Female decision-making in Drosophila flies requires the expression of a transcription factor in a small number of cholinergic neurons in discrete brain regions.
Abstract
Courtship is a widespread behavior in which one gender conveys to the other a series of cues about their species identity, gender, and suitability as mates. In many species, females decode these male displays and either accept or reject them. Despite the fact that courtship has been investigated for a long time, the genes and circuits that allow females to generate these mutually exclusive responses remain largely unknown. Here, we provide evidence that the Krüppel-like transcription factor datilógrafo (dati) is required for proper locomotion and courtship acceptance in adult Drosophila females. dati mutant females are completely unable to decode male courtship and almost invariably reject males. Molecular analyses reveal that dati is broadly expressed in the brain and its specific removal in excitatory cholinergic neurons recapitulates the female courtship behavioral phenotype but not the locomotor deficits, indicating that these are two separable functions. Clonal analyses in female brains identified three discrete foci where dati is required to generate acceptance. These include neurons around the antennal lobe, the lateral horn, and the posterior superior lateral protocerebrum. Together, these results show that dati is required to organize and maintain a relatively simple excitatory circuit in the brain that allows females to either accept or reject courting males.
Author Summary
Males of the fruit fly Drosophila melanogaster generate a series of courtship displays that convey visual, auditory, and olfactory information that females must decode in order to accept or reject mating. Despite the central role of female decision in sexual selection, relatively little is known about how genes and neural circuits generate this behavior. Here we show that the transcription factor datilografo (dati) is required to organize and maintain the neural circuitry required for acceptance in the central brain. Strikingly, dati is required in an excitatory circuit involving few neurons that express acetylcholine as their neurotransmitter and are located in the olfactory lobe, the first entry point for odor processing in the brain. In addition, dati is required in two other brain centers: a region where olfaction and presumably other senses are integrated and a novel region. Together these results show that a complex behavior can be generated by very few excitatory neurons, suggesting that the sharp cutoffs between acceptance and rejection may involve different thresholds of stimulation as postulated decades ago.
Introduction
Animals are capable of a staggering array of complex behaviors and many of them rely on innate abilities to compare different scenarios and generate specific and appropriate responses. For instance, most animals can determine with ease whether the best option is to confront or retreat from a predator or opponent. Risk assessment and similar mutually exclusive behaviors are likely to rely on neural circuits that collect information, remove irrelevant and noisy information, and quickly determine a course of action.
Courtship rituals are ancient forms of communication that allow animals to identify and rank potential mates in the midst of a noisy and usually complex environment. Thus, it is not surprising that courtships usually deploy a series of displays that involve bright colors, unusual sounds, and rhythmicities. The recipients of these displays, which in many species are females, evaluate their quality and generate the mutually exclusive behaviors of accepting or rejecting courtship.
One of the most fascinating aspects of the ability to generate courtship and respond with a decision is the fact that both behaviors are largely genetically encoded; that is, animals are capable of executing them perfectly with minimal practice and no instruction every generation. Pioneering work has established clear associations between individual male courtship behaviors with specific genes and alleles in Drosophila [1], and even led to the mapping of foci in the central nervous system required to generate discrete behaviors [2]–[5]. However, little is known about how females interpret and integrate aspects of the male's displays and decide if and when to accept male courtship [6]. This is a longstanding question of significance not only to our understanding of the molecular mechanisms of reproductive behavior but also to any comprehensive understanding of how neural circuits generate mutually exclusive decisions.
In Drosophila, males show their interest in females by making wing displays, singing a courtship song, dispersing airborne and contact pheromones, and physically contacting them [7]–[9]. In response to these cues, receptive females slow their movement and allow the male to proceed, to finally posture themselves to allow the male to mount them for copulation. In contrast, a disinterested or unreceptive female will engage in a number of rejection behaviors, such as fleeing, kicking the male, extruding her ovipositor, and raising or curling her abdomen [7]. Early studies have shown that no single sensory modality alone determines acceptance or rejection in mature females. Instead, the likelihood of acceptance or rejection relies on different sensory modalities that individually contribute to the final behavioral output [10]–[13]. Genes and alleles that either enhance or inhibit female receptivity have been isolated [14]–[17]. Mutations in these genes provide a unique opportunity to determine the genetic contribution to cell organization and physiological responses required to generate female mate choice [18]. In addition to mutations, somatic mosaics have been employed to determine the regions of the brain underlying female behavior [18],[19]. Nevertheless, critical information about the neural circuitry involved in female decision-making behavior and the genes that pattern these circuits is still sorely lacking. Here, we describe dati, a neural-specific transcription factor that is required for female courtship acceptance and locomotion, and use it to begin probing the nature of the circuit by which females integrate the signals they receive from courting males to reach the correct behavioral output.
Materials and Methods
Mutants, Transgenes, Genotypes, and Stocks
Flies were reared at 25°C or 22°C on standard cornmeal–molasses–yeast media (http://flystocks.bio.indiana.edu/Fly_Work/media-recipes/molassesfood.htm). The descriptions of fly stocks used and mutations therein can be found at (http://flybase.bio.indiana.edu/), unless otherwise stated. The mutant l(4)102CDd 2 [20] was molecularly mapped in this study. The single breakpoints of the chromosomal deletions Df(4)C1-7A, Df(4)B6-4A, and Df(4)B6-2D and the compound chromosome C(4)DRA-1 were described previously [21],[22]. The mutations KG02689 (dati 1) and KG01667 (dati 2) were also described previously [22],[23] and correspond to single P element insertions in the second intron and 300 bp upstream of the first exon of CG2052, respectively. dati F11.4 was generated by the excision of the P-element KG02689 inserted in dati 1 and corresponds to a precise excision as revealed by direct sequencing of the region that flanks the insertion. UAS-dati F32D is a leaky UAS transgene containing the cDNA of dati (GH06573, CG2052-PA) that rescues the embryonic lethality of l(4)102CDd 2 up to the first instar in the absence of a driver and its insertion was determined to be on the second chromosome in the neurally expressed gene Mmp2. The D. melanogaster stocks used were as follows: wild-type Canton-S (CS), w 1118, y w, y w; Df(4)B6-4A/ci D spap ol, y w; Df(4)B6-2D/ci D spap ol, y w; Df(4)C1-7A/C(4)DRA-1, y w; Df(4)BH/C(4)DRA-1, y w; C(4)DRA-1/dati1, y w; datiF11.4, y w; SM/Cha-Gal4 UAS-GFP; C(4)DRA-1/dati1, y w hs-FLP; SM/FRT42D Actin-Gal4; ci D spa/dati 1, y w hs-FLP; SM/FYGal80T; spa pol [23], w; UAS-mCherry.NLS, w; dati RNAi/dati RNAi [FBst0472372], w UAS-dcr-2; dati RNAi, y w hs-FLP; SM/FRT42D Actin-Gal4; C(4)DRA-1/dati 1, elav-Gal4 [FBst0008760], w; TM3, Sb/repo-Gal4 [FBst0007415], w; CyO/Cha-Gal4 UAS-GFP [FBst0006793], w; ple-Gal4 [FBst0008848], w; Ddc-Gal4 [FBst0007009], w; Gad-Gal4 (gift of T. Sakai) [17], Ubi-GFP.NLS [FBst0005626], Ubi-RFP.NLS [FBst0035496], and FYT/GAF [23].
Experimental crosses were raised at 25°C, unless otherwise specified. Both males and females used for mating experiments were collected as pupae and aged 3–6 d posteclosion before mating tests. All mating tests were performed at 22°C, between 1 and 4 pm EST.
Recording of Mating Behavior
Mating tests were performed in small arenas made by superimposing two sliding sheets of transparent polycarbonate containing 24 wells each (2.54 cm in diameter and 1.27 cm depth) [24]. Each well was divided in half by a thin removable sheet of plastic. Canton-S males and experimental females 3 to 6 d old were loaded into opposing sides of each chamber without anesthesia with a manual aspirator. Once all wells were loaded, the thin plastic sheet was removed and all pair matings began simultaneously. The chamber was lit from below by an Artograph LED LightPad (Artograph Inc.), and mating behavior was recorded using a Sanyo FH1-A (Sanyo Inc.) camcorder for 1 h. For each experimental group, we calculated the courtship acceptance rate, defined as the number of pairs that successfully copulated in the 1 h observation period divided by the number of pairs observed. The average Courtship Index (CI) was calculated for each experimental group. CI is defined as the fraction of time a male spent courting in a given observation period [25]. Male courtship for each pairing was observed for 10 min, starting at the onset of courtship. CIs for each pair mating in an experimental group were then aggregated into an average CI. Sample sizes are shown in the corresponding figure in results.
Quantification of Discrete Female Behaviors
Females 3 to 5 d old of experimental and control genotypes were pair-mated to Canton-S males in the mating chamber described above and video recorded for 1 h. For each pair mating, female behavior was analyzed for 10 min from the onset of courtship or until mating occurred. For this time period, every time a male initiated a step of courtship, the female reaction to courtship was recorded. The following six discrete rejection behaviors were quantified: fleeing, kicking, extruding ovipositor, jumping, flicking wings, and standing still. For each female, a Behavioral Index (BI) was obtained by calculating the frequency of each behavior displayed over the frequency of all behaviors, and these indices were averaged for each genotype.
Analysis of Locomotor Behavior
Locomotor behavior was analyzed using an adaptation of the negative geotaxis assay [26]. Five to eight flies of 3 to 5 d old were placed in a 15 mL Falcon tube without anesthesia and allowed to acclimate for 5 min. After this period of acclimation, the Falcon tube was inverted and rapped sharply against a fly transfer pad three times to knock flies to the bottom of the tube. The tube was then placed in front of the camcorder and flies were allowed to climb the walls. The heights reached by each of the flies after 5 s was assessed from the camcorder footage. Over 30 flies were analyzed for each experimental group. For each Falcon tube of flies, this assay was repeated for a total of five trials, spaced 30 s apart, and the heights of all flies from each trial averaged together.
Antibodies and Immunostaining
Antibody staining for brains was performed according to standard protocols [27]. Primary antibodies used were chicken anti-GFP (1∶1,000, Invitrogen), rabbit anti-RFP (1∶1,000, Invitrogen), mouse anti-nc82 (1∶1,000, DSHB), Guinea pig anti-Dati (1∶1,000, gift from T. Isshiki [35]), rat anti-Elav (1∶1,000, DSHB), and mouse anti-FasII (1∶100, DSHB). Secondary antibodies used were donkey anti-mouse 647 (1∶500), goat anti-rat 555 (1∶500, Invitrogen), donkey anti-guinea pig 647 (1∶500, Invitrogen), donkey anti-chicken 488 (1∶500, DyLight), and donkey anti-rabbit 555 (1∶500, Invitrogen). All samples were mounted in SlowFade (Invitrogen) and scanned on a Zeiss LSM 700 confocal microscope. Images generated from Z-stacks taken at 1 or 2 µm intervals are displayed as maximum intensity projections using Zeiss Zen 2009 or as orthogonal projections/surface projections using Image J.
Automated Image Analysis and Cell Counting
Automated cell counting was performed on confocal slices using Fiji software [28]. Briefly, a two-channel stack stained for dati (green) and Cha-Gal4 UAS-RFP.NLS (red) was converted to RGB, and the yellow overlap was segmented with white color using “Threshold Color” function. The blue channel containing the segmented nuclear overlaps was extracted and the noise removed by filtering the stack with the function “Despeckle.” Three-dimensional segmentation counts were generated by the plugin “3D Object Counter” [29]. Due to the large size of posterior brain stacks, they were stitched together using the plugin “Pairwise Stitching” before segmentation [30].
Clonal Analyses
Clonal analyses were performed using the FYT (FLP-recombinase recognition target site-yellow+-Translocation) system previously described [23]. After clone induction, third instar larvae containing GFP+ clones were handpicked, placed in a single vial, and allowed to develop up to adults 3 to 6 d old. Single females carrying clones were tested with single Canton-S male in the courtship arenas described above and video recorded for 1 h. After this time the number of couples that mated was recorded and the Courtship Indices determined. In the next day, the females that rejected males were retested with new Canton-S males for rejection, and only those that passed in the double rejection test were analyzed further [19]. Females that accepted and rejected males were referenced to specific wells and had their brains dissected. Each clone was located in a grid that divides the brain in 40 anterior and 40 posterior sectors. Because each brain may vary slightly in size or in the way it is mounted, the grid was manually stretched to find the best fit for each sample. In total, 491 clones from 83 brains were analyzed.
Olfactory Behavior Assays
In these experiments, we used a T-Maze [31] with 2 µl of benzaldehyde in one of the ends. Individual flies were loaded into the elevator of the apparatus and immediately lowered to the level containing the two ends with and without odor. After 10 s, the number of flies that moved away from the aversive odorant or towards it was recorded.
Mushroom Body and Antennal Lobe Image Analysis
Brains of different genotypes were dissected and stained for the mushroom body marker FasII and imaged as z-stacks at 1 µm intervals. Selected z-stacks containing the gamma lobe were manually segmented using the Fiji plugin “Segmentation Editor.” Measurement of γ lobe morphological defects was done in Fiji. A similar procedure was done to segment the antennal lobe, except that the limits of the segmented structures were defined by the expression of GFP in the pattern of CHA.
Statistical Analysis
All statistical analyses were performed using MiniTab 16.1.0 (Minitab Inc.). For all comparisons of courtship acceptance rate between control and experimental groups, a 2-Proportion Test was performed, and Fisher's exact p test value was used for the determination of significance level between two groups, unless otherwise indicated. CI data were arcsine transformed prior to statistical analysis as previously described [32] and analyzed by one-way ANOVA. The difference in climbing ability in locomotor tests was analyzed by one-way ANOVA. Behavioral indices (Figure S2) were analyzed by Mann–Whitney U Test. All other tests are two sample t tests unless otherwise noted.
Results
Identification of dati on the Drosophila Fourth Chromosome
We previously generated a series of molecularly mapped terminal deletions on the fourth chromosome that define relatively small genomic intervals that can be used to map mutations [22],[33]. These deletions were then used to map a collection of mutants available for this chromosome, to later test for locomotion and other behavioral abnormalities. One of them was l(4)102CDd2, an unmapped embryonic lethal mutation isolated nearly 50 years ago by Ben Hochman [20].
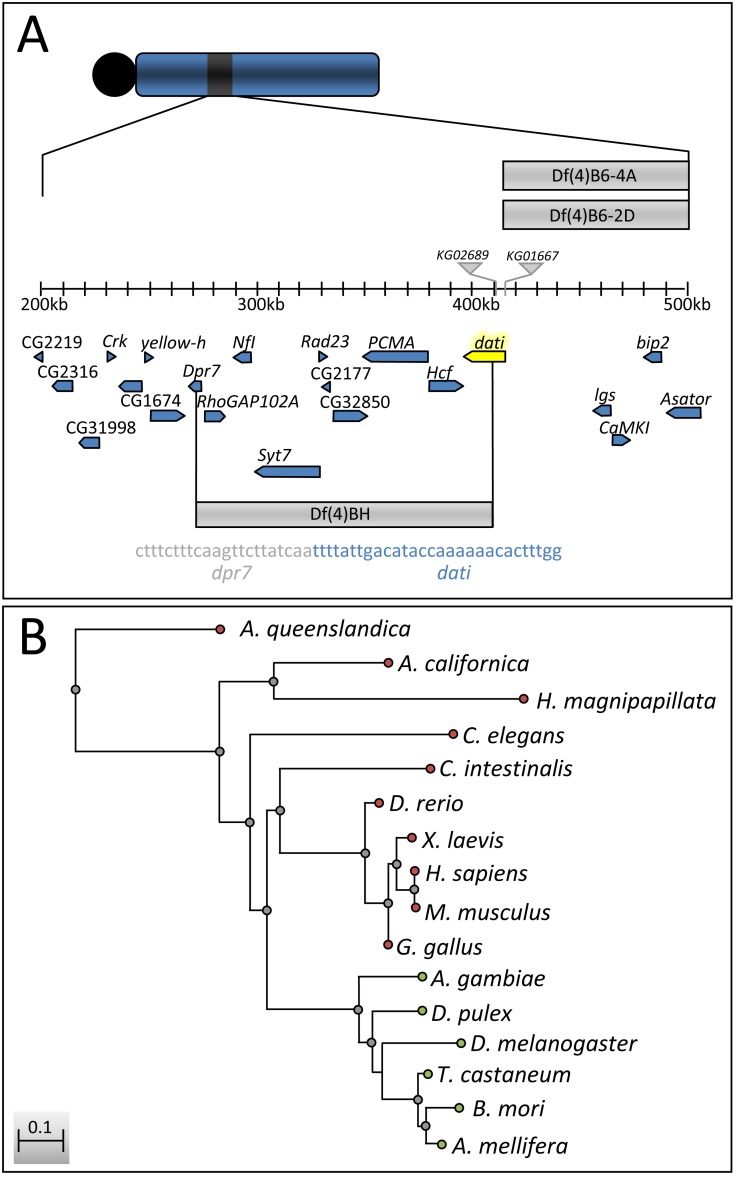
While mapping l(4)102CDd2 we found that 5%–8% of the heterozygotes between this mutation and two deletions (Df(4)B6-2D and Df(4)B6-4A) escaped the lethality of l(4)102CDd2 and exhibited a phenotype of uncoordinated movements, which becomes stronger with age (compare Movies S2 and S3). Due to the tapping of the forelegs of these genotypes, we named the mutation datilógrafo (dati) [34], which means typist in Portuguese. Subsequent analyses revealed that mutations in dati also render females completely unable to accept male courtship, as will be shown later. We located molecularly the mutation in l(4)102CDd2, which corresponds to a deletion that disrupts dpr7 and CG2052 plus eight other genes in between, and renamed it as deficiency on the fourth chromosome of Ben Hochman [Df(4)BH] (Figure 1A and Text S1). Because two single fourth chromosome P-element insertions localized at the breakpoint of these deletions in the CG2052 gene (KG02689 and KG01667) exhibited the same phenotypes as homozygotes or heterozygotes for Df(4)BH, we focused our analyses on the insertion KG02689 (dati 1), the strongest of these two alleles. dati encodes a zinc finger transcription factor closely related to rotund (rn) and squeeze (sqz) with homologs in several species, including humans (Figure 1B).
Figure 1. Molecular mapping of dati1 and conservation of dati across species.
(A) Physical position of dati on the fourth chromosome (highlighted in yellow). The P-elements KG02689 (dati 1) and KG01667 (dati 2) are represented by grey triangles. Deficiencies causing lethality in conjunction with dati1 are represented by gray bars. Note that the Deficiency of Ben Hochman [Df(4)BH] spans a region of 10 genes, between dpr7 and dati. Indicated below Df(4)BH is the sequence of the breakpoint in dpr7 (gray) and dati (blue). (B) Neighbor-joining distance tree with Kimura two-parameter distances of dati sequences across multiple species. Branches of the tree with green termini represent orthologs of dati, whereas branches with red termini represent homologs.
Consistent with its reported requirement in specifying late born neurons during embryogenesis [35], dati is specifically expressed in the central nervous system in embryos (Figure S1A). In larval stages, dati is expressed in the brain and ventral nerve cord (Figure S1B) but not in other larval tissues (e.g., wing, leg, eye, and antennal discs; unpublished data). In adults, dati is broadly expressed in the brain (Figure S1C).
dati Mutant Females Are Courted Normally But Fail to Accept Male Courtship
dati 1 mutants usually stand still for long periods of time, but when courted by males, they can flee at considerable speed. In addition, when cornered by a courting male, they engage in a series of rejection behaviors that include kicking and curling their abdomen (Movies S1, S2, S3) [7],[36]. To investigate how the behavior of dati 1 females departs from the wild type, we quantified six discrete behaviors normally displayed by wild-type females in response to male courtship (i.e., fleeing, kicking, extruding ovopositor, jumping, flicking wings, and standing still). dati females display all of the aforementioned behaviors but spend more time kicking and less time standing still than the wild type (Figure S2).
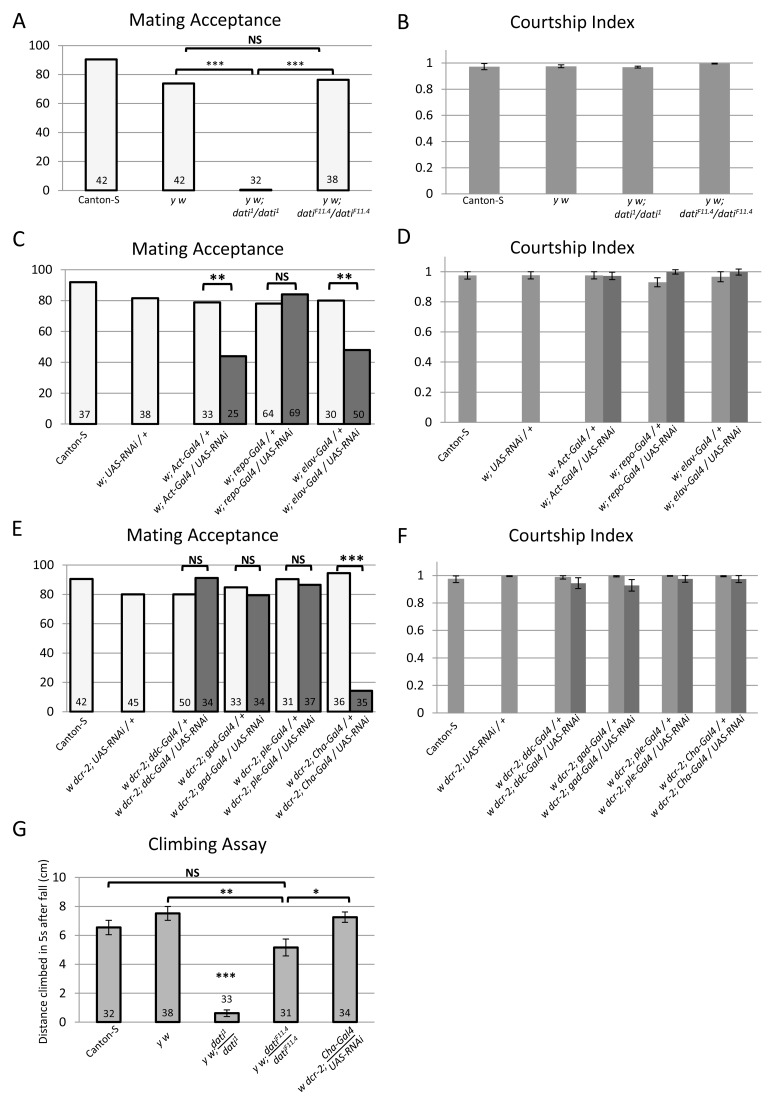
To further quantify the abnormal mating behavior of dati 1 mutant females, we compared their mating success with that of wild-type Canton-S, y w, and dati precise excision revertant females. From these data it becomes evident that the behavior of dati1 is significantly different from the wild-type Canton-S, y w and the revertant datiF11.4 females, which exhibit normal acceptance rates (Figure 2A). To test whether the deficit in matings was exclusively due to the female rejection, we assessed the sex appeal of dati 1 homozygous females using the CI (Figure 2B) [25],[37]. These experiments reveal that males respond to dati1 females normally, with courtship indices indistinguishable between all four groups.
Figure 2. Results of pair mating and locomotor experiments using dati1 and UAS-dati-RNAi.
(A, C, E) Percentage of females that accepted wild-type Canton-S male courtship within 1 h in pair mating assays. (B, D, F) Courtship indices of Canton-S males towards females tested in (A), (C), and (E), respectively. Courtship indices were arcsin transformed and subject to one-way ANOVA, and in all cases they are not significantly different from each other (p>.05). Error bars represent ±SEM of the measurements of each genotype. (A–F) Genotypes of females tested are indicated (“UAS-RNAi” means UAS-dati-RNAi). Dark and light bars in (C–F) indicate experimentals and controls, respectively. (A, B) dati 1 homozygous females strongly reject males, despite being courted vigorously by Canton-S males. This rejection phenotype is reverted in revertant females homozygous for the precise excision datiF11.4 allele. Controls with Canton-S and y w females were used, as revertants lose both y+ and w+ transgenes. (C, D) Ubiquitous expression of UAS-dati-RNAi driven by Act-Gal4 or expression in postmitotic neurons driven by elav-Gal4 causes rejection of male courtship. (C) Dark bars indicate the percentage of females expressing UAS-dati-RNAi driven by different Gal4 drivers that accepted male courtship. Light bars indicate results with control females (Canton-S females and females with either UAS-dati-RNAi or a Gal4 driver). Expression of dati-RNAi in glial cells (repo-Gal4) does not cause rejection. (E, F) Expression of dati-RNAi specifically in cholinergic neurons causes the female rejection behavior. (E) Percentage of courtship acceptance in females expressing UAS-dati-RNAi driven by neuron-specific Gal4 drivers (dark bars) versus control females with either Gal4 drivers or UAS-dati-RNAi only (light bars). Ddc-Gal4 is a driver of both serotonergic and dopaminergic neurons, ple-Gal4 of dopaminergic neurons, Gad-Gal4 of GABAergic neurons, and Cha-Gal4 of cholinergic neurons. (G) Courtship acceptance and locomotor deficits in dati mutant females are separable phenotypes. The graph shows the distance climbed by females of various genotypes within 5 s after being knocked to the bottom of a vial. Measurements shown are an average of five replicates from each group; error bars represent the mean ± the standard deviation. Sample size numbers are indicated inside bars in (A, D, E, G). Graphs (B, D, F) use the same datasets as (A, C, E), respectively. Statistical significance of differences in (A, C, E, G) was evaluated by the Fisher's exact probability test (***p<0.001; **p<0.01; *p<0.05).
The rejection of dati mutants was tested over a longer time by measuring the frequency of females that produced progeny with a wild-type male in 6 d. The difference between the two groups is not significant (dati 1 6 d = 2/14 versus dati 1 1 h = 0/32, p = 0.08), but both are significantly different than Canton-S (dati 1 6 d versus Canton-S 6 d = 29/30, p<0.0001 and Figure 2A). This result is consistent with the fact that females that fail to accept males within 30 min are unlikely to mate afterwards [38].
dati Is Required In Neurons for Normal Acceptance and Locomotion
To determine in which tissues dati is required for normal courtship behavior and locomotion, we knocked down its expression using RNAi and UAS-dcr-2 to enhance the knockdown. The knockdown of dati with the ubiquitous Actin-Gal4 [23] at 25°C resulted in few adult individuals that died shortly after eclosion with extreme locomotor abnormalities (unpublished data). To obtain a less severe phenotype more similar to dati 1 homozygotes, the UAS-dcr-2 construct was removed from the genotype and the flies were reared at 18°C. Under these conditions, females expressing the dati RNAi from Actin-Gal4 showed defects in acceptance and locomotion (Figure 2C,D; unpublished data). Similarly, the knockdown of dati with elav-Gal4 caused rejection and locomotor defects (Figure 2C). elav is a bona fide postmitotic marker, except for a transient embryonic expression in glial cells and neuroblasts in thoracic and abdominal segments [39]. However, we show that the knockdown of dati in glial cells using repo-Gal4 produced no effect (Figure 2), indicating that the courtship behavioral phenotypes are not generated in these cells. In addition, we later provide evidence that the behavioral effects of dati knockdown with elav-Gal4 are not associated with neuroblasts of the embryonic ventral nerve cord.
The Removal of dati in Cholinergic Neurons Impairs Normal Female Acceptance But Not Locomotion
Because our previous results suggested that dati might be required in some capacity in neurons, we next asked whether a specific neuronal population could phenocopy the mating deficit observed. The fly brain employs several neurotransmitters including dopamine, acetylcholine, GABA, glutamate, serotonin, histamine, octopamine, and tyramine [40]–[45]. To begin an unbiased search for specific neuronal populations, we first knocked down the expression of dati by RNAi using four Gal4 drivers of genes involved in the synthesis of different neurotransmitters (Dopa decarboxylase, pale, Choline Acetyltransferase, and Glutamic acid decarboxylase 1) (Figure 2E,F) to later test other neuronal types if necessary. Out of the four drivers tested, Choline Acetyltransferase Gal4 (Cha-Gal4) produced a strong and significant reduction in courtship acceptance (Figure 2E,F). Thus, the inability of dati females to accept males affects a particular neuronal type.
Interestingly, the removal of dati in cholinergic neurons does not impair locomotion as can be observed from “negative geotaxis” escape response tests [26],[46]. In these tests, dati 1 homozygous females normally achieve a much lower mean height 5 s after being knocked to the ground compared to wild-type Canton-S females (Figure 2G). Revertants also have a significantly better climbing ability than dati homozygotes (Figure 2G). However, their climbing ability was not completely restored to the levels of y w, indicating that although most of the climbing deficits can be ascribed to the mutation in dati, other genes in the genetic background contribute to the locomotor deficits observed. In contrast, the climbing abilities of dati RNAi knockdowns with the Cha-Gal4 driver were not different from wild-type Canton-S (Figure 2G), indicating that the male rejection behavior of dati 1 mutants is separable from the locomotor deficits.
dati Mutants Exhibit Abnormal γ-Lobes of the Mushroom Bodies, But These Defects Do Not Cause Female Rejection
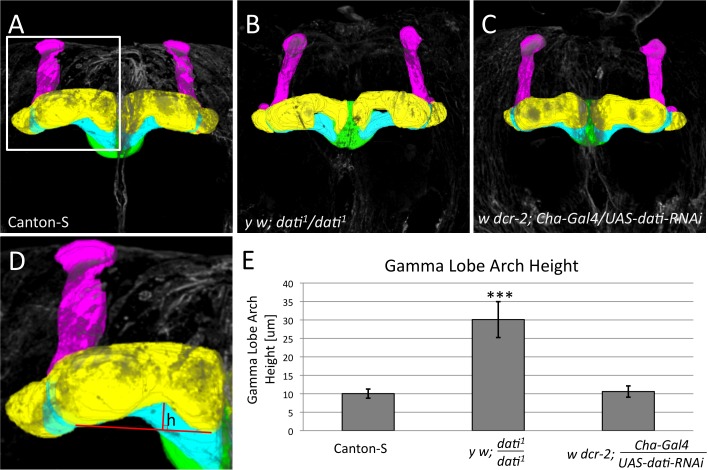
The results above revealed that the acceptance deficits of the dati 1 mutant are generated in cholinergic neurons. Because the mushroom bodies in Drosophila express CHA and have been implicated in memory formation, learning, and olfactory processing, we initially tested whether this neuropile was abnormal in dati 1 mutants [47],[48]. The alpha and beta lobes of dati 1 mutants appear indistinguishable from the wild-type mushroom bodies, but the gamma lobes are malformed with a generally withered appearance (Figure 3A,B,D) and have significantly different curvature (Figure 3E). To determine if the gamma lobe defects could be responsible for the behavioral rejection, we asked whether the knockdown of dati expression in CHA+ cells could recapitulate the morphological defect in the gamma lobe and behavioral phenotypes observed. These experiments revealed that although Cha-Gal4 UAS-dati-RNAi females reject males, the gamma lobe is not affected (Figure 3C,E). Together these experiments allowed us to conclude that although the loss of dati disrupts the gamma lobe neuropile, the focus of dati-mediated courtship acceptance lies elsewhere in the brain.
Figure 3. dati 1 homozygote females exhibit mushroom body defects, whereas Cha-Gal4/UAS-dati-RNAi females do not.
(A–D) 3D segmentations of mushroom bodies visualized by anti-FasII staining of (A) wild type, (B) dati1 homozygous, and (C) Cha-Gal4/UAS-dati-RNAi females. The α lobe (magenta), β lobe (blue), γ lobe (yellow), and ellipsoid body (green) are indicated. Note that the γ lobes in (B) are more curved than the gamma lobes in (A) and (C). Note that the genotype in (C) causes rejection like dati1 homozygous females (B), but not locomotor defects (Figure 1G). (D) Mushroom body shown in (A, box) and measurement of the morphological defect of the γ lobe. Red lines show the base (horizontal) and height (vertical) of the mushroom body arch. (E) Graph shows that there is a significant increase in the height (h) of the mushroom body arch in comparison to wild-type and Cha-Gal4<UAS-dati-RNAi females. All images are 3D reconstructions of confocal stacks rendered in Fiji. Error bars are ±standard deviation. Significance was tested by a two-sample t test and *** indicates p<0.001. The 3D reconstructions of the mushroom bodies are shown in Movie S4. Sample sizes are: Canton-S (N = 12), dati 1 (N = 12), and Cha-Gal4>UAS-dati-RNAi (N = 14).
dati Is Expressed in a Large Set of Neurons But in a Small Subset of the Cholinergic Neurons
To narrow the region where dati is required for female acceptance, we asked whether DATI- and CHA-positive neurons corresponded to a smaller subset than CHA neurons. DATI is broadly expressed in a complex pattern that involves a few thousand neurons. Automated cell counts indicate that there are around 2,400 neurons of the central brain that express dati, which corresponds roughly to 6.6% of all neurons of the fly's central brain (Figure S3) [49]. The overlap between CHA- and DATI-positive neurons is much smaller, comprising 345±55.3 (mean ± s.d) neurons of the anterior central brain (N = 5), and 1,049±134 neurons of the posterior central brain (N = 8). Based on these cell counts, DATI- and CHA-positive cells (i.e., cells that cause rejection with RNAi) correspond to a modest 4% of the total neurons in the central brain. Besides reducing the complexity of the neural circuit required for acceptance, these experiments revealed that dati is not required to determine cholinergic cell identity. Instead, dati appears to specify a subtype of neuronal identity that is presumably shared by neurons that express different neurotransmitters.
Mapping Brain Regions Where dati Is Required to Generate Acceptance Reveals Discrete Brain Foci
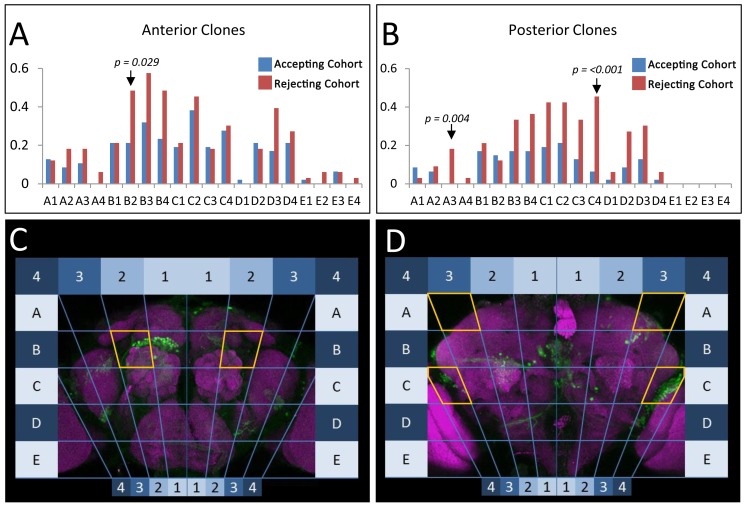
To determine the brain regions that mediate acceptance, we performed a clonal analysis using a new genetic tool we developed that allows for the systematic and efficient generation of somatic clones of fourth chromosome mutants, named the FYT system (Figure 4) [23]. In these experiments, we randomly removed dati in different positions in the brain, tested whether females accepted or rejected males, and located the position of each clone within a grid that divides the brain in 80 sectors (Figure 5A–D). By compiling a collection of 491 clones in the brain of females that either produce acceptance or double rejection (i.e., rejection in 2 consecutive days), it becomes clear that some regions in the brain produce significant deficits in acceptance while others do not. In the anterior brain, a single statistically significant region was identified in anterior sector B2 (AntB2, p = 0.029, Figure 5A). In the posterior brain, two regions stood out as highly significant (PosA3, p = 0.004 and PosC4, p≤0.001) (Figure 5B).
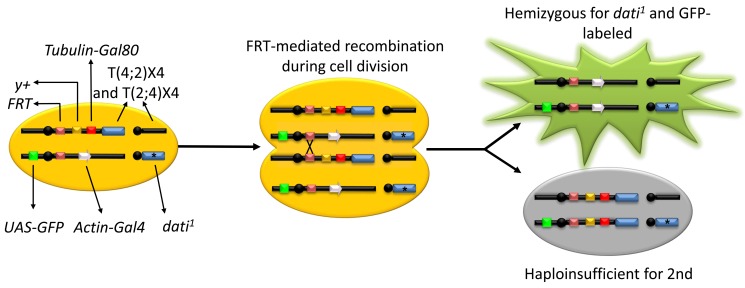
Figure 4. The FYT system.
The FYT system works in a manner similar to traditional MARCM, but with a reciprocal translocation bringing the fourth chromosome to the end of the second chromosome containing a centromeric FRT site. In this system, each cell has a heat shock-inducible source of Flippase on the X chromosome (not shown), plus two copies of the fourth chromosome (blue boxes), one carrying a mutant allele dati1 (asterisk) and the other carrying a wild-type copy translocated to the second chromosome. The second chromosome with the appended fourth chromosome contains an FRT site for somatic recombination, a y+ transgene to mark external tissues, and a ubiquitous source of the Gal4 repressor, Gal80 (red box). The homologous second chromosome carries an FRT site and a ubiquitous source of Gal4 (grey arrow) driving UAS-GFP (green box), which is repressed by Gal80. The induction of Flippase by heat shock (not shown) in a dividing cell removes the Gal80 repressor and generates two types of daughter cells. One that expresses GFP (green cell) and is hemizygous for a fourth chromosome bearing the mutation of interest, and another cell that dies due to aneuploidy (grey cell) [23].
Figure 5. Mapping regions of the brain where dati is required for normal acceptance using the GAF/FYT system.
(A and B) Frequency of marked dati1 clones in the brains of rejecting (red) and accepting (blue) females. Analysis of brains is divided into anterior (A) and posterior (B), and subdivided by regions corresponding to the maps seen in (C, D). The statistical significance of differences between accepting and rejecting clones was evaluated by the Fisher's exact test. The p values are indicated in the figure. Brain images are maximum intensity projections of confocal stacks rendered in Zen 2009.
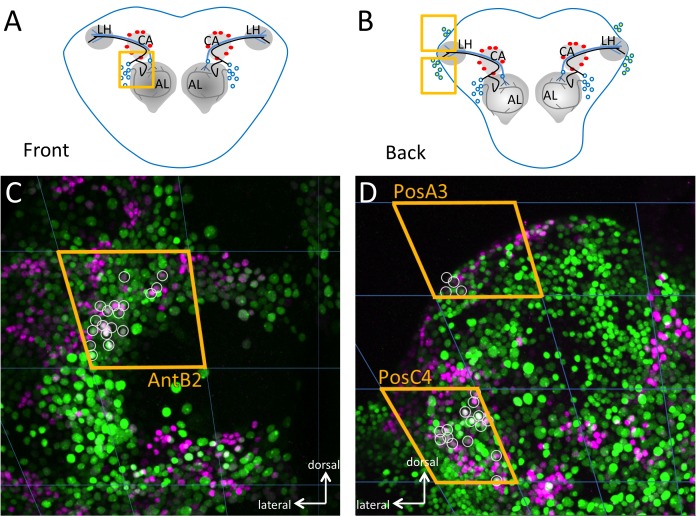
The anterior region AntB2 encompasses the first focus identified for female acceptance behavior using gynandromorphs [19] and also a region populated by extensively characterized local neurons (LNs) that express Sex lethal [50],[51]. The posterior region PosA3 is located in the posterior superior lateral protocerebrum (pslpr) immediately above the lateral horn. In contrast, PosC4 spans over the ventral part of the lateral horn, the edge of the posterior inferior lateral protocerebrum (pilpr), and posterior lateral protocerbrum (plpr) (Figure 5D). Together, these results show that dati is required in discrete neurons along a known olfactory path [52],[53], which involves second-order olfactory neurons and also third-order neurons located around the lateral horn. Interestingly, the ventral lateral horn has been recently identified as the region that processes pheromones [54],[55]. In contrast, PosA3 appears to be a novel focus implicated in female receptivity.
Rejection Foci Contain Few dati-Positive, Cholinergic Neurons
To narrow down the position of the neurons in each sector, we analyzed the neurons that express CHA and DATI within these regions. In the anterior brain, within the region AntB2, we can discern 13.8±2 neurons per hemi-antennal lobe (N = 16, Figure 6A and C). The posterior brain regions PosA3 and PosC4 that produced the most significant acceptance deficits also have very few DATI CHA neurons. Indeed, in these two regions we can identify 16.82±2.4 neurons that are positive for DATI and CHA (N = 15, Figure 6B and D). In the PosA3 sector (pslpr), we found 3.64±1.18 neurons (Figure 6D), and in the ventral lateral horn and posterior inferior lateral protocebrum (pilpr), there are 13.17±2.52 DATI CHA-positive neurons (N = 15). These results suggest that a strikingly small number of DATI CHA neurons are essential for female acceptance.
Figure 6. Detailed view of DATI/CHA double-positive cells in the three foci implicated in female acceptance as revealed by clonal analysis.
(A) A schematic frontal view of the brain where the antennal lobes (AL), the calyx (CA), and the lateral horn (LH) are indicated. The square indicates the position of the focus AntB2 indicated in (C). The trajectories of two projection neurons towards the lateral horn (black and blue line) and a projection of an LN are shown (grey line). The cell bodies around the antennal lobe are depicted as empty circles and the Kenyon cells as red circles. (B) Schematic rear view of the brain with the same three neuropiles and the Kenyon cells indicated in (A), the position of the foci PosA3 (upper square) and PosC4 (lower square). Cell bodies in the lateral horn are shown as empty circles. (C) Frontal maximum intensity projection of the antennal lobes as depicted in (A) with DATI protein stained in magenta, Cha-Gal4 UAS-mCherry.NLS in green, and the overlap between the two in light magenta or white (small circles). To determine the overlaps in a single image, the beginning and end of the double-labeled nuclei were identified in the z-stacks and a circle was drawn around the nucleus of the maximum intensity projections. In most cases the overlaps can be seen in the maximum intensity projection, but in a few cases this is not evident. The lateral and dorsal orientation of the neuropiles in (C) and (D) are indicated in the figure. Quantification of cell numbers and sample sizes are indicated in text.
dati Is Required to Generate a Subtype of Cholinergic Neurons
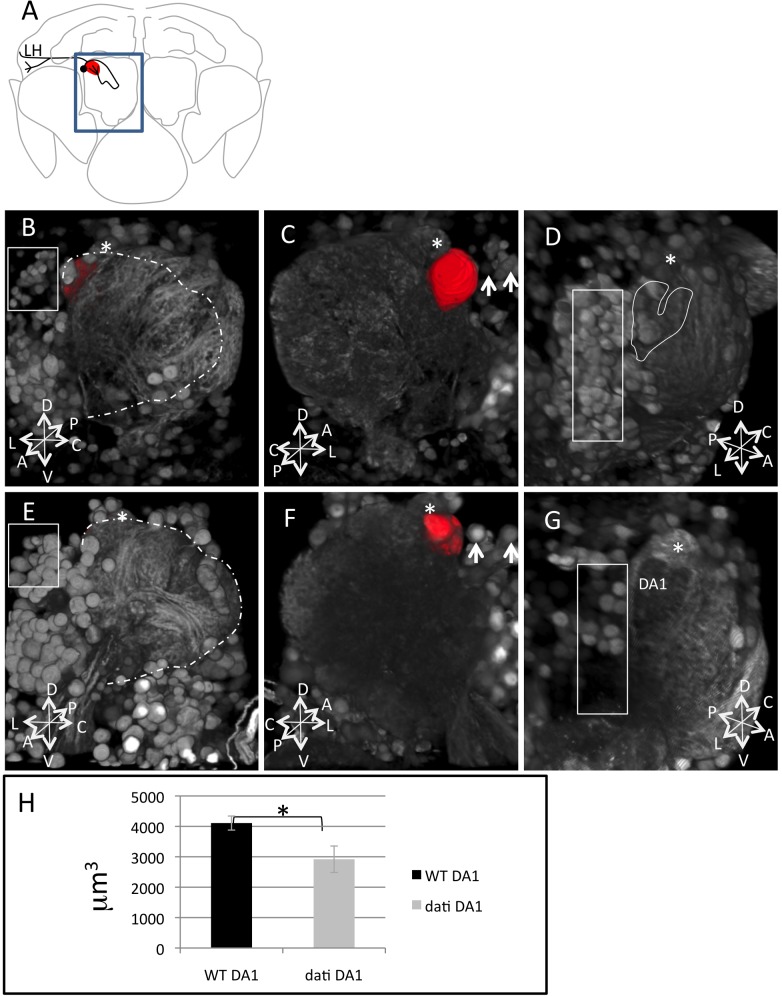
Because we had observed that the removal of dati in olfactory neurons in the region AntB2 impairs female acceptance and dati is required in the specification of late born neurons [35], we expected that the mutant might fail to specify a neuronal subtype DATI CHA. To begin addressing this issue, we compared the GFP expression patterns of Cha-Gal4 in wild-type and dati1 homozygotes (Figure 7A–G). These experiments revealed severe abnormalities in the cholinergic tracts of the antennal lobes (Figure 7B–G). A closer examination reveals that the population of dorsal lateral neurons in the region AntB2 are either reduced or transformed to cholinergic neuronal types with a distinct morphology than those normally found in this region (Figure 7C and F). These transformations within antennal lobe neurons affect several glomeruli, which include DA1, the target of the male pheromone cis-vaccenyl acetate (cVA) (Figure 7D–H) [56].
Figure 7. Homozygosity for dati1 causes loss of cholinergic projection neurons in the antennal lobe.
(A) The location of the antennal lobe in the brain (blue square). The cell body of a single lateral cholinergic projection neuron is indicated (black circle). Some cholinergic neurons in this position connect to the antennal lobe neuropile, which include the glomerulus DA1 (red, also shown in B and C) and the lateral horn (line in black). (B) Frontal view of a wild-type antennal lobe with the glomerulus DA1 highlighted in red by 3D segmentation and the glomerulus DL3 (asterisk). The dashed lines indicate the area occupied by the neuropile of the mutant in (E) and the square lateral neurons expressing CHA. (C) The same antennal lobe in (B) viewed from the brain outwards. The DA1 glomerulus (red) and the glomerulus DL3 (asterisk) are indicated. Arrows point to lateral neurons. (D) Side view of a wild-type antennal lobe. The square indicates lateral neurons expressing CHA, the asterisk the position of glomerulus DL3 (not visible from this angle), and the heart shape outlines the position of DA1 shown in (G). (E) Frontal view of a dati mutant brain, highlighting the antennal neuropile (dashed lines), the position of the glomerulus DL3 (asterisk), the glomerulus DA1 (also segmented in red to the left of DL3, but not visible from this angle in the 3D rendering), and the lateral neurons (square). Note that the antennal lobe neuropile in (E) is smaller than in (B), which indicates defects in innervation. Also note that the neurons in the square in (B) are smaller than those in (C). (F) Mutant antennal lobe in (E) viewed from the brain outwards. Asterisks indicate glomerulus DL3, red indicates glomerulus DA1, and arrows point to lateral neurons. Note that the size of DA1 in (F) is smaller than in (C) and that the lateral neurons are larger in (F) than in (C) (arrows). (G) Side view slightly from the top of another antennal lobe mutant for dati 1. The DL3 (asterisk), DA1 (red), and lateral neurons (square) are indicated. Note that the neuropile where DA1 is indicated in (G) appears darker than other parts of the antennal lobe and also darker than the corresponding region in (D) (heart-shaped outline), indicating that this region is less dense. Also note that the density of lateral neurons in (G) (square) is smaller than in (D) (square). Images are 3D projections of confocal stacks rendered in Fiji. (H) Quantification of DA1 volume in the wild type and mutant. Error bars indicate SEM. * indicate p<0.05, and the samples sizes are Canton-S, N = 6 and dati 1, N = 6. The coordinates P (posterior), A (anterior), D (dorsal), V (ventral), L (Lateral), and C (Central) are indicated.
The experiments above revealed that the loss of dati disrupts olfactory glomeruli. To test whether these disruptions lead to olfactory deficits, we assayed the performance of dati mutant females in a T-Maze in which flies are tested for moving away or towards an aversive odor. In this test, only 3% of the Canton-S flies (1 out of 30) moved towards the aversive odor compared to 32% of the dati mutant females (11 out of 34), indicating that olfactory behavior is indeed impaired in dati mutants (wild type versus dati, p = 0.001).
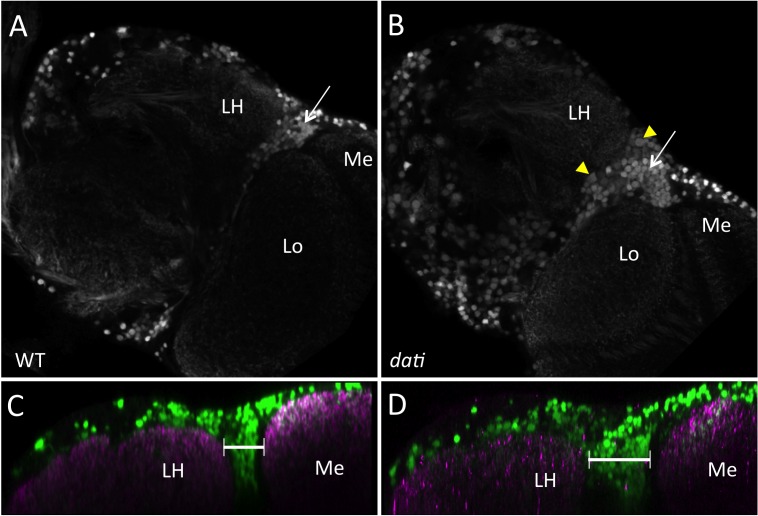
In the lateral horn, the loss of dati leads to a reduction of approximately 10% of the lateral horn neuropile area (Figure 8C and D; dati, N = 8; WT, N = 10) and the cholinergic projections from the antennal lobe towards the lateral horn are also affected (Figure S4). Like in the antennal lobe, we note the presence of larger neurons in the lateral horn of dati mutants, which are not present in the wild type (Figure 8A and B). Furthermore, there are more CHA-positive cells around the lateral horn, suggesting that in the absence of dati some neuronal precursors can proliferate to later assume a cholinergic fate or, alternatively, that in the absence of dati some cells assume a cholinergic fate (Figure 8A–D). Together, these results show that dati is required in postmitotic neurons as well as in the precursors of these cells.
Figure 8. Homozygosity for dati1 causes abnormal distribution of cholinergic neurons and improper innervation of the lateral horn.
(A) Wild-type female brain expressing GFP under the control of Cha-Gal4. The image is a single frontal confocal slice at the level of the lateral horn. The lateral horn (LH), the medulla (Me) and lobula (Lo) are indicated. Note the density of cholinergic neurons in the region between the lateral horn and optic lobe (arrow). (B) Frontal confocal slice of a dati 1 female brain expressing GFP under the control of Cha-Gal4 in approximately the same position shown in (A). Note increased numbers of CHA+ cells (arrow) and enlarged cells (yellow arrowheads). (C, D) Orthogonal view of the region shown in (A, B). CHA+ cells are labeled in green, and the neuropile is labeled with nc82 antibody (magenta). (C) Wild-type. (D) dati 1 mutant female. Note again the excess of CHA+ cells in (D) compared to (C, brackets). Image rendering in orthogonal views in (C, D) were done in Image J.
Nuclear Bar Coding Reveals That DATI CHA Neurons Mediate Short- and Long-Range Connections
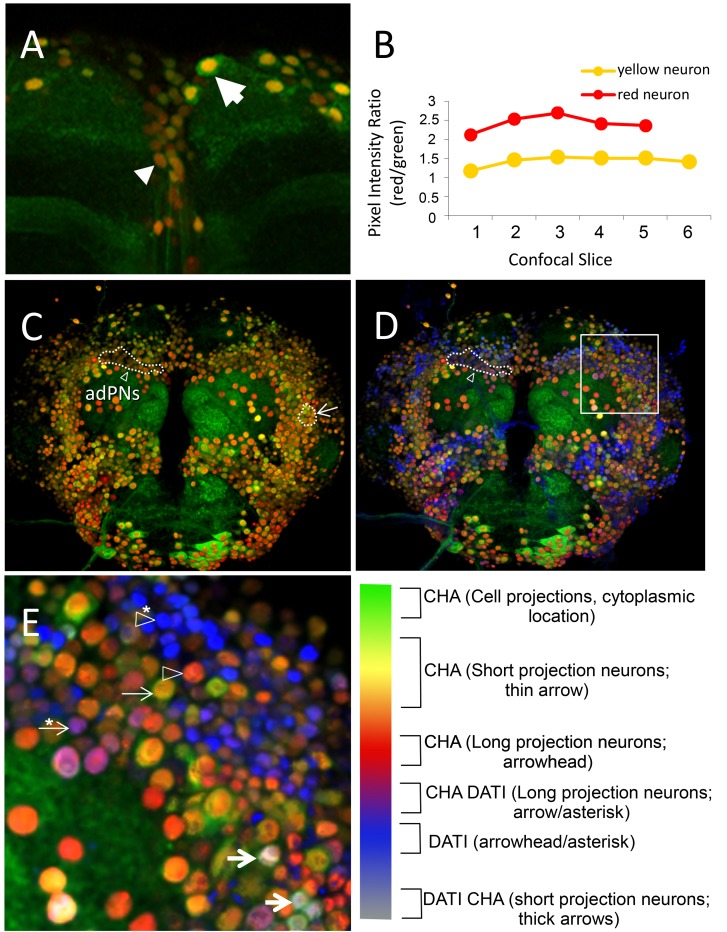
From the previous experiments, we found evidence that dati specifies a subpopulation of cholinergic neurons that project into the antennal olfactory glomeruli. Olfactory neurons in the antennal lobe descend from few neuroblast lineages that generate remarkably different neurons within and across lineages [50], and it has been suggested that morphologically different neurons are dedicated to specific neurocomputations [57]. This heterogeneity has been traditionally investigated in great detail in clones of single or few neurons using Gal4 drivers that reveal discrete neuronal populations [58]–[60]. However, we are often confronted with the opposite problem, which is to estimate whether a selected neuronal population makes simple, complex, or both simple and complex connections when a discrete Gal4 driver for these neurons is not available. This distinction is important to determine whether dati intrinsically modifies cell shapes or other aspects of neuronal physiology [61]. To that end, we developed a simple system of nuclear bar coding that distinguishes different DATI CHA neurons by color. Nuclear Bar Coding (NBC) consists of labeling nuclei of neurons with small or large volumes with different colors by expressing a localized nuclear RFP (mCherry.NLS) and GFP-S65T (nuclear and cytoplasmic) under the control of a Gal4 driver (in this case Cha-Gal4). Cells expressing the two fluorescent proteins from the same promoter are expected to be produced and degraded at comparable rates and result in nuclei with an overlay of two colors (Figure S5) [62],[63]. Assuming that these two proteins are not subject to a different regulation, the overlay of two colors should vary depending on the cellular volume. In cells with long or more intricate processes, GFP-S65T should be expected to fill up the cellular processes and shift the overlay of the two signals in the cell bodies towards that of the localized nuclear fluorescence (i.e., red color from RFP). Evidence for this shift was obtained in comparisons between cells with short and long cell processes (Figure 9A–C). Conversely, when both GFP and RFP are targeted to the nucleus, the shifts of nuclear bar coding are abolished (Figure S5). If, to this simple bar coding, we add a third color that detects DATI-positive cells (Figure 9D), then we can globally assess whether dati cholinergic neurons have simple or more complex projections. NBC allowed us to easily identify the descending neurons (Figure 9A), as well as long projection neurons located immediately above the antennal lobe, known as anterior–dorsal projection neurons (adPNs; Figure 9C,D), and LNs imbedded in antennal lateral neurons (Figure 9C–E). In addition, the NBC method reveals that the DATI CHA neurons within the region AntB2 make both short and long connections (Figure 9C–E). Thus, we conclude that dati does not specify only one type of cell shape, like other transcription factors that specify particular neurons [61].
Figure 9. dati is expressed in neurons that have small and large volumes.
(A) Maximum intensity projection of a confocal stack showing an antibody staining of a wild-type brain co-expressing nuclear RFP (red) and GFP-S65T (green) under Cha-Gal4 control. Neurons with large volumes (i.e., with long-range projections) appear with a gradation of dark orange to red (arrowhead; descending neuron), whereas neurons with small volumes (i.e., with short-range projections) appear as yellow to light orange (arrow). (B) Ratios of pixel intensity of the RFP and GFP channels of individual nucleus of the red and yellow neurons seen in (A) across all confocal slices encompassing their nuclei. Note the higher ratio of red/green from the red neuron with long processes, compared to the yellow neuron across each slice, indicating that the levels of GFP in the nuclei of long-range neurons are lower than in short-range neurons. Compare with results of the control experiment shown in Figure S4. (C) Overall view of the central brain. Anterior–dorsal projection neurons appear as dark red (adPNs, dotted line indicated by arrow), consistent with their long projections. LNs appear as light orange/yellow (dotted line indicated by arrowhead). (D) Superimposition of the channel detecting signal for anti-DATI staining (blue) to the image in (C). Note that adPNs appear in purple, indicating that these cells with long projections also express DATI (dotted line indicated by arrowhead). (E) High magnification of AntB2 region (box in D). DATI is expressed in cells with both small and large volumes, as indicated by color bar legend shown on the right. Neuron cell types are indicated by arrows and arrowheads, as explained in color bar legend. 3D image rendering was done in Image J.
Discussion
dati Encodes a Conserved ZNF Transcription Factor Related to Rotund/Squeeze and ZNF384 Required for Female Decision Making and Locomotion
Here we described DATI, a zinc finger transcription factor related to the Drosophila Rotund and Squeeze and the vertebrate ZNF384, one of the three genes known to be involved in acute lymphoblastic leukemia (ALL) [64],[65]. A survey of the sequences related to dati suggests that it descends from a Krüppel/rotund prototype present in cnidarians (e.g., Nematostella, gb|ABAV01025004.1|). Later this prototype evolved to become the rotund-like found in nematodes (e.g., C. elegans, Lin29) and mollusks (e.g., M. galloprovincialis, gb|GAEN01018610.1|) and was inherited by both vertebrates and invertebrates. Due to its similarity with Lin29, dati was previously referred to as Dmel/Lin29. However, orthology tests show that the ortholog of the C. elegans Lin29 is rotund, not dati. The first true ortholog of dati is found in marine arthropods (e.g., Daphnia pulex, Dpdati, gb|ACJG01001740.1|), which appeared in the Cambrian some 540 Mya [66].
Like its vertebrate homolog, dati is expressed in the nervous system and required for stem cell development [35],[67]–[69]. During embryogenesis, dati is one of the last genes to be activated in a serial activation of transcription factors that determines the identity of specific neuronal lineages in the ventral nerve cord [35]. The present study shows that dati is later required to specify regions of the central brain required for appropriate female acceptance.
dati mutant flies are moderately uncoordinated and almost invariably reject male courtship (Figure S1C and Figure 2). This rejection is so intense and persistent that it does not seem to be due to the mere loss of single sensory modalities, which inhibit but do not abolish acceptance [36]. Because of this strong rejection, we expected that dati might impair either more than one path required to generate acceptance in the brain or an area in which sensory information converges. In addition, we also tested if the locomotor and decision-making defects were associated or separable.
Locomotor Defects Are Separable From the Inability to Make Decisions
The mapping of foci by clonal analyses revealed individuals with clones that exhibited rejection but not locomotor defects (unpublished data). Conversely, we also found individuals with locomotor defects that were perfectly capable of accepting courtship and mating properly (unpublished data). Further evidence that locomotion and female behavior are separable was obtained in the experiments in which dati was knocked down in neurons that express different neurotransmitters (Figure 2). In this case, we found that none of the four drivers used (Ddc, Gad, ple, and Cha-Gal4) produced locomotor defects like those observed using either a ubiquitous driver or the neuronal driver elav-Gal4, but the removal of dati in CHA neurons resulted in strong female behavior deficits. Thus, we conclude that the locomotor defects and female acceptance map to different brain regions and distinct cells that express specific neurotransmitters.
DATI Adds an Additional Layer to the Identity of Cholinergic Neurons That Is Shared by Noncholinergic Neurons
Our results suggest dati has two roles in the nervous system—one developmental and another constitutive—both affecting female behavior. The over/underproliferation of cholinergic neurons in dati homozygotes suggests a requirement in neuronal precursors, which is consistent with the previous study that showed dati is transiently expressed in developing ganglion mother cells [35]. However, there is a requirement in neurons, as the courtship behavioral phenotype is recapitulated when dati is removed in postmitotic neurons. Further evidence for this requirement in adult neurons is the fact that dati is indeed expressed in neurons well into adulthood, and in fact, we identified a small group of neurons that only initiates expression of dati in adult neurons (unpublished data). Together these results suggest that dati may be required to maintain a neuronal identity. Because not all dati-positive neurons are cholinergic, and vice versa, it is unlikely that its primary role would be to determine the expression of this neurotransmitter. The Nuclear Bar Coding analysis suggests that dati does not evidently define any specific cell morphology either. We speculate that dati specifies a type of neuronal identity that allows neurons to respond to neurotransmitters that other cholinergic neurons without dati cannot. In this scenario, it is easy to see that removing dati from mature neurons would deprive them from the appropriate receptor(s) needed to receive input from their synaptic partners, and consequently silence female receptivity. Future tests should resolve whether dati indeed regulates channels/receptors to generate courtship acceptance.
The Regions Where dati Is Required Agree with Previous Mapping and Suggest the Existence of a Core Circuit for Female Decision Making
Different mutants and experimental approaches, including gynanders, spinster mosaics, mapping of cVA processing neurons, and the use of dati mosaics, here have identified some common and other distinct foci for female decision making. For instance, the first focus AntB2 that we identified maps to Sp11, the first brain region identified for female acceptance using mosaic gynandromorphs [19]. AntB2 also maps within the Spin-D site identified by mosaics of spinster [18], a gene also required for female behavior. In addition, the two other highly significant regions, PosC4 and PosA3, flank the lateral horn, and we note that the focus PosC4 co-maps with regions previously implicated in pheromonal processing in the female brain [52],[54]. Notably, the lateral horn may have a larger role in sensory integration, as it receives projections from centers that process visual and mechanosensory information [52]. Thus, the picture that emerges from previous work and the present study suggests that female decision making in Drosophila is modulated by a core circuit involving the antennal lobe and the lateral horn. However, we note that there are regions with ratios of acceptance and rejection that intuitively may appear to be relevant but that failed to reach statistical significance. In particular, there are three regions in the anterior brain (AntB3, AntB4, and AntD3) and seven regions in the posterior brain (PosB3, PosB4, PosC1, PosC2, PosC3, PosD2, and PosD3). We believe that these regions are unlikely foci for female receptivity, as our sample had resolution to identify the great significance of a relatively small focus like PosA3. Also, a similar study that analyzed a larger sample of Spinster foci for female receptivity also found brain regions that did not reach statistical significance but had ratios that could be intuitively interpreted as almost significant like ours. Like us, these authors disregarded these data as significant [18].
dati's Requirement in Few Excitatory Neurons in Three Discrete Brain Foci Reveals a Simple, Yet Fundamental, Mechanism of Female Decision Making in Drosophila
Besides providing the locations where courtship acceptance decisions are generated in the brain and the type of neurotransmitter involved, our results also reveal a significant neural mechanism at play. The DATI-CHA neurons mapped in the antennal lobe correspond to a subset of extensively studied cholinergic population known as the excitatory dorsal lateral Projection Neurons (ePNs) and excitatory lateral neurons (eLNs) [70]–[75]. The central role of excitatory cholinergic neurons revealed by our study and the localization of a region where sensory information is integrated constitute a nearly perfect cellular and molecular representation of the “Summation Hypothesis,” elaborated by Manning and others several decades ago based on behavioral inference [38],[76],[77]. This hypothesis states that acceptance of courtship involves the convergence of multiple excitatory stimulations provided by different sensory modalities until the stimulation reaches a critical threshold point that generates acceptance [76]. Most importantly, the Summation Hypothesis predicts that the two opposite female responses (i.e., rejection or acceptance) are not the result of opposing neural activities (e.g., excitation and inhibition) but rather the result of two different levels of excitation. Until now, there was no molecular and cellular evidence in support of this prediction. In this regard, our results are in agreement with this prediction, as the absence or presence of DATI in an excitatory circuit generates either complete rejection or overwhelming acceptance, respectively.
Corroborating our results, recent findings show that pheromone processing is not subject to the inhibitory mechanisms that apply to the processing of other odors [78]. Taken altogether, our results suggest that few dozen excitatory neurons converging in as few as three brain foci make the core components to generate a mating decision in Drosophila. Given that dati-related genes are present in a wide variety of organisms, it is likely that their common ancestor had the same or a similar mechanism of female acceptance.
Supporting Information
Embryonic and larval expression of dati . (A) Antibody staining of a wild-type late stage embryo for DATI (green) and ELAV (magenta). Note the presence of DATI in neurons of each hemisegment of the ventral nerve cord. Image is from a single slice of a confocal image stack. (B) Antibody staining of a wild-type L3 larval brain for DATI (gray). (C) Adult female brain stained with anti-DATI (green). The images are maximum intensity projections of confocal stacks.
(TIF)
Quantification of discrete responses to male courtship displayed by dati mutant females versus wild-type females. Female response to male courtship was quantified for 10 min after initiation of courtship by wild-type males. Bars show BI of each discrete response type of control group (Canton-S females, dark bars) and experimental (dati homozygous females, light bars) (see Materials and Methods for details). dati females are capable of displaying the same array of rejection behaviors to courtship as wild-type females (i.e., fleeing, kicking, extruding ovipositor, jumping, and flicking wings). Compared to wild-type females, dati females spend more time kicking males. In contrast, dati females spend significantly less time standing still, which is considered an accepting behavior displayed by wild-type females after being courted for some time. The statistical significance of differences was evaluated by the Mann–Whitney U test (***p<0.001; **p<0.01), and error bars represent ±SEM. The sample sizes are Canton-S, N = 12 and dati 1, N = 10.
(TIF)
Cell counts of neurons expressing DATI and both DATI and CHA. Cell counts of two different classes of neurons in Canton-S adult flies: DATI CHA double-positive cells and DATI-only positive cells. Light grey bars represent counts from scans made from the front of the central brain; black bars represent counts from scans made from the rear of the central brain. Automated cell counts were performed in Fiji as described in Materials and Methods.
(TIF)
dati mutants exhibit defects in the trajectory of projection neurons. (A) A wild-type brain and (B) a dati 1 mutant brain viewed from the brain neuropile towards the rear surface of the brain. In both images, 3D-rendered images were superimposed to 3D segmentation of the major cholinergic tracts (magenta). The lateral horn (LH) is indicated by the dashed circle. (C and D) Isolated segmentations of the major cholinergic tracts of the brain shown in (A and B, respectively) viewed from the rear brain surface. Note the thickness and complexity of the cell projection coming from the antennal lobe in (C) (red bracket) and the thinner and ill-defined projections in dati 1mutants (D) (red bracket). The coordinates P (posterior), A (anterior), D (dorsal), V (ventral), L (lateral), and C (central) are indicated.
(TIF)
Nuclear GFP and nuclear RFP have comparable rates of degradation. Fluorescent signals of nuclear GFP and nuclear RFP driven by the same ubiquitin promoter in the adult brain were captured and quantified. (A) A high-magnification confocal slice of the antennal lobe. The GFP and RFP pixel intensity values were collected along the white arrow. (B) Greyscale view of GFP.NLS expression from (A). (C) Greyscale view of RFP.NLS expression from (A). (D) Quantification of signals captured along the line shown in (A). Note that the levels of RFP and GFP are similar across the intensity peaks and valleys in contrast to when nuclear bar coding is performed (Figure 9).
(TIF)
Movie of a 2-d-old Canton-S female being courted by a Canton-S male. Note that although the female flees to some degree, she does so slowly and eventually stops to fully take in the males courtship display, and eventually allows him to mount.
(MP4)
Movie of a 2-d-old dati homozygous female being courted by a Canton-S male. Note that the female flees quickly from the male, and when she is caught up to by the male, she engages in rejection behaviors such as kicking.
(MP4)
Movie of a 7-d-old dati female being courted by a Canton-S male. Note the more severe incoordination of this female in comparison to younger female shown in Movie S2.
(MP4)
A 3D rendering of segmented images of the mushroom body of wild-type, Cha -Gal4 UAS- dati -RNAi and dati1 females labeled with anti-FasII. Labels for each genotype are shown in the movie. 3D rotations showing α (red), β (blue), and γ (yellow) lobes. Note that the γ lobe has a hammer-like shape in wild-type and Cha-Gal4<UAS-dati-RNAi females, whereas in dati1 mutants the γ lobe has an accentuated curvature in the center. This morphological defect is quantified in Figure 3D.
(MP4)
Determination of the molecular limits of l(4)102CDd2.
(TIF)
Acknowledgments
We thank Bloomington Stock Center for fly stocks. The anti-nc82, rat anti-Elav, and mouse anti-FasII antibodies developed by E. Buchner, G.M. Rubin, and C. Goodman, respectively, were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology.
Abbreviations
- adPN
anterior–dorsal projection neuron
- ALL
acute lymphoblastic leukemia
- ANOVA
analysis of variance
- BI
Behavioral Index
- Cha
Choline acetyltransferase
- CI
Courtship Index
- CS
Canton-S
- cVA
cis-vaccenyl acetate
- dati
datilógrafo
- dcr
dicer
- ddc
dopa decarboxylase
- elav
embryonic lethal abnormal vision
- eLNs
excitatory lateral neurons
- ePN
excitatory dorsal–lateral projection neurons
- FRT
flippase recognition target
- FYT
FRT-yellow-translocation
- Gad
Glutamic acid decarboxylase 1
- GFP
green fluorescent protein
- h
hour
- iPN
inhibitory projection neurons
- LH
lateral horn
- Lo
lobula
- Me
medulla
- min
minute
- MYA
million years ago
- NBC
nuclear bar coding
- NLS
nuclear localization sequence
- pilpr
posterior inferior lateral protocerebrum
- ple
pale
- plpr
posterior lateral protocerebrum
- pslpr
posterior superior lateral protocerebrum
- repo
reversed polarity
- RFP
red fluorescent protein
- rn
rotund
- s
second
- SEM
standard error of the mean
- spin
spinster
- sqz
squeeze
- WT
wild-type
Funding Statement
This work was funded by the grants from National Institutes of Health (R21EB016535) to RSN and CMM and National Science Foundation (IOS-1051662) to CMM. JMS was supported by a pre-doctoral GAANN fellowship from the US Department of Education. RLG was supported by Ohio First Scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Villella A, Hall JC (2008) Neurogenetics of courtship and mating in Drosophila. In: Hall J, Dunlap J, Friedmann T, van Heyningen V, editors. Advances in genetics. Oxford, UK: Academic Press, Vol. 62: . pp. 67–184. [DOI] [PubMed] [Google Scholar]
- 2. Hall JC (1979) Control of male reproductive behavior by the central nervous system of drosophila: dissection of a courtship pathway by genetic mosaics. Genetics 92: 437–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sakai T, Kitamoto T (2006) Differential roles of two major brain structures, mushroom bodies and central complex, for Drosophila male courtship behavior. J Neurobiol 66: 821–834 10.1002/neu.20262 [DOI] [PubMed] [Google Scholar]
- 4. Yu JY, Kanai MI, Demir E, Jefferis GSXE, Dickson BJ (2010) Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol 20: 1602–1614 10.1016/j.cub.2010.08.025 [DOI] [PubMed] [Google Scholar]
- 5. Dauwalder B (2011) The roles of fruitless and doublesex in the control of male courtship. Int Rev Neurobiol 99: 87–105 10.1016/B978-0-12-387003-2.00004-5 [DOI] [PubMed] [Google Scholar]
- 6. Jean-François F (2010) Drosophila female courtship and mating behaviors: sensory signals, genes, neural structures and evolution. Curr Opin Neurobiol 20: 764–769 10.1016/j.conb.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 7. Spieth HT (1974) Courtship behavior in drosophila. Annu Rev Entomol 19: 385–405 10.1146/annurev.en.19.010174.002125 [DOI] [PubMed] [Google Scholar]
- 8. Hall JC (1994) The mating of a fly. Science 264: 1702–1714. [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto D, Jallon JM, Komatsu A (1997) Genetic dissection of sexual behavior in Drosophila melanogaster. Annu Rev Entomol 42: 551–585. [DOI] [PubMed] [Google Scholar]
- 10. Von Schilcher F (1976) The function of pulse song and sine song in the courtship of Drosophila melanogaster. Anim Behav 24: 622–625 10.1016/S0003-3472(76)80076-0 [DOI] [Google Scholar]
- 11. Sakai T, Isono K, Tomaru M, Fukatami A, Oguma Y (2002) Light wavelength dependency of mating activity in the Drosophila melanogaster species subgroup. Genes Genet Syst 77: 187–195. [DOI] [PubMed] [Google Scholar]
- 12. Talyn BC, Dowse HB (2004) The role of courtship song in sexual selection and species recognition by female Drosophila melanogaster. Anim Behav 68: 1165–1180 10.1016/j.anbehav.2003.11.023 [DOI] [Google Scholar]
- 13. Grillet M, Dartevelle L, Ferveur J-F (2006) A Drosophila male pheromone affects female sexual receptivity. Proc Biol Sci 273: 315–323 10.1098/rspb.2005.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carhan A, Allen F, Armstrong JD, Hortsch M, Goodwin SF, et al. (2005) Female receptivity phenotype of icebox mutants caused by a mutation in the L1-type cell adhesion molecule neuroglian. Genes Brain Behav 4: 449–465 10.1111/j.1601-183X.2004.00117.x [DOI] [PubMed] [Google Scholar]
- 15. Juni N, Yamamoto D (2009) Genetic analysis of chaste, a new mutation of Drosophila melanogaster characterized by extremely low female sexual receptivity. J Neurogenet 23: 329–340 10.1080/01677060802471601 [DOI] [PubMed] [Google Scholar]
- 16. Suzuki K, Juni N, Yamamoto D (1997) Enhanced mate refusal in female Drosophila induced by a mutation in the spinster locus. Appl Entomol Zool 32: 235–243. [Google Scholar]
- 17. Sakai T, Kasuya J, Kitamoto T, Aigaki T (2009) The Drosophila TRPA channel, Painless, regulates sexual receptivity in virgin females. Genes Brain Behav 8: 546–557 10.1111/j.1601-183X.2009.00503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakurai A, Koganezawa M, Yasuyama K-I, Emoto K, Yamamoto D (2013) Select interneuron clusters determine female sexual receptivity in Drosophila. Nat Commun 4: 1825 10.1038/ncomms2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tompkins L, Hall JC (1983) Identification of brain sites controlling female receptivity in mosaics of Drosophila melanogaster. Genetics 103: 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hochman B, Gloor H, Green MM (1964) Analysis of chromosome 4 in Drosophila Melanogaster. I. Spontaneous and X-Ray-induced lethals. Genetica 35: 109–126. [DOI] [PubMed] [Google Scholar]
- 21. Sousa-Neves R (2000) The synthesis of a double fourth chromosome marker with y+. Dros Inf Serv 83: 8–10. [Google Scholar]
- 22. Sousa-Neves R, Lukacsovich T, Mizutani CM, Locke J, Podemski L, et al. (2005) High-resolution mapping of the Drosophila fourth chromosome using site-directed terminal deficiencies. Genetics 170: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sousa-Neves R, Schinaman JM (2012) A novel genetic tool for clonal analysis of fourth chromosome mutations. Fly (Austin) 6: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drapeau M, Long A (2000) The Copulatron, a multi-chamber apparatus for observing Drosophila courtship behaviors. Dros Inf Serv 83: 194–196. [Google Scholar]
- 25. Tompkins L, Hall JC, Hall LM (1980) Courtship-stimulating volatile compounds from normal and mutant Drosophila. J Insect Physiol 26: 689–697 10.1016/0022-1910(80)90042-6 [DOI] [Google Scholar]
- 26. Gargano JW, Martin I, Bhandari P, Grotewiel MS (2005) Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol 40: 386–395 10.1016/j.exger.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 27. Sweeney ST, Hidalgo A, de Belle JS, Keshishian H (2012) Antibody staining of the central nervous system in adult Drosophila. Cold Spring Harb Protoc 2012: 235–238 10.1101/pdb.prot067397 [DOI] [PubMed] [Google Scholar]
- 28. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolte S, Cordelières FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 30. Preibisch S, Saalfeld S, Tomancak P (2009) Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tully T, Quinn WG (1985) Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A 157: 263–277. [DOI] [PubMed] [Google Scholar]
- 32. Van Swinderen B, Hall JC (1995) Analysis of conditioned courtship in dusky-Andante rhythm mutants of Drosophila. Learn Mem 2: 49–61. [DOI] [PubMed] [Google Scholar]
- 33. Podemski L, Sousa-Neves R, Marsh JL, Locke J (2004) Molecular mapping of deletion breakpoints on chromosome 4 of Drosophila melanogaster. Chromosoma 112: 381–388. [DOI] [PubMed] [Google Scholar]
- 34. Sousa-Neves R, Mizutani CM, Lukacsovich T, Purcell J, Marsh JL (2005) datilografo (dati) encodes a zinc finger transcription factor required for proper locomotor activity in Drosophila. 46th A Dros Res Conf 46 Suppl: 1044C. [Google Scholar]
- 35. Tsuji T, Hasegawa E, Isshiki T (2008) Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development 135: 3859–3869 10.1242/dev.025189 [DOI] [PubMed] [Google Scholar]
- 36. Bastock M (1956) A gene mutation which changes a behavior pattern. Evolution (N Y) 10: 421–439 10.2307/2407002 [DOI] [Google Scholar]
- 37. Chu Y, Yang E, Schinaman JM, Chahda JS, Sousa-Neves R (2013) Genetic analysis of mate discrimination in Drosophila simulans. Evolution 67: 2335–2347 10.1111/evo.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manning A (1967) The control of sexual receptivity in female Drosophila. Anim Behav 15: 239–250. [DOI] [PubMed] [Google Scholar]
- 39. Berger C, Renner S, Lüer K, Technau GM (2007) The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn 236: 3562–3568 10.1002/dvdy.21372 [DOI] [PubMed] [Google Scholar]
- 40. Dierick HA, Greenspan RJ (2007) Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet 39: 678–682 10.1038/ng2029 [DOI] [PubMed] [Google Scholar]
- 41. Bicker G (1999) Introduction to neurotransmitter histochemistry of the insect brain. Microsc Res Tech 45: 63–64 doi:;10.1002/(SICI)1097-0029(19990415)45:2<63::AID-JEMT1>3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- 42. Littleton JT, Ganetzky B (2000) Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron 26: 35–43. [DOI] [PubMed] [Google Scholar]
- 43. Budnik V, White K (1987) Genetic dissection of dopamine and serotonin synthesis in the nervous system of Drosophila melanogaster. J Neurogenet 4: 309–314. [PubMed] [Google Scholar]
- 44. Daniels RW, Gelfand MV, Collins CA, DiAntonio A (2008) Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol 508: 131–152 10.1002/cne.21670 [DOI] [PubMed] [Google Scholar]
- 45. Waddell S (2010) Dopamine reveals neural circuit mechanisms of fly memory. Trends Neurosci 33: 457–464 10.1016/j.tins.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe TK, Anderson WW (1976) Selection for geotaxis in Drosophila melanogaster: heritability, degree of dominance, and correlated responses to selection. Behav Genet 6: 71–86. [DOI] [PubMed] [Google Scholar]
- 47. Balling A, Technau GM, Heisenberg M (1987) Are the structural changes in adult Drosophila mushroom bodies memory traces? Studies on biochemical learning mutants. J Neurogenet 4: 65–73. [PubMed] [Google Scholar]
- 48. McBride SM, Giuliani G, Choi C, Krause P, Correale D, et al. (1999) Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron 24: 967–977. [DOI] [PubMed] [Google Scholar]
- 49. Ito K, Awasaki T (2008) Clonal unit architecture of the adult fly brain. Adv Exp Med Biol 628: 137–158 10.1007/978-0-387-78261-49 [DOI] [PubMed] [Google Scholar]
- 50. Das A, Sen S, Lichtneckert R, Okada R, Ito K, et al. (2008) Drosophila olfactory local interneurons and projection neurons derive from a common neuroblast lineage specified by the empty spiracles gene. Neural Dev 3: 33 10.1186/1749-8104-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, et al. (2002) GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34: 58–61 10.1002/gene.10137 [DOI] [PubMed] [Google Scholar]
- 52. Tanaka NK, Awasaki T, Shimada T, Ito K (2004) Integration of chemosensory pathways in the Drosophila second-order olfactory centers. Curr Biol 14: 449–457 10.1016/j.cub.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 53. Masse NY, Turner GC, Jefferis GSXE (2009) Olfactory information processing in Drosophila. Curr Biol 19: R700–R713. [DOI] [PubMed] [Google Scholar]
- 54. Jefferis GSXE, Potter CJ, Chan AM, Marin EC, Rohlfing T, et al. (2007) Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell 128: 1187–1203 10.1016/j.cell.2007.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, et al. (2010) A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature 468: 686–690 10.1038/nature09554 [DOI] [PubMed] [Google Scholar]
- 56. Couto A, Alenius M, Dickson BJ (2005) Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 15: 1535–1547 10.1016/j.cub.2005.07.034 [DOI] [PubMed] [Google Scholar]
- 57. Sachse S, Rueckert E, Keller A, Okada R, Tanaka NK, et al. (2007) Activity-dependent plasticity in an olfactory circuit. Neuron 56: 838–850 10.1016/j.neuron.2007.10.035 [DOI] [PubMed] [Google Scholar]
- 58. Yu H-H, Kao C-F, He Y, Ding P, Kao J-C, et al. (2010) A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biol 8 10.1371/journal.pbio.1000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jefferis GS, Marin EC, Stocker RF, Luo L (2001) Target neuron prespecification in the olfactory map of Drosophila. Nature 414: 204–208 10.1038/35102574 [DOI] [PubMed] [Google Scholar]
- 60. Okada R, Awasaki T, Ito K (2009) Gamma-aminobutyric acid (GABA)-mediated neural connections in the Drosophila antennal lobe. J Comp Neurol 514: 74–91 10.1002/cne.21971 [DOI] [PubMed] [Google Scholar]
- 61. Parrish JZ, Emoto K, Kim MD, Jan YN (2007) Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 30: 399–423 10.1146/annurev.neuro.29.051605.112907 [DOI] [PubMed] [Google Scholar]
- 62. Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, et al. (2009) A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol 394: 732–746 10.1016/j.jmb.2009.09.050 [DOI] [PubMed] [Google Scholar]
- 63. Yen H-CS, Elledge SJ (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322: 923–929 10.1126/science.1160462 [DOI] [PubMed] [Google Scholar]
- 64. Martini A, La Starza R, Janssen H, Bilhou-Nabera C, Corveleyn A, et al. (2002) Recurrent rearrangement of the Ewing's sarcoma gene, EWSR1, or its homologue, TAF15, with the transcription factor CIZ/NMP4 in acute leukemia. Cancer Res 62: 5408–5412. [PubMed] [Google Scholar]
- 65. La Starza R, Aventin A, Crescenzi B, Gorello P, Specchia G, et al. (2005) CIZ gene rearrangements in acute leukemia: report of a diagnostic FISH assay and clinical features of nine patients. Leukemia 19: 1696–1699. [DOI] [PubMed] [Google Scholar]
- 66. Braun A, Chen J, Waloszek D, Maas A (2007) First early cambrian radiolaria. Geol Soc London, Spec Publ 286: 143–149 10.1144/SP286.10 [DOI] [Google Scholar]
- 67. Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, et al. (2006) Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics 7: 67–80 10.1007/s10048-006-0032-6 [DOI] [PubMed] [Google Scholar]
- 68. Lukk M, Kapushesky M, Nikkilä J, Parkinson H, Goncalves A, et al. (2010) A global map of human gene expression. Nat Biotechnol 28: 322–324 10.1038/nbt0410-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nakamoto T, Shiratsuchi A, Oda H, Inoue K, Matsumura T, et al. (2004) Impaired spermatogenesis and male fertility defects in CIZ/Nmp4-disrupted mice. Genes Cells 9: 575–589 10.1111/j.1356-9597.2004.00746.x [DOI] [PubMed] [Google Scholar]
- 70. Gu H, O'Dowd DK (2006) Cholinergic synaptic transmission in adult Drosophila Kenyon cells in situ. J Neurosci 26: 265–272 10.1523/JNEUROSCI.4109-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenböck G (2007) Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell 128: 601–612 10.1016/j.cell.2006.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Olsen SR, Bhandawat V, Wilson RI (2007) Excitatory interactions between olfactory processing channels in the Drosophila antennal lobe. Neuron 54: 89–103 10.1016/j.neuron.2007.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Silbering AF, Okada R, Ito K, Galizia CG (2008) Olfactory information processing in the Drosophila antennal lobe: anything goes? J Neurosci 28: 13075–13087 10.1523/JNEUROSCI.2973-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang J, Zhang W, Qiao W, Hu A, Wang Z (2010) Functional connectivity and selective odor responses of excitatory local interneurons in Drosophila antennal lobe. Neuron 67: 1021–1033 10.1016/j.neuron.2010.08.025 [DOI] [PubMed] [Google Scholar]
- 75. Parnas M, Lin AC, Huetteroth W, Miesenböck G (2013) Odor discrimination in Drosophila: from neural population codes to behavior. Neuron 79: 932–944 10.1016/j.neuron.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bastock M, Manning A (1955) The courtship of Drosophila melanogaster. Behaviour 8: 85–111. [Google Scholar]
- 77. Ewing AW (1964) The influence of wing area on the courting behavior of Drosophila melanogaster. Anim Behav 12: 316–320. [Google Scholar]
- 78. Liang L, Li Y, Potter CJ, Yizhar O, Deisseroth K, et al. (2013) GABAergic projection neurons route selective olfactory inputs to specific higher-order neurons. Neuron 79: 917–931 10.1016/j.neuron.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Embryonic and larval expression of dati . (A) Antibody staining of a wild-type late stage embryo for DATI (green) and ELAV (magenta). Note the presence of DATI in neurons of each hemisegment of the ventral nerve cord. Image is from a single slice of a confocal image stack. (B) Antibody staining of a wild-type L3 larval brain for DATI (gray). (C) Adult female brain stained with anti-DATI (green). The images are maximum intensity projections of confocal stacks.
(TIF)
Quantification of discrete responses to male courtship displayed by dati mutant females versus wild-type females. Female response to male courtship was quantified for 10 min after initiation of courtship by wild-type males. Bars show BI of each discrete response type of control group (Canton-S females, dark bars) and experimental (dati homozygous females, light bars) (see Materials and Methods for details). dati females are capable of displaying the same array of rejection behaviors to courtship as wild-type females (i.e., fleeing, kicking, extruding ovipositor, jumping, and flicking wings). Compared to wild-type females, dati females spend more time kicking males. In contrast, dati females spend significantly less time standing still, which is considered an accepting behavior displayed by wild-type females after being courted for some time. The statistical significance of differences was evaluated by the Mann–Whitney U test (***p<0.001; **p<0.01), and error bars represent ±SEM. The sample sizes are Canton-S, N = 12 and dati 1, N = 10.
(TIF)
Cell counts of neurons expressing DATI and both DATI and CHA. Cell counts of two different classes of neurons in Canton-S adult flies: DATI CHA double-positive cells and DATI-only positive cells. Light grey bars represent counts from scans made from the front of the central brain; black bars represent counts from scans made from the rear of the central brain. Automated cell counts were performed in Fiji as described in Materials and Methods.
(TIF)
dati mutants exhibit defects in the trajectory of projection neurons. (A) A wild-type brain and (B) a dati 1 mutant brain viewed from the brain neuropile towards the rear surface of the brain. In both images, 3D-rendered images were superimposed to 3D segmentation of the major cholinergic tracts (magenta). The lateral horn (LH) is indicated by the dashed circle. (C and D) Isolated segmentations of the major cholinergic tracts of the brain shown in (A and B, respectively) viewed from the rear brain surface. Note the thickness and complexity of the cell projection coming from the antennal lobe in (C) (red bracket) and the thinner and ill-defined projections in dati 1mutants (D) (red bracket). The coordinates P (posterior), A (anterior), D (dorsal), V (ventral), L (lateral), and C (central) are indicated.
(TIF)
Nuclear GFP and nuclear RFP have comparable rates of degradation. Fluorescent signals of nuclear GFP and nuclear RFP driven by the same ubiquitin promoter in the adult brain were captured and quantified. (A) A high-magnification confocal slice of the antennal lobe. The GFP and RFP pixel intensity values were collected along the white arrow. (B) Greyscale view of GFP.NLS expression from (A). (C) Greyscale view of RFP.NLS expression from (A). (D) Quantification of signals captured along the line shown in (A). Note that the levels of RFP and GFP are similar across the intensity peaks and valleys in contrast to when nuclear bar coding is performed (Figure 9).
(TIF)
Movie of a 2-d-old Canton-S female being courted by a Canton-S male. Note that although the female flees to some degree, she does so slowly and eventually stops to fully take in the males courtship display, and eventually allows him to mount.
(MP4)
Movie of a 2-d-old dati homozygous female being courted by a Canton-S male. Note that the female flees quickly from the male, and when she is caught up to by the male, she engages in rejection behaviors such as kicking.
(MP4)
Movie of a 7-d-old dati female being courted by a Canton-S male. Note the more severe incoordination of this female in comparison to younger female shown in Movie S2.
(MP4)
A 3D rendering of segmented images of the mushroom body of wild-type, Cha -Gal4 UAS- dati -RNAi and dati1 females labeled with anti-FasII. Labels for each genotype are shown in the movie. 3D rotations showing α (red), β (blue), and γ (yellow) lobes. Note that the γ lobe has a hammer-like shape in wild-type and Cha-Gal4<UAS-dati-RNAi females, whereas in dati1 mutants the γ lobe has an accentuated curvature in the center. This morphological defect is quantified in Figure 3D.
(MP4)
Determination of the molecular limits of l(4)102CDd2.
(TIF)