Abstract
In this work, we review the process of protein unfolding characterized by a solid-state nanopore based device. The occupied or excluded volume of a protein molecule in a nanopore depends on the protein’s conformation or shape. A folded protein has a larger excluded volume in a nanopore thus it blocks more ionic current flow than its unfolded form and produces a greater current blockage amplitude. The time duration a protein stays in a pore also depends on the protein’s folding state. We use Bovine Serum Albumin (BSA) as a model protein to discuss this current blockage amplitude and the time duration associated with the protein unfolding process. BSA molecules were measured in folded, partially unfolded, and completely unfolded conformations in solid-state nanopores. We discuss experimental results, data analysis, and theoretical considerations of BSA protein unfolding measured with silicon nitride nanopores. We show this nanopore method is capable of characterizing a protein’s unfolding process at single molecule level. Problems and future studies in characterization of protein unfolding using a solid-state nanopore device will also be discussed.
Keywords: Bovine Serum Albumin (BSA), current blockage, excluded volume, protein unfolding, solid-state nanopore, translocation time
1. Introduction
Protein molecules are made up of long chains of amino acids. The native (functional) state of most protein molecules is in a tightly folded three-dimensional conformation. Within cells, unfolding and refolding of proteins occurs on a continuous basis. Fluctuations between native protein structure and partially or fully unfolded states affect key biological processes such as binding, translocation across membranes, secretion, or degradation [1]. To be able to characterize the folding-unfolding process of proteins is of fundamental importance not only for basic science but also in biotechnological applications. Owing to the physiological implications, many bulk based experimental methods as well as theoretical approaches have been employed for analyzing the conformational changes accompanying the folded-unfolded process: Fluorescence [2], NMR [3], and molecular dynamics simulations [4] are a few such techniques. In recent years, it has been demonstrated that ion channels (protein pores) suspended in lipid membranes [5–8] and later solid-state nanopores [9] are capable of characterizing protein unfolding at single molecule level [10].
Polypeptides and proteins measured by protein pores
Protein pores or ion channels suspended in lipid membranes were first shown to be capable of sensing single polypeptide molecules [11, 12] or proteins [5–8, 13–15]. Protein pores such as αhemolysin have reproducible geometry, well-defined structure and dimensions. However, due to their small fixed pore diameter, ~1.5 nm, only polypeptides or denatured proteins are able to pass through, which limits protein pores being used in measuring the process of protein unfolding. In addition, protein pores become unstable under protein denaturing conditions.
Protein unfolding measured by solid-state nanopores
Solid-state nanopores fabricated with desired geometry and dimensions, have been demonstrated to be capable of detecting protein molecules both in their native (folded) [16–25] and in their denatured (unfolded) states [9, 26, 27]. Motivated to develop a solid-state nanopore based device to identify proteins by measuring them both in their folded and unfolded states, we have studied the process of protein unfolding with sold-state nanopores. The protein molecules we have studied are: β-lactoglobulin (βLGa) and Hpr [9], Laminin [10], Bovine Serum Albumin (BSA), and two sets of protein markers (NativeMark from Invitrogen, Protein Marker from NBL) in their native and denaturing conditions. Because the complexity of protein unfolding process and the sophistication of current signals observed for each protein, in this review we will mainly focus on one protein, BSA unfolding, as an example to describe the experiments, results, data analysis, and problems of using solid-state nanopores to characterize the process of protein unfolding.
2. Principles of measuring protein unfolding by a solid-state nanopore
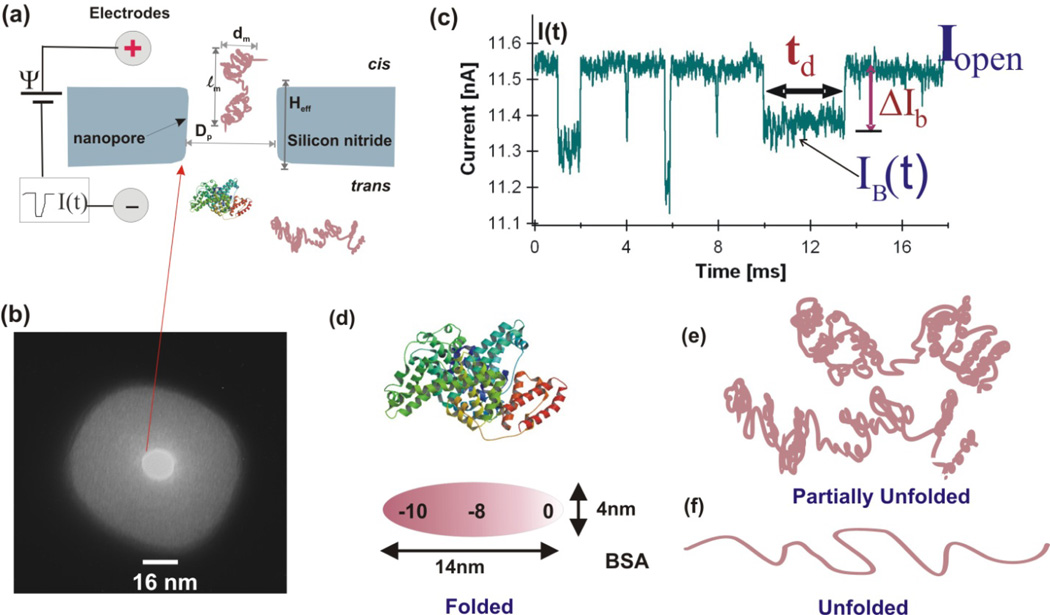
The center part of a solid-state nanopore device is a single nanopore fabricated in an insulating membrane (Fig. 1a and 1b) which separates two PDMS chambers filled with salt solution: the cis chamber in which protein samples is added, and the trans chamber where protein molecules move into after they translocate through the nanopore. A pair of silver chloride electrodes is embedded in the chambers. The electrodes are used to apply a constant voltage Ψ across the membrane and to measure the ionic current flow through the nanopore. The cis chamber is often electrically grounded and the trans is positively or negatively biased depending on the charge of the protein to be measured. The nanopores used for protein unfolding experiments reported so far are fabricated by a Focused Ion Beam (FIB) [26] or by a combination of FIB and low energy noble gas ion beam [28–30]. The nanopores are often 10–30 nm in thickness and 2–25 nm in diameter depending on the size of proteins.
Figure 1.
(a) Schematic diagram of a nanopore experiment setup. (b) A TEM image of a ~16 nm pore used for BSA measurement. (c) Several recorded BSA current blockage events in partially denatured condition (SDS+DTT+45 °C at pH 7 and 1M KCl measured with the nanopore shown in (b). (d) Illustration of one of the possible conformations of a BSA protein at native state (PDB, 3v03). (e) Possible partially denatured form of BSA. (f) completely unfolded form of a BSA molecule.
2.1. Parameters to be measured: current drop amplitude ΔIb and time duration td
A protein molecule of ~102 amino acids in a nanopore will partially block the flow of ions in the pore, increase the pore resistance, and cause a transient current drop event from the open pore current I0 as shown in Fig. 1c. Each current drop event in Fig. 1c represents a protein molecule interacting with or translocating through the pore. The current drop magnitude caused by a translocating protein is a function of time, ΔIb(t). Analysis of current drop events caused by protein molecules has been and is still a challenging task. For initial event sorting, we often calculate the mean current drop amplitude ΔIb and the dwell time td (Fig. 1c). The dwell time td of an event is defined as the time at half height of ΔIb. Below we describe how these two parameters are connected to the protein unfolding process.
ΔIb(t) is proportional to the excluded volume Λ (t) of a protein molecule in a nanopore
Approximately, the mean current drop amplitude ΔIb is determined by the pore geometry, conformation or shape of a protein molecule, and the solution conductivity. If we idealize a nanopore as a cylinder with diameter Dp and length Heff, assuming a protein molecule occupies a volume Λ(t) in the pore at time t (the instantaneous excluded volume), and further we assume that a translocating protein molecule in a nanopore driven by an applied voltage Ψ obeys Ohm’s law, the relationship between ΔIb(t) and Λ(t) can be written as [31–34]:
| (1) |
Where Io=Ψ/R0 is the open pore current, R0= Heff σ/Ap is the pore resistance, σ is the solution conductivity, Heff is the effective thickness of the pore taking into account the access resistance region on both sides of the nanopore [35–37], Ap is the area of the pore with an average diameter Dp, Vp = HeffAp the volume of the pore, and dm and lm are the diameter and length of the protein molecule. f(dm/Dp, lm/Heff) is a correction factor for the relative size and shape of the pore and the protein molecule and is often ignored for simplicity. For a folded protein molecule that can fit entirely inside a nanopore, f is less than one and contributes little to the current drop [32]. For a peptide chain that is much longer than the pore, f(dm/Dp, lm/Heff) ≈ 0, therefore ΔIb ≈ I0Λ/Vp, or Λ ≈ (ΔIb/I0)Vp. This relates the relative current drop amplitude, ΔIb(t)/I0, to the excluded volume of the protein molecule Λ(t).
The simple model described in Eq. (1) shows that if a nanopore’s volume Vp is kept a constant, the relative current drop magnitude, ΔIb(t)/I0, is directly proportional to a protein’s excluded volume, Λ, in the nanopore. For a completely unfolded protein going through a nanopore as a linear peptide chain with ℓm>Heff, only a segment of the protein is in the pore at any time, the excluded volume of the local segment is expected to be smaller than the whole protein at its 3D form, ΛlocalSeg< Λfolded,, therefore a smaller ΔIb(t)/I0 is expected to be measured for the unfolded protein than its 3D folded conformation [9]. Below we use BSA as an example to illustrate this point more quantitatively.
BSA excluded volume Λ in a nanopore and its conformation state
BSA belongs to the most abundant (with a typical concentration of ~77 mM) serum albumin protein family which is essential to maintain many important body functions in mammals. The BSA (66,430 D, Sigma) used in this work has 583 amino acid residues. One of the possible shapes and dimensions of a native state BSA shown in Fig. 1d is a prolate ellipsoid [38]. BSA is water soluble, monomeric, globular shape, and negatively charged at pH 7.4. Its isoelectric point at 25 °C is around 5.0 depending on the solution and salt concentration. BSA has 17 disulphide (covalent) bonds that make it as one of the most stable proteins. BSA is one of the most widely studied proteins and is used in numerous biochemical applications due to its stability.
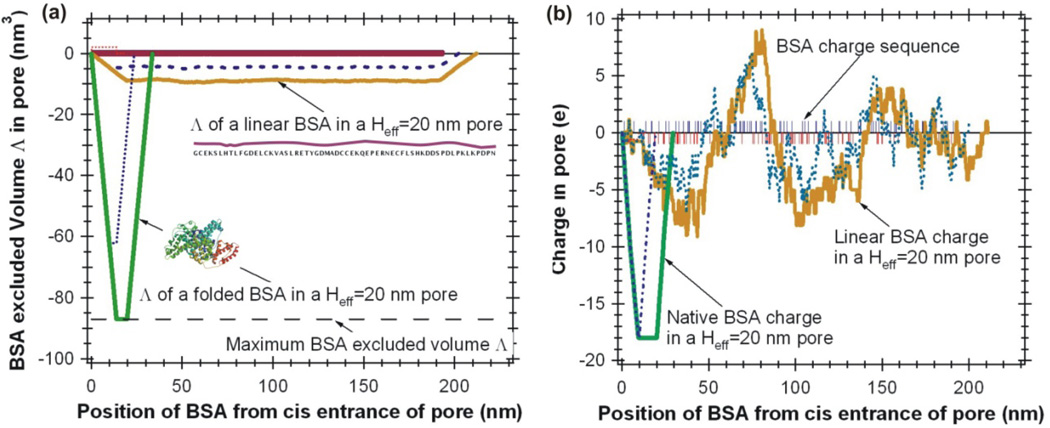
The conformation of a BSA molecule can be a folded (Fig. 1d), partially (Fig. 1e), and fully unfolded (Fig. 1f) states. By adding the atomic volume of all 583 amino acids [39, 40], the total atomic volume of a BSA molecule is estimated to be Λ= 87.0 nm3. The estimated contour length of a completely unfolded BSA is about ℓm ≈ 200 nm. When a BSA molecule translocates a nanopore (Heff< ℓm) as a linear peptide chain, only a segment of the amino acid chain will be inside the nanopore. As illustrated in Figure 2a, for a nanopore with Heff = 20 nm, the maximum excluded volume of a BSA molecule in its folded form is Λfold=87.0 nm3, as a linear peptide chain, Λunfold≈9.0 nm3. For a Heff = 10 nm pore, the maximum values are Λfold=62.2 nm3 and Λunfold≈4.5 nm3 (Fig. 2a, dotted lines). The ratio of Λunfold : Λfold = 1: 9.7, this suggests that the ΔIb(t)/I0 measured for a BSA molecule in a nanopore is expected to be 10 times smaller for a linear unfolded BSA chain comparing to a globular shaped folded BSA molecule.
Figure 2.
(a) Predicted excluded volume of BSA in a nanopore at folded globular state and unfolded linear shape in a Heff=20 nm pore (solid lines) and Heff=10 nm pore (dotted lines). (b) Predicted electrical charge of a BSA in a nanopore at folded globular state and unfolded linear shape (without SDS) in a Heff=20 nm pore (solid lines) and Heff=10 nm pore (dotted lines). The BSA protein sequence was obtained from: http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=3V03.
The excluded volume calculated by adding the atomic volumes of all amino acids in a nanopore provides an easy way to estimate the relative ratio of Λunfold/ Λfold. However, this estimation has ignored complicated issues such as the space inside a folded protein, hydration layer and territory bound water and ions on protein surface, the real excluded volume of a protein molecule in a nanopore is expected to be larger. For example, here we calculate Λfold=87.0 nm3 for a folded BSA, this is smaller than the value reported by Freedman et al. [25] calculated by formula Λ=(4/3)πa2b =117 nm3 using the dimensions shown in Fig. 1d, and it is also smaller than experimentally measured value166±29 nm3.
In conclusion, if the same nanopore is used, a BSA molecule’s excluded volume is expected to be Λ (folded) > Λ (partially unfolded) > Λ (unfolded). During a nanopore experiment, the measured relative current drop magnitudes are expected to have: ΔIb/I0(folded) > ΔIb/I0 (partially unfolded) > ΔIb/I0 (unfolded). Thus a solid-state nanopore device can be used to characterize the process of protein unfolding.
Protein unfolding states and the time duration td
The time duration of a current blockage event, td, is also related to a protein’s conformation state. When a BSA molecule in a nanopore is folded, it is expected to behave like a particle with a total charge Q= −18e at pH7 as shown in Fig. 2b. Under the electrical field strength of E=Ψ/ Heff, the electric force exerted on a BSA molecule is Fe=QE=QΨ/Heff, which is opposed by a viscous drag force Fdrag=ηCfυ assuming a BSA molecule is moving with a terminal velocity υ=Heff/td, then td can be written as
| (2) |
Here ηCf is the friction coefficient [41], η is the solution viscosity, and Cf is a constant. In this case, the time duration td is expected to be inversely proportional to the applied voltage Ψ, and the time distribution histogram for td is expected to be a Gaussian [9, 10, 41]. This equation also works for a uniformly charged amino acid chain, which is expected to behave like a DNA molecule with υ=(lm+Heff)/td.
When a protein is completely unfolded and going through a nanopore as a linear amino acid chain, the time distribution is expected to depend on the protein’s charge sequence [9, 10]. As shown in Fig. 2b for an unfolded BSA, the net charge inside a nanopore is a function of translocation position of the amino acid chain; and the net charge could be positive, neutral, and negative. The net electrical driving force would change direction when the net charge of the local segment changes sign, which could drive the protein back and forth near the electrically neutral charged region. The polypeptide chain could be transiently trapped in the nanopore due to a protein’s heterogeneous charge sequence, the translocation process could be thermally activated and the time duration distribution is expected to be multiple exponentials [9, 10] instead of a Gaussian for a folded BSA molecule.
Pore-protein interactions affect the dynamics of protein translocation
The above discussions on ΔIb and td have ignored the interactions between a nanopore surface with a passing protein molecule. Reports on protein translocation in solid-state nanopores have shown that if the size of a protein and the diameter of the pore are close, the pore-protein interactions could dominate a protein’s translocation dynamics [9, 19]. The pore-protein interactions could be controlled by functionalizing a nanopore’s surface to adjust its charge, surface hydrophobicity/hydrophilicity, and by performing a nanopore experiment at different salt concentrations. Although these studies are critically important, understanding and controlling pore-protein interactions are still challenging tasks today. To reduce the pore-protein interactions, nanopores several nanometers larger in diameter than a protein is preferred.
2.2 Methods to unfold proteins for solid-state nanopore analysis
Most native-state proteins have a well-defined 3D structure, which can be disrupted by heat, denaturants, and extreme pH [42]. The task to completely unfold a native state protein to a linear amino acid chain and measure it with a solid-state nanopore has been a challenge. Here we briefly summarize several denaturing agents used in our lab: Guanidine HCl, urea, and SDS plus DTT.
Guanidine hydrochloride
CH6ClN3 or [CH6N3]+Cl− in solution, is one of the strongest denaturants used in studies of protein unfolding. Since it contains Cl− ions, it can be used with or without additional Cl− containing salt that is required for a nanopore experiment using AgCl electrodes. In 6M guanidine chloride solution, all proteins lose their well-ordered structures, and most of them become randomly coiled.
Urea
CO(NH2)2, has two NH2 groups joined by a carbonyl (C=O) functional group. It is highly soluble in water and non-toxic. In 8M urea, most proteins are denatured. However, since urea is electrically neutral, adding 8M urea decreases solution conductivity significantly [9].
Sodium dodecyl sulfate
(SDS) +DTT, CH3(CH2)11OSO3Na, is an anionic surfactant and is a common component used in cleaning products. SDS can denature a protein and coat a layer of negative charge on a peptide chain. This would be ideal because such a negatively charged peptide chain would go through a nanopore like a uniformly charged DNA molecule. However, attempts at adding SDS directly to the cis and trans chambers had not been successful in our lab because SDS can form bubbles easily. Dithiothreitol (DTT), C4H10O2S2, is an unusually strong reducing agent and is often used to break disulfide bonds.
3. Measuring BSA unfolding with solid-state nanopores
3.1. BSA in SDS + DTT + temperature
The experiment
Below we use BSA as an example to demonstrate how a solid-state nanopore can be used to characterize the unfolding process of a protein with disulfide bonds. To compare ΔIb/I0 signal of folded BSA with partially and completely unfolded, the protein needs to be measured at least in 3 conformations, folded, partially denatured, and completely unfolded. This requires a nanopore that is large enough to be occupied by a native state globular shaped protein. One model of a BSA molecule has a dimension of dm=4 nm and lm=14 nm (Fig. 1d), hence a pore larger than 16 nm is preferred. The nanopore used for this BSA unfolding experiment was about ~16 nm (Fig. 1b). The electrolyte solution contains 1M KCl with 10 mM Tris and 1mM EDTA at pH 7. BSA protein (Sigma-Aldrich) was first dissolved to make a stock solution of 1 mg/ml (~ 15 µM) in ~150 mM KCl TE buffer. The trans chamber was positively biased to drive the negatively charged BSA (pH 7) to pass through the nanopore. The ionic current signal was recorded using an Axopatch 200B (Molecular Devices) in event driven and voltage-clamp mode. The low pass Bessel filter in the Axopatch 200B was set to 10 kHz or 100 kHz as indicated.
Results
After the BSA sample was added to the cis chamber to a final concentration of ~12 nM, current blockage events were observed. Several examples of these events are shown in Fig. 3a. After calculating the average current drop amplitude ΔIb and the dwell time td for each event, we plotted the ΔIb vs td in Fig. 3b. Fig. 3b shows the event distribution of BSA measured in 1M KCl at room temperature ~22°C without denaturants.
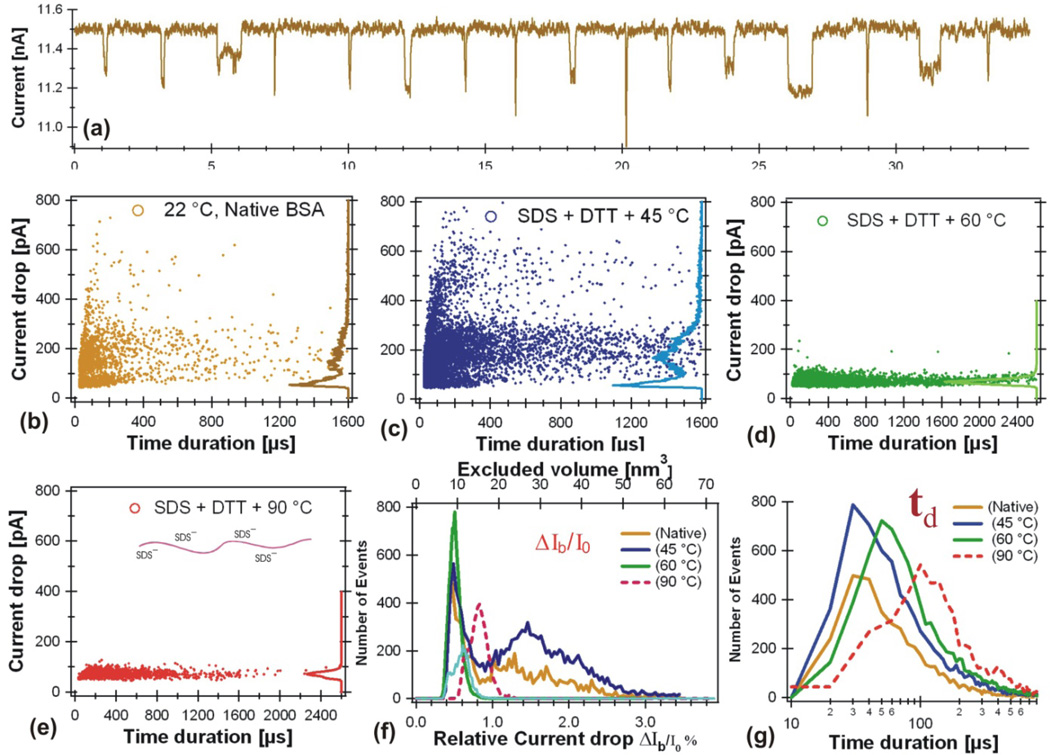
Figure 3.
BSA unfolding states measured in a ~16 nm pore (Fig. 1b) in 1M KCl at pH 7. (a) Current drop events observed for BSA in 1M KCl at pH 7 with no denaturant. The stock BSA sample contains 1.4 mg SDS (4.85 mM) and 2mM DTT with 1 mg BSA (15µM) in 1ml 1M KCl solution were kept for 5 minutes at the higher temperature indicated in the figures, than immediately cooled down in a water bath kept at room temperature. The BSA protein was then added to the cis chamber. The bias voltage was Ψ=120 mV. The open pore current was ΔI0=11.5 (b), 11.5 (c), 13.4 (d), and 8.7 (e) nA respectively. (f) Number of events distributions vs ΔIb/I0 for data shown in panels b, c, e, and d. (g) Number of events distributions vs td for data shown in panels b, c, e, and d. The final concentration of the BSA protein in the cis chamber was about 12 nM for all the measurements. The final concentrations of SDS and DTT were 3.9 µM and 1.6 µM respectively.
To denature the BSA, 1.4 mg SDS (3.9 mM) with 2 mM DTT was added to the stock BSA (1mg BSA in 1ml) solution. This mixture was heated at 45° for 5 minutes, then immediately cooled down in a water bath at room temperature. We then added this BSA treated with SDS +DTT and heated at 45° to the cis chamber. The resulting event distribution is shown in Fig. 3c. The same experiment was repeated for the BSA heated at higher temperatures are shown in Fig. 3d (preheated at 60° C) and Fig. 3e (preheated at 90° C).
For native BSA at room temperature, the current drop ΔIb histogram on the right axis of Fig. 3b shows approximately two clusters of events, consistent with our earlier observations [16]. One cluster has a smaller current drop peak at ΔIb =60 ± 10 pA, the other cluster has a broader ΔIb = 100–300 pA (Fig. 3b). For the BSA protein preheated at 45° C, the plot of ΔIb vs td in Fig. 3c shows features similar to the native state BSA data displayed in Fig. 3b, except more events have larger ΔIb and longer td. Preheated at 60°C, the ΔIb vs td plot only shows one peak (Fig. 3d) at ΔIb =60 ± 10 pA. Preheated at 90 °C, the same one peak at ΔIb =60 ± 10 pA (Fig. 3e) was observed.
Discussion
The disappearance of the broader peak at ΔIb = 100–300 pA observed for BSA in Figure 3b and 3c suggests that under the conditions of SDS + DTT preheated above 60 °C (Fig. 3d and 3e), the BSA molecules were unfolded and all BSA protein molecules have lost their 3D structure. Note that the process of protein folding-unfolding is dynamic and the time scale is in the order of milliseconds. The observed one peak at ΔIb =60 ± 10 pA suggests that the BSA preheated above 60 °C remained denatured measured by the nanopore, possibly the heating treatment had permanently unfolded the protein. This also suggests that the small current blockage peak observed at ΔIb ≈ 60 pA at all temperatures likely belongs to the denatured BSA translocation events. This conclusion is also consistent with our earlier studies on other protein unfolding measured with solid-state nanopores [9] and proteins and peptides measured with protein nanopores [5, 11, 12]. In the work by Oukhaled at el. and Merstorf et al., the authors concluded that the small ΔIb and short td blockages were due to the passage of completely unfolded proteins, since their frequency increased as the concentration of the denaturing agent increased. This frequency dependency has also been observed with an increase of the temperature [8]. However, from this experiment, we can’t exclusively conclude that all the events in the small ΔIb = 60 pA peak represented unfolded BSA translocation, some short events could be produced by the collision of BSA molecules with the nanopore without translocation.
The broader peak with greater ΔIb =100–300 pA in Fig. 3b implies larger excluded volume of protein molecules in the pore, possibly produced by folded BSA. The same larger ΔIb =100–300 pA peak in Fig. 3c indicates that most BSA molecules were still having larger excluded volumes (folded state), but could have some degree of denaturation and aggregation. Since some events have larger ΔIb (400–600pA) and very longer td, an increased pore-protein interactions at this condition was also possible. It is puzzling why the native state BSA in a nanopore produced such a broad peak in ΔIb, implying a broad distribution of the excluded volume Λ and translocation configurations. Broad range of ΔIb distributions were also observed by previous reports studying BSA and other protein translocation in solid-state nanopores [9, 16–19, 21, 25–27], suggesting some protein molecules going through solid-state nanopores with multiple conformations, some could be partially denatured.
Fig. 3f shows that the relative current drop amplitude ΔIb/I0 is close to 2% for foplded BSA and about 0.5% for unfolded. Assuming Vp doesn’t change, the ratio of ΔIb/I0 for unfolded/folded measured in this experiment is about 1:4, not the ratio Λunfold: Λfold ≈ 1:10 predicted in Fig. 2a. Two possibilities could explain this smaller ratio: 1) if the single peak in Fig. 3d represents the completely unfolded BSA peptide chain in the pore, then events in the broader ΔIb peak were produced by partially denatured BSA under the experimental condition; 2) if the broader peak at ΔIb = 100–300 pA represents the intact folded BSA protein translocation in the nanopore, then the peak at ΔIb = 60 pA represents unfolded but looped translocation configuration, not the linear amino acid chain imagined in Fig. 2a.
The time duration histograms in Fig. 3g show that the peak values of td were similar for BSA in its native state and for BSA pretreated at 45°C. The tds had increased and the distributions were broader for the unfolded BSA pretreated 5 minutes at 60 °C and 90 °C. It is not clear if the negatively charged SDS had affected the td measured in this experiment.
This example demonstrates that by measuring the relative current drop magnitude ΔIb/I0, a solid-state nanopore device can characterize a protein’s unfolding process. Next, we show two more examples: 1) BSA in 6M GuHCl that BSA can only be partially denatured; 2) BSA voltage dependence without chemical denaturants to see if a native state BSA behaves like a rigid charged particle.
3.2. BSA in 6M Guanidine Chloride
Proteins can be completely or partially (if disulphide bonds exist) denatured in 6M Guanidine Chloride (GuHCl). The conductivity of 6M Guanidine hydrochloride solution was approximately σ=179.3 ms/cm at room temperature (22 °C) at pH=7.0 (σ=114 for 1M KCl). Using a pore with Dp ~18 nm (TEM image shown in Fig. 4) in 6M GuHCl solution at Ψ=120 mV, an open pore current I0=12.9 nA was measured. BSA protein was first dissolved in 6M GuHCl, and then added into the cis chamber. The BSA concentration in the cis chamber was ~100 nM. The trans chamber was positively biased.
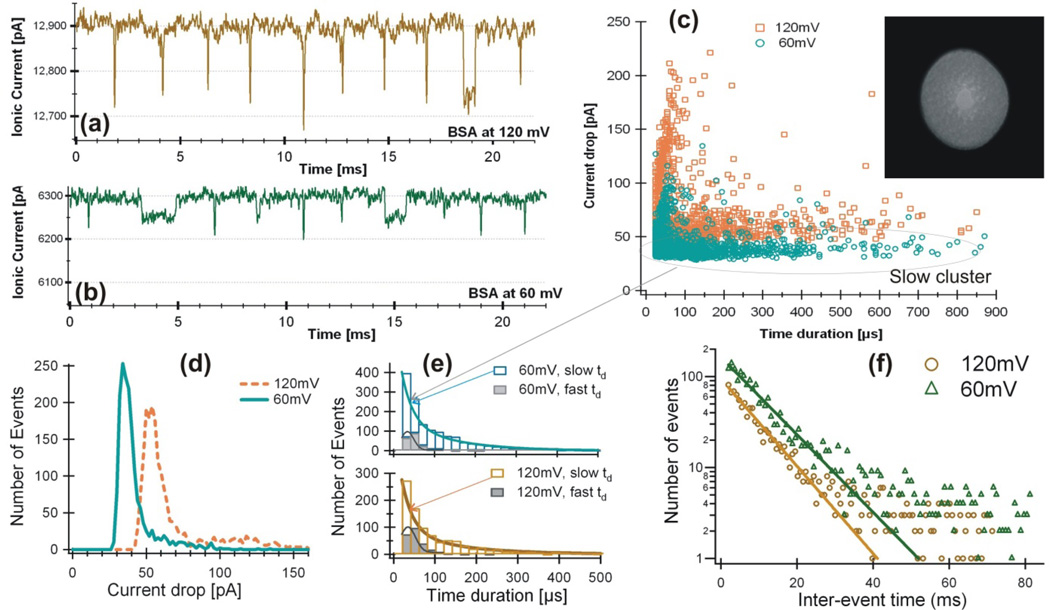
Figure 4.
BSA protein in 6M Guanidine Hydrochloride solution measured by a pore of Dp=18±2 nm. Examples of current blockage events at 120 mV (a, I0=12.9 nA), 60 mV (b, I0=6.3 nA), and ΔIb vs I0 (c). The nanopore used for this experiment is shown in the insert of (c). Histogram distributions are shown for current blockage ΔIb (d), time duration td (e), and the interval between events (f).
Examples of current blockage events produced by the BSA sample are shown for Ψ=120 mV (Fig. 4a) and for Ψ=60 mV (Fig. 4b). Comparing with the completely denatured BSA which only showed one cluster of events in Figure 3c and 3d, the plot of ΔIb vs td (Fig. 4c) shows two clusters of events at both voltages, ~80% of the events are in the cluster of longer td with smaller ΔIb, indicating not all BSA molecules are completely denatured in 6M GuHCl. Figure 4d shows that the peak values of current drop amplitudes are: ΔIb = 34±6 pA and the relative blockage ΔIb/I0=0.54% at Ψ=60 mV, Ib = 54±8 pA and ΔIb/I0=0.42% at Ψ=120 mV. The ratio of ΔIb/I0 is decreased at Ψ=120 mV compared to Ψ=60 mV. To see the behavior of the two clusters of events more clearly, the time duration histograms are shown separately in Figure 4e at both voltages. Fitting with a Gaussian to the events of larger ΔIb and short td, the peak values are: tdp = 33.6 ± 0.7 µs for Ψ=60 mV and tdp = 34.7 ± 1.4 µs for Ψ=120 mV. Two exponential fittings to the events of smaller ΔIb and longer td, the exponential decal constants are tau1 =26.7 ± 5 µs and tau2 = 125 ± 37 µs for 60mV, tau1 =26.1 ± 4 µs and tau2 = 130 ± 40 µs for 120 mV. The histograms of the waiting time between events at both of these voltages in Figure 4f shows that the inter-event times are longer at Ψ=60 mV than Ψ=120 mV.
Discussion
This measurement shows that BSA molecules in 6M GuHCl were not completely unfolded, which is consistent with that guanidine is not capable of breaking the disulphide bonds in a protein. The ratio of ΔIb/I0 decreased from 0.54% at Ψ=60 mV to 0.42% at Ψ=120 mV, this suggests a BSA molecule might become more stretched at 120 mV so it blocked less amount of current flow, consistent with recent observations reported by Cressiot et al on studying a maltose binding protein [27]. The same ratio of ΔIb/I0 ≈ 0.5% was observed here suggesting that the peak at ΔIb = 60 pA in Figure 3 is most likely from looped BSA translocation configuration. The events in the slow cluster with short tau1≈26 µs could be produced by collision of BSA molecules with the pore, the longer td were most likely produced by partially unfolded BSA. With tau2 = 125 µs for 60mV and tau2 = 130 µs for 120 mV, the dwell times of these events were longer at Ψ=120 mV, suggesting that these events with longer tds could be produced by BSA linear translocation as shown in Figure 2B. The translocation process could be thermally activated. Another possibility is that BSA molecules become more stretched at higher voltage that it takes longer time for them to translocate a nanopore. However, without a reference molecule like a DNA that doesn’t change its shape in a pore, the BSA excluded volume in the pore can’t be estimated precisely, therefore we can’t determine if the BSA molecules were in linear or looped configuration in a pore. The inter-event histogram (Fig. 4f) shows that the current drop frequency increased as the applied voltage increased from 60 mV to 120 mV, consistent with earlier report by Cressiot et al. [27].
3.3. Voltage dependence of BSA in 1M KCl without denaturant
Recent measurements with solid-state nanopores have shown that the electric field in a nanopore can change protein structure: folded protein molecules could be partially denatured and partially folded proteins could be further stretched. Using β-lactoglobulin, our previous experiment demonstrated that a fraction of β-lactoglobulin seemed partially denatured without any denaturing agent [9]. Freedman et al. had shown that the electric field in a nanopore did denature folded BSA [25]. Oukhaled et al. [26] and Cressiot et al. [27] had demonstrated that unfolded protein molecules could be further stretched in nanopores with increased applied voltage. Here we show an example of the electric field effects to folded BSA and double stranded DNA molecules by nanopore experiment and computer simulation.
Results
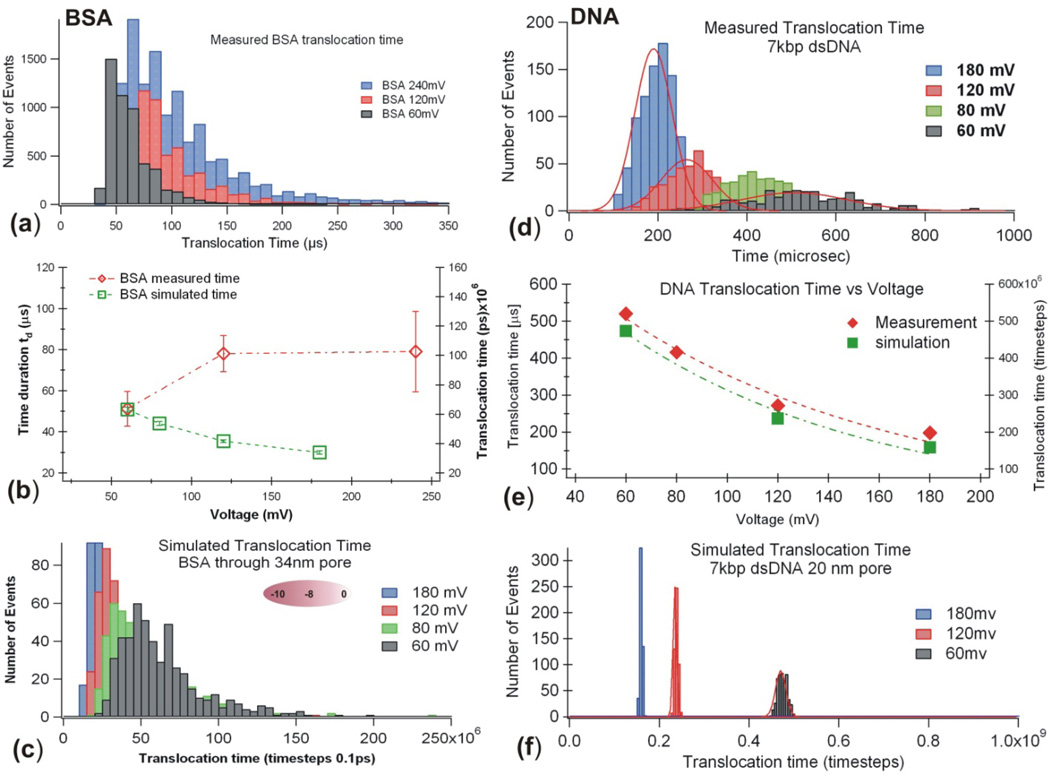
To probe if the strong electric field in a nanopore, E ≈ Ψ /Heff = 105V/cm, will change the conformation of a very stable protein like BSA in folded state, we measured voltage dependence of BSA in 1M KCl solution without any denaturing agent. Figure 5a shows that when the applied voltage Ψ was increased from, 60 mV, to 120 mV and 240 mV, the time duration td was slightly increased (Fig. 5b, open diamond ◇). The same trend was also observed when a set of native proteins (NativeMark from Invitrogen, contains IgM, Apoferritin, Lactate Dehydrogenase, BSA, etc.) were measured with the same nanopore (data not shown). If the BSA molecules were rigid charged particles as shown in Figure 1d, the translocation time td would be expected to follow Eq. 2, therefore td would be shorter as increased. To aid visualization, by assuming BSA molecules are rigid charged particles (with position profile shown in Fig. 2) driven through a nanopore by voltage Ψ, but simultaneously moves randomly under thermal motion, we have simulated BSA translocation times (Fig. 5b, open squares □). The simulated BSA distributions (Fig. 5c) show that the particle like BSA translocation times decreased as voltage Ψ increased. To further validate the computer simulation program (see details in Appendix), a rigid uniformly charged 7k dsDNA translocation time was also simulated and the results are compared with the data measured in a nanopore (Fig. 5 def). For both folded BSA and uniformly charged DNA molecules, as voltage Ψ increases, the time durations simulated decrease and the time distributions are Gaussians (Fig. 5d and 5f).
Figure 5.
For the native state BSA, I0=13.2 nA in 1M KCl at 120 mV during the measurement. Voltage dependence of the time duration distributions measured by the nanopore (a), the most probable values of td as a function of voltages (b, ◇), Computer simulated time histograms vs voltage (c). For the 7k dsDNA, I0=11.5±1.5 nA in 1M KCl at 120 mV during the experiment. Voltage dependence of the time duration distributions measured by the nanopore (d), the most probable values of td as a function of voltages (e), Computer simulated time durations vs voltage (f). Error bars were the standard deviation from Gaussian fit to the data.
Discussion
The study in Figure 5 suggests that the highly charged native state BSA molecules can’t be treated as rigid particles during a nanopore translocation experiment. Highly charged protein molecules could be partially denatured upon entering a nanopore at the voltages used, consistent with previous observation using β-lactoglobulin [9] and more recent report using BSA [25]. This assumption is reasonable considering that some proteins unfold at pulling forces larger than ~5 pN [43, 44]. The electrical field strength in a nanopore is E ≈ ψ /Heff = 105V/cm. The electric force experienced per charge is estimated to be eE ≈ 1.6 pN/e. A BSA molecule has about 100 negative and positive residues each at pH 7.0. When a BSA molecule is entering a nanopore, opposite charges in the protein will be driven in opposite directions by the electric field (see Fig. 2b.) and this electric force is estimated to be much larger than the strength of the hydrogen bonds and will reduce the attractive electric force between these charges that hold a protein in a folded shape, thus a BSA could be partially denatured. In addition, the heat produced by current flow in a pore may also contribute to protein denaturing. This electric field effect is not an advantage for the purpose of protein identification; it could cause a previous unfolded protein entering a nanopore with a charged region, not one end of a peptide chain, increase the chance of looped protein translocation configuration.
Conclusion
From a very limited number of reports on studying protein unfolding with solid-state nanopores, we conclude that a solid-state nanopore device is capable of characterizing the process of protein unfolding at single molecule level by measuring a protein’s relative current drop amplitude that is directly proportional to the protein’s excluded volume in a nanopore. A protein in a folded state occupies a larger excluded volume in a nanopore therefore blocks more ionic current flow than the protein in unfolded or random coil state. The technique of solid-state nanopore is not only sensitive enough to distinguish the folding state of a protein; it also gives the distribution of possible states.
The disadvantage of this method is that the electric field used to drive a protein molecule through a nanopore and to produce an ionic current signal can change protein structure, partially unfold a native state protein or stretch an unfolded peptide chain. When an unfolded protein molecule enters a nanopore, due to its heterogeneous charge distribution, a negatively or positively charged region may enter first depending on the voltage polarity, which could cause looped or multiple peptide chain translocation configurations. This looped translocation configuration will not affect the determination of a protein’s folding state, however, this will be a key problem for future applications such as identifying proteins by their linear amino acid chain translocation signal in a nanopore.
Perspective of studying protein unfolding by solid-state nanopores
Characterization of protein unfolding using solid-state nanopores is a new research field that only started several years ago and is still at its initial developing stage. To progress further in this field, experimental and theoretical studies are needed to understand how the translocation dynamics and configurations of unfolded protein molecules in a nanopore are related to protein structure, nanopore geometry, and pore-protein interactions.
Acknowledgements
The authors thank the funding support of this research provided by NIH R21HG003290, NIH R21HG004776, and ABI0710. The authors also thank Professor Jene Golovchenko and the Harvard nanopore group for their assistance on nanopore fabrication.
Appendix
Translocation time simulation
To better understand protein translocation time distribution, we have simulated td in figure 5. Below is a brief description of the computer simulation. We model a BSA molecule as rigid with charge distribution shown as in Figure 1d. When this BSA molecule is pulled through a nanopore by the electric field in a nanopore, the BSA molecule also moves randomly due to Brownian motion along the centerline of the pore. The motion of the BSA molecule can be modeled by a 1-D Langevin equation:
Where ν is the speed of the molecule, Fe(x)=Eq(x) is the electric force, q(x)=∑Qin is the total charge of a BSA in the pore, f is the drag coefficient, related to the diffusion coefficient by f= kbT/D, κ is defined by the fluctuation-dissipation theorem, and W(t) is a ‘noise term’ or Wiener process with zero mean that represents the random thermal forces on the molecule. The variable × is the position of the first part of the molecule that enters the pore.
To calculate time durations td, we simulated ensembles of hundreds of molecules travelling through the pore using the Euler-Murayama numerical methods to solve the one dimensional Langevin equation, which includes the Wiener process term, ΔW, which adds a normally distributed random number with mean zero for each timestep Δt. The change in speed is calculated by
And the change in position is calculated by
The translocation time td is calculated by adding all the timesteps it takes for a BSA or DNA molecule to pass through the nanopore.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Eftink MR. The Use of Fluorescence Methods to Monitor Unfolding Transitions in Proteins. Biophys. J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meinhold DW, Wright PE. Measurement of protein unfolding/refolding kinetics and structural characterization of hidden intermediates by NMR realaxation dispersion. PNAS. 2011 May 11; doi: 10.1073/pnas.1105682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daggett V, Levitt M. Protein Unfolding Pathways Explored through Molecular Dynamics Simulations. J. Mol. Biol. 1993;(232):600–619. doi: 10.1006/jmbi.1993.1414. [DOI] [PubMed] [Google Scholar]
- 5.Oukhaled G, Mathe J, Biance A-L, Bacri L, Betton J-M, Lairez D, Pelta J, Auvray L. Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys. Rev. Lett. 2007;98 doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 6.Stefureac R, Waldner L, Howard P, Lee JS. Nanopore Analysis of a Small 86-Residue Protein. Small. 2008;4(1):59–63. doi: 10.1002/smll.200700402. [DOI] [PubMed] [Google Scholar]
- 7.Merstorf C, Cressiot B, Pastoriza-Gallego M, Oukhaled A, Betton JM, Auvray L, Pelta J. Wild Type, Mutant Protein Unfolding and Phase Transition Detected by Single-Nanopore Recording. ACS Chem. Biol. 2012;7(4):652–658. doi: 10.1021/cb2004737. [DOI] [PubMed] [Google Scholar]
- 8.Payet L, Martinho M, Pastoriza-Gallego M, Betton J-M, Auvray L, Pelta J, Mathé J. Thermal Unfolding of Proteins Probed at the Single Molecule Level Using Nanopores. Anal. Chem. 2012;84(9):4071–4076. doi: 10.1021/ac300129e. [DOI] [PubMed] [Google Scholar]
- 9.Talaga D, Li J. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 2009;131:9287–9297. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledden B, Fologea D, Talaga DS, Li J. Sensing Single Protein Molecules with Solid-state Nanopores, in Nanopores: Sensing and Fundamental Biological Interactions. In: Iqbal SM, Bashir R, editors. New York: Springer; 2011. pp. 129–150. [Google Scholar]
- 11.Sutherland TC, Long Y-T, Stefureac R-I, Bediako-Amoa I, Kraatz H-B, Lee JS. Structure of Peptides Investigated by Nanopore Analysis. Nano Lett. 2004;4(7):1273–1277. [Google Scholar]
- 12.Movileanu L, Schmittschmitt JP, Scholtz JM, Bayley H. Interactions of peptides with a protein pore. Biophys. J. 2005 Aug;89:1030–1045. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefureac R, Long Y, Kraatz H, Howard P, Lee J. Transport of alpha-helical peptides through alpha-hemolysin and aerolysin pores. Biochemistry. 2006;45(30):9172–9179. doi: 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- 14.Pastoriza-Gallego GGM, Thiebot B, Betton J-M, Pelta J. Polyelectrolyte and unfolded protein pore entrance depends on the pore geometry. Biochimica et Biophysica Acta - Biomembranes. 2009;1788:1377–1386. doi: 10.1016/j.bbamem.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, Goot FGvd, Auvray L, Betton J-M, Pelta J. 1.Dynamics of Unfolded Protein Transport through an Aerolysin Pore. J. AM. CHEM. SOC. 2011;133(9):2923–2931. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- 16.Fologea D, Ledden B, McNabb DS, Li J. Electrical Characterization of Protein Molecules in a Solid-State Nanopore. Appl. Phys. Lett. 2007;91 doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han A, Creus M, Schurmann G, Linder V, Ward TR, Rooij NFd, Staufer U. Label-Free Detection of Single Protein Molecules and Protein-Protein Interactions Using Synthetic Nanopores. Anal. Chem. 2008;80:4651–4658. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]
- 18.Han A, Schurmann G, Mondin G, Bitterli RA, Hegelbach NG, de Rooij NF, Staufer U. Sensing protein molecules using nanofabricated pores. Appl. Phys. Lett. 2006;88:093901–093903. [Google Scholar]
- 19.Firnkes M, Pedone D, Knezevic J, Döblinger M, Rant U. Electrically Facilitated Translocations of Proteins through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano Lett. 2010;10(6):2162–2167. doi: 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- 20.Stefureac RI, Trivedi D, Marziali A, Lee JS. Evidence that small proteins translocate through silicon nitride pores in a folded conformation. j. Phys. Condens. Matter. 2010;17(22):454133. doi: 10.1088/0953-8984/22/45/454133. [DOI] [PubMed] [Google Scholar]
- 21.Niedzwiecki DJ, Grazul J, Movileanu L. Single-Molecule Observation of Protein Adsorption onto an Inorganic Surface. Journal of the American Chemical Society. 2010;132(31):10816–10822. doi: 10.1021/ja1026858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Controlling the translocation of proteins through nanopores with bioinspired fluid walls. Nature Nanotechnology. 2011 doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yusko EC, Prangkio P, Sept D, Rollings RC, Li J, Mayer M. Single-Particle Characterization of Aβ Oligomers in Solution. ACS nano. 2012;6(7):5909–5919. doi: 10.1021/nn300542q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sexton LT, Mukaibo H, Katira P, Hess H, Sherrill SA, Horne LP, Martin CR. An Adsorption-Based Model for Pulse Duration in Resistive-Pulse Protein Sensing. J Am Chem Soc. 2010;132:6755–6763. doi: 10.1021/ja100693x. [DOI] [PubMed] [Google Scholar]
- 25.Freedman KJ, Jürgens M, Prabhu A, Ahn CW, Jemth P, Edel JB, Kim MJ. Chemical, Thermal, and Electric Field Induced Unfolding of Single Protein Molecules Studied Using Nanopores. Anal. Chem. 2011;83:5137–5144. doi: 10.1021/ac2001725. [DOI] [PubMed] [Google Scholar]
- 26.Oukhaled A, Cressiot B, Bacri L, Pastoriza-Gallego M, Betton J-M, Bourhis E, Jede R, Gierak J, Auvray L, Pelta J. Dynamics of Completely Unfolded and Native Proteins through Solid-State Nanopores as a Function of Electric Driving Force. Acs nano. 2011;5:3628–3638. doi: 10.1021/nn1034795. [DOI] [PubMed] [Google Scholar]
- 27.Cressiot B, Oukhaled A, Patriarche G, Pastoriza-Gallego M, Betton J, Auvray L, Muthukumar M, Bacri L, Pelta J. Protein transport through a narrow solid-state nanopore at high voltage: experiments and theory. Acs nano. 2012;6(7):6236–6243. doi: 10.1021/nn301672g. [DOI] [PubMed] [Google Scholar]
- 28.Cai Q, Ledden B, Krueger E, Golovchenko JA, Li J. Nanopore Sculpting with Noble Gas Ions. J. Appl. Phys. 2006;100 doi: 10.1063/1.2216880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001 Jul;412(12):166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 30.Stein D, Li J, Golovchenko JA. Ion-Beam Sculpting Time Scales. Phys. Rev. Lett. 2002;89(27):276106-1-4. doi: 10.1103/PhysRevLett.89.276106. [DOI] [PubMed] [Google Scholar]
- 31.Bezrukov SM. Ion Channels as Molecular Coulter Counters to Probe Metabolite Transport. J. Membr. Biol. 2000;174:1–13. doi: 10.1007/s002320001026. [DOI] [PubMed] [Google Scholar]
- 32.DeBlois RW, Bean CP. Counting and Sizing of Submicron Particles by the Resistive Pulse Technique. Rev. Sci. Instrum. 1970;41(7):909–916. [Google Scholar]
- 33.Gregg EC, Steidley KD. Electrical Counting and Sizing of Mammalian Cells in Suspension. Biophys. J. 1965;5:393–405. doi: 10.1016/S0006-3495(65)86724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henriquez RR, Ito T, Sun L, Crooks RM. The Resurgence of Coulter Counting as a Nanoscale Analytical Method. Analyst. 2004;129:478–482. doi: 10.1039/b404251b. [DOI] [PubMed] [Google Scholar]
- 35.Hall JE. Access Resistance of a Small Circular Pore. J.Gen. Physiol. 1975;66:531–532. doi: 10.1085/jgp.66.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King GM, Golovchenko JA. Probing Nanotube-Nanopore Interactions. Phys. Rev. Lett. 2005;95(21) doi: 10.1103/PhysRevLett.95.216103. [DOI] [PubMed] [Google Scholar]
- 37.Hyun C, Rollings R, Li J. Probing Access Resistance of Solid-State Nanopores with a Scanning-Probe Microscope Tip. small. 2012;8(3):385–392. doi: 10.1002/smll.201101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters TJ. Serum Albumin. Adv. Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 39.Sirimulla S, Lerma M, Herndon WC. Prediction of Partial Molar Volumes of Amino Acids and Small Peptides: Counting Atoms versus Topological Indices. J. Chem. Inf. Model. 2010;50:194–204. doi: 10.1021/ci900318c. [DOI] [PubMed] [Google Scholar]
- 40.Zamyatnin AA. AMINO ACID, PEPTIDE, AND PROTEIN VOLUME IN SOLUTION. Ann. Rev. Biophys. Bioeng. 1984;13:145–165. doi: 10.1146/annurev.bb.13.060184.001045. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Talaga DS. The distribution of DNA translocation times in solid-state nanopores. J. Phys. Condens. Matter. 2010;22 doi: 10.1088/0953-8984/22/45/454129. p. 454129 (8pp) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocco AG, Mollica L, Ricchiuto P, Baptista AM, Gianazza E, Eberini I. Characterization of the Protein Unfolding Process Induced by Urea and Temperature. Biophys. J. 2008;94:2241–2251. doi: 10.1529/biophysj.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lellermayer MSZ, Smith SB, Granzer HL, Bustamante C. Folding-Unfolding Transitions in Single Titin Molecules Characterized with Laser Tweezers. Science. 1997;276:1112. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 44.Bechtluft P, Leeuwen RGHv, Tyeman M, Tomkiewicz D, Nouwen N, Tepper HL, Driessen AJM, Tans SJ. Direct Observation of Chaperone-Induced Changes in a Protein Folding Pathway. Science. 2007 Nov;318(30):1458–1461. doi: 10.1126/science.1144972. [DOI] [PubMed] [Google Scholar]