Abstract
The definitive diagnosis of the type of epilepsy, if it exists, in medication-resistant seizure disorder is based on the efficient combination of clinical information, long-term video-electroencephalography (EEG) and neuroimaging. Diagnoses are reached by a consensus panel that combines these diverse modalities using clinical wisdom and experience. Here we compare two methods of multimodal computer-aided diagnosis, vector concatenation (VC) and conditional dependence (CD), using clinical archive data from 645 patients with medication-resistant seizure disorder, confirmed by video-EEG. CD models the clinical decision process, whereas VC allows for statistical modeling of cross-modality interactions. Due to the nature of clinical data, not all information was available in all patients. To overcome this, we multiply-imputed the missing data. Using a C4.5 decision tree, single modality classifiers achieved 53.1%, 51.5% and 51.1% average accuracy for MRI, clinical information and FDG-PET, respectively, for the discrimination between non-epileptic seizures, temporal lobe epilepsy, other focal epilepsies and generalized-onset epilepsy (vs. chance, p<0.01). Using VC, the average accuracy was significantly lower (39.2%). In contrast, the CD classifier that classified with MRI then clinical information achieved an average accuracy of 58.7% (vs. VC, p<0.01). The decrease in accuracy of VC compared to the MRI classifier illustrates how the addition of more informative features does not improve performance monotonically. The superiority of conditional dependence over vector concatenation suggests that the structure imposed by conditional dependence improved our ability to model the underlying diagnostic trends in the multimodality data.
I. Introduction
The diagnosis of seizure disorder is challenging, and relies on the effective integration of multiple streams of information, or modalities. Clinicians must combine clinical information, obtained through the clinical interview, with various technological modalities including, but not limited to, scalp electroencephalography (EEG), structural and diffusion magnetic resonance imaging (MRI), and fluoro-deoxyglucose positron emission tomography (PET). Each modality provides incomplete but complementary information upon which a diagnosis can be built, and each modality has its own limitations. Clinical information depends typically upon accurate reporting from patients and/or caregivers who are untrained observers, and some work has shown that their reports are no more accurate than random guessing [1]. Neuroimaging relies on the development of observable structural and/or metabolic abnormalities that are associated, but not necessarily by cause or effect, with epileptogenic regions. Based on analysis of these factors, clinicians are able to provide effective treatment for two-thirds of patients with seizure disorder.
When a patient has failed two or more antiepileptic drugs (AEDs), or the etiology of the seizures is unclear, they are admitted for long-term video-EEG monitoring. During these admissions, 20 to 30% of patients with medication-resistant seizure disorder are found to have non-epileptic seizures [4]. For those patients with epilepsy, the goal of long-term monitoring is to determine if the seizures have focal or generalized onset and, if the seizures have focal onset, determine where the focus is and if it is surgically resectable [3]. Each of these determinations leads to changes in the treatment plan to target the cause of the seizures more effectively.
Our objective in designing computer-aided diagnostic tools (CADTs) is to improve diagnostic accuracy and certainty by providing information complementary to clinicians' judgment. This has the potential to decrease the cost of and time to diagnosis by providing clinicians' information that they would not otherwise have access to. Due to the inherently multimodal nature of the diagnosis of epilepsy, we focus on how to develop effective multimodal CADTs using the information available to clinicians.
In this manuscript, we assess the efficacy of two methods of multimodal learning: vector concatenation (VC) and conditional dependence (CD), with simplified data from clinical information (CI), MRI and PET. Vector concatenation represents a purely information theory perspective that relies on algorithms to discover the relationships between modalities. For other applications, VC has resulted in decreased performance relative to single modality models, likely due to overfitting and the “curse of dimensionality.” CD attempts to overcome these limitations by considering each modality sequentially [7]. CD also models clinical practice, where clinicians make a preliminary diagnosis based on the clinical interview, then look to technological data to modify that initial impression.
II. Methods
All 645 selected patients with medication-intractable seizures were admitted to the University of California, Los Angeles adult (age 13-88) video-EEG epilepsy monitoring unit (UCLA EMU) between the years of 2006 and 2013. Patients were split according to their definitively diagnosed etiology: temporal lobe epilepsy (TLE, n=235), other focal-onset epilepsy (OFE, n=109), generalized-onset epilepsy (Gen, n=50), unspecified epilepsy (UES, n=81) and non-epileptic seizures (NES, n=170). Patients diagnosed with unspecified epilepsy had confirmed epilepsy, but the seizure onset zone was not determined. Definitive diagnosis was based on consensus panel review of long term scalp video-EEG, MRI, FDG-PET, clinical history, physical and neurologic exam, and/or neuropsychiatric testing. Not all patients underwent all studies. Patients with prior neurosurgery, those with inconclusive video-EEG results, and events suspicious for mixed NES and epilepsy seizure disorder were excluded from analysis (n=219). This work was approved by the UCLA Institutional Review Board and was consistent with the Helsinki declaration. Written informed consent was obtained from all patients (or guardians of patients).
Our analysis focused on three modalities: CI, MRI and PET. All data were acquired as part of the patients' clinical care according to the resources available at the time of care. Simple clinical information was extracted, including age, gender, duration of seizure disorder prior to neuroimaging, seizure frequency and a history of clinically suspected stroke, febrile seizures, focal or generalized neurotrauma, and neuroinfection. For patients with multiple neuroimages, only the most recent, pre-operative scan of each modality was included. Neuroimaging results were based on review of clinical records written primarily, but not exclusively, by Dr. Noriko Salamon, who is an expert in the interpretation of neuroimaging for the diagnosis and pre-surgical assessment of epilepsy. The MRI findings were simplified into binary indicator variables for ex-tratemporal FLAIR or T2 hyperintensities, evidence of mesial temporal sclerosis, mass/space occupying lesion, encephalomalacia, cavernoma/hemangioma/angioma, cortical dysplasia, ischemic changes, gliosis, grey or white matter heterotopia, diffuse atrophy, focal extratemporal atrophy, meningioma, encephalocele, non-specific tumor, edema, vascular abnormality, cortical thickening, tuberosclerosis, unspecified lesion, cerebellar tonsil ectopia, abnormal gyration/sulcus structure, neurocystocercosis, hydrocephalus, and other MRI finding. The PET findings were simplified into indicators for hypo- or hyper-metabolism in the temporal lobe, frontal lobe, occipital lobe, parietal lobe, insula, diffuse cerebral cortex, cerebellum or whole brain diffuse hypometabolism, as well as foci of abnormal metabolism (i.e. high metabolism in white matter). Both neuroimaging modalities also included an aggregate indicator of abnormal findings.
Our data were extracted entirely from real-world clinical archives; not all data values were available for all patients. For the purposes of data imputation, we split the missing data into two groups. Duration of seizure disorder (0.5% missing) and seizure frequency (7% missing) were considered to be missing completely at random (MCAR), because these variables clearly are defined for every patient, and there was no trend in percent missing in any diagnostic subgroup. In contrast, if the clinical notes did not mention a historical factor (i.e., neurotrauma), we assumed that the patient had no history of this factor because the clinician is biased to report a historical factor if it exists. Overall 624 (97%) and 486 (75%) patients had MRI and PET records, respectively. The presence or absence of neuroimaging was not a significant predictor of diagnosis, when other clinical factors were taken into account (data not shown). Therefore, we assumed that this data was MCAR. We multiply imputed the data 20 independent times using the mi package in R [6]. Based on their theoretical and observed distribution, duration and seizure frequency were log transformed to maintain linearity. For the neuroimaging, there was insufficient information to impute each individual abnormality, therefore only the aggregate abnormality indicator for each modality was imputed. Separate analysis was conducted on each imputed dataset and results were aggregated with respect to the within and between imputation variance [8].
All classifications were based on C4.5 decision trees in Weka [5] with leave-one-out cross-validation (LOOCV), and performance was compared to chance distributions determined by permutation tests. Briefly, at each node, the C4.5 finds the feature and threshold that maximizes the normalized information gain. In LOOCV, one patient is excluded from all training. Once the decision tree is built, its performance is assessed on this “unseen” patient. For each method, we evaluated the overall accuracy, sensitivity for each diagnostic class (TLE, OFE, Gen, UES, NES). UES patients were considered correctly classified if they were predicted to have any type of epilepsy, but not NES. All other patients were considered misclassified if they were predicted to have UES. This penalty was reflected in the cost matrix of the C4.5 classifier. To compare multiple classifiers head-to-head we calculated the paired performance change, where the difference in accuracy is paired within patient, then averaged across patient because the performance on each patient cannot be assumed to be independent across classifiers. The null distribution for all performance measures was calculated by conducting the same analysis (imputation, training, LOOCV and aggregating results across imputed datasets as in [8]) on data with permuted diagnostic labels, without replacement. At least 100 permutations were done on each imputed dataset. The rank order of performance measures from the permutations were used as as empirical markers for the 1% quantile bins of each chance, or null, distribution used to determine significance, because the permuted labels had no relation to the underlying diagnostic class.
We compared VC and CD. VC ignores the modality structure and treats all features as components of one large model. CD, otherwise known as “stacking” [7], classifies each patient into discrete, multivariate classes based on each modality individually in a specified order. Intuitively, for each test case the classifier gives a preliminary diagnosis based on the first modality. Then, a second layer classifier is learned from all training samples that also were classified as that same preliminary diagnosis, either correctly or incorrectly. To frame this theoretically, Bayes theorem states that:
where Dx and Data indicate the diagnosis and data, respectively. In CD, we factor P(Dx|DataM1,M2) by each modality to get:
where M1 and M2 indicate two modalities, in order. Therefore, P(DataM2|Dx) is conditionally dependent on DataM2. Although we have described two-modality CD, this reasoning can be extended to apply to m modalities for any positive integer m. The final predicted diagnosis is the diagnosis that maximizes this likelihood, given the data and the classification model used to estimate the probabilities.
III. Results
The LOOCV accuracy and per-class sensitivity, taking into account the multiple imputations [8], of the single and multi-modality classifiers is illustrated in Figure 1. The accuracy of the single modality classifiers was 53.1%, 51.5%, and 51.1% fo MRI, CI, and PET, respectively. The accuracy of VC was 39.2% and 37.7% using MRI+PET+CI and just MRI+CI, respectively. The accuracy of CD was 58.7%, 56.6%, 52.9%, and 51.8% when modalities were considered in the order MRI→CI, CI→MRI, MRI→PET→CI, and CI→MRI→PET, respectively. All accuracies were significantly better than chance (p<0.01) except the MRI+CI, MRI→PET→CI, and MRI→CI (p>0.1). All pairwise comparisons revealed that all classifiers were superior to vector concatenation (p<0.01), but no other pairwise comparisons were significant (p>0.08).
Fig. 1.
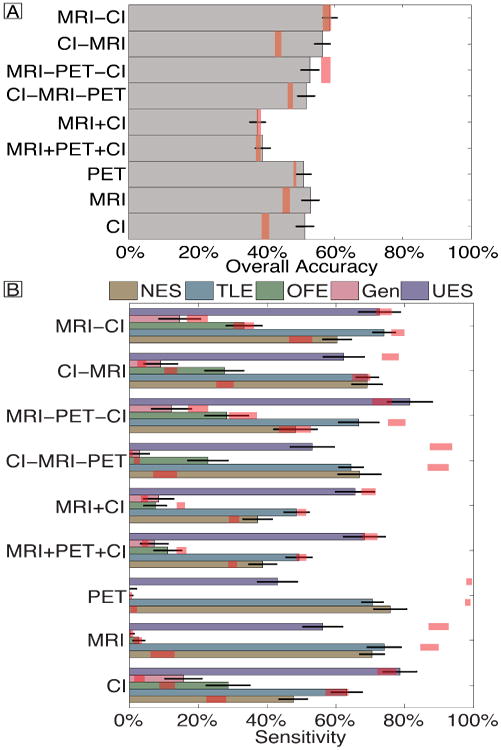
Overall accuracy (A) and per-class sensitivity (B) of each classifier. Error bars reflect binomial theoretical standard error bars, with multiple imputation. Red shading reflects the 95% quantile bounds from permutation tests. Vector concatenation and conditional dependence are indicated by + and -, respectively. For conditional dependence, the order of modalities is read from left to right. Abbreviations: Clinical information (CI).
Table I illustrates the distribution of the considered diagnostic features, except for the long list of neuroimaging indicators, by diagnostic class. All trees were more than 10 nodes deep and were too large for display.
Table I.
Summary of the most prevalent features in each diagnostic group, prior to multiple imputation.
| mean (standard error of the mean) | TLE | OFE | Gen | UES | NES |
|---|---|---|---|---|---|
|
| |||||
| Female (%) | 51 (3) | 60 (5) | 53 (7) | 49 (6) | 71 (3) |
| Age (years) | 38.1 (0.8) | 33.5 (1.4) | 32.3 (2.0) | 34.5 (1.7) | 38.4 (1.2) |
| Duration Seizure Disorder (log10year) | 1.074 (0.033) | 1.065 (0.044) | 1.002 (0.072) | 0.959 (0.082) | 0.464 (0.066) |
| Seizure Frequency (log10Seizures/month) | 0.787 (0.049) | 0.988 (0.083) | 0.789 (0.132) | 0.807 (0.100) | 1.148 (0.069) |
| History of Stroke (%) | 3 (1) | 5 (2) | 6 (3) | 8 (3) | 9 (2) |
| History of Febrile Seizures (%) | 16 (3) | 18 (4) | 12 (6) | 13 (4) | 9 (3) |
| History of Neurotrauma (%) | 35 (3) | 31 (4) | 24 (6) | 25 (5) | 36 (4) |
| History of Neuroinfection (%) | 16 (3) | 8 (3) | 3 (3) | 8 (4) | 16 (3) |
| Abnormal MRI (%) | 68 (3) | 56 (5) | 41 (7) | 49 (6) | 24 (3) |
| Abnormal PET (%) | 71 (3) | 48 (5) | 36 (8) | 43 (6) | 26 (6) |
| Mesial Temporal Sclerosis (%) | 68 (3) | 34 (5) | 27 (6) | 35 (5) | 10 (2) |
| Other MRI Findings (%) | 45 (3) | 48 (5) | 37 (7) | 41 (6) | 22 (3) |
| Temporal Hypometabolism (%) | 60 (3) | 29 (3) | 33 (8) | 37 (6) | 18 (5) |
| Other PET Findings (%) | 27 (3) | 27 (5) | 13 (5) | 13 (4) | 11 (4) |
Abbreviations: Temporal Lobe Epilepsy (TLE), Other Focal Epilepsy (OFE), Generalized-onset epilepsy (Gen), Unspecified-onset Epileptic Seizures (UES), Non-Epileptic Seizures (NES), Magnetic Resonance Imaging (MRI), fluoro-deoxyglucose Positron Emission Tomography (PET).
IV. Discussion
In real-world applications, combining information from multiple modalities does not always improve accuracy; this combination must consider the statistical and practical limitations inherent in modeling high dimensional data. Conditional dependence (CD) was superior to vector concatenation (VC) in overcoming these limitations, but did not result in a significant improvement over the single best modality classifier: the MRI.
The efficacy of CD relies on efficiently splitting the patients into more homogenous subgroups. The curse of dimensionality states that as the number of dimensions increases the number of samples needed to achieve the same sampling density increases exponentially. This curse can be overcome if the data truly exist in a lower dimensional subspace. This can occur when there are subgroups of patients within each diagnostic class that are more similar to each other, and therefore are distributed over a relatively limited region of feature space. These subgroups can be discovered using hypothesis-driven methods like CD, or through data-driven “committee-of-experts” methods that we will examine in the future. We hypothesize that, when applied in the most efficient order (neuroimaging first), CD identifies subgroups of patients with similar etiology. The relatively simple clinical variables then can identify if the clinical presentation of this etiology matches with the expected presentation of patients with similar etiologies. In particular, this order is interesting because it is the opposite of how clinicians diagnose patients. This illustrates how the ideal structure of automated computer analysis may differ from how clinicians' diagnose, due to the relative strengths of each analysis method. This reflects our belief that CADTs cannot, and should not, replace clinicians' expertise.
Even though neuroimaging-first produced higher accuracies than CI-first, this was not significantly higher than the accuracy on permuted diagnostic labels. Variation of chance between 36% (nTLE/ntotal) and 49% (nTLE +nUES/ntotal) was expected due to the latent structure of the data and classifiers naively diagnosing all patients as the most common class (TLE), which also was considered correct for patients with UES. However, chance accuracies of 58% for the neuroimaging-first CD classifiers seem inflated, for a number of reasons that can and should be explored. For instance, latent structure of the data could have been used to identify coherent subclasses that the randomly permuted diagnostic labels did not break up. This exploration is outside the scope of this short manuscript.
While most of our diagnostic accuracies were significantly above chance, they were too low to be readily applicable to clinical medicine. We expect that CADT performance would improve by including more detailed clinical information, including ictal semiology and co-morbidity profile; as well as integrating in automated MRI- and/or PET-based CADTs that utilize features not appreciated by radiologists (i.e. [9]–[12]). However, the addition of these other diagnostic features could magnify the problem of the curse of dimensionality. We, therefore, chose to focus first on simplified, high-salience features to assess multimodal classification methods.
To develop this CADT, we relied solely on archived clinical data from a tertiary epilepsy center, which has its benefits and limitations. The primary benefit is that the information we used reflects the information that would be available in clinic. This ensures that the CADT performance on this data is more similar to how the CADT would perform when applied in a similar setting, at the cost of accurately describing the underlying pathology [13]. As discussed above, the clinical information may be misreported, and radiologists cannot determine the epileptogenic region in all patients. Therefore, even though our CADTs may be clinically applicable, these observed trends may or may not reflect the true pathologic process of disease.
Archived clinical data often are limited because some data are missing. In this case, we multiply-imputed the missing durations, seizure frequency and neuroimaging results based on multilinear trends in all of the other included variables. This allowed the imputed missing data points to contribute to the MRI- and PET-based classifiers. While we expect the variance and, therefore the uncertainty, of each diagnosis to increase with the amount of missing data, in the case of our CADT, multiple imputation has the additional benefit of allowing us to apply one unified model to all patients, irrespective of what data has been collected.
V. Conclusion
Conditional dependence resulted in a more clinically-applicable CADT compared to vector concatenation. The imposed structure of conditional dependence improved performance. The opposite order of modalities in our analysis suggests that computers view the data differently from clinicians and could provide a non-redundant, complementary perspective on the data that could improve diagnostic accuracy and certainty, when combined with clinicians' expertise.
Acknowledgments
This work was supported by the National Institutes of Health (T32 GM08042, T32 GM008185, T90 DA023422, R33 DA026109), the William M. Keck Foundation, and the UCLA Department of Biomathematics.
References
- 1.Syed TU, LaFrance WC, Jr, Kahriman ES, Hasan SN, Rajasekaran V, Gulati D, Borad S, Shahid A, Fernandez-Baca G, Garcia N, Pawlowski M, Loddenkemper T, Amina S, Koubeissi MZ. Can semiology predict psychogenic nonepileptic seizures? A prospective study. Ann Neurol. 2011;69:997–1004. doi: 10.1002/ana.22345. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert DL, Sethuraman G, Kotagal U, Buncher R. Meta-analysis of EEG test performance shows wide variation among studies. Neurology. 2003;60:564–570. doi: 10.1212/01.wnl.0000044152.79316.27. [DOI] [PubMed] [Google Scholar]
- 3.Sauro KM, Macrodimitris S, Rkassman C, Wiebe S, Pillay N, Federico P, Murphy W, Jette N. Quality indicators in an epilepsy monitoring unit. Epilepsy & Behavior. 2014;33:7–11. doi: 10.1016/j.yebeh.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Kerr WT, Anderson A, Lau EP, Cho AY, Xia H, Bramen J, Douglas PK, Braun ES, Stern JM, Cohen MS. Automated diagnosis of epilepsy using EEG power spectrum. Epilepsia. 2012;53(11):e189–e192. doi: 10.1111/j.1528-1167.2012.03653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explorations. 2009;11(1):10–18. [Google Scholar]
- 6.Yu-Sung S, Gelman A, Hill J, Yajima M. Multiple imputation with diagnostics (mi) in R: Opening windows into the black box. J Stat Softw. 2011;45(2):1–31. [Google Scholar]
- 7.Wolpert DH. Stacked generalization. Neural networks. 1992;5:241–259. [Google Scholar]
- 8.Rubin DB. Multiple imputation after 18+ years (with discussion) JASA. 1996;91:473–489. [Google Scholar]
- 9.Kerr WT, Nguyen ST, Cho AY, Lau EP, Silverman DH, Douglas PK, Reddy NM, Anderson A, Bramen J, Salamon N, Stern JM, Cohen MS. Computer aided diagnosis and localization of lateralized temporal lobe epilepsy using interictal FDG-PET. Front Neurol. 2013;4:31. doi: 10.3389/fneur.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farid N, Girard HM, Kemmotsu N, Smith ME, Magda SW, Lim WY, Lee RR, McDonald CR. Temporal lobe epilepsy: quantitative MR volumetry in detection of hippocampal atrophy. Radiology. 2012;264(2):545–550. doi: 10.1148/radiol.12112638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Focke NK, Yogarajah M, Symms MR, Gruber O, Paulus W, Duncan JS. Automated MR image classification in temporal lobe epilepsy. NeuroImage. 2012;59(1):356–362. doi: 10.1016/j.neuroimage.2011.07.068. [DOI] [PubMed] [Google Scholar]
- 12.Keihaninejad S, Heckemann RA, Gousias IS, Hajnal JV, Duncan JS, Alijabar P, Rueckert D, Hammers A. Classification and lateralization of temporal lobe epilepsies with and without hippocampal atrophy based on whole-brain automatic MRI segmentation. PLoS One. 2012;7(4):e33096. doi: 10.1371/journal.pone.0033096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr WT, Cho AY, Anderson A, Douglas PK, Nguyen ST, Reddy NM, Lau EP, Hwang E, Raman K, Trefler A, Silverman DH, Cohen MS. 3rd International Workshop Pattern Recognition in Neuroimging. Philadelphia: Conference Publishing Services; 2013. Balancing clinical and pathological relevance in the machine learning diagnosis of epilepsy. [DOI] [PMC free article] [PubMed] [Google Scholar]


