Abstract
Soybean is extensively cultivated worldwide and is the largest source of biologically fixed nitrogen among legumes. It is nodulated by both slow and fast growing rhizobia. Indigenous soybean rhizobia in Vertisols of central India were assessed for utilization of 35 carbon sources and intrinsic resistance to 19 antibiotics. There was greater utilization of trehalose and raffinose by fast growers (87 and 73 % by fast vs. 35 and 30 % by slow growers); but slow growers had higher ability to utilize glucosamine (75 % by slow vs. 33 % by fast growers). A larger proportion of slow growers were resistant to vancomycin, polymyxin-B and rifampicin (70, 65 and 55 %) compared to fast growers (13, 7 and 7 % each). Among the two 16S rRNA sequence types in the slow growers, those belonging to Bradyrhizobium spp. utilized glucosamine while those belonging to Rhizobium radiobacter did not. All the fast growers had 16S rRNA homology to R. radiobacter and majority could not utilize glucosamine. It is suggested that during initial isolations and screening of rhizobia in strain selection programmes, using carbon sources like glucosamine and antibiotics like vancomycin, polymyxin-B and rifampicin in the media may provide a simple way of distinguishing Bradyrhizobium strains from R. radiobacter among the slow growers.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-013-0430-z) contains supplementary material, which is available to authorized users.
Keywords: Biological nitrogen fixation, Carbon utilization, Intrinsic antibiotic resistance, Microbial inoculants, Nodulation
Introduction
There are growing concerns about the adverse effects associated with the manufacture and use of nitrogenous fertilizers and a renewed focus on benign alternatives like biological nitrogen fixation (BNF). Soybean is the most important legume fixing 16 Tg N/year amounting to 77 % of global crop legume BNF [1]. It is nodulated by slow (Bradyrhizobiumjaponicum), extra-slow growing (B. liaoningense and B. yuanmingense) and fast growing rhizobia. The fast growers belong to Sinorhizobium fredii as well as saprophytic bacteria resembling Agrobacterium spp. reported in effectively nodulated soybean fields in South America [2, 3]. In India, large scale cultivation of soybean began in late 1960s with the import of yellow seeded modern soybean varieties from USA along with their slow growing symbiont—B. japonicum [4]. Later, the import of soybean inoculants gave way to indigenous inoculants prepared from native isolates of Indian soils. The selection of such native strains adapted to the tropical conditions consisted of inoculation of an area, followed by re-isolation of bacteria from nodules of soybean trap plants. The effects of inoculation during early years of soybean introduction in India, with the imported bradyrhizobia were very convincing. But the responses declined gradually with time [4] due to the naturalization of the introduced slow growing strains (evolution of native or indigenous strains) similar to that observed in other soybean growing countries like Brazil [2].
The rhizobial population structure in soils is a resultant of the interactions among the bacteria, host plant and environmental factors [5]. Large diversities of soybean rhizobia (in countries for which soybean was an exotic crop) could have arisen due to transfer of symbiotic genes from the inoculant strains to indigenous non-symbiotic rhizobia [2]. In earlier studies in India, effective nodulation of soybean was reported to be mostly by slow growing bradyrhizobia; nodulation by fast growing rhizobia was to a much lesser extent but ineffective [6]. But in central India, a majority of the soybean isolates reported recently were found to be fast growing [7]. Knowledge of the biodiversity of rhizobia is thus important for the design of successful inoculation strategies. Lindstrom et al. [8] argued in favour of linking the knowledge of the molecular diversity of rhizobia to the practical requirement of searching for competitive inoculants of rhizobia especially in situations where there is extensive adaptation to the local conditions leading to evolution of native rhizobia due to transference of symbiotic genes. There is thus a need to comprehensively study the diverse ecological attributes of the indigenous slow and fast growing soybean rhizobial strains in major soybean growing zones and to distinguish Bradyrhizobium strains among them by simple biochemical tests that can be routinely deployed in inoculant production facilities. Therefore, we assessed the native soybean rhizobia in Vertisols of Central India and other regions for phenotypic attributes like catabolic versatility and intrinsic antibiotic resistance (IAR) and made an attempt to link the observed differences to the strain identity in an effort to contribute to simple screening methods for strain selections.
Materials and Methods
The root nodules were sampled from healthy soybean plants in experimental and farmers’ fields in Vertisols in Madhya Pradesh, Central India. Soils were also taken from other soybean growing areas in India and rhizobia isolated using soybean as the ‘trap plant’. The details of sites, soil pH, electrical conductivity and organic carbon status are given in Supplementary Material 1. Rhizobia were isolated from healthy pink nodules in Congo-red yeast extract agar (CRYEMA) medium and characterized by morphological, physiological and nodulation tests as per standard procedures [9]. Reference strains—B. japonicum USDA 110 (R27), B. japonicum USDA 122 (R28) and B. elkanii USDA 31 (R29) were included for comparison. The details of strains, morphological and physiological tests, and nodulation testing in green house are given in Supplementary Material 1.
Hi-carbohydrate kit (Himedia laboratories, India) containing 35 carbon sources belonging to mono-, di- and trisaccharides and their derivatives, polysaccharides and some other sources was used. 5 μl of the rhizobial suspensions (0.5 OD at 620 nm) were inoculated and the strips incubated at 28 °C for 48 h to observe change of color. Nineteen antibiotics of different groups were used for assessing IAR. They were: β-lactams (Ampicillin, 10 μg, Carbenicillin—100 μg), aminoglycosides (Streptomycin 25 μg, Kanamycin 30 μg, Gentamicin 10 μg, Neomycin 30 μg, Amikacin 30 μg, Novobiocin 30 μg), tetracyclines (Tetracycline 30 μg), macrolides (Erythromycin 15 μg), polypeptides (Polymyxin 300 units), rifampin (Rifampicin 5 μg), glycopeptides (Vancomycin 30 μg), quinolones (Ciprofloxacin 5 μg, Nalidixic acid 30 μg), sulfonamides (Trimethoprim 5 μg), aminocyclitol (Spectinomycin 100 μg) and others (Chloramphenicol 30 μg, Co-trimoxazole 25 μg). 24 h old cultures of Rhizobium were used to make bacterial lawns on CRYEMA plates and impregnated antibiotic discs (Himedia labs) were pressed onto the agar surface. They were incubated at 28 °C; clearing zone around the discs (2–5 days for fast growers and 7–10 days for slow growers) were measured.
The carbon utilization (35 sources) and the IAR (19 antibiotics) of the rhizobial strains was used for cluster analysis using Similarity for Qualitative data (SIMQUAL) subroutine of Numerical taxonomic and multivariate analysis system (NTSYS). The isolates were grouped by the unweighted paired group method using arithmetic means (UPGMA) and depicted as dendrograms. The data on the 16S rRNA gene sequence of 17 out of 35 strains (7 slow and 10 fast growing strains representing areas of major soybean cultivation in different geographical areas) obtained in a parallel study (NCBI accession nos. given in Supplement 1) were used to confirm strain identity and any relationship with growth rate.
Results
The characteristics of the 40 rhizobial strains are given in Supplementary Material 1. Twenty strains were slow or very slow growing with 1–2 mm size colonies appearing after 7–10 days. The very slow growers had white opaque morphology whereas the slow growers had white opaque or watery translucent morphology. These were from three locations in Central (Geelakhedi, Dt. Rajgarh; Tikamgarh, Vidisha), one in Western (Parbhani) and three in northern India (Pantnagar, Palampur, Ludhiana). Fifteen strains were fast growers with 4–8 mm size colonies appearing after 2–3 days with watery translucent colonies, all were from ten locations in Central India (Anuppur, Betul, Chhindwara, Dewas, Dhar, Harda, Narsinghpur, Seoni, Vidisha, Indore). Five strains had intermediate growth with 2–5 mm size colonies appearing after 7–10 days, exhibited pink colonies and were from three locations in Central India (Bhopal, Chhatarpur and Indore). All the strains with intermediate growth rate exhibited pink colonies. Because the majority of strains are slow or fast, the intermediate category was ignored for the present purpose since the intention was to bring out the contrasting differences between slow and fast growers.
Most of slow or very slow growers (14 out of 20) were alkali producers whereas most of the fast growers (13 out of 15) were acid producers. 29 out of the 40 strains showed poor growth on glucose peptone agar. Of the 10 that grew, 4 were slow growing, 1 was intermediate and 6 were fast growing. None of the rhizobia grew on ketolactose agar. A significant number, 30 strains out of 40 (12 slow, 5 intermediate and 13 fast growing) grew on Hoffer’s alkaline broth at pH 11. Thus, out of the four physiological tests routinely deployed for rhizobia [9] only ketolactose test showed consistency as a distinguishing criteria. All the slow and fast growers nodulated soybean both in sterile sand culture and in soil (Supplementary Material 1).
Among the 20 monosaccharides, mannose, l-arabinose and ribose were utilized by maximum strains (28–29) (see Supplementary Material 2). However all the monosaccharides were utilized by one or the other strain (Fig. 1). Out of 8 disaccharides, sucrose was utilised by maximum number (28) while others were utilized by one or other strain. Out of 5 polysaccharides, esculin was utilised by maximum number of strains (22). All of the 35 carbon sources were not utilized by any single strain of the slow or fast growing rhizobia. The number of sources utilized by any one strain ranged from 0 to 29 (mean 15 sources).
Fig. 1.
Utilization of various carbohydrates by slow and fast growing soybean rhizobia
Among the 20 slow growers there were 19 different carbon utilization patterns while among the 15 fast growers there were 14 patterns. The slow growers had greater catabolic versatility; one or the other strain could utilize any one of the 35 carbon sources. The sources that were utilized by the greatest number of slow growers were glucosamine (75 %), ribose (55 %), mannose (55 %) and l-arabinose (55 %). On the other hand the fast growers could not utilize xylitol, α methyl d-mannoside, sorbose, sodium gluconate and melezitose. The carbon sources that were utilized by the greatest number of fast growers were mannose (93 %), melibiose (93 %), ribose (87 %), l-arabinose (87 %), xylose (87 %), trehalose (87 %) and sucrose (87 %). The extent of carbon utilization is more in fast growing soybean rhizobia but a greater number of carbon sources were utilized by slow growers (Fig. 1).
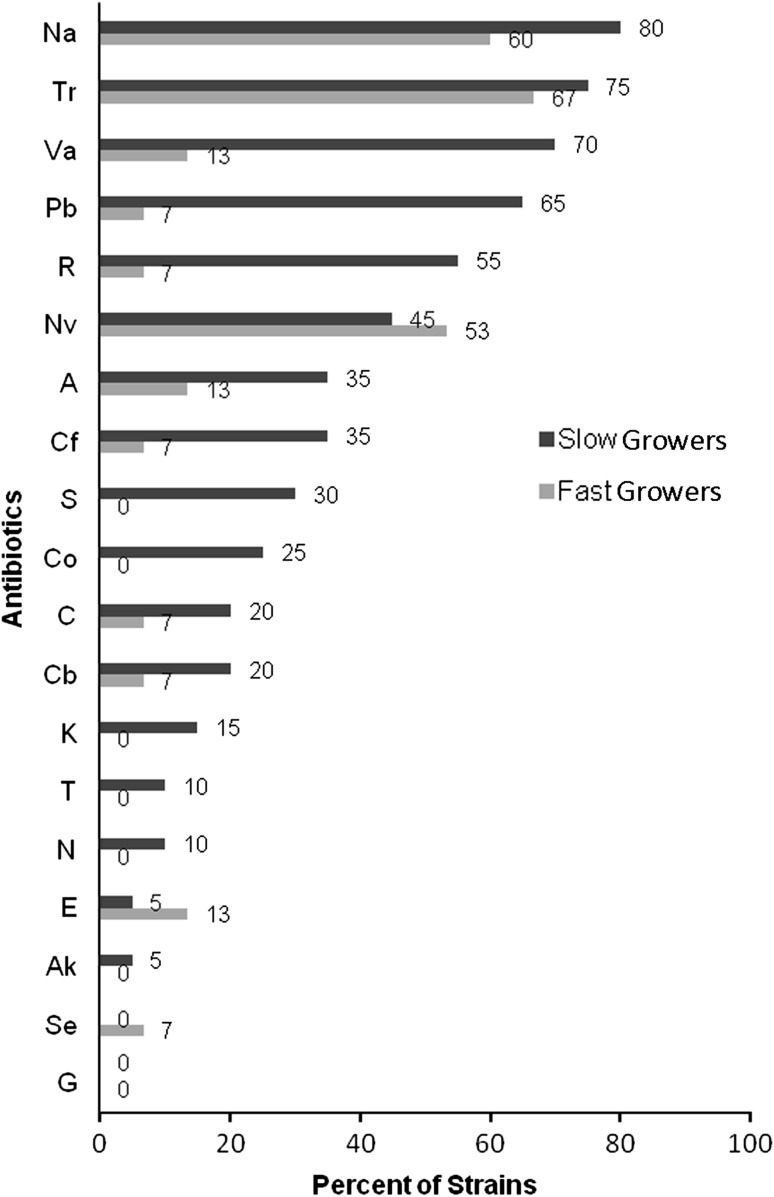
Of the 20 slow growers, 15 were resistant to trimethoprim, 16 to nalidixic acid, 13 to polymyxin B, 14 to vancomycin and 9 to Novobiocin (see Supplementary Material 3). Of the 15 fast growing rhizobia, 10 were resistant to trimethoprim, 9 to nalidixic acid and 8 to novobiocin. Thus the slow growers were more resistant to antibiotics (Fig. 2). Both slow and fast growers showed maximum resistance to nalidixic acid and trimethoprim (slow growers—Na 80 % and Tr 75 %, fast growers—Na 60 % and Tr 67 %). All the slow and fast growing rhizobia were sensitive to gentamicin. In addition, the slow growers were also sensitive to spectinomycin. The fast growers were more sensitive to six antibiotics namely amikacin, neomycin, tetracycline, kanamycin, co-trimoxazole and streptomycin. A larger proportion of slow growers were resistant to rifampicin and polymyxin-B (55 and 65 %) as compared to fast growers (7 % each). There was only one slow growing (R26 from Indore 9b) and two fast growing strains (R18 from Seoni, R24 from Indore 7.1) that were completely sensitive to all the 19 antibiotics.
Fig. 2.
IAR of slow and fast growing soybean rhizobia
Discussion
Among the routine cultural methods that are used to characterize rhizobia, it was found that with a few exceptions, all the slow growers were alkali producers and all the fast growers were acid producers. All the slow and fast growers were also negative for ketolactose test. The presence of fast growing soybean nodulating rhizobia has been reported in Indian soils but in low frequency (5 out of 60 isolates) [6] but these were non-nodulators or ineffective. In the present study, ten new strains isolated during the study were used (along with others isolated earlier or from other collections); of these, four were soil isolates (R50–R53) and six were from freshly uprooted soybean plants in the field (R1–R2 and R33–R36). Among these, nine were slow growers and one was of intermediate growth. However, fast growers were also encountered during isolations from these sites, albeit in low frequency (2–3 colonies out of 32 isolations made from each of the sites).
The slow and fast growing rhizobia fell in distinct clusters in the dendrogram based on carbon utilization pattern (Supplementary Material 4). There were 24 clusters at 70 % level of similarity and 6 clusters at 25 % level. A majority of the slow growers clustered at the bottom and were separated from the fast growers which clustered at the top. The dendrogram based on IAR (Supplementary Material 5) was also similar; most of the slow growers clustered together at the bottom of the dendrogram; clusters 3, 4 and 5 were exclusively represented by slow growers. All the fast growers except one fell in cluster 1 at the top of the dendrogram. These distinct differences led us to classify all the attributes studied by us into two groups of slow and fast growth rate so as to infer strain differences from simple biochemical criteria.
Similarity of the sequences of the 16S rRNA gene to the type strains in the Ribosomal Data Project, RDP 10 showed that slow growers belonged to both B. japonicum as well as Rhizobiumradiobacter—three slow growers, R16 (JQ514080), R33 (JQ665274) and R34 (JQ514083, Supplementary Material 1) showed highest homology with B. japonicum group (B. japonicum, B. liaoningense and B. yuanmingense) (96.4–98.1 %) while rest of the four slow growers, R19, R32, R50 and R51 (JQ514081, JQ665273, JQ514084, JQ514085 respectively) had highest similarity to R. radiobacter (86.5 % for R19 and 94.4–98.3 % for the rest). But all the fast growers R3–R5, R8–R12, R14 and R22 (JQ514071-79, JQ665272) belonged to only R.radiobacter (93.8–98.0 % homology). This meant that while all the fast growers were R. radiobacter, the slow growers could belong to B. japonicum as well as to R. radiobacter and thus simple tests other than growth rate would be required in routine quality checks to screen-out non-bradyrhizobia.
In the earlier studies one of the distinguishing features of soybean rhizobia reported was the inability of the slow growers to utilize sucrose, trehalose and raffinose [10, 11]. However, in our study, these sources were utilized by slow growers also, but to a lesser extent; utilization by fast and slow growers for sucrose was 87 and 50 %, trehalose 87 and 35 %, raffinose 73 and 30 %. Another distinguishing feature was either the inability of fast growers to utilize arabinose or grow very slowly on it [10]. In our study the proportion of fast growers utilizing d-arabinose was 7 % while that of slow growers was 20 %. But the proportion of l-arabinose utilizers among fast and slow growers was 87 and 55 % respectively.
Utilization of glycerol, lactose, maltose, sucrose, trehalose and cellobiose are usually associated with fast growers [10, 12]. In our study the fast growers could not utilize xylitol, α methyl d-mannoside, sorbose, sodium gluconate and melezitose while a small proportion of slow growers utilized them. A new finding in our study was the greater utilization of glucosamine by slow growers (33 % by fast vs. 75 % by slow) and its utility as a distinguishing criteria. It is significant that the carbon utilization pattern of the two 16S rRNA sequence types among the slow growers in Indian soils showed a distinct diverging trend. While all the three slow growing Indian strains belonging to Bradyrhizobium spp. (R16, R33, R44) could utilize glucosamine, the other three Indian strains belonging to R. radiobacter (R19, R50, R51) could not utilize it. The sole exception was the slow growing commercial isolate from USA-R32 that utilized glucosamine). Glucosamine can thus be used to select for bradyrhizobia among the slow growers in initial isolations from root nodules. It is also pertinent to note that of the ten fast growing rhizobia (R. radiobacter) sequenced, except one-R8, none could utilize glucosamine (Supplementary Materials 1, 2).
Intrinsic resistance to low concentrations of many antibiotics allows identification of strain differences while avoiding the selection of mutants with high tolerance. In our study among the 20 slow growers there were 20 different IAR patterns while among the 15 fast growers there were 9 patterns. Both slow and fast growing rhizobia showed maximum resistance to nalidixic acid and trimethoprim. Resistance to nalidixic acid was very high in fast (60 %) and slow growers (80 %). Most of the soybean rhizobia of central India were resistant to nalidixic acid, chloramphenicol and trimethoprim [7]. There was no antibiotic to which only the fast growers were resistant and to which slow growers were sensitive. In our study both slow and fast growing soybean rhizobia showed resistance towards much lower concentrations (85- to 333-fold less) of chloramphenicol, nalidixic acid, rifampin and vancomycin. The slow growing rhizobia were also resistant to kanamycin, neomycin, streptomycin, tetracycline and co-trimaxazole whereas all the fast growers were sensitive to these five antibiotics.
A majority of the effective fast growing soybean rhizobia in Brazil [13] were resistant to low level of erythromycin, kanamycin and rifampicin, and to higher levels of chloramphenicol, erythromycin, gentamicin, kanamycin, rifampicin and tetracycline. Resistance to streptomycin was low. In a study in Africa, 40 % Bradyrhizobium spp were resistant to rifampicin and 60 % to erythromycin [14]. We are not aware of any report on the complete sensitivity of the slow growers to gentamicin, erythromycin and spectinomycin and also the complete sensitivity of fast growers to gentamicin that we found in our present study. A larger proportion of slow growers were resistant to vancomycin (70 % by slow vs. 13 % by fast), polymyxin-B (65 % by slow vs. 7 % by fast) and rifampicin (55 % by slow vs. 7 % by fast) as compared to fast growers and thus they can be included in selective media to enrich for slow growing bradyrhizobia in isolations from root nodules.
To understand loss of inoculant effectiveness, it is necessary to distinguish how introduced bradyrhizobia (slow growing) and indigenous adapted rhizobia (slow as well as fast growing) differ and whether simple biochemical methods could be used to distinguish them. Santos et al. [2] reported how soybean rhizobia that were native, genetically very distant from parental strains, became good nodulators and N2 fixers, possibly via transfer of nodulation, nitrogen fixation and other competition-related genes from the inoculum strain. In our study there was close similarity of some of the slow and all fast growing rhizobia to R. radiobacter (Agrobacterium radiobacter type strain ATCC 19358). A. radiobacter was re-classified as R. radiobacter [15] and is a bonafide species [16].
Our study on indigenous adapted soybean rhizobia in central India suggests that for the purpose of enriching Bradyrhizobium strains among the slow growers during initial isolations from nodules, using carbon sources like glucosamine and antibiotics like vancomycin, polymyxin-B and rifampicin in the media would provide an easy way of distinguishing them from other slow growers belonging to R. radiobacter.
Electronic supplementary material
Acknowledgments
We are grateful to the Indian Council of Agricultural Research, New Delhi for funding this investigation under the network project on ‘Application of Microorganisms in Agriculture and Allied sectors’ (AMAAS) of the National Bureau of Agriculturally Important Microorganisms (NBAIM), Mau, U.P. India. We are grateful to Director, IISS, Bhopal for providing the facilities for this investigation. We thank Dr. Sushil K. Sharma and Dr. M. P. Sharma, Directorate of Soybean Research, Indore, M.P., India for supply of the rhizobial strains (strain nos. R21–29, Online Resource 1).
References
- 1.Herridge DF, Peoples MB, Boddey RM. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil. 2008;311:1–18. doi: 10.1007/s11104-008-9668-3. [DOI] [Google Scholar]
- 2.Santos MA, Vargas MAT, Hungria M. Characterization of soybean Bradyrhizobium strains adapted to the Brazilian savannas. FEMS Microbiol Ecol. 1999;30:261–272. doi: 10.1111/j.1574-6941.1999.tb00654.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen LS, Figueredo A, Villani H, Michajluk J, Hungria M. Diversity and symbiotic effectiveness of rhizobia isolated from field-grown soybean nodules in Paraguay. Biol Fertil Soils. 2002;35:448–457. doi: 10.1007/s00374-002-0493-1. [DOI] [Google Scholar]
- 4.Rawat AK, Rao DLN, Sahu RK. Effect of soybean inoculation with Bradyrhizobium and wheat inoculation with Azotobacter on their productivity and N turnover in a vertisol. Arch Agron Soil Sci. 2012 [Google Scholar]
- 5.Zhang YM, Li Y, Chen WF, Jr, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX. Biodiversity and biogeography of rhizobia associated with soybean plants grown in the north china plain. Appl Environ Microbiol. 2011;77:6331–6342. doi: 10.1128/AEM.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annapurna K, Balakrishnan N, Vital L. Verification and rapid identification of soybean rhizobia in Indian soils. Curr Microbiol. 2007;54:287–291. doi: 10.1007/s00284-006-0423-9. [DOI] [PubMed] [Google Scholar]
- 7.Sharma MP, Srivastava K, Sharma SK. Biochemical characterisation and metabolic diversity of soybean rhizobia isolated from Malwa region of central India. Plant Soil Environ. 2010;56:375–383. [Google Scholar]
- 8.Lindstrom K, Murwira M, Willems A, Altier N. The biodiversity of beneficial microbe-host mutualism: the case of rhizobia. Res Microbiol. 2010;16:453–463. doi: 10.1016/j.resmic.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Somasegaran P, Hoben HJ. Handbook for rhizobia: methods in legume-Rhizobium technology. New York: Springer; 1994. [Google Scholar]
- 10.Sadowsky MJ, Keyser HH, Bohlool BB. Biochemical characterization of fast and slow growing rhizobia that nodulate soybeans. Int J Syst Bacteriol. 1983;33:716–722. doi: 10.1099/00207713-33-4-716. [DOI] [Google Scholar]
- 11.Young CC, Chang JY, Chao CC. Physiological and symbiotic characteristics of Rhizobium fredii isolated from subtropical-tropical soils. Biol Fertil Soils. 1988;5:350–354. doi: 10.1007/BF00262145. [DOI] [Google Scholar]
- 12.Stowers MD, Eaglesham ARJ. Physiological and symbiotic characteristics of fast-growing Rhizobium japonicum. Plant Soil. 1984;77:3–14. doi: 10.1007/BF02182807. [DOI] [Google Scholar]
- 13.Hungria M, de O Chueire LM, Coca RG, Megias M. Preliminary characterization of fast growing rhizobial strains isolated from soyabean nodules in Brazil. Soil Biol Biochem. 2001;33:1349–1361. doi: 10.1016/S0038-0717(01)00040-2. [DOI] [Google Scholar]
- 14.Abaidoo RC, Keyser HH, Singleton PW, Borthakur D. Comparison of molecular and antibiotic resistance profile methods for the population analysis of Bradyrhizobium spp. (TGx) isolates that nodulate the new TGx soybean cultivars in Africa. J Appl Microbiol. 2002;92:109–117. doi: 10.1046/j.1365-2672.2002.01518.x. [DOI] [PubMed] [Google Scholar]
- 15.Young JM, Kuykendall LD, Martınez-Romero E, Kerr A, Sawada H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int J Syst Evol Microbiol. 2001;51:89–103. doi: 10.1099/00207713-51-3-945. [DOI] [PubMed] [Google Scholar]
- 16.Lindstrom K, Young JPW. International Committee on Systematics of Prokaryotes, Subcommittee on the taxonomy of Agrobacterium and Rhizobium, minutes of the meeting, 7 September 2010, Geneva, Switzerland. Int J Syst Evol Microbiol. 2011;61:3089–3093. doi: 10.1099/ijs.0.036913-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.