Abstract
ω-3 and ω-6 Polyunsaturated fatty acids (PUFAs) play a role in the pathogenesis of colon cancer. Upon oxidation, PUFAs generate α,β-unsaturated aldehydes or enals, such as acrolein (Acr) and (E)-4-hydroxy-2-nonenal (HNE), which can form cyclic adducts of deoxyguanosine (Acr-dG and HNE-dG, respectively) in DNA. Both Acr-dG and HNE-dG adducts have been detected in human and animal tissues and are potentially mutagenic and carcinogenic. In vivo levels of Acr-dG in DNA are at least two orders of magnitude higher than those of HNE-dG. In addition to the facile reaction with Acr, the higher levels of Acr-dG than HNE-dG in vivo may be due to a lower rate of repair. Previous studies have shown that HNE-dG adducts are repaired by the NER pathway (Choudhury et al., Biochemistry 43 (2004) 7514–7521). We hypothesize that Acr-dG adducts are repaired at a slower rate than HNE-dG and that HNE-dG in DNA may influence the repair of Acr-dG. In this study, using a DNA repair synthesis assay and a LC-MS/MS method, we showed that Acr-dG in a plasmid DNA is repaired by NER proteins, but it is repaired at a much slower rate than HNE-dG in human colon cell extracts, and the slow repair of Acr-dG is likely due to poor recognition/excision of the lesions in DNA. Furthermore, using a plasmid DNA containing both adducts we found the repair of Acr-dG is significantly inhibited by HNE-dG, however, the repair of HNE-dG is not much affected by Acr-dG. This study demonstrates that the NER repair efficiencies of the two major structurally-related in vivo cyclic DNA adducts from lipid oxidation vary greatly. More importantly, the repair of Acr-dG can be significantly retarded by the presence of HNE-dG in DNA. Therefore, this study provides a mechanistic explanation for the higher levels of Acr-dG than HNE-dG observed in tissue DNA.
Keywords: Acrolein, 4-hydroxy-2-nonenal, DNA adduct, nucleotide excision repair (NER), repair kinetics, human colon cells
1. Introduction
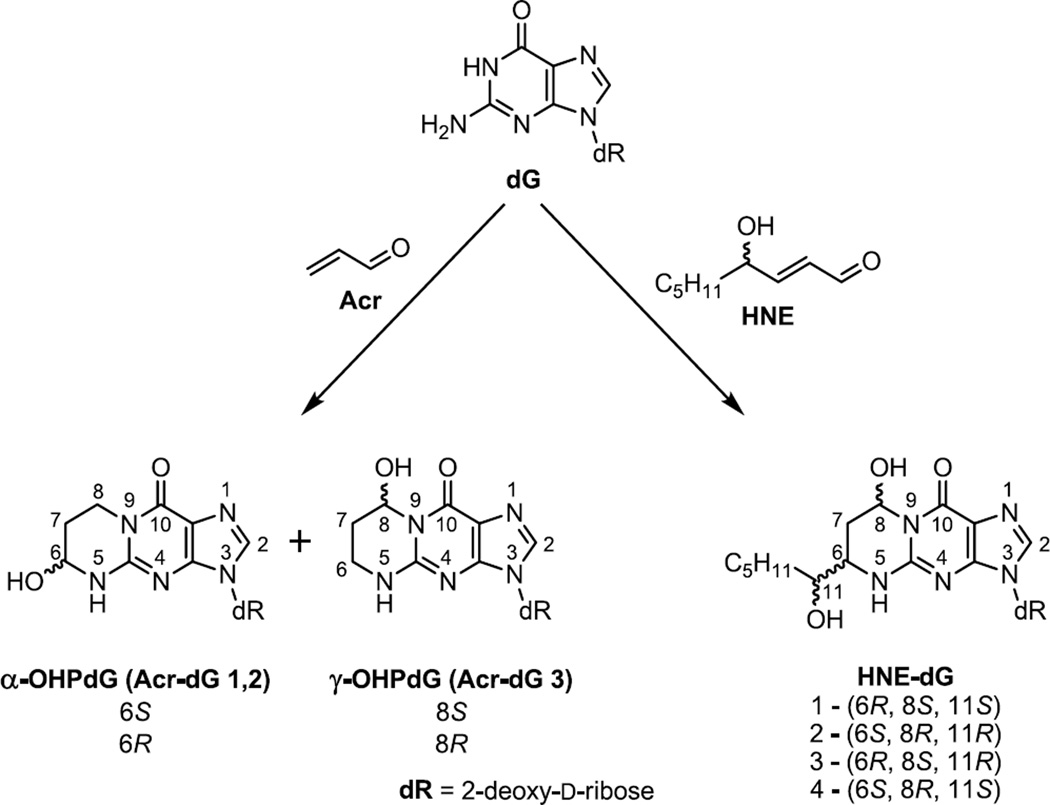
Dietary fats, particularly ω-3 and ω-6 PUFAs, are implicated in the pathogenesis of colon and rectum cancers [1–5]. Studies in animal models and of cell cultures suggest that ω-6 PUFAs may have tumor promoting effects, whereas ω-3 PUFAs seem to protect against the progression of colon cancer [6–10]. Both ω-3 and ω-6 fatty acids are genotoxic via the lipid peroxidation pathway [11,12]. PUFAs can be oxidized by reactive oxygen species to generate electrophilic α,β-unsaturated aldehydes (enals), notably acrolein Acr and HNE, which are capable of forming protein and DNA adducts that may trigger apoptotic and mutagenic responses in cells, depending on the levels of modification, repair, and mutagenic potency [13–15]. Acr and HNE adducts of deoxyguanosine are two major cyclic 1,N2-propano-dG adducts ubiquitously detected in rodent and human tissues as background lesions [16–18]. Both Acr-dG and HNE-dG possess several stereo and regio-isomers (Acr-dG 1,2 and Acr-dG 3, also designated as α-OHPdG and γ-OHPdG, respectively, and HNE-dG 1, 2, 3 and 4; Fig. 1) [19–21]. Among the regio-isomers, Acr-dG 3 is invariably detected as the predominant adduct in vivo. Acr-dG adducts are derived primarily from ω-3 and, to a lesser extent, from ω-6 PUFAs, whereas HNE-dG adducts are a product of oxidized ω-6 PUFAs [12]. Acr-dG can also be formed as a result of environmental exposure, such as cigarette smoke which contains a relatively high amount of Acr [22].
Figure 1.
Chemical Structures of isomeric Acr-dG and HNE-dG adducts.
A previous study showed that Acr-dG adducts, upon reaching a threshold level in DNA, may induce apoptosis in colon cells [13]. But the role of Acr-dG in mutagenesis remains unsettled, as the results of the site-specific mutagenesis studies seem to depend on the assay conditions and its regio-chemistry [23–31]. However, importantly studies on binding and mutations in the p53 gene by Acr and HNE provide evidence that supports a mutagenic role of these cyclic dG adducts in human cancers [32–34]. The basal levels of Acr-dG detected in vivo are estimated to be in the range of adduct per 107 bases, whereas levels of HNE-dG are adduct per 109 bases, i.e. approximately two orders of magnitude lower than Acr-dG [16,17]. While the difference is remarkable, the reason remains unclear. The levels and persistence of adducts in tissues are determined by rates of formation, repair, and DNA replication. It is plausible that, in addition to its facile formation, the low repair rate of Acr-dG may contribute to the difference. Cyclic adducts are repaired either by base excision repair (BER) or nucleotide excision repair (NER) mechanisms. The etheno adducts, which can be generated by epoxides of enals or other products of oxidative metabolism, are mainly repaired by the BER pathway, initiated by a specific DNA glycosylase, N-methylpurine DNA-glycosylase (MPG) and thymine DNA glycosylase, present throughout the phylogeny, including humans [35–40]. However, 1,N2-propano-dG (P-dG), a model bulky cyclic propano adduct, is repaired by the NER pathway [41]. In E. coli, Acr- or other enal-derived cyclic propano DNA adducts, like the model adduct, are known to be repaired by NER [30]. In human cells, HNE-dG adducts are shown to be repaired by the NER pathway [42]. More recently it was reported that NER is also involved in the repair of Acr-dG [26]. However, the repair rate of Acr and HNE adducts has not been directly compared. This is important because it may shed light onto the difference in Acr-dG vs. HNE-dG levels observed in vivo. The purpose of this study is to examine and compare the repair kinetics of Acr- and HNE-dG in human cell extracts using a combination of the repair synthesis assay and the LC-MS/MS method, the former assay is to determine the total repair activity (recognition/excision/ gap synthesis) and the latter is to measure the recognition/excision activity.
2. Materials and methods
2.1. Chemicals
HPLC grade acetonitrile (ACN) and methanol (MeOH) were purchased from EMD (EMD Chemicals, Gibbstown, NJ). Ammonium formate was acquired from Fluka (Fluka Chemie GmbH, Switzerland). Alkaline phosphatase grade I from calf intestine was purchased from Roche (Roche Applied Science, Indianapolis, IN). Acr was purchased from Alfa Aesar (Alfa Aesar, Ward Hill, MA). HNE was a generous gift from Dr. Shantu Amin of Penn State University. [13C10,15N5]-2′-deoxyguanosine was obtained from Spectra Stable Isotopes (Cambridge Isotope Laboratories, Andover, MA). 2′-deoxyguanosine monohydrate, 2′-deoxyguanosine, deoxyribonuclease I from bovine pancreas Type II (DNase I) and purified phosphodiesterase I from Crotalus adamanteus venom were purchased from Sigma (Sigma-Aldrich Corp., St. Louis, MO). All other reagents used were analytical or HPLC grade.
2.2. Cell culture
The NER-proficient cell line GM000637 (Coriell, Camden, NJ) and NER deficient human XPA cells (kindly provided by Dr. Randy Legarski, MD Anderson Cancer Center, Houston, TX) were grown in DMEM and Minimum Essential Medium (MEM; Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and penicillin/streptomycin (Gibco/Invitrogen, Carlsbad, CA) in a 5% CO2 humidified incubator. XAN1cells, derivative of XPA cells stably transfected with XPA minigene [43], kindly provided by Dr. J. Christopher States, University of Louisville School of Medicine, Louisville, KY, were grown in MEM (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and penicillin/streptomycin. The NER-proficient colon cancer cell line HT-29 was grown under the same conditions as for GM000637.
2.3. Preparation of nuclear extracts for the in vitro NER assay
The nuclear extracts were prepared from HT-29, XPA, XAN1 and GM000637 cells using an NE-PER kit from PIERCE/Thermo Fisher Scientific (Rockford, IL) with a protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN) and stored at −80° C in small aliquots. The typical yield was 4–5.5 mg of protein from a single 10-cm plate, and the concentration ranged from 4 to 6 mg/mL. Each aliquot was thawed only once for the in vitro NER activity assay to avoid inactivation due to repeated freeze-thaw cycles.
2.4. Preparation of plasmid substrates containing HNE-dG and Acr-dG
HNE was synthesized according to the method previously described [44]. The Acr was purchased from Sigma-Aldrich Co., St. Louis, MO. The pBluescript (pBSII) plasmid DNA was received from Recombinant DNA Laboratory core facility, UTMB, Galveston, TX, and used for subsequent Acr and HNE modifications. The plasmid DNA was modified with HNE as described by Hu et al. [32]. Purified pBSII (10 µg) in TE buffer (10 mM Tris, 1 mM EDTA, pH 7.2) was incubated with a final concentration of 15 mg/mL of HNE (stock solution, 100 mg/mL in methanol) at 37° C for 20 h. Control pBSII was treated with methanol only and used as a HNE-untreated substrate in subsequent NER assays. The unreacted HNE was removed by repeated phenol/chloroform extraction. The Acr-modified plasmid DNA substrate was prepared by treating purified pBSII (11.2 µg) in 100 µL with a final concentration of 8 mg/mL Acr in PBS, pH 7.2 at 37° C for 10 min. Control pBSII was treated with the buffer only and used as Acr-untreated substrate in subsequent NER assays. The unreacted Acr was removed by repeated phenol/chloroform extraction. The dual-modified plasmid DNA substrate was prepared by treating purified pBSII (11.2 µg) in 100 µL with a final concentration of 15 mg/mL of HNE in PBS, pH 7.2 at 37° C for 20 h, and then Acr was added at a final concentration of 8 mg/mL to the same reaction mixture for further 10 min incubation at 37° C. The unreacted HNE and Acr were removed by repeated phenol/chloroform extraction. All the treated plasmids were then precipitated with ethanol, dissolved in TE buffer (pH 7.2), and used to quantify the HNE-dG and Acr-dG level in both single- and dual-modified plasmids by LC-MS/MS.
2.5. Repair synthesis assay by [32P] incorporation/agarose gel electrophoresis
In typical 20 µL reaction mixtures, 300 ng each of HNE- and Acr- modified pBSII, or untreated pBSII closed circular plasmids were incubated at 30° C for stipulated times with HT-29, XPA, XAN1 or GM000637 cell nuclear extracts (60 µg) in the presence of 74 kBq of α-[32P] dCTP (110 TBq/mmol; Amersham Pharmacia Biotech, Piscataway, NJ); 13 mM HEPES-KOH (pH 7.9); 50 mM KCl; 7.4 mM MgCl2; 1 mM DTT; 2 mM ATP; 50 mM each dGTP, dATP, dTTP, and 10 mM dCTP; 40 mM creatine phosphate; 0.5 µg of creatine phosphokinase (Type I; Sigma-Aldrich Co., St. Louis, MO); and 6.4 µg of BSA as described [42]. The reaction was stopped by adding 25 mM EDTA, subjected to digestion with RNAse and Proteinase K as described [45]. DNA was purified using a QIAquick Nucleotide Removal kit (Qiagen, Valencia, CA). The purified plasmid DNA was then linearized with EcoRI and electrophoresis was performed on a 1% agarose gel containing 0.5 µg/mL ethidium bromides. It was then dried on DE-81 filter paper, and exposed to Phosphor Imager (Amersham Pharmacia Biotech, Piscataway, NJ) for visualization and quantification of radioactivity in the bands. A standard curve was generated by spotting a known amount of α-[32P] dCTP on the DE-81 paper, which was then processed along with the dried gel.
2.6. Repair Excision Assay by Detection of HNE-dG and Acr-dG Adducts in Plasmid DNA by the LC-MS/MS method
A LC-MS/MS method was applied to directly measure HNE-dG adducts in plasmid DNA before and after the repair-synthesis assay. To yield sufficient DNA for this assay, repair synthesis was carried out under proportionally scaled-up conditions as compared to the methods described earlier. The assay was performed with 2 µg of HNE-dG-containing and Acr-dG-containing pBSII DNA, and DNA modified with both HNE and Acr, or untreated DNA, 100 µg HT-29 cell nuclear extracts and similar concentrations of dNTPs (no α-[32P] dCTP) and other essential ingredients described earlier. The plasmid DNA from repair reactions was purified using a combination of QIAquick Nucleotide Removal kit (Qiagen, Valencia, CA) and agarose gel electrophoresis and then analyzed for remaining HNE-dG and Acr-dG adducts by the LC-MS/MS method. The details of the latter method are given below.
2.7. Synthesis of Acr-dG, [13C10,15N5]-Acr-dG, HNE-dG, and [13C10,15N5]-HNE-dG standards
Acr-dG was prepared as described previously [19] using arginine to improve the reaction yield [46]. HNE-dG was prepared as before [20]. Both [13C10,15N5]-Acr-dG and [13C10,15N5]-HNE-dG as an internal standard were prepared by the same methods as their parent compounds but using [13C10,15N5]-dG instead of dG. Detailed conditions for all reactions and compound characterization data can be found in the supporting materials.
2.8. Plasmid DNA hydrolysis for quantification of Acr-dG and HNE-dG by LC-MS/MS
DNA (0.1–10 µg) was dissolved in 125–150 µL of 5 mM magnesium chloride and spiked with internal standards. Samples for Acr-dG quantification were spiked with 100 fmol of [13C10,15N5]-α-Acr-dG and 50 fmol of [13C10,15N5]-γ-Acr-dG. Samples for HNE-dG quantification were spiked with 100 fmol of mixture of [13C10,15N5]-HNE-dG isomers 1 and 2, 50 fmol of [13C10,15N5]-HNE-dG isomer 3, and 50 fmol of [13C10,15N5]-HNE-dG isomer 4. For quantification of both Acr-dG and HNE-dG adducts together samples were spiked with 200 fmol of [13C10,15N5]-α-Acr-dG, 100 fmol of [13C10,15N5]-γ-Acr-dG, 200 fmol of a mixture of [13C10,15N5]-HNE-dG isomers 1 and 2, 100 fmol of [13C10,15N5]-HNE-dG isomer 3, and 100 fmol of [13C10,15N5]-HNE-dG isomer 4. Enzymatic hydrolysis was performed by incubation with 65 units of DNase I at 37° C for 30 min followed by a second addition of 65 units of DNase I. After an additional 10 min at 37° C, 19 units of Alkaline phosphatase and 0.003 units of phosphodiesterase I were added, and the mixture was incubated for 1 h. After hydrolysis, 25 µL of hydrolysate was frozen and kept at −80° C for further dG quantification. The rest of the hydrolysate was purified by solid phase extraction (SPE) using C18, 200 mg, Bond Elut columns (Varian Inc., Lake Forest, CA). The columns were conditioned by ACN (3 × 1 mL) and water (3 × 1 mL). After sample loading, columns were washed with water (2 × 1 mL), and samples were collected with 20% ACN (2 × 1 mL) when used only for Acr-dG adduct quantification, or by 30% ACN (2 × 1 mL) if the samples were used for HNE-dG or HNE-dG and Acr-dG adducts together. After SPE, the eluants were dried over a vacuum using a SpeedVac centrifugal concentrator, re-dissolved in 1:1 ACN:water (2 × 400 µL), moved to autosampler vials, dried, and kept at −80° C. For quantification, assay samples were reconstituted in 50 µL of water, and 37 µL of sample was injected onto LC-MS/MS.
2.9. Acr-dG and HNE-dG quantification by LC-MS/MS
Acr-dG and HNE-dG adducts were quantified by similar methods as previously reported [18,47,48]. Briefly, mass spectrometry quantification was carried out on an AB SCIEX API 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX, Framingham, MA) coupled with a Waters ACQUITY UPLC liquid chromatography equipped with Waters ACQUITY UPLC BEH C18, 50 × 2.1 mm, 1.7 µm particle size column (Waters Corporation, Milford, MA). The separation of Acr-dG and HNE-dG adducts was performed by isocratically eluting with 3% and 12.5%, respectively ACN in 1 mM ammonium formate buffer over 2.5 and 10 min, respectively using a 0.5 mL/min flow rate at 40° C, followed by a 100% ACN wash. The ESI source was used in positive mode for both adducts. Adducts were quantified using multiple reaction monitoring mode (MRM). Ion transitions of 324.2→208.1 m/z (Acr-dG) and 339.2→218.1 m/z ([13C10,15N5]-Acr-dG) with a collision energy (CE) of 20 eV were used for quantitation, and those of 324.2→190.1 m/z (Acr-dG) and 339.2→200.1 m/z ([13C10,15N5]-Acr-dG) with a CE of 47 eV were used for structural conformation of Acr-dG. For HNE-dG, ion transitions of 424.2→308.2 m/z (HNE-dG) and 439.2→318.2 m/z ([13C10,15N5]-HNE-dG) with CE of 19 eV were used for quantitation, and those of 424.2→290.2 m/z (HNE-dG) and 439.2→300.2 m/z ([13C10,15N5]-HNE-dG) with a CE of 31 eV were used for structural conformation. A more detailed description of this assay can be found in the supporting materials.
2.10. dG quantification by UV-HPLC
The levels of adduct in DNA are expressed as number of adduct/dG. dG in DNA was analyzed by reversed–phase HPLC using enzymatically hydrolyzed DNA samples. A more detailed description of this assay is given in the supporting materials.
2.11. Statistical analysis
Statistical analyses were performed on data collected from at least three independent experiments. Student’s t test (one-tailed) was used to evaluate the statistical difference, and the differences were considered statistically significant when P<0.05.
3. Results
3.1. Acr-dG is repaired by the NER pathway in human cell free extracts, but its repair is less efficient than HNE-dG
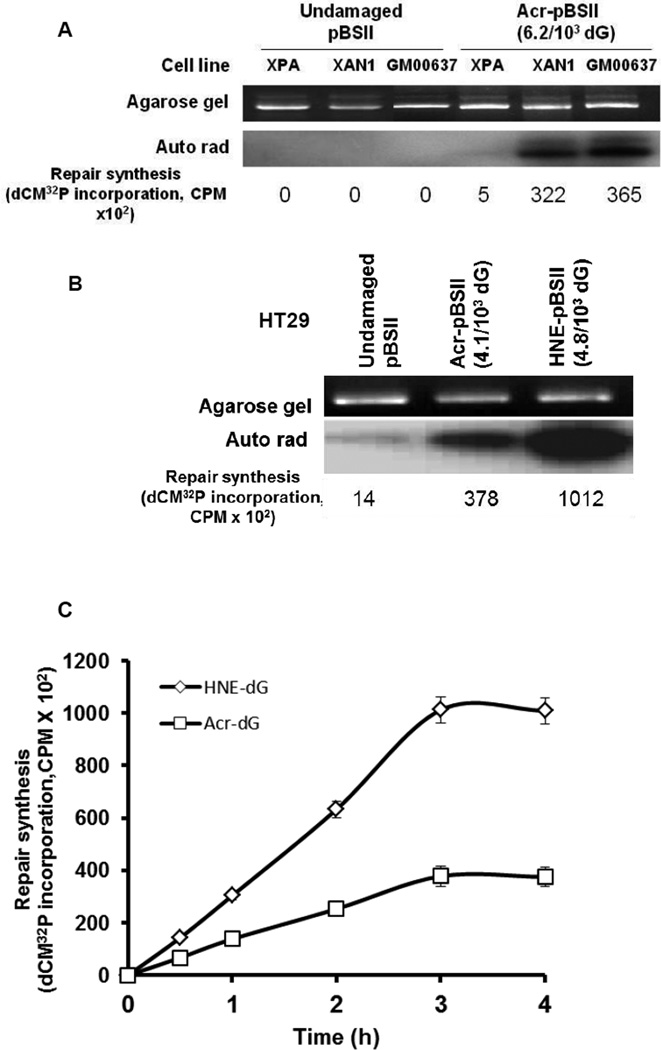
To determine and verify that the NER pathway is involved in the repair of Acr-dG, we used the NER-proficient and NER-deficient (XPA) human cells in a cell-free assay similar to that described previously for HNE-dG [42]. We found that the extracts from the normal fibroblast line (GM000637) has a substantial DNA repair synthesis capacity for Acr-dG in pBluescript II (pBSII; 4.1 adducts/103 dG), whereas the extracts from the NER-deficient line (XPA) show very little or no repair. However, extracts from XAN1, a derivative of XPA cells stably transfected with XPA minigene and expressing active XPA protein [43], restored the repair synthesis of Acr-dG (Fig. 2A). After establishing the role of NER in Acr-dG repair in XPA cells, we next compared the repair efficiencies of Acr-dG and HNE-dG adducts using human colon cell extracts. The pBSII plasmid DNA containing HNE-dG or Acr-dG was used as a substrate. The results indicate that at comparable levels of modification, HNE-dG (4.8/103 dG) appears to be more efficiently repaired than Acr-dG (4.1/103 dG) through the NER pathway (Fig. 2B). Because of the small quantity of plasmid DNA (300 ng) used, it was necessary to use the highly modified substrates for reliable quantification of adducts by the LC-MS/MS method, especially in the post-repair samples (see section 3.3). A time study was then carried out to ascertain that the difference in repair is not observed at only one time point. The kinetics showed that repair synthesis for both adducts increases steadily as time progresses and reaches a maximum between 3–4 h (Fig. 2C). Thus, the repair of Acr-dG and HNE-dG in human cell nuclear extracts occurs in a time-dependent manner and it is also clear that Acr-dG is repaired at a significantly slower rate than HNE-dG (P<0.05).
Figure 2.
DNA Repair synthesis of Acr-dG or HNE-dG adducts in plasmid DNA. (A) To confirm the NER’s role in Acr-dG repair, DNA Repair synthesis assays were carried out by incubating the plasmid DNA (pBSII) with (Acr-pBSII) and without (Undamaged) Acr-dG modification with cell extracts from XPA (NER–deficient), XAN1 (NER-proficient) and GM00637 (NER-proficient) (60 µg) for 3 h at 30° C. (B) Comparison of DNA repair synthesis of Acr- or HNE-dG adducts in pBSII DNA upon incubations with HT-29 cell nuclear extracts. The ethidium bromide stained gel is shown in the upper panel and autoradiogram of the dried gel in the lower panel. The DNA Repair synthesis activity for recognition/excision and gap synthesis is expressed as radioactivity (CPM) for dCM32P incorporation. (C) The kinetics of DNA Repair synthesis of Acr- or HNE-dG adducts in pBSII DNA incubated with HT-29 cell nuclear extracts. The reaction conditions are the same as described in Fig. 2 A. The repair synthesis is expressed as CPM for dCM32P incorporation. The data are the mean value derived from three independent experiments with standard errors. P<0.05 by Student’s t test (one-tailed).
3.2. Excision repair of Acr-dG and HNE-dG in plasmid DNA determined by LC-MS/MS
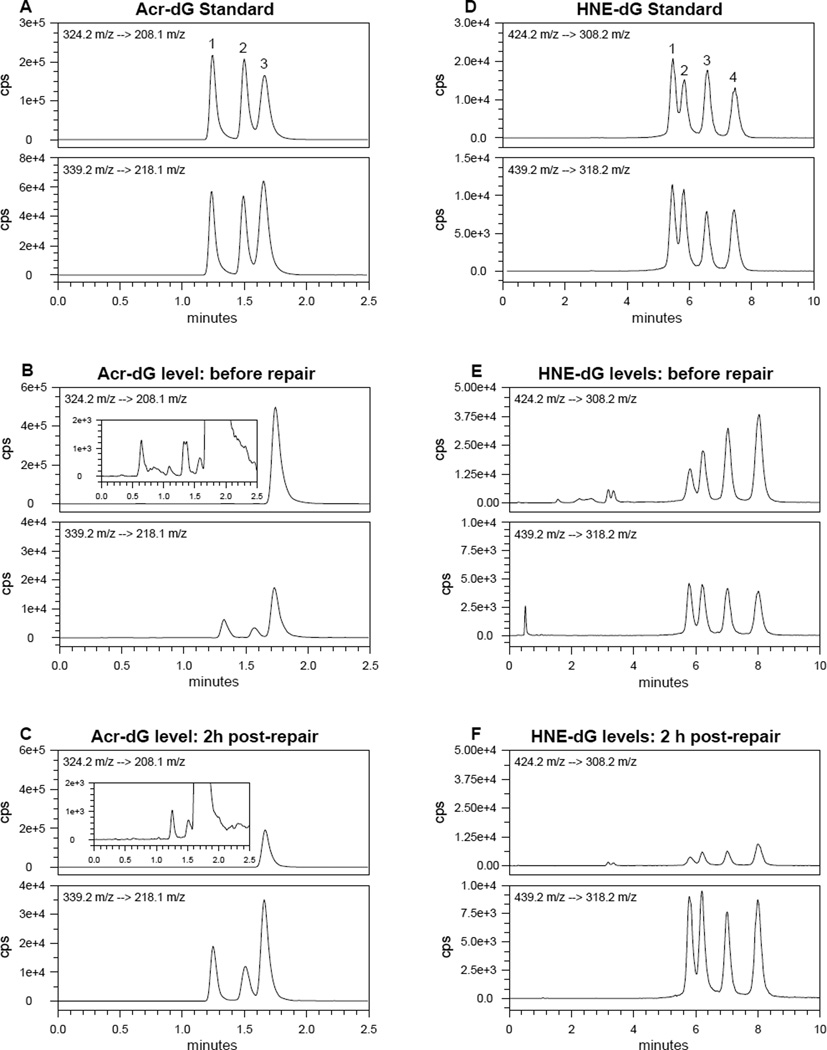
To examine whether Acr-dG and HNE-dG adducts are specifically removed from plasmid DNA we used the LC-MS/MS method. Unlike the in vitro incorporation assay described above that determines complete repair activity (excision and gap-synthesis), the LC-MS/MS method can specifically quantify the Acr-dG and HNE-dG adducts remaining in the substrates after repair, namely the excision steps of the repair process. In the Acr-modified plasmid DNA, Acr-dG3 (γ-OHPdG) was detected as by far the predominant isomeric adduct (Fig.3A and B). This is expected based on previous studies showing the regio-selective formation of Acr-dG 3 in DNA in Tris-containing buffer [49]. It was found that ~30% of Acr-dG (Fig 3A, B and C) vs. 70–80% of HNE-dG (Fig 3D, E and F) were excised in nuclear extracts of human colon HT29 cells within 2 h at 30° C. Therefore, the results of the LC-MS/MS assays corroborate those from the repair synthesis assay by gel electrophoresis and demonstrate specifically that Acr-dG and HNE-dG in the modified DNA can be excised and repaired by human colon cells and that HNE-dG adducts are repaired significantly more efficiently than Acr-dG.
Figure 3.
Multiple reaction monitoring mass chromatograms for Acr-dG or HNE-dG adducts. (A) Acr-dG adduct standards (upper panel, 324.2→208.1 m/z) and [13C10,15N5]-Acr-dG internal standards (lower panel, 339.2→218.1 m/z). The lower panel is internal standards. (B) Acr-dG levels in pBSII DNA (upper panel) before repair. The lower panel is internal standards. Acr-dG 3 is the major isomer and the insert with a different scale shows small amounts of Acr-dG 1 and 2 are present in the plasmid DNA. (C) Acr-dG levels in pBSII DNA (upper panel) after incubating with HT-29 cell nuclear extracts in the DNA repair experiments for excision activity. The lower panel is internal standards. (D) HNE-dG standards (upper panel, 424.2→308.2 m/z) and [13C10,15N5]-HNE-dG internal standards (lower panel, 439.2→318.2 m/z). (E) HNE-dG levels in pBSII DNA (upper panel) before repair and internal standards (lower panel). (F) HNE-dG levels in pBSII DNA (upper panel) after incubating with HT-29 cell nuclear extracts in the DNA repair experiments for excision activity. The lower panel is internal standards.
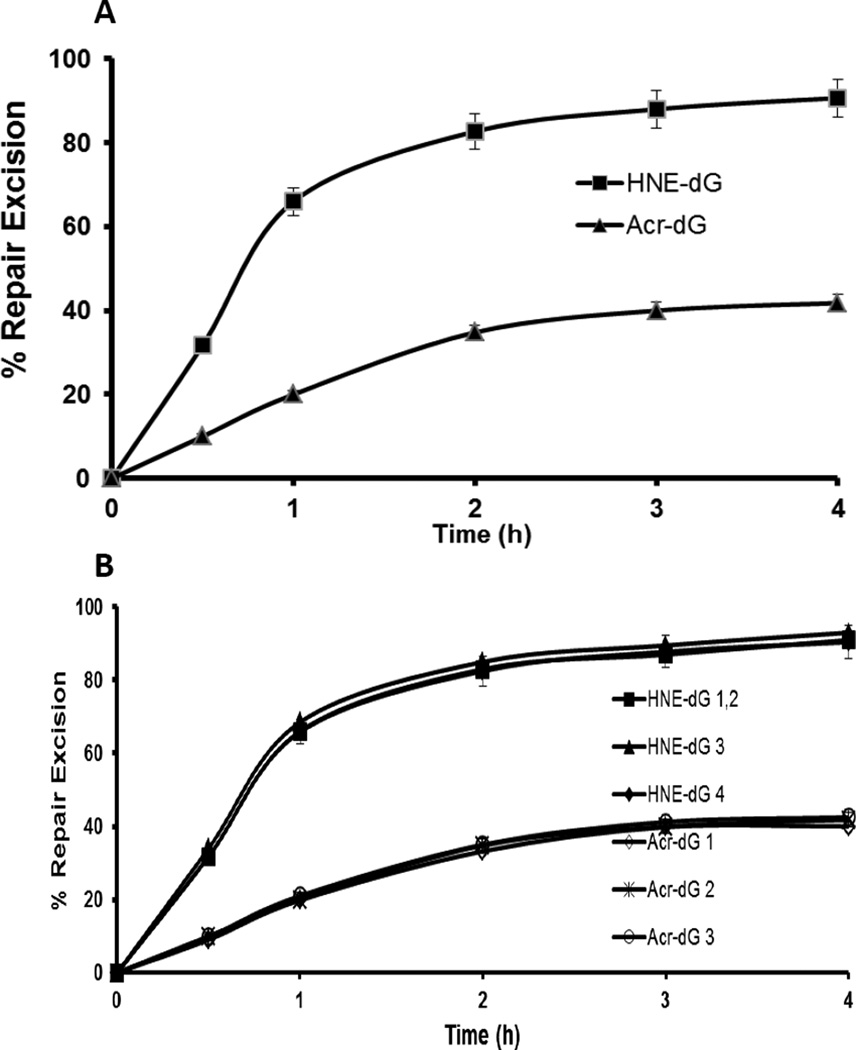
3.3. Acr-dG and its isomers are excised following a slower kinetics than HNE-dG and its isomers by human colon cell extracts
We then determined by the LC-MS/MS method the repair kinetics of Acr and HNE dG under similar assay conditions by measuring the residual adducts in the substrates after repair at various time points up to 4 h. The slower excision rate of Acr-dG than that of HNE-dG is statistically significant (Fig 4A, P<0.01). The results showed that the rate of excision of both Acr-dG and HNE-dG in the plasmid DNA reached a maximum between 2–4 h (Fig. 4A). Whereas more than 60% of HNE-dG in DNA was excised within one h, the excision of Acr-dG did not exceed 50% even at 4 h. Taken together, the results from the complete repair synthesis assay by dCM32P incorporation (Fig. 2) and of the similar kinetics shown by the two methods suggest that the slower rate of Acr-dG repair compared to HNE-dG in plasmid DNA is most likely a result of inefficient excision, instead of the incorporating step (gap synthesis) during repair. Moreover, based on the levels of each Acr-dG isomers detected, it was concluded that the rates of repair of all three Acr-dG isomers are nearly identical (Fig 4B). Similar to the previous findings, the four HNE-dG isomers were repaired at almost equal efficiency under the assay conditions [42]. All the isomers of Acr-dG are also excised at a significantly (P<0.01) slower rate than the isomers of HNE-dG (Fig. 4B).
Figure 4.
The kinetics of DNA Repair excision activity of (A) Acr- or HNE-dG adducts in pBSII DNA and (B) their respective isomers. The plasmid DNA containing 4.2 Acr-dG/103 dG or 2.3 HNE-dG/103 dG was separately incubated with HT-29 cell nuclear extracts under the conditions as described in section 2.5. The repair kinetics of Acr and HNE dG under similar assay conditions was determined by the LC-MS/MS method by measuring the residual adducts in the substrates after repair at various time points up to 4 h. The data are the mean value derived from three independent experiments and presented with standard errors. A, P<0.01 and B, P<0.01 by Student’s t test (one-tailed).
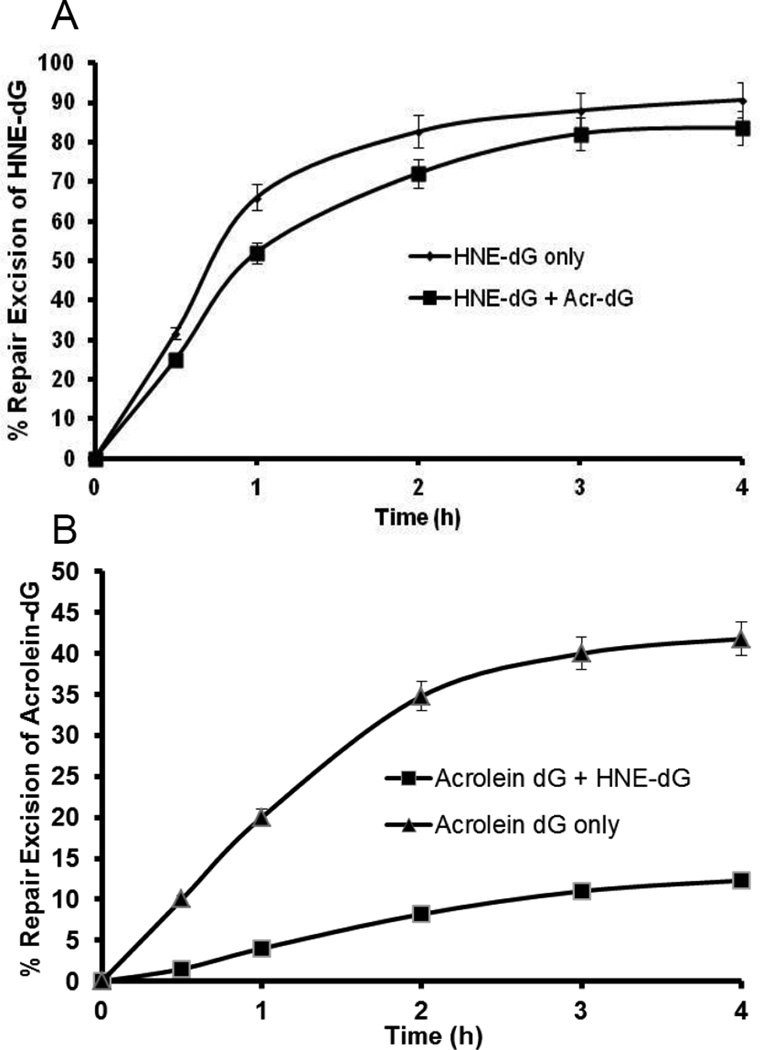
3.4. HNE-dG in DNA retards the repair of Acr-dG, but not vice versa
Next, we determined to what extent Acr-dG and HNE-dG influence each other in repair. This is an important question because cellular DNA is modified by more than one type of adduct and it is expected that the repair machineries have to process multiple classes of adducts simultaneously. However, only limited data is available, primarily due to a lack of specificity of the dCM32P incorporation assay to detect and quantify specific adducts in DNA. The LC-MS/MS method capable of detecting the specific adducts provides a means to address this fundamental question. Using the plasmid DNA modified by both Acr and HNE with levels of Acr-dG and HNE-dG at 4.5/103 dG and 2.5/103 dG, respectively in the same DNA, the repair kinetics of HNE-dG and Acr-dG in human colon cell extracts were then determined. The data showed that the presence of Acr-dG does not have much effect on the repair excision of HNE-dG (Fig. 5A P>0.05). On the other hand, the repair excision of Acr-dG was significantly inhibited at all time points by the presence of HNE-dG in DNA (Fig. 5B, P<0.01).
Figure 5.
The kinetics of DNA Repair excision activity of pBSII DNA containing both Acr- and HNE-dG adducts. The dual-modified plasmid DNA containing 4.5 Acr-dG/103 dG and 2.5 HNE-dG/103 dG was incubated with HT-29 cell nuclear extracts as described in section 2.5. The LC-MS/MS method was used to determine (A) Repair kinetics of HNE–dG in dual-modified vs. single-modified plasmid DNA and (B) Repair kinetics of Acr-dG in dual-modified vs. single-modified plasmid DNA. The data are the mean value derived from three independent experiments and presented with standard errors. A, P>0.05 and B, P<0.01 by Student’s t test (one-tailed).
4. Discussion
This study demonstrated that, like HNE-dG, Acr-dG is repaired by NER proteins, but its repair follows a significantly slower overall kinetics than HNE-dG in extracts from human colon cells and that the slow repair of Acr-dG is likely due to poor excision efficiency. It also showed that when both adducts are present in DNA the HNE-dG repair was not greatly affected by the presence of Acr-dG in DNA, whereas the repair of Acr-dG was significantly inhibited by HNE-dG. These results provide a mechanistic explanation for the high levels of Acr-dG observed in rodent and human DNA [16,17].
It is generally recognized that the repair rate of DNA adducts can be influenced by their structures. For example, although the small oxidative adducts, such as 8-oxoguanine and thymine glycol, are primarily repaired by base excision repair pathway, the UV-damaged bulky DNA adducts and the small DNA oxidative adducts can be both repaired by NER. However, these oxidative small adducts are repaired with a significantly lower efficiency than the UV-damages [50, 51]. Our previous study also showed that HNE-dG is repaired via NER preferentially to UV-lesions [42]. It is expected that because of the structural similarity of Acr-dG to P-dG, a model compound without the OH group in the propano ring moiety that is shown to be repaired by NER, the same pathway should also be involved in the repair of the Acr adducts. Interestingly, the presence of the OH group at either the 6- or 8-position of Acr-dG does not affect its repair rate (Fig. 4B), but the presence of a long bulky alkyl group at the 6-position in HNE-dG seems to harbor NER activity significantly (Figs. 2 and 4). By directly comparing the repair of Acr and HNE adducts this study provides concrete evidence that Acr-dG adducts are repaired by NER at a much slower rate than HNE-dG adducts.
Our data further suggest that the lower rate for the repair of Acr-dG than that of HNE-dG may lie in the recognition/excision steps of the NER pathway. Recognition/excision has been found to be the slowest and thus the rate-limiting step of NER for UV-adducts [52]. It is reasonable to assume that the rates of gap-synthesis would be similar for both HNE-dG and Acr-dG once they are excised from DNA. Thus, the slower rate of recognition/excision of Acr-dG (Fig. 4A) is the main reason underlying the slower repair of Acr-dG by NER (Figs. 2B and C). However, both HNE and Acr are known to generate various DNA adducts other than the modifications of dG [53,54]. The presence of these other adducts, albeit at lower levels, may have effect in repair synthesis kinetics (Fig. 2C) obtained by the repair synthesis assay, and may account for differences in synthesis kinetics from excision kinetics measured by LC-MS/MS method for either Acr-dG or HNE-dG only (Fig. 4). It is not known why Acr-dG is not efficiently recognized and/or excised compared to HNE-dG, as the X-ray crystallography study of the three-dimensional structures of protein complexes with HNE-dG or Acr-dG adducted DNA is not as yet available. It is conceivable that, compared to Acr-dG, HNE-dG adducts are better recognized and/or excised, thus NER proteins are preoccupied with the repair of HNE-dG in DNA. This is analogous to the observation that 3,N4-ethenocytosine (eC) inhibits the excision activity of another structurally-related adduct, 1,N6-ethenoadenine (eA) by a BER DNA-glycosylase, MPG, because MPG seems to bind tightly to eC, but does not excise eC [55–57]. Surprisingly, the repair synthesis of Acr-dG is not only slower than HNE-dG, but it also plateaued at a much lower level (Fig. 2C). The same is true for the excision activity towards these lesions (Fig. 4). Thus, it seems that some Acr-dG lesions are completely refractory to repair, possibly due to poor recognition and it is isomer-independent (Fig. 4B). This phenomenon may also account for the greater accumulation of Acr-dG than HNE-dG in the genome. It is generally acknowledged that NER proteins in mammalian cells recognize different types of DNA lesions with different binding affinities, which determine the relative kinetics of the repair of different lesions. For example, the 6-4 photoproducts are recognized and repaired more rapidly than the cyclobutane pyrimidine dimer (CPD) in human and rodent cells [58,59]. Therefore, the difference in the ability of NER complex to locate and incise HNE-dG and Acr-dG adducts in DNA may contribute to the differential in repair efficiency for HNE-dG vs. Acr-dG. It is also possible that Acr and HNE adduction may affect DNA helical structure in different ways; consequently, they are recognized and excised at different rates. Indeed, the lesion recognition by NER proteins does not depend solely on the chemical structure of the DNA lesion itself, but lesion-induced alterations in DNA helical structure, such as local unwinding of a few DNA bases around the damaged site, may also play a critical role in the recognition step. During this unwinding process, the DNA bends resulting in further unwinding by NER enzymes. Additionally, the DNA lesions opposite a mismatch become even better substrates for NER proteins [60]. Human colon HT29 cells were used in this study because ω-3 and ω-6 PUFAs, the endogenous sources of the cyclic adducts, are implicated in colon carcinogenesis [1–5]. Intriguingly, the HT29 cell extracts seem more efficient and robust than HeLa cell extracts in excision and repair synthesis reactions. As previously shown, HeLa extracts could excise ~50% of total HNE-dG from DNA in 2.5 h [42], however, HT29 cell extracts excised the similar amount of adducts in less than an hour (Fig. 4A). In fact, ~90% of total HNE-dG adducts are removed in 3–4 h by HT29 cell extracts.
The steady-levels of adducts in tissue DNA are the net outcome of their formation and removal, and they also depend on the rate of cell proliferation. The results of this study, although using plasmids with much higher modification than that found in vivo, showed that the higher basal levels of Acr-dG in tissue DNA may be due in part to its slow repair, especially when HNE-dG adducts are present in the DNA. Our findings, as a proof of principle, suggest that the interactions among different adducts in DNA with respect to their repair could be an important factor for their ultimate levels in vivo. As the formation of DNA adduct is a critical determinant of how cells respond to the damage, a recent study showed that Acr-dG after reaching a threshold in the DNA of human colon cells may contribute to the apoptosis induced by docosahexaenoic acid [13]. This observation raises an intriguing question that is whether the accumulation of Acr-dG in cells due to its poor repair could play a role in the cancer protective effect of ω-3 polyunsaturated fatty acids.
Supplementary Material
Highlights.
The repair kinetics of two common cyclic DNA adducts derived lipid peroxidation, Acr-dG and HNE-dG, in human colon cells was studied
This study is the first to measure DNA repair rates by both DNA repair synthesis and LC-MS/MS methods
Acr-dG, like HNE-dG, is repaired by NER pathway, but it is repaired at a much slower rate compared to HNE-dG
HNE-dG can inhibit the repair of Acr-dG if both are present in the same DNA
These results provide an explanation for the higher levels of Acr-dG than HNE-dG observed in vivo
Acknowledgements
We thank the Proteomics & Metabolomics Shared Resource of Lombardi Comprehensive Cancer Center (supported partially by NIH/NCI grant P30-CA051008) for providing resources for DNA adducts analysis. We also thank Dr. Shantu Amin of Penn State University for providing us HNE, and Dr. J. Christopher States of University of Louisville School of Medicine for providing us XAN1 cells.
Role of the funding source
The work was supported by NIH/NCI grants RO1 CA113447 (R. Roy) and RO1 CA43159 (F.-L. Chung). The funding sources had no involvement in study design, data collection, data analysis and interpretation, the writing of the article or the decision to submit the article for publication.
Abbreviations
- ACN
acetonitrile
- Acr
acrolein
- BEH
bridged ethyl hybrid
- BER
base excision repair
- CE
collision energy
- ESI
electrospray ionization
- HNE
(E)-4-hydroxy-2-nonenal
- HNE-dG
HNE-derived 1,N2-deoxyguanosine adducts
- HPLC
high performance liquid chromatography
- LC
liquid chromatography
- MRM
multiple reactions monitoring mode
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- NER
nucleotide excision repair
- OHPdG (Acr-dG)
α- and γ-OH-1,N2-propanodeoxyguanosine
- P-dG
1,N2-Propano-dG
- PUFA
polyunsaturated fatty acid
- UPLC
ultra performance liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Appendix A. Supplementary data
References
- 1.Caygill CP, Hill MJ. Fish, n-3 fatty acids and human colorectal and breast cancer mortality. Eur. J. Cancer Prev. 1995;4:329–332. doi: 10.1097/00008469-199508000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Caygill CP, Charlett A, Hill MJ. Fat, fish, fish oil and cancer. Br. J. Cancer. 1996;74:159–164. doi: 10.1038/bjc.1996.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howe GR, Aronson KJ, Benito E, Castelleto R, Cornée J, Duffy S, Gallagher RP, Iscovich JM, Deng-ao J, Kaaks R, Kune GA, Kune S, Lee HP, Lee M, Miller AB, Peters RK, Potter JD, Riboli E, Slattery ML, Trichopoulos D, Tuyns A, Tzonou A, Watson LF, Whittemore AS, Shu Z. The relationship between dietary fat intake and risk of colorectal cancer: evidence from the combined analysis of 13 case-control studies. Cancer Causes Control. 1997;8:215–228. doi: 10.1023/a:1018476414781. [DOI] [PubMed] [Google Scholar]
- 4.Kojima M, Wakai K, Tokudome S, Suzuki K, Tamakoshi K, Watanabe Y, Kawado M, Hashimoto S, Hayakawa N, Ozasa K, Toyoshima H, Suzuki S, Ito Y, Tamakoshi A. Serum levels of polyunsaturated fatty acids and risk of colorectal cancer: a prospective study. Am. J. Epidemiol. 2005;161:462–471. doi: 10.1093/aje/kwi066. [DOI] [PubMed] [Google Scholar]
- 5.Kimura Y, Kono S, Toyomura K, Nagano J, Mizoue T, Moore MA, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Yasunami Y, Maekawa T, Takenaka K, Ichimiya H, Imaizumi N. Meat, fish and fat intake in relation to subsite-specific risk of colorectal cancer: The Fukuoka Colorectal Cancer Study. Cancer Sci. 2007;98:590–597. doi: 10.1111/j.1349-7006.2007.00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy BS, Narisawa T, Vukusich D, Weisburger JH, Wynder EL. Effect of quality and quantity of dietary fat and dimethylhydrazine in colon carcinogenesis in rats. Proc. Soc. Exp. Biol. Med. 1976;151:237–239. doi: 10.3181/00379727-151-39181. [DOI] [PubMed] [Google Scholar]
- 7.Reddy BS. Omega-3 fatty acids in colorectal cancer prevention. Int. J. Cancer. 2004;112:1–7. doi: 10.1002/ijc.20320. [DOI] [PubMed] [Google Scholar]
- 8.Schønberg SA, Lundemo AG, Fladvad T, Holmgren K, Bremseth H, Nilsen A, Gederaas O, Tvedt KE, Egeberg KW, Krokan HE. Closely related colon cancer cell lines display different sensitivity to polyunsaturated fatty acids, accumulate different lipid classes and downregulate sterol regulatory element-binding protein 1. FEBS J. 2006;273:2749–2765. doi: 10.1111/j.1742-4658.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 9.Reddy BS. Nutritional factors and colon cancer. Crit. Rev. Food Sci. Nutr. 1995;35:175–190. doi: 10.1080/10408399509527698. [DOI] [PubMed] [Google Scholar]
- 10.Larsson SC, Kumlin M, Ingelman-Sundberg M, Wolk A. Dietary long-chain n-3 fatty acids for the prevention of cancer: a review of potential mechanisms. Am. J. Clin. Nutr. 2004;79:935–945. doi: 10.1093/ajcn/79.6.935. [DOI] [PubMed] [Google Scholar]
- 11.Chung F-L, Chen HJ, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 12.Pan J, Chung F-L. Formation of cyclic deoxyguanosine adducts from omega-3 and omega-6 polyunsaturated fatty acids under oxidative conditions. Chem. Res. Toxicol. 2002;15:367–372. doi: 10.1021/tx010136q. [DOI] [PubMed] [Google Scholar]
- 13.Pan J, Keffer J, Emami A, Ma X, Lan R, Goldman R, Chung F-L. Acrolein-derived DNA adduct formation in human colon cancer cells: its role in apoptosis induction by docosahexaenoic acid. Chem. Res. Toxicol. 2009;22:798–806. doi: 10.1021/tx800355k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L, Wang J, Sekhar KR, Yin H, Yared NF, Schneider SN, Sasi S, Dalton TP, Anderson ME, Chan JY, Morrow JD, Freeman ML. Novel n-3 fatty acid oxidation products activate Nrf2 by destabilizing the association between Keap1 and Cullin3. J. Biol. Chem. 2007;282:2529–2537. doi: 10.1074/jbc.M607622200. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs AT, Marnett LJ. Heat shock factor 1 attenuates 4-Hydroxynonenal-mediated apoptosis: critical role for heat shock protein 70 induction and stabilization of Bcl-XL. J. Biol. Chem. 2007;282:33412–33420. doi: 10.1074/jbc.M706799200. [DOI] [PubMed] [Google Scholar]
- 16.Nath RG, Chung F-L. Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung F-L, Nath RG, Ocando J, Nishikawa A, Zhang L. Deoxyguanosine adducts of t-4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: detection and potential sources. Cancer Res. 2000;60:1507–1511. [PubMed] [Google Scholar]
- 18.Zhang S, Villalta PW, Wang M, Hecht SS. Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2007;20:565–571. doi: 10.1021/tx700023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung F-L, Young R, Hecht SS. Detection of cyclic 1,N2-propanodeoxyguanosine adducts in DNA of rats treated with N-nitrosopyrrolidine and mice treated with crotonaldehyde. Carcinogenesis. 1989;10:1291–1297. doi: 10.1093/carcin/10.7.1291. [DOI] [PubMed] [Google Scholar]
- 20.Sodum RS, Chung F-L. 1,N2-ethenodeoxyguanosine as a potential marker for DNA adduct formation by trans-4-hydroxy-2-nonenal. Cancer Res. 1988;48:320–323. [PubMed] [Google Scholar]
- 21.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J. Am. Chem. Soc. 2003;125:5687– 5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 22.Nath RG, Ocando JE, Guttenplan JB, Chung F-L. 1-,N2-propanodeoxyguanosine adducts: potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 1998;58:581–584. [PubMed] [Google Scholar]
- 23.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J. Biol. Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 24.Kawanishi M, Matsuda T, Nakayama A, Takebe H, Matsui S, Yagi T. Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutat. Res. 1998;417:65–73. doi: 10.1016/s1383-5718(98)00093-x. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived alpha-HOPdG and gamma-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 26.Wang H-T, Hu Y, Tong D, Huang J, Gu L, Wu X-R, Chung F-L, Li G-M, Tang M. Effect of carcinogenic acrolein on DNA repair and mutagenic susceptibility. J. Biol. Chem. 2012;287:12379–12386. doi: 10.1074/jbc.M111.329623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, gamma -hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J. Biol. Chem. 2003;278:784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 28.Yang I-Y, Miller H, Wang Z, Frank EG, Ohmori H, Hanaoka F, Moriya M. Mammalian translesion DNA synthesis across an acrolein-derived deoxyguanosine adduct. Participation of DNA polymerase eta in error-prone synthesis in human cells. J. Biol. Chem. 2003;278:13989–13994. doi: 10.1074/jbc.M212535200. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Pfeifer GP, Besaratinia A. Lack of mutagenicity of acrolein-induced DNA adducts in mouse and human cells. Cancer Res. 2007;67:11640–11647. doi: 10.1158/0008-5472.CAN-07-2528. [DOI] [PubMed] [Google Scholar]
- 30.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 31.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct gamma -OH-1,-N2-propano-2′-deoxyguanosine. J. Biol. Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 32.Hu W, Feng Z, Eveleigh J, Iyer G, Pan J, Amin S, Chung F-L, Tang M-S. The major lipid peroxidation product, trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at codon 249 of human p53 gene, a unique mutational hotspot in hepatocellular carcinoma. Carcinogenesis. 2002;23:1781–1789. doi: 10.1093/carcin/23.11.1781. [DOI] [PubMed] [Google Scholar]
- 33.Feng Z, Hu W, Hu Y, Tang M. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15404–15409. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C, Hohler T, Galle PR, Phillips DH, Markin R, Marrogi AJ, Harris CC. Increased p53 mutation load in nontumorous human liver of wilson disease and hemochromatosis: oxyradical overload diseases. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singer B, Antoccia A, Basu AK, Dosanjh MK, Fraenkel-Conrat H, Gallagher PE, Kuśmierek JT, Qiu ZH, Rydberg B. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dosanjh MK, Chenna A, Kim E, Fraenkel-Conrat H, Samson L, Singer B. All four known cyclic adducts formed in DNA by the vinyl chloride metabolite chloroacetaldehyde are released by a human DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1024–1028. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dosanjh MK, Roy R, Mitra S, Singer B. 1,N6-ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase) Biochemistry. 1994;33:1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]
- 38.Roy R, Biswas T, Lee JC, Mitra S. Mutation of a unique aspartate residue abolishes the catalytic activity but not substrate binding of the mouse N-methylpurine-DNA glycosylase (MPG) J. Biol. Chem. 2000;275:4278–4282. doi: 10.1074/jbc.275.6.4278. [DOI] [PubMed] [Google Scholar]
- 39.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matijasevic Z, Sekiguchi M, Ludlum DB. Release of N2,3-ethenoguanine from chloroacetaldehyde-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9331–9334. doi: 10.1073/pnas.89.19.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson KA, Fink SP, Marnett LJ. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. J. Biol. Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 42.Choudhury S, Pan J, Amin S, Chung F-L, Roy R. Repair kinetics of trans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 2004;43:7514–7521. doi: 10.1021/bi049877r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myrand SP, Topping RS, States JC. Stable transformation of xeroderma pigmentosum group A cells with an XPA minigene restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis. 1996;17:1909–1917. doi: 10.1093/carcin/17.9.1909. [DOI] [PubMed] [Google Scholar]
- 44.Esterbauer H, Weger W. Über die Wirkungen von Aldehyden auf gesunde und maligne Zellen, 3. Mitt.: Synthese von homologen 4-Hydroxy-2-alkenalen, II. Monatsh. Chem. 1967;98:1994–2000. [Google Scholar]
- 45.Kohno K, Shimamoto T. Nucleotide excision repair assay in Drosophila melanogaster using established cell lines. Methods Mol. Biol. 1999;113:337–346. doi: 10.1385/1-59259-675-4:337. [DOI] [PubMed] [Google Scholar]
- 46.Sako M, Inagaki S, Esaka Y, Deyashiki Y. Oxidative hydrolysis of a cyclic 1,N2-propano-2′-deoxyguanosine, an adduct of 2′-deoxyguanosine with acetaldehyde or crotonaldehyde. Tetrahedron Lett. 2003;44:7303–7306. [Google Scholar]
- 47.Liu X, Lovell MA, Lynn BC. Development of a method for quantification of acrolein-deoxyguanosine adducts in DNA using isotope dilution-capillary LC/MS/MS and its application to human brain tissue. Anal. Chem. 2005;77:5982–5989. doi: 10.1021/ac050624t. [DOI] [PubMed] [Google Scholar]
- 48.Liu X, Lovell MA, Lynn BC. Detection and quantification of endogenous cyclic DNA adducts derived from trans-4-hydroxy-2-nonenal in human brain tissue by isotope dilution capillary liquid chromatography nanoelectrospray tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:710–718. doi: 10.1021/tx0502903. [DOI] [PubMed] [Google Scholar]
- 49.Chung F-L, Wu MY, Basudan A, Dyba M, Nath RG. Regioselective formation of acrolein-derived cyclic 1,N2-propanodeoxyguanosine adducts mediated by amino acids, proteins, and cell lysates. Chem. Res. Toxicol. 2012;25:1921–1928. doi: 10.1021/tx3002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protić M, Hübscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 53.Kowalczyk P, Cieśla JM, Komisarski M, Kuśmierek JT, Tudek B. Long-chain adducts of trans-4-hydroxy-2-nonenal to DNA bases cause recombination, base substitutions and frameshift mutations in M13 phage. Mutat. Res. 2004;550:33–48. doi: 10.1016/j.mrfmmm.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 55.Gros L, Maksimenko AV, Privezentzev CV, Laval J, Saparbaev MK. Hijacking of the human alkyl-N-purine-DNA glycosylase by 3,N4-ethenocytosine, a lipid peroxidation-induced DNA adduct. J. Biol. Chem. 2004;279:17723–17730. doi: 10.1074/jbc.M314010200. [DOI] [PubMed] [Google Scholar]
- 56.Lingaraju GM, Davis CA, Setser JW, Samson LD, Drennan CL. Structural basis for the inhibition of human alkyladenine DNA glycosylase (AAG) by 3,N4-ethenocytosine-containing DNA. J. Biol. Chem. 2011;286:13205–13213. doi: 10.1074/jbc.M110.192435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu D, Samson LD. Direct repair of 3,N4-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair (Amst) 2012;11:46–52. doi: 10.1016/j.dnarep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nairn RS, Mitchell DL, Adair GM, Thompson LH, Siciliano MJ, Humphrey RM. UV mutagenesis, cytotoxicity and split-dose recovery in a human-CHO cell hybrid having intermediate (6-4) photoproduct repair. Mutat. Res. 1989;217:193–201. doi: 10.1016/0921-8777(89)90071-2. [DOI] [PubMed] [Google Scholar]
- 59.Szymkowski DE, Lawrence CW, Wood RD. Repair by human cell extracts of single (6-4) and cyclobutane thymine-thymine photoproducts in DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9823–9827. doi: 10.1073/pnas.90.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.