Abstract
The application of machine learning to epilepsy can be used both to develop clinically useful computer-aided diagnostic tools, and to reveal pathologically relevant insights into the disease. Such studies most frequently use neurologically normal patients as the control group to maximize the pathologic insight yielded from the model. This practice yields potentially inflated accuracy because the groups are quite dissimilar. A few manuscripts, however, opt to mimic the clinical comparison of epilepsy to non-epileptic seizures, an approach we believe to be more clinically realistic. In this manuscript, we describe the relative merits of each control group. We demonstrate that in our clinical quality FDG-PET database the performance achieved was similar using each control group. Based on these results, we find that the choice of control group likely does not hinder the reported performance. We argue that clinically applicable computer-aided diagnostic tools for epilepsy must directly address the clinical challenge of distinguishing patients with epilepsy from those with non-epileptic seizures.
Keywords: controls, FDG-PET, epilepsy, non-epileptic seizures, machine learning, neuroimaging
I. Introduction
Machine learning (ML) has proven clinically useful in many aspects of the diagnosis and pathologic characterization of epilepsy. For decades, seizure detection and prediction algorithms have been applied to scalp and intracranial electroencephalography (EEG) to characterize each patient's seizures [1, 2]. The seizure detection algorithms are integrated regularly into the clinical software for EEG review. More recently, ML has been applied to scalp EEG, intracranial EEG, structural and diffusion MRI and FDG-PET to diagnose epilepsy compared to both patients with non-epileptic seizures (PWN) and seizure-naïve, neurologically normal patients (NNPs) [3-9]. These applications helped to provide meaningful pathological insight into the features of epilepsy. They likely also have the potential to become computer-aided diagnostic tools (CADTs) that can be applied readily in clinical medicine. The choice of which “control” group to compare patients with epilepsy (PWE) against is not uniform. In this manuscript, we provide evidence to suggest that that the optimal control group depends upon whether an ML tool is intended to elucidate facets of the neurophysiology of epilepsy, or to estimate the reliability of the tool to diagnose patients in a clinical setting.
Non-epileptic seizures are primarily psychiatric events. In greater than 90% of cases, these seizure-like attacks are a symptom of conversion disorder in which patients' psychological challenges present as physical symptoms [10, 11]. These attacks tend to mimic seizures that the patient has seen or heard about; they are therefore difficult to distinguish from epileptic seizures. The diagnosis of non-epileptic seizures is based on ruling out all organic causes for the attacks; therefore these patients are frequently exposed to antiepileptic medication and other neurologic interventions prior to definitive diagnosis. There are no neuropathologic changes that predispose these patients to have attacks and the electrophysiological mechanism for the attacks is dissimilar from the mechanism for epileptic seizures. Therefore, antiepileptic medication and other neurologic interventions will not control these attacks, though their side effect profile remains the same. Although a small minority of PWN also suffer from epileptic seizures, it is important to distinguish PWN from PWE so that they can receive the treatment appropriate to the underlying cause of their seizures.
The pathologic benefit of comparing NNPs to PWE is clear. Using our knowledge of normal physiology, this comparison has been used to describe how the complexity of EEG recording during seizure (ictus) is less than that of interictal EEG [1, 2]; how epileptic lesions are associated with focal cortical atrophy and hyperintensity on MRI [12]; and how focal interictal hypometabolism in FDG-PET can indicate the seizure onset zone [13]. These differences are thought to be macro-scale features that confirm our understanding of epilepsy on a neural level.
The decreased complexity in EEG reflects hypersynchronized activity of neurons in the epileptic network, coupled with inhibited activity in the surrounding tissue [14]. Focal cortical atrophy and MRI signal hyperintensity are MRI-based signs of increased cell death of inhibitory cells and the ensuing gliosis [15]. The cause for focal hypometabolism is defines less clearly, but it has been shown that neurons within the epileptic network have altered metabolic activity [16]. Cell death and gliosis also may play a role in interictal hypometabolism [16]. While these findings help us to understand the neuropathologic features of epilepsy, there are, unfortunately, important caveats to their interpretation.
Patients with seizures are exposed to environmental factors that may artificially increase physicians' ability to discriminate NNPs from PWE. The mechanism of action of many antiepileptic drugs (AEDs) is to decrease the synchronicity and excitability of neural networks [14], thereby potentially increasing the baseline complexity of EEG so that the contrast with seizure activity is enhanced. Similarly, some AEDs have psychiatric side effects that appear similar to the psychiatric co-morbidities of PWN [17, 18]. In contrast with NNPs, PWN frequently are treated with these AEDs before their seizures are determined to be non-epileptic, therefore the use of PWN as a control group more accurately controls for the potential effect of AEDs.
Even though the diagnoses of non-epileptic seizures and epilepsy are distinct, many of their risk factors are shared. PWNs model their seizures after those they have seen or heard about before, therefore the relationship with family history is difficult to describe [19, 20]. Similarly, both PWN and PWE are associated with traumatic brain injury (TBI), albeit PWN are more associated with mild TBI [10, 19]. The presence of psychiatric comorbidities increases the suspicion for non-epileptic seizures, but epilepsy also has been shown to be associated with significant psychiatric challenges, potentially as a side effect of AEDs, or to the loss of independence and the stigma associated with the disease [21] Therefore, in order to assess reliably if ML models can detect signs of the underlying pathology associated with epilepsy, it is useful to compare PWE to PWN.
Lastly, and potentially most importantly, the comparison to PWN mirrors the clinical question at hand. Physicians only question if a patient has epilepsy if they present with seizure-like events. They do not, as suggested by the comparison with NNPs, consider epilepsy in all patients they encounter.
As a result of these concerns, we seek to study in our FDG-PET database which control group results in a more accurate diagnosis of lateralized temporal lobe epilepsy (TLE). This allows us to measure the potential effect of the confounding factors discussed above. We also inspect if there were detectable and/or interpretable differences between PWN and NNP.
II. Methods
A. Dataset
All of the 105 patients with seizures included in our analysis were admitted to the University of California, Los Angeles (UCLA) Seizure Disorder Center's video-EEG Epilepsy Monitoring Unit between 2005 and 2012. Each patient's diagnosis was based on a consensus panel review of their clinical history, physical and neurological exam, neuropsychiatric testing, video-EEG, interictal FDG-PET, ictal FDG-PET, structural and diffusion MRI and/or CT scan. This multimodal assessment is the gold standard for epilepsy diagnosis, and for localization of the epileptic focus. The patients included in this analysis were chosen because they had an FDG-PET; had no history of penetrative neurotrauma, including neurosurgery; were determined by consensus diagnosis to have a single, lateralized epileptogenic focus and had no suspicion of mixed non-epileptic and epileptic seizure disorder. These patients were diagnosed with either left temporal lobe epilepsy (LTLE, n=39), right temporal lobe epilepsy (RTLE, n=34) or non-epileptic seizures (PWN, n=32). PET images were determined to be interictal by clinical findings and concurrent scalp EEG. Neurologically normal, seizure naïve patients (NNP, n=30) were scanned for other reasons on the same clinical scanners and were age matched to the PWN. Details of PET acquisition and feature extraction using NeuroQ (Syntermed, CA) are described in [4].
B. Machine Learning Details
The Multilayer Perceptron was implemented to discriminate between either PWN or NNP versus RTLE or LTLE with default parameters in Weka [22] using the protocol described in [4] where the clinical implications of the PWN versus TLE discrimination are discussed. The minimum redundancy-maximum relevancy (mRMR) toolbox for MATLAB [23, 24] was used to generate a ranked list of the 47 ROI metabolisms (features) based on the training set. We used a random field theory correction (RFTC) to correct for the bias in selecting the maximum cyclical leave-one-out cross validation (CL1OCV) accuracy after testing multiple numbers of ROIs that contributed to the model [4]. Weka [22] was used to implement CL1OCV of a cost-sensitive MLP that was weighted to maximize balanced accuracy, defined by the mean of sensitivity and specificity.
III. Results
A. Cross-Validation Accuracy Differences
Using either control group, we diagnosed lateralized TLE effectively with greater than 81% CL1OCV accuracy (RFTC z-test of proportions versus naïve classifier, z>5.8, p<10-8; Fig. 1). There was no significant difference between the CL1OCV accuracies when NNP or PWN was used when diagnosing RTLE or LTLE (two sample z-test of proportions, |z|<1.5, p>0.16). We discriminated between PWN and NNP with 77% CL1OCV accuracy which was significantly better than chance (RFTC z-test of proportions versus naïve classifier, z=4.9, p<10-5).
Fig. 1.
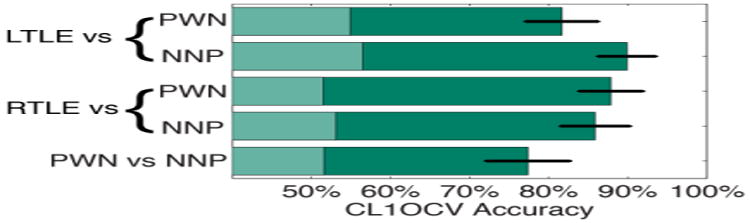
CL1OCV accuracy of our computer-aided diagnostic tool using each control group. Error bars indicate standard error from the mean. Shading indicates performance of a naïve classifier. PWN: Patients with non-epileptic seizures; NNP: Neurologically normal patients; L or R TLE: Left or Right Temporal Lobe Epilepsy
B. Insight into Focality of the Epilepsies
Fig. 2 illustrates the number of features that produced the random field theory corrected CL1OCV accuracy. Using the PWN as a control, the RTLE comparison required fewer ROIs than the LTLE comparison. Using the NNPs as a control, the same trend was seen, albeit with different mRMR feature rankings (Table 1).
Fig. 2.
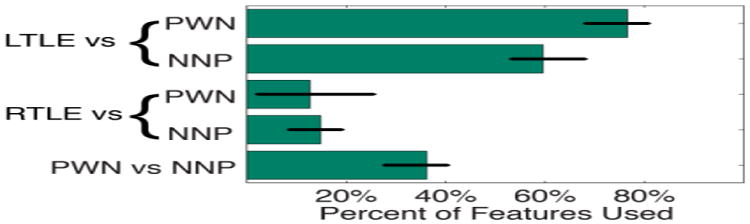
Percent of the 47 ROIs that contributed to the best CL1OCV accuracy. Error bars indicate accuracies within the same significant random field theory cluster.
Table I. ROI mRMR Ranking.
| ROI Rank | Region of Interest | |||
|---|---|---|---|---|
| LTLE vs PWN | LTLE vs NNP | RTLE vs PWN | RTLE vs NNP | |
| 1 | Midbrain | R Ass Vis | R ila Temp | R ila Temp |
| 2 | L ilp Temp | R pm Temp | L SM | R ilp Temp |
| 3 | R ilp Temp | L i Frontal | R ilp Temp | L Lentiform |
| 4 | L Ass Vis | R s Parietal | L sl Temp | L SM |
| 5 | L Broca's | L SM | R Thal | R sl Temp |
The mRMR Rank of the top 5 ROIs based on the full data. Note that these rankings utilize the full dataset and therefore do not necessarily coincide with any individual training set, each of which are missing data from just one patient. The preceding L and R refer to left and right respectively. The lower case i, l, a, p, s, and m stand for inferior, lateral, anterior, posterior, superior, and medial respectively. The other abbreviations are for temporal cortex (Temp), thalamus (Thal), associative visual cortex (Ass Vis), and Sensorimotor cortex (SM). The temporal, parietal and frontal ROIs are all cortical ROIs. Colors indicate repeat ROIs.
IV. Discussion
Even though there are substantial differences in the resting state neural metabolism of PWNs and NNPs, the choice of control group did not substantively affect our ability to diagnose PWEs, nor did it provide different pathologic insight into the difference between LTLE and RTLE. This suggests that comparing PWE to NNPs did not artificially increase our discriminative ability, contrary to our hypothesis. Our reliable discrimination between PWNs and NNPs, and the difference in feature rankings however, indicate that the multilayer perceptron may harness separate pathologic findings depending on the control group used. Interested readers are directed to [4] for an in depth description of this clinical and pathologic insight.
This suggests that there may be no control group that a priori will result in higher performance. The comparison to NNPs might describe better the combination of pathologic insults that results from, and/or causes, epilepsy. In our case, the lack of temporal ROIs in the top 5 rankings for LTLE suggests that the most salient pathologic consequences, and/or initiating factors, may lie outside the epileptogenic focus whereas the opposite may be true for RTLE. In contrast, the comparison to PWN demonstrates directly how the algorithm would perform in the clinic. In addition to utilizing the neurometabolic changes associated with epilepsy, this model may also harness the neurometabolic changes associated with PWN. Therefore, all of the observed differences cannot be attributed directly to epilepsy. As discussed above, depression was associated with hippocampal volume loss [25, 26]. Therefore, conversion disorder may also have characteristic FDG-PET findings.
These results reveal the challenge of developing a CADT to diagnose patients effectively. Just detecting epilepsy is not enough; we must also discriminate it reliably from disorders whose presentation is similar. The ideal CADT for epilepsy would effectively rule out transient ischemic attacks, confusion episodes, syncope, drug abuse and other disorders on the differential diagnosis for seizures (for full differential diagnosis see [27]). This presents a clear challenge: effectively recruiting and scanning enough patients with each of these disorders is prohibitively expensive in both time and money. Therefore, when planning experiments, we believe that one must choose the control group(s) that reflects the desired balance of clinical relevance to pathologic relevance.
There are a few limitations in the generalizability of these findings to the diagnosis of epilepsy and other disorders. In patients who present with their first seizure, the clinical question is not merely if the seizures are epileptic or non-epileptic: the patient also needs to know if they are at risk for future seizures. This clinical comparison may be better served by the contrast between NNPs and PWE. In addition, PWN are more frequently misdiagnosed as frontal lobe or generalized epilepsy instead of TLE. Therefore, there are also some caveats to using PWN as the control group for TLE diagnosis.
The challenge of identifying the proper control group to train CADTs is not unique to epilepsy. For example, many CADTs for Alzheimer's disease frequently are controlled both by NNPs and patients with mild cognitive impairment [28, 29]. Few studies, however, consider the full differential diagnosis for dementia, including Parkinson's dementia, fronto-temporal lobe dementia, Lewy-body disease, and other dementias. Similarly, much work has been done in discriminating patients with schizophrenia from NNPs even though antipsychotic medication is associated with substantial neurologic changes [30].
We believe that there may be two divergent goals for machine learning in clinical populations: the pathologic description of disorders and the development of clinically applicable tools. Therefore, when describing the underlying pathophysiology of disease, the goal of machine learning is not necessarily to optimize classification accuracy. It is instead to pose a biologically plausible model that reflects trends seen in the data. This is related, but potentially separate, from the ultimate goal of using machine learning to maximize the clinical utility of computer-aided diagnostic tools. We argue here that to maximize clinical applicability, one must mimic the clinical question at hand by carefully selecting the control group.
Acknowledgments
The authors give thanks to Maunoo Lee and Cheri Geist for data management of the PET records and organizational support. This work was supported by the University of California (UCLA)-California Institute of Technology Medical Scientist Training Program (NIH T32 GM08042), the Systems and Integrative Biology Training Program at UCLA (NIH T32 GM008185), NIH R33 DA026109 (to M.S.C.), the W.M. Keck foundation and the UCLA Department of Biomathematics. M.S.C. is a member of the California NanoSystems Institute.
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare.
Contributor Information
Wesley T. Kerr, Email: WesleyTK@UCLA.edu, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA.
Andrew Y. Cho, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA
Ariana Anderson, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA.
Pamela K. Douglas, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA
Edward P. Lau, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA
Eric S. Hwang, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA
Kaavya R. Raman, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA
Aaron Trefler, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA.
Mark S. Cohen, Laboratory of Integrative Neuroimaging Technology University of California, Los Angeles Los Angeles, USA
Stefan T. Nguyen, Ahmanson Translational Imaging Division University of California, Los Angeles Los Angeles, USA
Navya M. Reddy, Ahmanson Translational Imaging Division University of California, Los Angeles Los Angeles, USA
Daniel H. Silverman, Email: DSilver@UCLA.edu, Ahmanson Translational Imaging Division University of California, Los Angeles Los Angeles, USA.
References
- 1.Osorio I, Lyubushin A, Sornette D. Automated seizure detection: unrecognized challenges, unexpected insights. Epilepsy & behavior : E&B. 2011;22(Suppl 1):s7–17. doi: 10.1016/j.yebeh.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Sackellares JC. Seizure prediction. Epilepsy Currents. 2008;8(3):55–9. doi: 10.1111/j.1535-7511.2008.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr WT, Anderson A, Lau EP, Cho AY, Xia H, Bramen J, et al. Automated diagnosis of epilepsy using eeg power spectrum. Epilepsia. 2012;53(11):e189–92. doi: 10.1111/j.1528-1167.2012.03653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr WT, Nguyen ST, Cho AY, Lau EP, Silverman DH, Douglas PK, et al. Computer aided diagnosis and localization of lateralized temporal lobe epilepsy using interictal fdg-pet. Front Epilepsy. 2013 doi: 10.3389/fneur.2013.00031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farid N, Girard HM, Kemmotsu N, Smith ME, Magda SW, Lim WY, et al. Temporal lobe epilepsy: quantitative mr volumetry in detection of hippocampal atrophy. Radiology. 2012;264(2):542–50. doi: 10.1148/radiol.12112638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keihaninejad S, Heckemann RA, Gousias IS, Hajnal JV, Duncan JS, Aljabar P, et al. Classification and lateralization of temporal lobe epilepsies with and without hippocampal atrophy based on whole-brain automatic MRI segmentation. PloS one. 2012;7(4):e33096. doi: 10.1371/journal.pone.0033096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Focke NK, Yogarajah M, Symms MR, Gruber O, Paulus W, Duncan JS. Automated mr image classification in temporal lobe epilepsy. NeuroImage. 2012;59(1):356–62. doi: 10.1016/j.neuroimage.2011.07.068. [DOI] [PubMed] [Google Scholar]
- 8.Rosso OA, Mendes A, Berretta R, Rostas JA, Hunter M, Moscato P. Distinguishing childhood absence epilepsy patients from controls by the analysis of their background brain electrical activity (ii): a combinatorial optimization approach for electrode selection. Journal of neuroscience methods. 2009;181(2):257–67. doi: 10.1016/j.jneumeth.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Santiago-Rodriguez E, Harmony T, Cardenas-Morales L, Hernandez A, Fernandez-Bouzas A. Analysis of background eeg activity in patients with juvenile myoclonic epilepsy. Seizure : the journal of the British Epilepsy Association. 2008;17(5):437–45. doi: 10.1016/j.seizure.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Binder LM, Salinsky MC. Psychogenic nonepileptic seizures. Neuropsychology review. 2007;17(4):405–12. doi: 10.1007/s11065-007-9047-5. [DOI] [PubMed] [Google Scholar]
- 11.Patel H, Scott E, Dunn D, Garg B. Nonepileptic seizures in children. Epilepsia. 2007;48(11):2086–92. doi: 10.1111/j.1528-1167.2007.01200.x. [DOI] [PubMed] [Google Scholar]
- 12.Henry TR, Chupin M, Lehericy S, Strupp JP, Sikora MA, Sha ZY, et al. Hippocampal sclerosis in temporal lobe epilepsy: findings at 7 T. Radiology. 2011;261(1):199–209. doi: 10.1148/radiol.11101651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KK, Salamon N. [18F] Fluorodeoxyglucose–Positron-Emission Tomography and MR Imaging Coregistration for Presurgical Evaluation of Medically Refractory Epilepsy. AJNR American journal of neuroradiology. 2009;30:1811–6. doi: 10.3174/ajnr.A1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogawski MA, Loscher W. The neurobiology of antiepileptic drugs. Nature reviews Neuroscience. 2004;5(7):553–64. doi: 10.1038/nrn1430. [DOI] [PubMed] [Google Scholar]
- 15.Schneider-Mizell CM, Parent JM, Ben-Jacob E, Zochowski MR, Sander LM. From network structure to network reorganization: implications for adult neurogenesis. Physical Biology. 2010;7:1–11. doi: 10.1088/1478-3975/7/4/046008. [DOI] [PubMed] [Google Scholar]
- 16.Person C, Koessler L, Louis-Dorr V, Wolf D, Maillard L, Marie PY, editors. IEEE Eng Med Biol Soc. 2010. Analysis of the relationship between interictal electrical source imaging and PET hypometabolism. [DOI] [PubMed] [Google Scholar]
- 17.Hoppe C, Elger CE. Depression in epilepsy: a critical review from a clinical perspective. Nature reviews Neurology. 2011;7(8):462–72. doi: 10.1038/nrneurol.2011.104. [DOI] [PubMed] [Google Scholar]
- 18.Ettinger AB. Psychotropic effects of antiepileptic drugs. Neurology. 2006;67(11):1916–25. doi: 10.1212/01.wnl.0000247045.85646.c0. [DOI] [PubMed] [Google Scholar]
- 19.Vincentiis S, Valente KD, Thome-Souza S, Kuczinsky E, Fiore LA, Negrao N. Risk factors for psychogenic nonepileptic seizures in children and adolescents with epilepsy. Epilepsy & behavior : E&B. 2006;8(1):294–8. doi: 10.1016/j.yebeh.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Holmes GL, Sackellares JC, McKiernan J, Ragland M, Dreifuss FE. Evaluation of childhood pseudoseizures using EEG telemetry and video tape monitoring. The Journal of pediatrics. 1980;97(4):554–8. doi: 10.1016/s0022-3476(80)80008-4. [DOI] [PubMed] [Google Scholar]
- 21.Kanner AM, Schachter SC, Barry JJ, Hersdorffer DC, Mula M, Trimble M, et al. Depression and epilepsy, pain and psychogenic non-epileptic seizures: clinical and therapeutic perspectives. Epilepsy & behavior : E&B. 2012;24(2):169–81. doi: 10.1016/j.yebeh.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Hall M, Frank E, Holmes GL, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explorations. 2009;11(1):10–18. [Google Scholar]
- 23.Ding C, Peng HC. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005;3(2):185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- 24.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Analysis and Machine Intelligence. 2005;27(8):1226–38. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 25.Bremner JD, Narayan M, Anderson ER, Staib LH, Miller HL, Charney DS. Hippocampal volume reduction in major depression. The American journal of psychiatry. 2000;157(1):115–8. doi: 10.1176/ajp.157.1.115. [DOI] [PubMed] [Google Scholar]
- 26.Frodl T, Meisenzahl EM, Zetzsche T, Born C, Groll C, Jager M, et al. Hippocampal changes in patients with a first episode of major depression. The American journal of psychiatry. 2002;159(7):1112–8. doi: 10.1176/appi.ajp.159.7.1112. [DOI] [PubMed] [Google Scholar]
- 27.Mikati MA. In: Kliegman: Nelson Textbook of Pediatrics. 19th. Kliegman RM, Stanton BF, Geme JWI, Schor NF, Behrman RE, editors. Philadelphia: Saunders; 2011. [Google Scholar]
- 28.Dai Z, Yan C, Wang Z, Wang J, Xia M, Li K, et al. Discriminative analysis of early Alzheimer's disease using multi-modal imaging and multi-level characterization with multi-classifier (M3) NeuroImage. 2011 doi: 10.1016/j.neuroimage.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Cho Y, Seong JK, Jeong Y, Shin SY. Individual subject classification for Alzheimer's disease based on incremental learning using a spatial frequency representation of cortical thickness data. NeuroImage. 2012;59(3):2217–30. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isom AM, Gudelsky GA, Benoit SC, Richtand NM. Antipsychotic medications, glutamate, and cell death: A hidden, but common medication side effect? Medical hypotheses. 2013;80(3):252–8. doi: 10.1016/j.mehy.2012.11.042. [DOI] [PubMed] [Google Scholar]


