Abstract
Background
Infection with high-risk human papillomaviruses (hrHPV) is a frequent cause of cervical intraepithelial neoplasias and carcinomas. The recently developed p16/Ki67 dual-stain of cytologic preparations possesses superior specificity over current HPV testing for detecting moderate and high-grade dysplasias and can potentially be applied in routine cytology screening. Image analysis can possibly improve the efficiency of evaluating Pap smears, if dual stained nuclei are accurately localized and reliably distinguished from the background of other cells.
Methods
Here we describe a technique comprising color deconvolution, radial symmetry detector and a superpixel-based segmentation for computerized delineation of nuclei in Pap smears stained with p16/Ki67.
Results
The performance of the method was determined by the precision and recall in 99 images (n=19323 cells) and reached 0.952 and 0.958, respectively. The accuracy of delineation, assessed by the Jaccard index (n=1080 cells), was 0.794. In single cells the precision and recall was higher than in clumps (p = 0.005).
Conclusions
In summary, the new technique delineates large and small nuclei irrespectively of coloration with a significantly better performance than a method solely involving the radial symmetry detector. Therefore, it is suited to automatically define nuclear areas for quantification of nuclear biomarkers in smears.
Introduction
Cervical Pap smear is a commonly used test to detect cervical intraepithelial neoplasia (CIN), a precursor lesion of cervical cancer (1). Abnormal cells in Pap smears are diagnosed through light microscopy, which is a time consuming, labor intensive and costly process. In addition, their interpretation is confounded by significant intra- and inter-observer variability and occasional false positive and false negative diagnoses (2).
Nearly all cervical intraepithelial neoplasias (CIN) and carcinomas are caused by high-risk human papillomaviruses (hrHPV). Whereas the majority of low-grade (LG) CIN spontaneously regress without treatment, persistent infection with hrHPV is associated with progression to high-grade (HG) CIN and cervical cancer that requires treatment. P16 is a cyclin-dependent kinase inhibitor that is over-expressed in HG CIN and invasive cervical squamous carcinoma, and is clinically helpful in distinguishing LG from HG lesions (3,4). Ki67 is a proliferation marker that is also over-expressed in HG CIN (5). The two markers are combined in the CINtec® PLUS test, a dual immunostain used to resolve cytomorphological ambiguities and to improve diagnostic accuracy (6). According to the manufacturer’s guidelines the presence of ≥ 1 dual stained (CINtec® PLUS positive) cell constitutes a positive test result. CINtec® PLUS can be applied to routinely prepared and previously screened liquid-based Pap smears. Since interpretation of the stain does not rely on cytomorphology, the dual stain offers the potential for more objective and more reproducible evaluation of Pap smears and prediction of cancer risk (7).
Over-expression of p16 (p16 positivity) is visible as a brown reaction product in the nucleus and/or cytoplasm while expression of Ki67 (Ki67 positivity) is visible as a red reaction product confined to the nucleus. The coloration of p16/Ki67 dual-stained cells is distinct from either p16 or Ki67 positive cells and from negative cells. However, it is difficult to visually distinguish single dual-stained cells with co-localized brown and red colorations in the nucleus from the others.
Image analysis, when enhanced by machine learning tools, can automate selected tasks involving recognizing abnormal cells in Papanicolaou (8–10) and dual stained smears (11). However, the recognition of abnormal cells by image analysis will only succeed if nuclei of all cells on the slide are automatically found and delineated. This task is computationally intense, time consuming and prone to errors arising from variable size of cells and nuclei, coarse and irregular chromatin texture, variable nuclear to cytoplasmic area ratio, and cell clumping. Recently published advances are mostly dedicated to the computerized delineation of nuclei in images of Papanicolaou stained smears. Gentcav et al developed a multi-scale blind partitioning followed by a binary classification of partitioned regions to separate nuclei from cytoplasm (8). A seeded watershed-based method that automatically finds nuclear centroids was described by Plissiti et al (10). The same authors proposed to resolve overlapped cells or overlapped nuclei by a spatially adaptive active physical model (9). However, the coloration introduced by immunohistochemistry adds an additional level of complexity and computational challenges to the evaluation of Pap smears (11,12), and since the need for automated quantification of nuclei stained for overexpression of Ki67 and p16 is relatively new the available literature dedicated to this problem is scarce.
The demand for automated instrumentation that can reproducibly detect p16/Ki67 dual-stained cells is underscored by the increasing number of studies reporting the utility of the stain (4,6,13–15). Thus, our objective was to develop a methodology for delineation of cell nuclei of p16 and Ki67 positive cells in Pap smears. In the current study we integrate our previously reported radial symmetry transform detector (RSD) with superpixel segmentation and k-nearest neighbor (k-nn) classification to localize all nuclei and refine the boundaries between the nucleus and cytoplasm. The method can be applied irrespective of the immunostaining pattern and is not affected by the confluency of cells. To validate our approach, we compared the accuracy of the nuclear outline that is generated by the computer to the tracings of an experienced pathologist in clinical samples.
Materials
Selection of images and data acquisition
A protocol was approved by Institutional Review Board to obtain data from 10 liquid-based (SurePath™) cervical Pap smears (8 with and 2 without abnormal cells). The Papanicolaou stained smears were destained and then restained utilizing the CINtec® PLUS dual staining reagent kit (Ventana Medical Systems, Tuscon, AZ) as per the manufacturer’s recommendations and as previously described (4). To destain the Papanicolaou stained smears, our immunohistochemistry lab utilized the following protocol: Xylene (5min), Xylene (5min), 100% Ethanol (5min), 95% Ethanol (5min), 70% Ethanol (5min), 50% Ethanol (5min), distilled water, 1% HCl in 70% Ethanol to remove haematoxylin, wash in distilled water, followed by an incubation for 40 min in a 100° citrate buffer (mild antigen retrieval). Variants of this protocol are commonly used to destain cytopathology smears (16,17).
In each slide pathologists marked ≥5 and up to 20 areas with abnormal cells, which were selected for imaging. In the negative slides the selection of areas (n=10) was arbitrary. Imaging was performed on the whole slide imaging platform, Vectra 2 (Perkin-Elmer, CA), equipped with a scientific-grade CCD color camera and image acquisition software (Nuance). 20× objective, 4ms exposure time and 1×1 pixel binning were set to acquire a single RGB color image array with 1040×1392 pixels (pixel size = 0.5µm×0.5µm) in horizontal and vertical direction from each selected area. During the acquisition the images were flat-field corrected. A total of 99 color images containing approximately twenty thousand cells were acquired. Each image in a 0–255 scale (24-bit image file) was exported as an uncompressed TIFF format for subsequent analyses.
Ground truth nuclear outlines and training features (roundness and color)
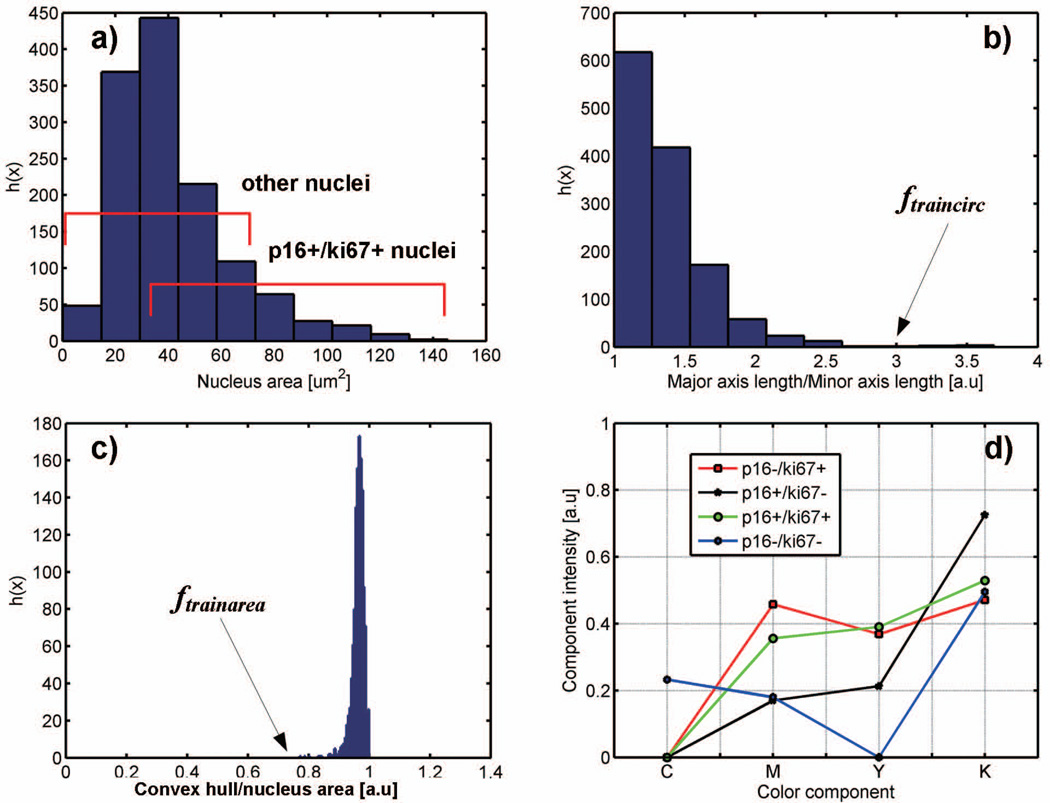
One of the goals was to devise a reliable method that can accurately capture the shape and coloration of nuclei. In ten images (5 with and 5 without abnormal cells) randomly selected from the set of 99 images a total of 1080 nuclei were manually outlined by a pathologist, and the nuclear areas were determined. These nuclei were used in the validation study. The rest of images were used to develop the segmentation algorithm. The first step in the development of the segmentation algorithm was to define thresholds of nuclear area and roundness. These parameters were arbitrarily established through boundaries of distributions of the nuclear area and roundness of annotated nuclei. The roundness was defined by: (1) major to minor nuclear axis ratio, and (2) by ratio of the convex hull to the full area of the nucleus, where the convex hull is a smallest possible area enclosing the nucleus by a non-concave polygon. Fig.1a-c show the distributions of nuclear area, major to minor axis ratio and ratio of the convex hull to the full nuclear area for 1080 nuclei. Nuclei with a major/minor axis length ratio ≤ 3 (named ftraincirc) and a convex hull/nuclear area > 0.75 (named ftrainarea) comprised 99.4% of all nuclei. Approximately 78% of these possessed the ratio of major to minor axis <1.5, which indicated a circular contour. The rest of the nuclei had elongated shapes. The ftrainarea and ftraincirc were used to separate shapes of candidate nuclei from non-nuclear shapes obtained during the segmentation process. The average nuclear area, Aavr=41µm2 (202.3 pixels), was derived from Fig. 1a and used to determine the window W for superpixel segmentation.
Figure 1.
Histograms of shape features in manually delineated nuclei: a) nuclear area, b) major axis length to minor axis length ratio and the ftraincirc cutoff, c) convex hull to nuclear area ratio and the ftrainarea cutoff, d) averaged CMYK intensities of nuclear immunoreactivity in 4 IHC staining patterns.
Modeling of chromogenic staining in CMYK (cyan/magenta/yellow/black) color space has been shown to be well suited for observer independent, reproducible and high-throughput image analysis tasks (18). We retrieved colorimetric features from 100 nuclei (a subset of 1080) and 25 cytoplasmic areas. Each feature was obtained by first converting colors of all pixels from the RGB space to the CMYK (cyan/magenta/yellow/black) space and then averaging across all pixels to obtain one CMYK pattern for one nucleus. Nuclear patterns were organized into 4 groups of 25 nuclei each: Ki67 positive (p16−/Ki67+)N, p16 positive (p16+/Ki67−)N, p16 and Ki67 dual positive (p16+/Ki67+)N, and dual negative (p16−/Ki67−)N. The 2 cytoplasmic staining patterns were derived from 10 (p16+/Ki67−)C and 15 (p16−/Ki67−)C cells in a similar manner. The selection of nuclear and cytoplasmic areas was arbitrary in order to take into account the diversity of staining intensities (lighter to darker colors) in groups of the nuclear and cytoplasmic p16/Ki67 patterns. Altogether 125 patterns formed a training set ftrainCMYK that allowed the classification of superpixels to nuclear or cytoplasmic compartments.
Methods
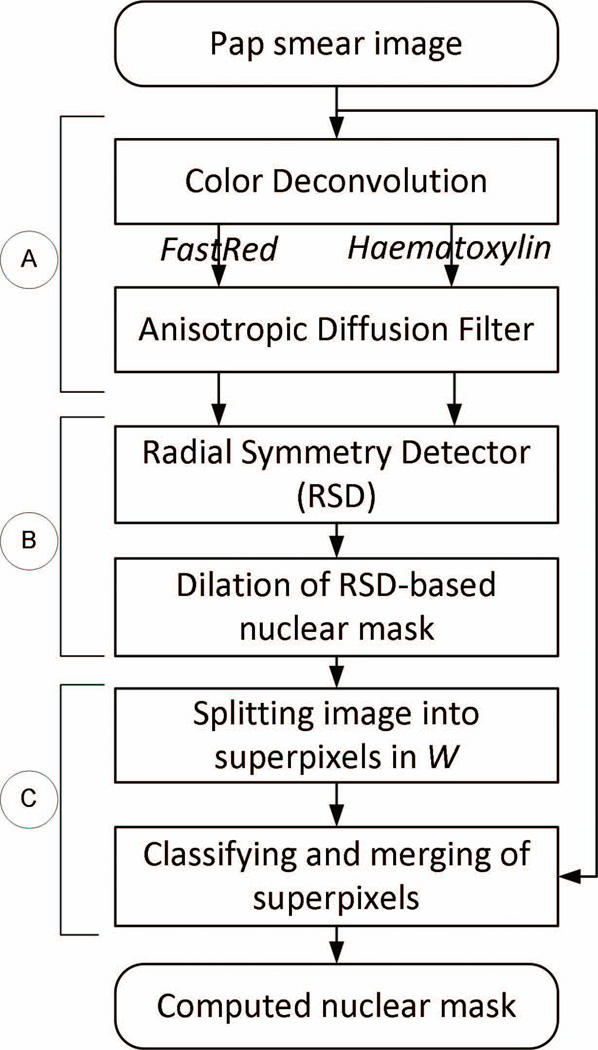
The overall approach to outline nuclei illustrated in Fig. 2, involved three sequential steps: (A) pre-processing with color deconvolution and anisotropic diffusion smoothing to enhance edges between nuclei and cytoplasm, (B) application of the radial symmetry-based detector to generate a preliminary mask of nuclei followed by mask dilation, and (C) simple linear iterative clustering (SLIC) to split small image regions into superpixels, and to combine superpixels of similar colorimetric patterns using the k-nn classification.
Figure 2.
Overview of analytical workflow. Images of nuclear stains deconvolved from the original image were smoothened (A), and then processed by RSD to obtain a preliminary mask of nuclei (B). SLIC superpixels were implemented to correctly determine the boundary between nucleus and cytoplasm (nuclear membrane). The SLIC was performed in windows W positioned around the nuclear membrane of artificially expanded (dilated) nuclei from step (B), followed by k-nn clustering to combine neighboring superpixels with colorimetric patterns of high similarity (C).
(A) Pre-processing and preliminary detection of nuclei
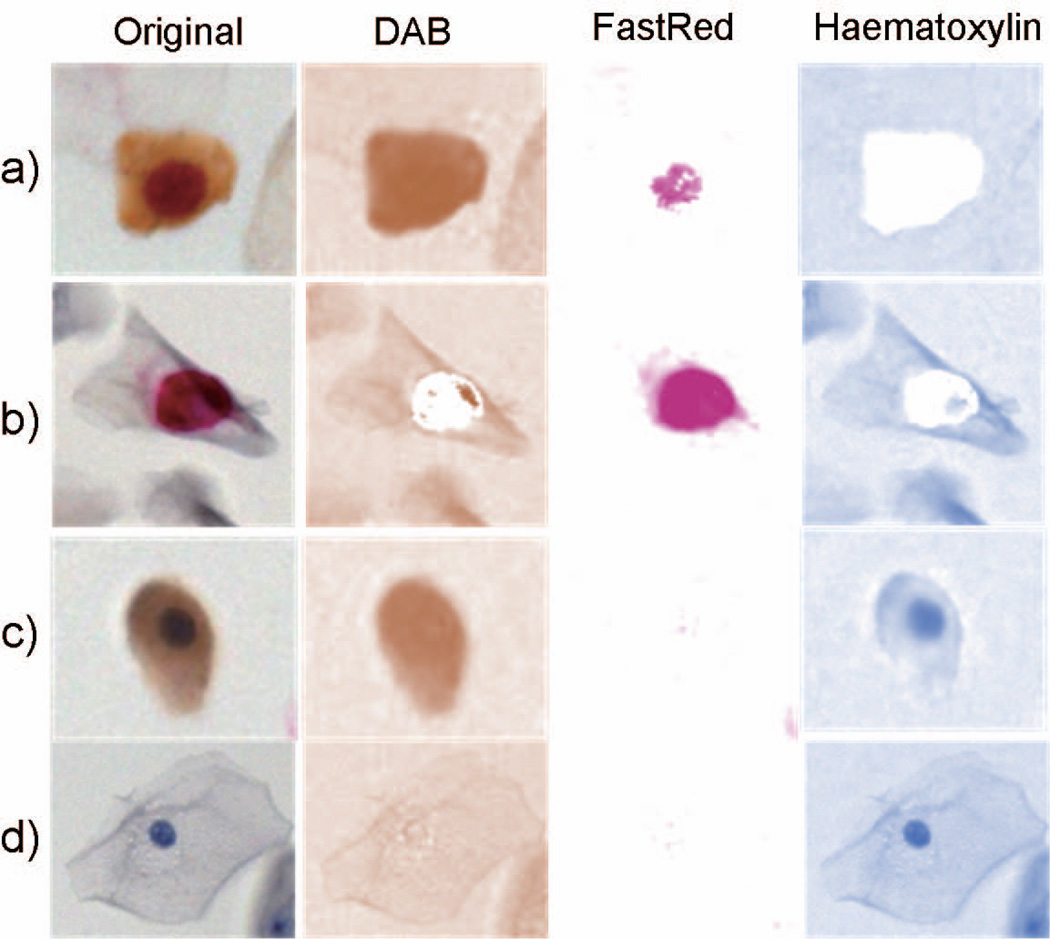
For the detection of p16 and Ki67, DAB and FastRed chromogens are converted to brown (p16) and red (Ki67) colors. The pre-processing began with a separation of original RGB images into monochromatic images of DAB, FastRed and haematoxylin through a color deconvolution algorithm (19) within the open source image manipulation platform (ImageJ) (20). Three chromogenic fingerprints that specify the RGB spectrum of DAB=[0.268, 0.570, 0.776], FastRed=[0.214, 0.851, 0.478], and Haematoxylin=[0.490, 0.769, 0.410] were used for the deconvolution. Haematoxylin and FastRed images were retained for further processing, while the DAB image was discarded due to lack of distinction between the nuclear and cytoplasmic signals (Fig. 3a and c in DAB column). The Perona-Malik anisotropic diffusion (11,21) was applied to improve the contrast between nucleus and cytoplasm and to smoothen the nuclear texture in FastRed and Haematoxylin images.
Figure 3.
Color deconvolution in images of epithelial cells with different staining patterns: a) p16+/Ki67+, b) p16−/Ki67+, c) p16+/Ki67−, and d) p16−/Ki67− . Respective columns labeled DAB and FastRed display localization of individual stains after deconvolution demonstrating that only FastRed and Haematoxylin images are suitable for nuclear segmentation.
(B) Application of radial symmetry detector and mask dilation
In the second step outlined in Fig. 2, we began by applying our previously published method to identify nuclei based on circularity [12], which utilized an adjusted radial symmetry operator for processing Pap smear images. This operator captured gray-level images with high intensities in nuclear areas and low intensities anywhere else. Nuclei were located by adaptive thresholding of the gray-level image to generate a binary mask and the mask was combined with the radial symmetry operator to form a radial symmetry detector (RSD). In this study we expand our previous method by embedding the RSD into the analytical workflow (Fig. 2) for preliminary detection of nuclei and concatenated the binary mask into a RSD-based nuclear mask.
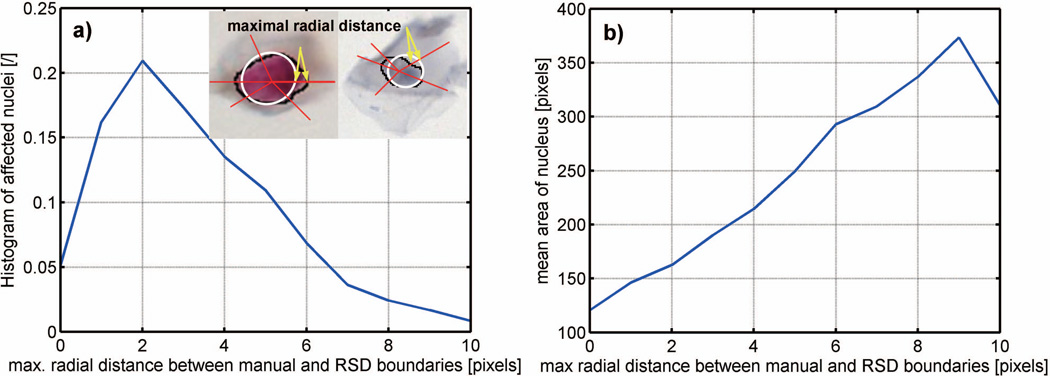
Although the success rate of RSD to localize nuclei was high, a discrepancy between the RSD-based and the ground-truth nuclear contours occurred in a population of nuclei that were poorly contrasted or had a low circularity. In order to provide a more reliable delineation of nuclei an additional processing step was introduced (Fig. 2, step C). Its main component is a superpixel-based analysis in a small window W around the RSD-based nuclear mask. In order to determine the size of W we first dilated the RSD-based nuclear mask to assure that it entirely covered the nucleus and also permitted the inclusion of adjacent cytoplasm. To prevent excessive dilation, the number of pixels added to the mask was controlled by a square structuring element which was empirically adjusted to a size of K. The borderline values of K were set based on the maximal radial error measured between the manual outline and outlines generated by RSD-based nuclear mask (Fig. 4). The error of the computed nuclear outline relative to the manual outline was determined along multiple lines originating at the center of the nucleus and crossing both the manual and the RSD outlines (Fig. 4 insert). The maximum error - the greatest difference between two outlines - was used as the ultimate error value for a given nucleus. In 50% of nuclei the maximal radial error was ≤ 3 pixels, and the error increased with nuclear size, reaching 10 pixels for the largest nuclei.
Figure 4.
Distribution of maximal radial distance error measured between RSD-based and pathologist outlines: (a) histogram of affected nuclei, and (b) maximal radial error as a function of mean nuclear area. The two nuclei in the insert depict the error between the RSD-based (white line) and pathologist (black line) outlines. Yellow arrows show where maximal distances occurred.
To prevent excessive dilation of the RSD-based mask the structuring element was set as: , where: round is the function returning nearest integer, A is the area under the RSD-computed mask, and Aavr is the average nuclear area obtained from manual outlines (Fig.1a). The window size was adjusted by expanding the bounding box (the smallest possible rectangle around the RSD-based nuclear mask) to include the entire nucleus and the adjacent cytoplasm by a factor of 2.5 or by a fixed value of 250 pixels in both directions whichever yielded a smaller window area.
(C) Classification and merging of superpixels
(C.1) Implementation of SLIC
Superpixels are clusters of pixels formed by combining neighboring pixels in an image into small regions of arbitrary shape. The borders of superpixels are defined by changes in coloration. The proximity and color information in a superpixel are used as descriptors. We utilized the SLIC technique (22) because of its good segmentation performance, low computational complexity (O(n)), and superior recognition of boundaries when compared with other superpixel-based methods. Briefly, SLIC uses multiple parameters defined by the color components and pixel coordinates to iteratively partition an image into a predetermined number of superpixels (Nsp). The initial partitioning and placement of the center of a superpixel was derived from color gradients and a predefined interval where: N is the total number of pixels in the region, and Nsp is the number of superpixels provided by the user. Boundaries and centers of a superpixel fluctuate until their positions remain unchanged in two consecutive iterations. An in-depth description of SLIC can be found in (23). In our approach SLIC was applied to the rectangular area W (Fig. 5d) centered on the RSD-based nuclear mask (Fig. 5a and b). An ideally partitioned W should contain a very small number of superpixels (ideally one) that entirely cover the nucleus and several additional extra-nuclear superpixels (Fig. 5d). In order to optimally determine the number of superpixels in W, the number of superpixels was set as the ratio of the number of pixels in W and the pixels in the area under the dilated RSD-based mask: Nsp = card(W)/Ak.
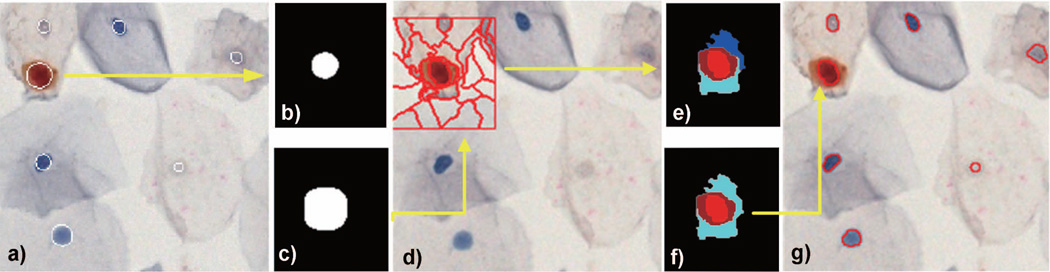
Figure 5.
Superpixels segmentation, classification and merging in Pap smear images: (a) computed RDS-based nuclear mask superimposed on the digital image, b) RSD-based mask of a p16+/Ki67+ nucleus, c) dilated RSD-based mask, d) window W around the dilated mask split into superpixels, e) the k-nn classification was used to assign superpixels to nuclear (red) and cytoplasmic (cyan, blue and brown) locations, f) non-circular and non-convex superpixels were removed and the boundary of the remaining superpixel were superimposed on the digital image (g). Note the difference between the RSD-based mask – nuclei approximated by circles in a) and the fine boundary of nuclear contour in g) which better represents the location of boundary between the nuclear envelope and the cytoplasm.
(C.2) Selection, classification and merging of superpixels
To accurately delineate nuclear superpixels within W, those that overlapped with the dilated nuclear mask were selected for further analysis, whereas the others were discarded. Intensities of each involved superpixel (Fig. 5e) were converted from the RGB to the CMYK color space, and then averaged across respective colors to form a feature vector, fCMYK. Prior to their classification ftrainCMYK vectors in the library were normalized to the zero mean and unit standard deviation in each color channel. The means and standard deviations were used to also normalize color components of fCMYK. A k-nn classifier was implemented to determine the nuclear or cytoplasmic origin of fCMYK vectors. To classify the fCMYK vectors, each vector was compared to the library of vectors ftrainCMYK with known cytoplasmic and nuclear origins (see Materials). The vectors in the library were ranked based on their similarity to the fCMYK of unknown origin. The highest frequency in subcellular origin (nucleus or cytoplasm) of the top 9 ranked vectors (k-nn neighborhood) was used to assign the unknown fCMYK vector either to either a nuclear or cytoplasmic location. Since the ranked vectors also contain intrinsic information about p16 and Ki67 status the comparison to the ftrainCMYK vectors provides additional information about the localization and expression of p16 and Ki67. p16 positive vectors can be either nuclear or cytoplasmic, while Ki67 positive or p16/Ki67 double positive vectors are exclusively nuclear.
After their classification, adjacent superpixels with the same colors were merged (Fig. 5f), and analyzed for roundness by calculating the major to minor axis ratio (fcirc) and the ratio of convex hull to the total area of the merged superpixels (farea). If fcirc≤ftraincirc and farea>ftrainarea, the merged region was accepted; otherwise it was discarded. If none of merged superpixels fulfilled the roundness criteria, the RSD-based was used as the final product of the computed nuclear mask (Fig. 5g).
Validation
The performance of the novel nuclear segmentation method to correctly outline nuclei was evaluated by the nuclear area agreement – a direct comparison of computed nuclear masks with masks from manual delineations. Further, the accuracy of nuclear detection was determined by using the precision and recall measures.
The accuracy of the computed nuclear outlines was measured in 99 images. A subset of 10 images was chosen to represent the spectrum of p16/Ki67 expression patterns and contained 1080 nuclei that were manually delineated by a pathologist. The nuclear area agreement between the computed mask (C) and manual ground truth tracings (G) was evaluated by the overlap measure: , and the Jaccard index: ; where ∩ and ∪ represent the intersection and union of binary images, with the summation of involved pixels across the window W. For perfectly overlapping masks both Ao and J reach 1.
The accuracy of nuclear detection was evaluated by the precision = TP/(TP + FP) and recall = TP/(TP + FN) (sensitivity) in the remaining 89 images, in which a pathologist identified TP, FP and FN nuclei. In addition, the Ao measure was used for nuclei that were partially outlined by the algorithm. If the outline encompassed < 50% of the manually outlined nuclear area, the result was counted as a FN, otherwise it was considered a TP. The evaluations of detection accuracy were only based on TP, FP and FN since TN rates are not required to assess the performance of a detector, which detects single events, in our case, the presence of a nucleus. Calculating TN rates would be required to manually delineate all objects that are not nuclei (including areas of cytoplasm, artifacts etc.), which is impractical. The chi-square test for categorical data involving TP and FN detections was applied in order to test the null hypotheses that no association exists between the detection performance of the newly proposed technique and (a) the nucleus size, (b) clumping of cells, and (c) our previously published nuclei detection technique.
Results
We developed a method that utilizes radial symmetries and superpixels combined with morphological and colorimetric characteristics to delineate epithelial cell nuclei in Pap smears. To develop and then validate the segmentation algorithm we used a set of ten (out of 99) images in which nuclear contours from 1080 cells, (327 immunoreactive and 753 non-immunoreactive), were outlined by a pathologist (See Materials). The same images (without outlines) were first used to determine the distributions of size, shape, and color CMYK characteristics (Fig. 1) for the development of the segmentation algorithm and the outlines of computed nuclear masks were compared to the manual outlines of a pathologist in the validation phase of the project.
To assess the accuracy of the segmentation algorithm in delineating nuclei, a quantitative comparison of computer generated outlines vs. manual outlines in the 10 images yielded averaged Ao = 0.889 and J=0.794 for all 1080 nuclei, and Ao = 0.901 and J = 0.807 for large nuclei - with areas > Aavr. (33.8% of all nuclei) (Table 1).
Table 1.
Performance of nuclei detection in the set of nuclei manually delineated by the pathologist.
| Measure | p16+/Ki67+ | p16+/Ki67− | p16−/Ki67+ | p16−/Ki67− | Non-clustered cells |
Clustered cells |
All cells |
|---|---|---|---|---|---|---|---|
| Ao | 0.891 | 0.88 | 0.914 | 0.898 | 0.915 | 0.868 | 0.889 |
| J | 0.832 | 0.810 | 0.835 | 0.820 | 0.815 | 0.772 | 0.794 |
The segmentation procedure was also tested on the remaining 89 images containing 19323 cells to determine the precisions and recalls of the nuclear detection and to juxtapose them to those obtained by our previously developed method (Table2). The precision and recall rates for all nuclei were 0.952 and 0.958. TP and FN rates, for combined single positive and negative nuclei were 0.957 and 0.049, respectively. In the next step we tested whether nuclear size and cell clumping affect the segmentation performance (Table. 2) TP and FN rates evaluated by the Chi-square test showed that the difference in the detection performances of small and large nuclei was not significant at the p=0.1 level, suggesting that the detection performance was not significantly affected by nuclear size. On the other hand, the difference in the detection performances of our previously published method and the proposed one were significant at the significance level p=0.005 both in individual and clumped cells. These suggest that the present method outperformed the previous one. (Table 2). Example nuclei segmentation results are shown in Figures 6 and 7.
Table 2.
Performance of nuclear detection in 89 Pap smear images containing 19323 cells.
| Nucleus category | TP | FP | FN | Precision | Recall |
|---|---|---|---|---|---|
| All nuclei (old method (Gertych et al., 2012)) | 3933 | 241 | 295 | 0.942 | 0.930 |
| All nuclei (current method) | 18512 | 934 | 811 | 0.952 | 0.958 |
| Large: A ≥ Aavr | 6108 | 297 | 265 | 0.953 | 0.958 |
| Small: A < Aavr | 12404 | 636 | 546 | 0.951 | 0.957 |
| In single cells | 10830 | 395 | 340 | 0.964 | 0.969 |
| In clumps | 7682 | 538 | 471 | 0.934 | 0.942 |
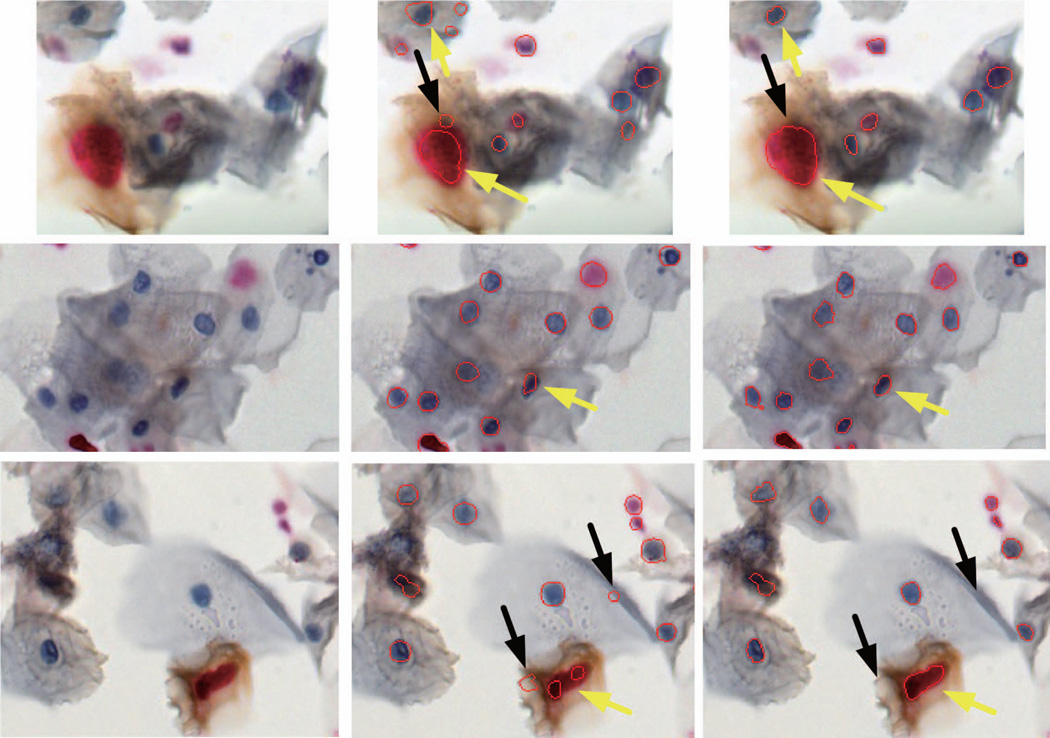
Figure 6.
Example segmentations of nuclei in Pap smears stained for Ki67 and p16. Boundaries of automatically delineated nuclei are marked in red. Left column - original images, middle column - results from RSD – our previously developed algorithm, and right column – final output of the proposed method involving RSD combined with superpixel analysis. Yellow arrows (in the right column) show contour detection improvement after implementation of superpixels. Black arrows point onto areas detected by the RSD that were removed during the superpixels analysis. Note the presence of individual and clumped cells, and differences in size, shape, coloration and chromatin texture of nuclei. Images were recorded for 20× magnification.
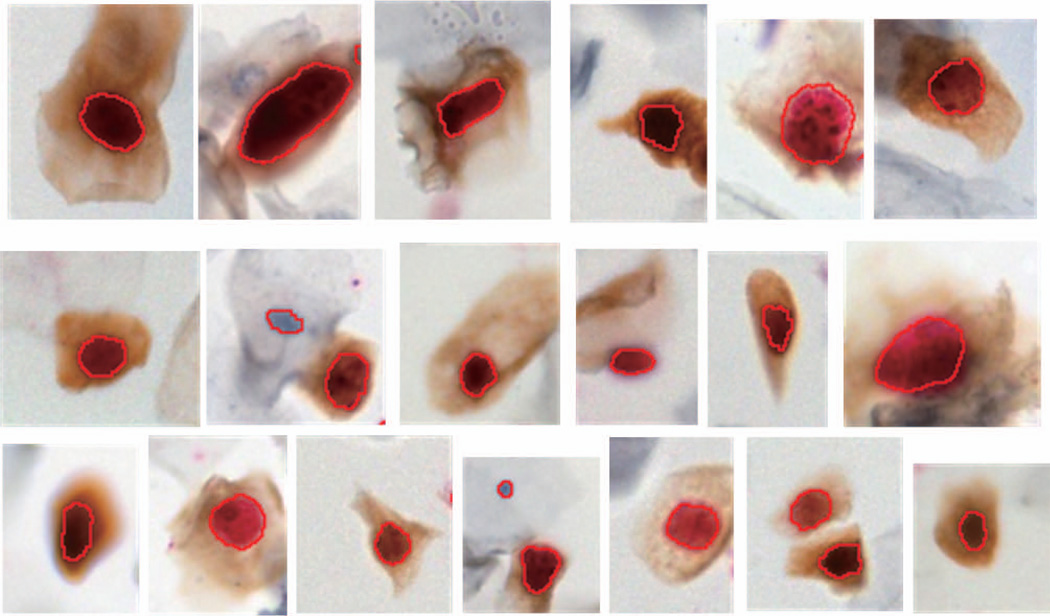
Figure 7.
Segmentations of p16+/Ki67+ positive cells. Note the diversity of staining intensities in the cytoplasm and nucleus and complexity of nuclear morphology.
Our algorithm was implemented in Matlab R13 (Matlab, Natick, MA) on a PC based computer with dual core 2.8GHhz processor. The computational cost of the analyses was evaluated by measuring time performance of processing steps outlined in Fig. 2. The average analysis time was 43sec for an image containing on average 215 nuclei. Of this time, 15% was attributed to the color deconvolution and anisotropic diffusion smoothing, 49% to RSD and the remaining 36% (0.06sec per nucleus) to RSD mask dilation, window W sizing, SLIC segmentation, and classification and merging in windows W. Since this lab-developed code was neither optimized for speed nor parallelized, we believe it is possible to obtain a much better processing performance through code parallelization and low-level programming.
Discussion
It has been shown that although the sensitivities of CINtec® PLUS and the screen for high-risk HPV genotypes are similar, the specificity for detecting CIN2+ is much higher for CINtec® PLUS. There are also several large studies supporting the potential use of p16/Ki-67 dual-stained cytology (7,15–17,24). CINtec® PLUS is already available for clinical use in Europe, Asia, Latin America, and Canada, and the manufacturer is planning clinical trials with the goal of obtaining FDA approval for clinical use of CINtec® PLUS in liquid based cytology in the U.S. Thus, we consider the likelihood of future clinical implementation of CINtec® PLUS in the U.S. to be high. Given that standard Pap smears can be, and in some laboratories already are, evaluated with the use of intelligent devices, it is also likely that smears stained for biomarkers (p16/Ki67 and/or others) could be analyzed using specifically designed hardware/software.
The goal of this study was to automate the segmentation of nuclei for enumeration of dual p16 and Ki67 positive cells in Pap smears. Pap smears from the archives of the Cedars-Sinai Medical Center Department of Pathology and Laboratory Medicine provided the image data to develop and test a novel algorithm. Our combined RSD-SLIC methodology outperformed the RSD algorithm reported in our previous study (11) and as shown in Figure 5 constitutes the main contribution in this paper. The precision and recall improved by as much as 6.7% and 7.1%, respectively, mainly through separation of the original image into Haematoxylin and FastRed images during the preprocessing steps. The improved precision and recall is equivalent to the decrease in FN and FP detections. Remaining FN detections were attributed to: (1) weak nuclear haematoxylin staining comparable to staining in cytoplasm, (2) strong p16 staining in nuclei and cytoplasm of p16+/Ki67− cells, which resulted in the delineation of whole cells instead of their nuclei, or (3) partially segmented nuclei in all types of cells. FP detections occurred in debris, corners or folded cell membranes mimicking circular shapes, fragments of nuclei, as well as in dense or thick clumps with several layers of cells.
The nuclear patterns were collected from cells with diverse color intensities spanning between extremes of bright and dark colors, and the training set was kept small – 125 randomly selected nuclear patterns to train the classifier. In contrast to many tissue-based analyses, our approach is not subject to classifier overfitting concerns because our testing set is more than tenfold larger than the training set. Although we did not perform any studies to measure the effect of training set selection on the overall algorithm performance, one should note that only a small and well defined set of nucleus-cytoplasm color pairs exists in the p16/Ki67 stained smears. These are: a) dark blue nucleus and light blue cytoplasm in p16−/Ki67− cells, b) dark brown nucleus over a light brown cytoplasm in p16+/Ki67− cells, c) red nucleus over blue cytoplasm in p16−/Ki67+ cells, and d) mixed red and brown colors in the nucleus with a variety of brown cytoplasm shades in p16+/Ki67+ cells. Of these, only the latter case can be challenging in distinguishing nuclear superpixels from cytoplasmic superpixels because the colorimetric patterns of p16+/Ki67+ and p16+/Ki67− are close in the CMYK color space. As shown in Fig 7 the training set dealt with mixed shades of brown and red colors in p16+/Ki67+ cells from different patients. Similar delineation performances were found in cells of the remaining three colorimetric patterns.
The parameters governing the superpixel segmentation such as the window size W and the number of superpixels Nsp determined by dilation kernel K are more critical to the accuracy of delineation than the selection of colorimetric patterns. Nearly all FN detections were attributed to an inaccurate split of W by superpixels which was followed by the delineation of nucleus and some adjacent cytoplasm. In order to avoid an ad hoc assignment of W, K, Nsp, Aavr, ftraincirc and ftrainarea, we empirically derived them from the ground truth cells. Given the involved number of slides for training and the complexity of the proposed analytical assay, our attention was mainly focused on the proper delineation of a large number of nuclei. Assuming that there are no two cells or two cell clusters alike, our current validation seems to be sufficient before our tool will be advanced to the next step involving a statistically significant number of cases. Currently the analysis of an image with moderate cell confluency (215 cells) takes 43s using our current version of the software. Since the circular area that contains the cellular specimen encompasses 342 FOVs per slide, at this cell confluency the analysis would require 245 min. However, after implementation of image analysis procedures in a low-level programming language, code parallelization and optimization, we anticipate that the analysis time for an image (and for a slide as well) could be decreased to 10% of the current time. The multispectral imager utilized in this study (Vectra II) acquires FOVs sequentially, requiring about 30 min to scan one slide. With the optimized software the system can analyze one FOV while another FOV is being scanned. Thus, analysis of a slide could be completed soon after the last FOV is captured.
Analysis of a large set of normal and abnormal cells showed that this novel semi-supervised image processing tool can delineate nuclei almost as well as the human observer. Compared to our previous study (11), this method improves the Jaccard index by as much as 10.1%. Even with the improved algorithms shown here, there are still discrepancies from the pathologists’ outlines, which can be attributed to small local contour variations. Achieving better performance of contour delineation by means of the proposed analytical framework, and perhaps other frameworks developed for this purpose, would be very challenging for two reasons: a) the imperfection and subjectivity of manual tracing itself does not always guarantee a perfect overlap with the underlying border between the nucleus and cytoplasm, and b) as imaging hardware automatically adjusts image focus for the entire field of view, it may introduce poor local contrast that will confuse both manual and machine-based raters.
Conclusions
In this paper we describe how we have improved the computerized delineation of nuclei in Pap smears stained with dual immunohistochemical stains. Experimental results suggest that our concept of linking radial symmetries and superpixels is well suited for automated profiling of dual p16/Ki67 overexpression and achieves an accuracy of detection close to that of humans. Based on the excellent performance in Pap smears, the method can aid in the screening of Pap smears for p16 and Ki67 positive cells in a high-throughput manner. Given the flexible framework, our new method can also serve as a starting point for the development of other new nuclear detection algorithms for cytologic preparations that are stained with two probes and a counterstain.
Acknowledgements
This work was supported in part by a grant from the Department of Surgery at Cedars-Sinai Medical Center, and in part by a NIH/NCI grant 5R21CA143618-02 (to AG). AW was supported by the Department of Pathology and Laboratory Medicine at Cedars-Sinai Medical Center. BSK was supported for this work by NIH/NCI grant 5R01CA131255-05 and by funding from the Departments of Biomedical Sciences and Pathology and Laboratory Medicine at Cedars-Sinai Medical Center.
Footnotes
Conflict of Interest
The authors declared no potential conflicts of interest with respect to the authorship and publication of this article.
References
- 1.Sirovich BE, Welch HG. The frequency of Pap smear screening in the United States. J Gen Intern Med. 2004;19:243–250. doi: 10.1111/j.1525-1497.2004.21107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nanda K, McCrory DC, Myers ER, Bastian LA, Hasselblad V, Hickey JD, Matchar DB. Accuracy of the Papanicolaou test in screening for and follow-up of cervical cytologic abnormalities: a systematic review. Ann Intern Med. 2000;132:810–819. doi: 10.7326/0003-4819-132-10-200005160-00009. [DOI] [PubMed] [Google Scholar]
- 3.Cuschieri K, Wentzensen N. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2008;17:2536–2545. doi: 10.1158/1055-9965.EPI-08-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loghavi S, Walts AE, Bose S. CINtec(R) PLUS dual immunostain: a triage tool for cervical pap smears with atypical squamous cells of undetermined significance and low grade squamous intraepithelial lesion. Diagn Cytopathol. 2013;41:582–587. doi: 10.1002/dc.22900. [DOI] [PubMed] [Google Scholar]
- 5.Nam EJ, Kim JW, Hong JW, Jang HS, Lee SY, Jang SY, Lee DW, Kim SW, Kim JH, Kim YT, et al. Expression of the p16 and Ki-67 in relation to the grade of cervical intraepithelial neoplasia and high-risk human papillomavirus infection. J Gynecol Oncol. 2008;19:162–168. doi: 10.3802/jgo.2008.19.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walts AE, Lechago J, Bose S. P16 and Ki67 immunostaining is a useful adjunct in the assessment of biopsies for HPV-associated anal intraepithelial neoplasia. Am J Surg Pathol. 2006;30:795–801. doi: 10.1097/01.pas.0000208283.14044.a9. [DOI] [PubMed] [Google Scholar]
- 7.Ikenberg H, Bergeron C, Schmidt D, Griesser H, Alameda F, Angeloni C, Bogers J, Dachez R, Denton K, Hariri J, et al. Screening for cervical cancer precursors with p16/Ki-67 dual-stained cytology: results of the PALMS study. J Natl Cancer Inst. 2013;105:1550–1557. doi: 10.1093/jnci/djt235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genctav A, Aksoy S, Onder S. Unsupervised segmentation and classification of cervical cell images. Pattern Recognition. 2012;45:4151–4168. [Google Scholar]
- 9.Plissiti ME, Nikou C. Overlapping cell nuclei segmentation using a spatially adaptive active physical model. IEEE Trans Image Process. 2012;21:4568–4580. doi: 10.1109/TIP.2012.2206041. [DOI] [PubMed] [Google Scholar]
- 10.Plissiti ME, Nikou C, Charchanti A. Combining shape, texture and intensity features for cell nuclei extraction in Pap smear images. Pattern Recogn. Lett. 2011;32:838–853. [Google Scholar]
- 11.Gertych A, Joseph AO, Walts AE, Bose S. Automated detection of dual p16/Ki67 nuclear immunoreactivity in liquid-based Pap tests for improved cervical cancer risk stratification. Ann Biomed Eng. 2012;40:1192–1204. doi: 10.1007/s10439-011-0498-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wentzensen N, Bergeron C, Cas F, Eschenbach D, Vinokurova S, von Knebel Doeberitz M. Evaluation of a nuclear score for p16INK4a-stained cervical squamous cells in liquid-based cytology samples. Cancer. 2005;105:461–467. doi: 10.1002/cncr.21378. [DOI] [PubMed] [Google Scholar]
- 13.Walts AE, Bose S. p16, Ki-67, and BD ProExC immunostaining: a practical approach for diagnosis of cervical intraepithelial neoplasia. Hum Pathol. 2009;40:957–964. doi: 10.1016/j.humpath.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Reuschenbach M, Clad A, von Knebel Doeberitz C, Wentzensen N, Rahmsdorf J, Schaffrath F, Griesser H, Freudenberg N, von Knebel Doeberitz M. Performance of p16INK4a-cytology, HPV mRNA, and HPV DNA testing to identify high grade cervical dysplasia in women with abnormal screening results. Gynecol Oncol. 2010;119:98–105. doi: 10.1016/j.ygyno.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Wentzensen N, Schwartz L, Zuna RE, Smith K, Mathews C, Gold MA, Allen RA, Zhang R, Dunn ST, Walker JL, et al. Performance of p16/Ki-67 immunostaining to detect cervical cancer precursors in a colposcopy referral population. Clin Cancer Res. 2012;18:4154–4162. doi: 10.1158/1078-0432.CCR-12-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birner P, Bachtiary B, Dreier B, Schindl M, Joura EA, Breitenecker G, Oberhuber G. Signal-amplified colorimetric in situ hybridization for assessment of human papillomavirus infection in cervical lesions. Mod Pathol. 2001;14:702–709. doi: 10.1038/modpathol.3880375. [DOI] [PubMed] [Google Scholar]
- 17.Ylitalo N, Bergstrom T, Gyllensten U. Detection of genital human papillomavirus by single-tube nested PCR and type-specific oligonucleotide hybridization. J Clin Microbiol. 1995;33:1822–1828. doi: 10.1128/jcm.33.7.1822-1828.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham NA, Morrison A, Schwock J, Aviel-Ronen S, Iakovlev V, Tsao MS, Ho J, Hedley DW. Quantitative image analysis of immunohistochemical stains using a CMYK color model. Diagn Pathol. 2007;2:8. doi: 10.1186/1746-1596-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perona P, Malik J. Scale-Space and Edge Detection Using Anisotropic Diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990;12:629–639. [Google Scholar]
- 22.Achanta R. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012;34:2274–2282. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 23.Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. SLIC Superpixels. 2010 doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 24.Wentzensen N, Fetterman B, Tokugawa D, Schiffman M, Castle PE, Wood SN, Stiemerling E, Poitras N, Lorey T, Kinney W. Interobserver reproducibility and accuracy of p16/Ki-67 dual-stain cytology in cervical cancer screening. Cancer Cytopathol. 2014 doi: 10.1002/cncy.21473. [DOI] [PubMed] [Google Scholar]