Abstract
A rapid and sensitive two-step RT-PCR protocol for simultaneous detection of major apple viruses, namely Apple mosaic virus (ApMV), Apple stem pitting virus (ASPV), Apple stem grooving virus (ASGV), Apple chlorotic leaf spot virus (ACLSV) and Apple scar skin viroid (ASSVd), was developed. Five specific primer pairs were tested and confirmed for these viruses and viroid together in a single tube, giving amplicons of ~198, ~330, ~370, ~547 and ~645 bp corresponding to ASGV, ASSVd, ASPV, ApMV and ACLSV, respectively. Using a guanidinium-based extraction buffer along with a commercial kit resulted in better quality RNA as compared to kit, suited for multiplex RT-PCR. A rapid CTAB method for RNA isolation from apple tissue was developed, which produce good yield and saves time. To the best of our knowledge, this is the first report on the simultaneous detection of five pathogens (four viruses and a viroid) from apple with NADH dehydrogenase subunit 5 (nad5) as an internal control.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-013-0431-y) contains supplementary material, which is available to authorized users.
Keywords: ApMV, ASGV, ACLSV, ASSVd, ASPV, mRT-PCR
Introduction
Apple is one of the most important and widely grown fruit crop, providing a living for farmers worldwide. India is ranked seventh in world apple production. Due to prevalent agricultural practices, the crop has been found infected by many viral and subviral pathogens across world, notably Apple stem pitting virus (ASPV) [12, 15], Apple chlorotic leaf spot virus (ACLSV) [13, 15], Apple mosaic virus (ApMV) [23, 24], Apple stem grooving virus (ASGV) [26] and Apple scar skin viroid (ASSVd) [5, 7, 16]. ACLSV, and ASGV belong to family Betaflexiviridae, genus Trichovirus, and Capillovirus, ASPV belongs to family Alphaflexivirideae, genus Foveavirus. Apart from these some other viruses and viroids are known to infect apple in other parts of world. All of these pathogens reduce fruit quality and quantity, and plant growth [3, 22, 27]. High level of infection or mixed infection causes severe losses especially in perennial crops such as apple. Planting virus-free materials is necessary to limit losses caused by viral and subviral pathogens. For routine diagnosis, molecular techniques such as PCR provide a highly sensitive and rapid method of detecting these pathogens over conventional bioassays and ELISA. However, use of PCR in diagnostic laboratories can be limited due to the cost of its reagents and only one pathogen is detected in one time. This can be circumvented with the development of multiplex RT-PCR protocols that detect multiple pathogens in single reaction, thus providing a quicker, sensitive, reliable and cost-effective routine diagnosis. Previously such type of protocol was developed for detection of apple viruses viz. in which two pathogens were detected in two tubes and four in one tube [8, 19, 20]. Present study focuses on development of mRT-PCR which can detect all major pathogens (five) including ASSVd viroid in one reaction, as mixed infections are very common in apple. ASSVd is one of the prevalent pathogen in apple orchards of India [17] and this study offers an advantage of previously reported ones.
Materials and Methods
Sample Collection
Samples were collected from Rohru, Hatkoti, Jubbal and Kotkhai areas in the Shimla district, the Rekong Peo, Kothi and Kalpa areas of the Kinnaur district, and Palampur in the Kangra district (based on visual viral symptom and without symptoms as well), of Himachal Pradesh located in Northern India. Field and tissue-culture samples from our institute (sample number 13–21; P1–P27 and 31–38, respectively, Table 1) were also collected.
Table 1.
Results of ELISA and multiplex RT-PCR of samples collected from different regions of Himachal Pradesh
| S. no. | Sample code | Virus detected by multiplex RT-PCR | Symptoms observed | ELISA | ||||
|---|---|---|---|---|---|---|---|---|
| ApMV | ASGV | ASPV | ACLSV | ASSVd | ||||
| 1 | T1/111 | + | − | − | − | + | Chlorosis | − |
| 2 | T2/106 | − | − | − | − | − | Symptomless | − |
| 3 | T3/111 | + | − | + | − | + | Curled and underdeveloped leaves | − |
| 4 | T4/106 | + | − | − | − | + | Mosaic | − |
| 5 | T5/106 | + | − | − | − | + | Curling | − |
| 6 | T6/106 | + | + | + | − | + | Severe chlorosis | − |
| 7 | T7/106 | + | − | − | − | + | Mosaic | − |
| 8 | T8/106 | − | − | − | − | − | Symptomless | − |
| 9 | T9/109 | + | + | + | + | + | Severe chlororsis | − |
| 10 | T10/109 | + | − | − | − | + | Mottled leaves | − |
| 11 | T11/106 | − | + | + | + | − | Severe mosaic | − |
| 12 | T12/106 | − | − | − | − | + | Symptomless | − |
| 13 | RC (Positive control) | + | + | + | + | + | Severe mosaic | + (for all four viruses) |
| 14 | RC-2 (Considered as negative control) | − | + | − | − | − | Symptomless | − |
| 15 | RC-3 (Negative control) | − | + | − | − | − | Symptomless | − |
| 16 | Rf -1 | + | − | − | − | + | Symptomless | − |
| 17 | Rd-1 | + | + | + | − | + | Deformed fruits, mosaic | − |
| 18 | Rd-2 | + | + | + | − | + | Deformed fruits and severe mosaic | − |
| 19 | Rd-3 | − | − | + | − | − | Chlorosis | − |
| 20 | Rd-4 | + | − | + | − | − | Symptomless | − |
| 21 | Rd-5 | − | + | − | + | + | Symptomless | − |
| 22 | Gd-1 | + | + | + | − | + | Mottled leaves, deformed fruits | − |
| 23 | Gd-2 | + | − | − | + | − | Mosaic pattern | − |
| 24 | Gd-3 | + | + | + | + | + | Severe chlorosis, deformed fruits | − |
| 25 | Gd-4 | + | + | + | − | + | Severe chlorosis, deformed fruits | − |
| 26 | Gd-5 | − | − | − | − | − | Symptomless | − |
| 27 | Gd-6 | + | + | + | + | − | Chlorosis | − |
| 28 | Gd-7 | + | + | + | + | + | Deformed, underdeveloped fruit and leaves, chlorosis | − |
| 29 | 793 | + | + | − | + | − | Mild mosaic | − |
| 30 | 106 | + | + | + | − | + | Mottling | − |
| 31 | M7 (Positive control) | + | + | + | − | + | Symptomless | − |
| 32 | Gd | + | − | + | − | + | Symptomless | − |
| 33 | Rf-2 | + | + | − | − | + | Symptomless | − |
| 34 | Ga-1 | + | + | + | − | + | Symptomless | − |
| 35 | Ga-2 | − | − | − | − | − | Symptomless | − |
| 36 | Ga-3 | + | − | − | + | − | Symptomless | − |
| 37 | Ga-4 | − | − | + | − | + | Symptomless | − |
| 38 | T.No.1 | − | + | + | + | − | Mild mosaic pattern | − |
| 39 | T.No.2 | − | + | + | + | − | Mild curling | − |
| 40 | T.No.3 | − | + | + | + | − | Mild chlorosis | − |
| 41 | T.No.4 | − | + | + | + | − | Mild chlorosis | − |
| 42 | V1 | − | + | + | + | − | Mild chlorosis | − |
| 43 | V2 | − | + | + | − | − | Mild mottling | − |
| 44 | V3 | − | + | + | − | − | Underdeveloped leaves | − |
| 45 | R1 | − | + | + | + | − | Mild chlorosis and curled leaves | − |
| 46 | R2 | − | + | + | + | − | Underdeveloped leaves | − |
| 47 | K-1 | − | + | + | + | − | Chlorotic and necrotic spots | − |
| 48 | K-2 | + | + | + | + | − | Mottling | − |
| 49 | K-3 | − | + | + | − | − | Mottling | − |
| 50 | K-4 | − | + | + | + | − | Chlorosis | − |
| 51 | Kw-1 | + | + | + | + | − | Mosaic and reduced leave size | − |
| 52 | Kw-2 | − | + | + | − | − | Reduced leave size | − |
| 53 | Kw- 3 | − | + | + | − | + | Curling and reduced leave size | − |
| 54 | Kw-4 | + | + | + | + | − | Severe mosaic | − |
| 55 | B1 | − | + | + | − | + | Symptomless | − |
| 56 | B2 | − | + | + | − | + | Symptomless | − |
| 57 | B3 | − | + | + | − | + | Mild chlorosis | − |
| 58 | B4 | − | + | + | + | + | Chlorosis | − |
| 59 | P1 | + | + | + | + | + | Mild chlorosis | + (for all four viruses) |
| 60 | P2 | + | + | − | − | + | Underdeveloped, mosaic pattern on leaves | − |
| 61 | P3 | + | − | + | − | + | Underdeveloped leaves | − |
| 62 | P4 | + | + | + | − | − | Severe mosaic | + (only for ApMV) |
| 63 | P5 | + | + | + | + | − | Symptomless | − |
| 64 | P6 | + | − | + | + | + | Underdeveloped leaves | + (for ApMV, ACLSV) |
| 65 | P7 | + | − | − | − | + | Underdeveloped, mottled | − |
| 66 | P8 | + | + | + | − | − | Underdeveloped and curled leaves | + (only for ApMV) |
| 67 | P9 | + | − | + | − | − | Chlorosis, deformed fruits | − |
| 68 | P10 | + | − | − | − | − | Symptomless | − |
| 69 | P11 | − | + | − | − | − | Mild mosaic | − |
| 70 | P12 | + | + | + | − | + | Mild mosaic, underdeveloped fruits | + (only for ApMV) |
| 71 | P13 | + | + | + | − | − | Yellowish leaves | − |
| 72 | P14 | + | + | − | − | + | Symptomless | − |
| 73 | P15 | + | + | − | − | − | Symptomless | − |
| 74 | P16 | + | + | + | − | + | Leaf curling and chlorosis | + (only for ApMV) |
| 75 | P17 | + | + | + | − | + | Severe mosaic, stunted plant, small fruit size | + (only for ApMV) |
| 76 | P18 | + | + | + | − | − | Chlorosis | − |
| 77 | P19 | + | + | + | − | Symptomless | − | |
| 78 | P20 | + | + | + | − | + | Symptomless | − |
| 79 | P21 | + | + | + | − | − | Symptomless | − |
| 80 | P22 | + | + | + | − | + | Symptomless | − |
| 81 | P23 | + | − | + | − | − | Underdeveloped leaves | − |
| 82 | P24 | + | + | + | − | − | Chlorosis and necrotic spots | − |
| 83 | P25 | + | + | + | + | + | Symptomless | + (for ApMV, ACLSV, ASPV) |
| 84 | P26 | + | + | + | − | − | Symptomless | − |
| 85 | P27 | + | − | + | + | + | Mild mosaic and underdeveloped leaves | + (for ApMV, ACLSV, ASPV) |
| 86 | P28 | + | − | + | − | + | Symptomless | − |
| 87 | P29 | + | + | + | − | + | Mild mosaic | − |
| 88 | Positive control provided with ELISA Kit | + | + | + | + | − | − | + |
| 89 | Negative control provided with ELISA Kit | − | − | − | − | − | − | − |
(+) Positive, (−) Negative
Samples description Samples 1–12: Hatkoti and Rohru, HP (T1 to T12 were tree no. having rootstocks mentioned); Samples 13–21, 56–59 and P1–P29: Palampur, HP (RC: Red Chief; Rf: Red Fuji; B1- B4 Golden delicious; P1–P29 varieties not known); Samples 22–30: Jubbal, HP (Gd: Golden delicious; 793and 106: root stocks); Samples 31–38: Tissue culture plants (109, M7: root stocks; Gd: Golden delicious; Rf-2: Red Fuji; Ga: Gala); Samples 39–55: Kinnaur, HP (T.No.1–T.No.4: Golden; R1 and R2: Royal delicious; K1–K4: Royal delicious; Kw-1 to Kw-4; Royal delicious)
Nucleic Acid Extraction
Total RNA was extracted from 100 mg leaf and bark tissue using commercial kit (RNA plant mini kit, Qiagen, Hilden, Germany). The extraction buffer (EB) provided with the kit was slightly modified to contain 4 M guanidine thiocyanate, 0.2 M sodium acetate pH 5.0, 25 mM EDTA and 2.5 % (w/v) PVP-40 [18] in 1:1 ratio. In modified CTAB method, 100 mg of tissue was crushed in liquid nitrogen and was transferred to 1 ml CTAB containing 1 % 2-β mercaptoethanol. This mixture was heated for 10 min at 60 °C. Plant debris was separated and supernatant was extracted with chloroform: isoamyl alcohol (24:1). Aqueous phase was recovered and incubated with 3units of DNase I (Fermentas, Vilnius, Lithuania) at 37 °C/30 min. To this 0.8 volume of isopropanol was added and mixed gently by inverting tubes. This mixture was incubated on ice/5 min and then centrifuged at 14,000 rpm/20 min at 4 °C. Pellet was washed with 80 % ethanol. Final pellet was resuspended in 40 μl of RNase free water and analyzed on nanodrop (Themo scientific) and on agarose gel (1 %). Similar protocols were followed for leaf and bark tissue.
Oligonucleotides
Viruses and viroid-specific primer pairs were used for the detection of ACLSV, ASGV, ApMV, ASPV and ASSVd (Supplementry Table 1), alongwith NADH dehydrogenase subunit 5 gene (nad5) as an internal control for plant mRNA [14, 19]. For primer designing coat protein gene was used for ACLSV, ASGV, ApMV, ASPV while in case of ASSVD whole genome was used. Amplicons were sequenced using an ABI Prism 310 with an ABI prism Big Dye-Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
Simplex RT-PCR
First-strand cDNA synthesis was carried out in 25 μl reaction containing 800 ng of total RNA, 1 μl (200 units) of reverse transcriptase enzyme (M-MLV; USB, USA), 5 μl (5×) RT buffer, 1 μl of 40 mM dNTP mix, and 1 μl (10 pmol/μl) of specific primers. Reaction was first denatured at 65 °C for 5 min along with primers and then chilled on ice/5 min followed by addition of other components. Reaction was incubated at 37 °C/75 min followed by incubation at 80 °C/5 min. PCR was carried out in a thermocycler (G-Storm thermal cycler GS2, Somerton Biotechnology Centre, Somerset, UK) using each 1 μl (10 pmol/μl) virus specific forward and reverse primers, 5 μl first strand cDNA, 5 μl (10x) taq buffer, 1.5 units of taq DNA polymerase (Genei Banglore), 1 μl of 2.5 mM dNTP mix in a total of 50 μl volume. Reaction was amplified for 30 cycles (30 s at 94 °C, 35 s at 58 °C, and 1 min at 72 °C). The PCR products were fractionated on 1 % agarose gel by electrophoresis for 1 h at 85volt and documented by gel doc (Syngene G: Box).
Multiplex RT-PCR
For RT, all conditions were the same except random hexamer primers 100 ng were used for cDNA synthesis and for multiplex PCR reaction 5 μl cDNA, 0.16 pmol of each primers pair, 1.2× Taq buffer, 0.3 mM dNTPs, 1.5 units of Taq DNA polymerase (Bangalore Genei) and 1.5 mM MgCl2 (additional) in final concentration was used. cDNA was amplified for 35 cycles (40 s at 94 °C, 75 s at 58 °C, and 90 s at 72 °C). Same reaction composition was carried out with Hot Start Taq DNA polymerase (Fermentas, Vilnius, Lithuania). Multiplex PCR was also carried out with a multiplexing kit (Qiagen, Hilden, Germany) along with Q-solution following the manufacturer’s instructions. Amplicons were fractionated on 3 % agarose gel and desired bands were excised from gel, eluted, cloned in pGMET-Easy vector (Promega) and sequenced using the ABI prism Big Dye™ Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, USA).
Optimization of Multiplex RT-PCR
Four primer pairs were used in this study (Supplementary Table 1) had never been used previously in multiplex RT-PCR assays. The multiplex RT-PCR assay was optimized with regular Taq (Bangalore Genei, Bengaluru, India), Hot Start Taq (Fermentas, Vilnus, Lithuania) DNA polymerase and multiplexing kit from Qiagen under the conditions described above with nad5 as an internal control.
ELISA
Leaf samples were tested by double-antibody sandwich ELISA (DAS-ELISA) for the targeted viruses ACLSV, ApMV, ASPV, and ASGV using a commercial kit (Bioreba, Reinach, Switzerland).
Results and Discussion
ELISA
Only few sample (10) found positive in DAS-ELISA. Of these positive samples, only RC, P-1, were found positive for all viruses. In P-6 (ApMV, ACLSV); P-25, P-27 (ApMV, ACLSV, ASPV); P-4, P-8, P-12, P-16 & P-17 (ApMV) found positive for mentioned viruses. Samples recollected in September (P-1 to P-29) were also analyzed but all were found negative in DAS-ELISA.
Amplification of the Various Viral Gene Fragments By Simplex RT-PCR
RNA was extracted from leaf and bark tissue and all pathogens were successfully detected from this preparation (Fig. 1). Both methods of RNA extraction were found to be reliable and rapid, giving good yield of RNA. Modified CTAB method is easy and quick to perform as all precipitation steps were omitted. Approximately within 2 h good quality RNA (~30–35 μg/100 mg tissue) was obtained. Also the quality is assessed during reaction as in this assay plant gene nad5 is incorporated as internal control. To check the specificity of the primer pairs, simplex RT-PCR was first carried out and validated to choose the best working primers. Amplicons of ~198, ~330, ~370, ~547 and ~645 bp corresponding to ASGV, ASSVd, ASPV, ApMV and ACLSV, respectively, were obtained by this method (Fig. 1). RC, Rd-1, Gd-2, Gd-3 (from Palampur and Jubbal) known viral sources were used first in the simplex and then in the multiplex RT-PCR assay (Table 1).
Fig. 1.
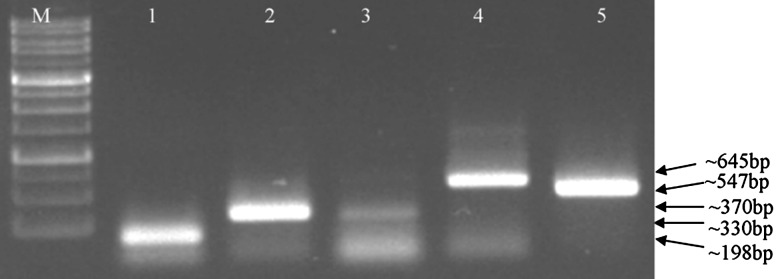
Gel electrophoresis of individual virus detection. Lanes 1–5 specific amplification for ASGV, ASPV, ASSVd, ACLSV and ApMV respectively; lane M 100 bp DNA marker
Optimization of Multiplex RT-PCR
Multiplex RT-PCR assay was optimized with regular Taq (Bangalore Genei, Bengaluru, India), Hot Start Taq (Fermentas, Vilnus, Lithuania) DNA polymerase and multiplexing kit from Qiagen under the conditions described above with nad5 as an internal control (Fig. 2 and Supplementary Fig. 1). Specific amplification products were obtained for all viral and subviral pathogens as well as nad5 following electrophoresis on a 3 % agarose gel and staining with EtBr (10 mg/ml). Specificity of the fragments obtained following electrophoresis was validated by sequencing the amplicons. On sequencing amplicons showed 198, 330, 370, 547 and 645 bp corresponding to ASGV, ASSVd, ASPV, APMV and ACSLV, respectively. An increase in MgCl2 concentration was found to increase band intensity. All samples showing virus-like symptoms (i.e. collected from different regions of Himachal Pradesh) were found positive and most of the samples were found infected with more than one virus (Table 1). Samples Rf-1, Rd-2, Rd-4 Rd-5, B1, B2 and tissue-culture plants (except Ga-2, T8/106, T2/106, Gd-5), which did not display any of the reported viral symptoms, were also found positive by multiplex RT-PCR. Further the method was validated on 87 samples collected from different areas of H.P.
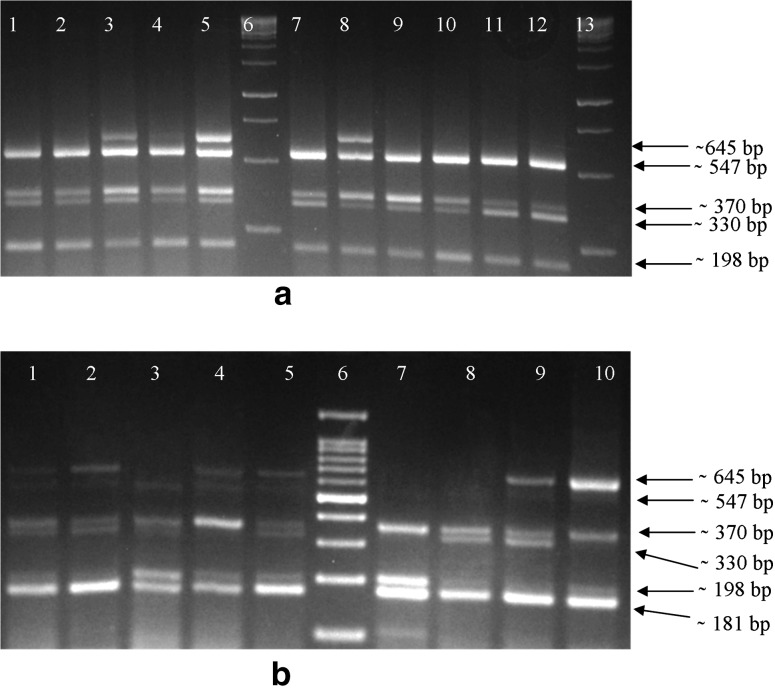
Fig. 2.
a Multiplex RT-PCR without internal control. Lanes 1 and 2 amplified product of M7 was loaded showing specific amplification of four all pathogens except ACLSV; lane 3 and 4 contains: product of RC and Gd-3; lane 5 and 8 contains amplicon of Gd-7; lane 6 and 13 100 bp marker (Genedirex); lane 7: product of sample 106; lanes 9 and 10 amplified product of Gd-1; lane 11 and 12 amplicons of T6/109 sample. b Multiplex RT-PCR with internal control (nad5). Lanes 1–5: amplicons of samples RC, T9/109, Rd-1, Gd-6 and Gd-7; lanes 7–10 amplicons of Kw-2, Kw-3, K-4 and Kw-4; lane 6 100 bp marker. All products were fractionated on a 3 % agarose gel
Sensitivity of Multiplex RT-PCR and comparison with ELISA
To test the detection sensitivity of the multiplex RT-PCR, amplification was carried with serially diluted cDNA along with other parameters such as the use of random hexamer primers versus specific primers, and amplification by Taq DNA polymerase, hot-start Taq DNA polymerase or multiplexing kit were compared (Supplementry Fig. 1a–c). To avoid non-specific annealing of primers and their dimer formation, primers that bind at high temperature were selected. To avoid non-specific binding and amplification, hot-start Taq DNA polymerase was used, and gave better results than the regular Taq DNA polymerase. To determine the sensitivity of the reaction, an initial 5 μl of cDNA was serially diluted from tenfold up to 10−6-fold. Multiplex RT-PCR was carried out with these dilutions using the hot-start Taq DNA polymerase and a multiplexing kit. Up to 10−1, all pathogens were detected with hot-start Taq, whereas with the multiplexing kit, sensitivity reached 10−2 (Fig. 3). At higher dilutions, all pathogens except ApMV were detected (Fig. 3), showing the high sensitivity and specificity of the protocol. To check the reproducibility of assay in dormant season when viral titre is low, field samples (P1-P29) collected twice in two seasons were analyzed by mRT-PCR. Similar results were obtained in multiplex RT-PCR in both seasons (Table 1), however there was variation in DAS-ELISA results (Table 1).
Fig. 3.
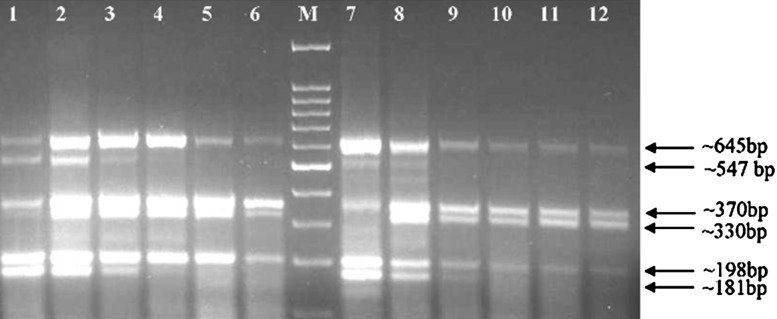
Sensitivity limits of multiplex RT-PCR. Lane 1 Amplification from tenfold diluted cDNA; lanes 2–6 amplicons from 10−2 to 10−6 fold diluted cDNA. Lanes 1–6 amplicons obtained with multiplexing kit; lanes 7–12 amplification with hot-start Taq DNA polymerase at same dilutions; lane M 100 bp marker
Validation of Amplicons Obtained by Multiplex RT-PCR
Amplicons were sequenced and compared to known sequences in the database and were found to be specific for the target viruses. As a cross-check, some samples were analyzed by simplex RT-PCR followed by multiplex RT-PCR. Initially, protocol is validated on known samples and now it is routinely used. The amplification protocol was tested on 88 field samples collected from different areas of the Kangra, Shimla and Kinnaur districts of Himachal Pradesh (Table 1). A reliable and highly sensitive method for viral and subviral pathogen detection is a pre-requisite for selecting trees for budwood (scions) and for raising virus-free rootstocks. As mixed infection is common in apple, development of a highly sensitive, reliable, rapid and economical method of detection is an absolute necessity, especially taking into consideration the ubiquitous presence of viruses and viroids in the field. ELISA is not very reliable and is time-consuming. A number of procedures have been developed for simultaneous detection of various viruses wherein workers [10] reported the detection of ASGV along with Cherry mottle leaf virus in apple. A multiplex RT-PCR assay was developed for diagnosis of ASPV and six viroids Citrusexocortisviroid (CEVd), Citrus bent leafviroid (CBLVd), Hop stuntviroid (HSVd), CitrusviroidIII (CVd-III), CitrusviroidIV (CVd-IV), CitrusviroidOS (CVd-OS) and Apple stem grooving virus (ASGV, synonym: Citrustatter leafvirus, CTLV) from citrus plants [9]. There have been many other efforts in this direction and reports have been published for many crops [1, 2, 4, 6, 10, 11, 21, 25]. PCR and multiplex RT-PCR are routinely used for virus diagnosis. Further, real time PCR and LAMP (loop mediated isothermal amplification) methods are useful for virus titre estimation (Real time-PCR) and diagnosis when titre of the virus is not a problem (LAMP). LAMP method offers ease of detection, but requires few sets of HPLC purified primers, which makes it costly. There is requirement of a turbidometer for final assessment. It has been rarely used for plant virus detection. Realtime-PCr on the other hand offers great sensitivity and exact quantification of pathogen, but there is high cost associated (probes or dyes) alongwith specialized equipment. In the present study, an rapid, improved, cost effective approach was adopted to develop multiplex RT-PCR assay for four major apple viruses and Apple scar skin viroid, using the plant gene nad5 as an internal control. Several parameters were optimized to develop a reliable multiplex assay. We obtained positive results with primer concentrations varying from 0.5 to 0.8 μl (10 pmol/μl stock), but best results were obtained with 0.8 μl of each primer, along with increased MgCl2 concentration up to 1.6 mM. Annealing temperature and extension time also play a critical role during multiplex RT-PCR detection. Generally, a higher annealing temperature, an increase in the number of cycles and a small extension of the annealing time provided reliable and specific results. The quality of the nucleic acids is a limiting step in multiplex RT-PCR, so it is important to choose the best method of nucleic acid extraction. Selection of an internal control is another important step to obtaining reliable results in PCR, as this gives preliminary information on the quality of the RNA and helps in interpreting negative results [19]. The two-step multiplex RT-PCR is more sensitive than its one-step counterpart [6]. Use of random hexamers for the RT and hot-start Taq DNA polymerase improved the cDNA quality and reaction specificity and resulted in detection at higher dilutions. Previously reported methods detected four major apple viruses (ACLSV, ASGV, ASPV, ApMV) with and without an internal control, in two separate tubes [19, 20] and in a single tube [8]. Here we report the simultaneous detection of five major economic pathogens of pome fruits in single tube (four major viruses ACLSV, ASGV, ASPV, ApMV and viroid), which are a real problem in the orchards. This mRT-PCR assay is reliable, sensitive, simple, rapid, cost-effective method to detect these pathogens in apple and could be opted in small labs in early screening of nursery raised plants, mother plants used for raising virus free planting material through tissue culture and other phytosanitary purposes. Approximately within 7–8 h one can get results from multiple samples starting from RNA extraction to gel analysis by using this multiplex RT PCR. Also this will be useful to quarantine and certification programs and virus surveys when large numbers of samples are tested. Results also confirm the presence of mixed infection in Indian apple orchards as also reported previously [17].
Electronic supplementary material
Supplementry fig. 1 Evaluation of first-strand cDNA primers, Taq DNA polymerase and commercially available multiplexing kit efficiency. a) Use of specific primers for cDNA synthesis with Genei Taq for amplification. Lanes 1–5: amplified products of RC, RC2 (healthy control), K2, Gd-7 and T11/106, respectively. Lane M: 100 bp marker. b) Use of random hexamer for cDNA synthesis and Hot-start Taq DNA polymerase for amplification. Lane 1-4 Fragments amplified from samples B1, B4, B3, B2 and lane 5 healthy control samples 109. c) Use of random hexamer for cDNA synthesis and multiplexing kit & Genei Taq DNA polymerase for PCR amplification. Lanes 1–4: amplified products of samples B1, B2, B3, B4, respectively amplified using multiplexing kit, lane 5: no template (control). Lanes 6–9 same samples amplified using Genei Taq DNA polymerase. Lane M: 100 bp marker. (TIFF 3609 kb)
Supplementary fig. 2 Samples analyzed by multiplex RT-PCR a). Lane 1, sample found postive only for ASGV & all other viruses, viroid was absent in it: lane2; ASGV, ASSVd; lane M 100 bp DNA marker b). Mix infection was observed when amplicons were fractionated on agarose gel by electrophoresis, except one sample present in lane 10. We are now using these samples (Fig. 2a; lane1, Fig. 2b; lane 10) as a negative control (their infection status is known for mentioned pathogens), as complete virus free material is not yet identified (its very difficult in perennial crops). Lane M contains 1 kb DNA ladder. (TIFF 412 kb)
Acknowledgments
The authors are thankful to the Director, Council of Scientific & Industrial Research, Institute of Himalayan Bioresource Technology, Palampur, Himachal Pradesh, India for providing the necessary facilities to carry out the work, and to Digvijay Singh for technical help with nucleotide sequencing. A fellowship to SK and financial support from the Department of Biotechnology, Govt. of India; grant number BT/PR/10594/PBD/16/760/2008 and Council of Scientific & Industrial Research are duly acknowledged. This is IHBT publication number 3274.
References
- 1.Alabi JO, Kumar PL, Naidu RA. Multiplex PCR for the detection of African cassava mosaic virus and East African cassava mosaic Cameroon virus in cassava. J Virol Methods. 2008;154:111–120. doi: 10.1016/j.jviromet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Bertolini E, Olmos A, Martinez MC, Gorris MT, Cambra M. Single-step multiplex RT-PCR for simultaneous and colourimetric detection of six RNA viruses in olive trees. J Virol Methods. 2001;96:33–41. doi: 10.1016/S0166-0934(01)00313-5. [DOI] [PubMed] [Google Scholar]
- 3.Campbell AI. The effect of some latent virus infections on the growth and cropping of apples. J Hortic Sci. 1963;38:15–19. [Google Scholar]
- 4.Deb M, Anderson M. Development of a multiplexed PCR detection method for Barley and Cereal yellow dwarf viruses, wheat spindle streak virus, wheat streak mosaic virus and soil-borne wheat mosaic virus. J Virol Methods. 2008;148:17–24. doi: 10.1016/j.jviromet.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Desvignes CJ, Grasseau N, Boye R, Cornaggia D, Aparicio F, Di Serio F, Flores R. Biological properties of apple scar skin viroid: isolates, host range, different sensitivity of apple cultivars, elimination, and natural transmission. Plant Dis. 1999;83:768–772. doi: 10.1094/PDIS.1999.83.8.768. [DOI] [PubMed] [Google Scholar]
- 6.Gambino G, Gribaudo I. Simultaneous detection of nine grapevine viruses by reverse transcription-polymerase chain reaction with coamplification of a plant RNA as internal control. Virol J. 2010;96:1223–1229. doi: 10.1094/PHYTO-96-1223. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto J, Koganezawa H. Nucleotide sequence and secondary structure of apple scar skin viroid. Nucleic Acids Res. 1987;15:7045–7052. doi: 10.1093/nar/15.17.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan M, Myrta A, Polak J. Simultaneous detection and identification of four pome fruit viruses by one tube pentaplex RT-PCR. J Virol Methods. 2006;133:124–129. doi: 10.1016/j.jviromet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Ito T, Ieki H, Ozaki K. Simultaneous detection of six citrus viroids and Apple stem grooving virus from citrus plants by multiplex reverse transcription polymerase chain reaction. J Virol Methods. 2002;106:235–239. doi: 10.1016/S0166-0934(02)00147-7. [DOI] [PubMed] [Google Scholar]
- 10.James D. A simple and reliable protocol for the detection of Apple stem grooving virus by RT-PCR and in a multiplex PCR assay. J Virol Methods. 1999;83:1–9. doi: 10.1016/S0166-0934(99)00078-6. [DOI] [PubMed] [Google Scholar]
- 11.Jarosova J, Kundu JK. Simultaneous detection of stone fruit tree viruses by one-step multiplex RT-PCR. Sci Hortic. 2010;125:68–72. [Google Scholar]
- 12.Jelkmann W. Nucleotide sequences of apple stem pitting virus and of the coat protein gene of a similar virus from pear associated with vein yellows disease and their relationship with potex- and carlaviruses. J Gen Virol. 1994;75:1535–1542. doi: 10.1099/0022-1317-75-7-1535. [DOI] [PubMed] [Google Scholar]
- 13.Jelkmann W. The nucleotide sequence of a strain of Apple chlorotic leaf spot virus (ACLSV) responsible for plum pseudopox and its relation to an apple and plum back split strain. Phylopathol. 1996;86:101. [Google Scholar]
- 14.Kato S, Shimamoto Y, Mikami T. The apple mitochondrial ATP9 gene: RNA editing and cotranscription with exons a and b of the nad5 gene. Physiol Plant. 1995;93:572–575. doi: 10.1111/j.1399-3054.1995.tb06860.x. [DOI] [Google Scholar]
- 15.Koganezawa H, Yanase H. A new type of elongated virus isolated from apple trees containing the stem pitting agent. Plant Dis. 1990;74:610–614. doi: 10.1094/PD-74-0610. [DOI] [Google Scholar]
- 16.Koganezawa H, Ito T. Apple fruit crinkle viroid. In: Hadidi A, Flores R, Randles JW, Semancik JS, editors. Viroids. Collingwood: CSIRO Publishing; 2003. pp. 150–152. [Google Scholar]
- 17.Kumar S, Singh RM, Ram R, Badyal J, Hallan V, Zaidi AA, Varma A. Determination of major viral and sub viral pathogens incidence in Apple orchards in Himachal Pradesh. Indian J Virol. 2012;23:75–79. doi: 10.1007/s13337-011-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKenzie DJ, Mclean MA, Mukerji S, Grenn M. Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Dis. 1997;81:222–226. doi: 10.1094/PDIS.1997.81.2.222. [DOI] [PubMed] [Google Scholar]
- 19.Menzel W, Jelkmann W, Maiss E. Detection of four apple viruses by multiplex RT-PCR assays with co-amplification of plant mRNA as internal control. J Virol Methods. 2002;99:81–92. doi: 10.1016/S0166-0934(01)00381-0. [DOI] [PubMed] [Google Scholar]
- 20.Menzel W, Zahn V, Maiss E. Multiplex RT-PCR-ELISA compared with bioassay for the detection of four apple viruses. J Virol Methods. 2003;110:153–157. doi: 10.1016/S0166-0934(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 21.Nie X, Singh RP. Detection of multiple potato viruses using an oligo(dT) as a common cDNA primer in multiplex RT-PCR. J Virol Methods. 2000;86:179–185. doi: 10.1016/S0166-0934(00)00140-3. [DOI] [PubMed] [Google Scholar]
- 22.Posnette AF, Cropley R, Ellenberger EC. The effect of virus infection on the growth and crop of apple, pear, and plum trees. Phytopathologia Mediterranea. 1963;2:158–161. [Google Scholar]
- 23.Rybicki EP. The Bromoviridae. In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD, editors. Virus taxonomy. Sixth Report of the International Committee on Taxonomy of Viruses. Vienna: Springer-Verlag; 1995. pp. 450–457. [Google Scholar]
- 24.Shiel JP, Berger H. The complete nucleotide sequence of apple mosaic virus (ApMV) RNA 1 and RNA 2: ApMV is more closely related to alfalfa mosaic virus than to other ilarviruses. J Gen Virol. 1999;81:273–278. doi: 10.1099/0022-1317-81-1-273. [DOI] [PubMed] [Google Scholar]
- 25.Thompson RJ, Wetzel S, Klerks MM, Vaskova D, Schoen CD, Spak J, Jelkmann W. Multiplex RT-PCR detection of four aphid-borne strawberry viruses in Fragaria spp in combination with a plant mRNA specific internal control. J Virol Methods. 2003;111:85–93. doi: 10.1016/S0166-0934(03)00164-2. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa N, Sasaki E, Kato M, Takahashi T. The nucleotide sequence of apple stem grooving capillovirus genome. Virol J. 1992;191:98–105. doi: 10.1016/0042-6822(92)90170-T. [DOI] [PubMed] [Google Scholar]
- 27.Zahn V. Obstvirustestung im Wandel der Zeit. Obstbau. 1996;21:547–550. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementry fig. 1 Evaluation of first-strand cDNA primers, Taq DNA polymerase and commercially available multiplexing kit efficiency. a) Use of specific primers for cDNA synthesis with Genei Taq for amplification. Lanes 1–5: amplified products of RC, RC2 (healthy control), K2, Gd-7 and T11/106, respectively. Lane M: 100 bp marker. b) Use of random hexamer for cDNA synthesis and Hot-start Taq DNA polymerase for amplification. Lane 1-4 Fragments amplified from samples B1, B4, B3, B2 and lane 5 healthy control samples 109. c) Use of random hexamer for cDNA synthesis and multiplexing kit & Genei Taq DNA polymerase for PCR amplification. Lanes 1–4: amplified products of samples B1, B2, B3, B4, respectively amplified using multiplexing kit, lane 5: no template (control). Lanes 6–9 same samples amplified using Genei Taq DNA polymerase. Lane M: 100 bp marker. (TIFF 3609 kb)
Supplementary fig. 2 Samples analyzed by multiplex RT-PCR a). Lane 1, sample found postive only for ASGV & all other viruses, viroid was absent in it: lane2; ASGV, ASSVd; lane M 100 bp DNA marker b). Mix infection was observed when amplicons were fractionated on agarose gel by electrophoresis, except one sample present in lane 10. We are now using these samples (Fig. 2a; lane1, Fig. 2b; lane 10) as a negative control (their infection status is known for mentioned pathogens), as complete virus free material is not yet identified (its very difficult in perennial crops). Lane M contains 1 kb DNA ladder. (TIFF 412 kb)