Abstract
Lymphoblastoid cell lines (LCLs) are commonly used in molecular genetics, supplying DNA for the HapMap and 1000 Genomes Projects, used to test chemotherapeutic agents, and informing the basis of a number of population genetics studies of gene expression. The process of transforming human B cells into LCLs requires the presence of Epstein-Barr virus (EBV), a double-stranded DNA virus which through B-cell immortalisation maintains an episomal virus genome in every cell of an LCL at variable copy numbers. Previous studies have reported that EBV alters host-gene expression and EBV copy number may be under host genetic control. We performed a genome-wide association study of EBV genome copy number in LCLs and found the phenotype to be highly heritable, although no individual SNPs achieved a significant association with EBV copy number. The expression of two host genes (CXCL16 and AGL) was positively correlated and expression of ADARB2 was negatively correlated with EBV copy number in a genotype-independent manner. This study shows an association between EBV copy number and the gene expression profile of LCLs, and suggests that EBV copy number should be considered as a covariate in future studies of host gene expression in LCLs.
Introduction
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus. Following primary infection EBV establishes lifelong persistent infection through latent infection of memory B cells where the virus genome is transcriptionally silent [1], [2]. Reactivation from latency is required for the production of infectious EBV, with such lytic EBV replication being under the control of host and virus factors. In particular, terminal differentiation of memory B cells into plasma cells can lead to EBV lytic reactivation [3]. The mechanisms of host induction of EBV lytic replication are incompletely understood, but periodic shedding of EBV in saliva [4] and variation in saliva virus load between people [5] suggest host genetic variation may contribute to EBV lytic cycle induction. Lymphoblastoid cell lines (LCLs) are human B cells immortalised in vitro by EBV and are a useful model of latent infection of B cells. Previous studies on LCLs have shown that when multiple LCLs are derived from the same individual, inter-individual variation in EBV copy number in LCLs is greater than intra-individual variation [6]. A study of the impact of EBV copy number on the gene expression profiles of 198 HapMap LCLs reported that expression of 125 human genes was significantly correlated with EBV copy number [7]. A comparison of Epstein-Barr virus copy number in 62 adult and paediatric LCLs found considerable inter-individual variation in EBV copy number that correlated with expression of immediate-early viral lytic genes BRLF1 and BZLF1, suggesting that spontaneous lytic reactivation is the cause of high EBV genome copy numbers in a subset of LCLs. After the addition of acyclovir, a drug which inhibits viral reactivation, Davies et al. showed EBV genome copy numbers fall in LCLs, and return to previous high levels after the removal of acyclovir [8]. This suggests that spontaneous lytic reactivation may be under the control of cell-intrinsic factors. When the viral gene expression profiles of LCLs were compared, using RNAseq data from multiple experiments from different laboratories, Arvey et al. [9] reported two major EBV gene expression profiles: latency type III and a lytic pattern of expression. There is evidence of both BZLF1 expression and virus particle production in some LCLs [10]. We have therefore hypothesised that high EBV copy number in LCLs is the result of poor host cell control of the EBV latent-lytic cycle switch, and may be under the control of host genetic factors.
Genome-wide association studies have been successfully used to identify the host genes involved in the pathogenesis of infectious disease [11], [12], [13], [14], [15]. Two genome-wide association studies of genetic control of antibodies to herpesviruses have been performed. A study of EBV antibody titres in ∼2000 individuals identified 15 loci exceeding genome-wide significance associated with either the quantitative or discrete trait of antibody titre [16]. By contrast, a similarly-sized study of cytomegalovirus (CMV) antibody response, a betaherpesvirus which also establishes lifelong latent infection in humans, did not find any genome-wide significant associations [17]. Other studies of host genetic response to herpesvirus infection and lytic reactivation have been limited to family linkage [18] and candidate gene [19] studies of herpes simplex virus-induced disease, and small studies of susceptibility to infection of chickens with an avian herpesvirus, Marek's disease virus [20], [21]. As yet, there have been no attempts to characterise common human genetic polymorphisms associated with cell-intrinsic response to EBV infection.
Here we describe a study to identify human genetic variants associated with Epstein-Barr virus genome copy number in the HapMap [22] and 1000 Genomes [23] LCLs, incorporating sequencing and genotyping data from the HapMap and 1000 Genomes projects. We also investigate differences in gene expression associated with EBV genome copy number using publicly available gene expression data for a subset of the HapMap 3 samples [24].
Results
Relative EBV copy number in LCLs and between-population comparisons
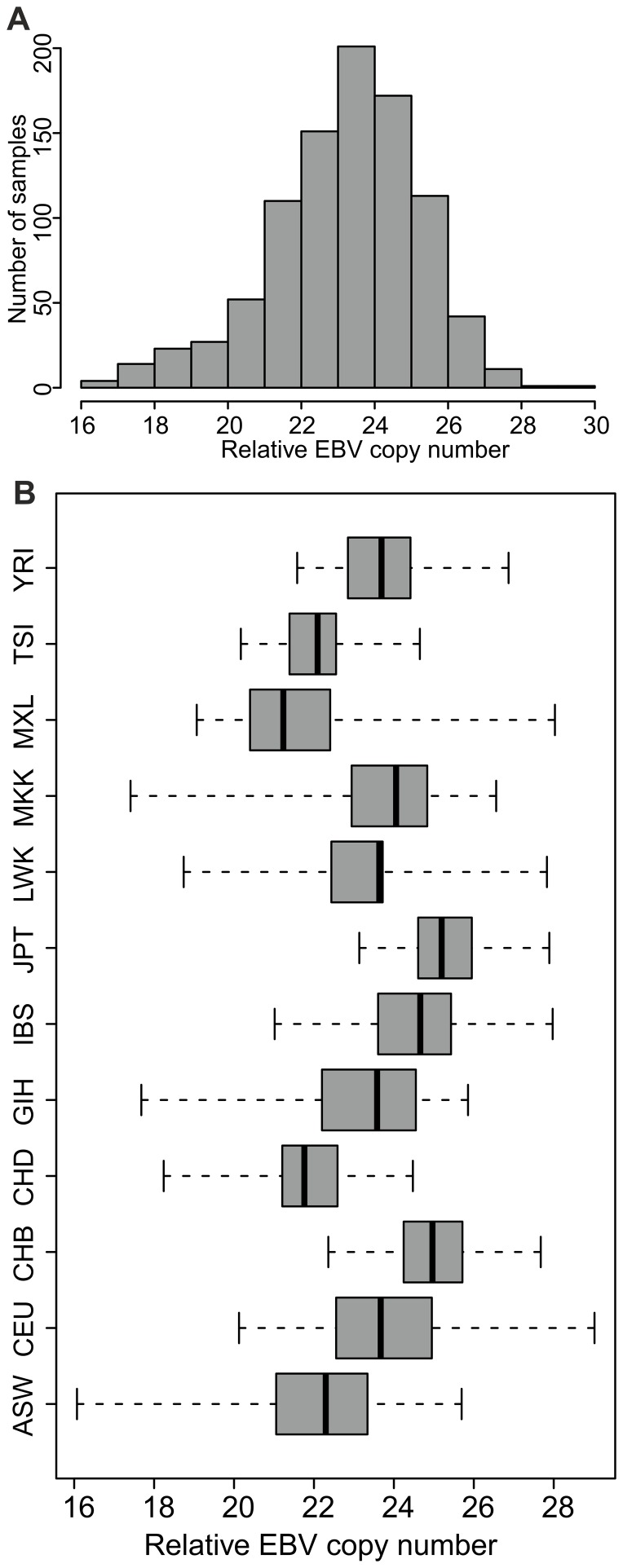
We determined the relative EBV genome copy number for 915 LCLs from the HapMap and 1000 Genomes populations using quantitative PCR (qPCR). The qPCR assay had an 8 log10 dynamic range from 1×109 to 1×102 copies/reaction (Figure S1) and an analytical sensitivity of 100 copies/reaction. The PCR efficiency was 99.4%. The samples assayed included individuals from 12 populations (Table 1); and EBV copy numbers of all samples were within the dynamic ranged of the qPCR. Across all populations the mean relative (to single copy host gene) EBV copy number per LCL is 23.36 with a range of 16.07–29.02 (SD±2.03), corresponding to an absolute range of 1 copy per cell to 350 copies per cell (Figure 1 A). We examined the trait of EBV copy number in different populations (Figure 1 B), and found significant differences in the mean EBV copy number between the populations (ANOVA p<2.2×10−16). Interestingly, apparently similar ethnic groups in different geographical areas have different mean EBV genome copy numbers. The Denver Han Chinese (CHD) and Beijing Han Chinese (CHB) populations have statistically significantly different means (difference = −3.23, 95% CI −4.34–−2.12), p = 1.17×10−8), although the CHB and Japanese from Tokyo (JPT) do not differ from one another significantly. Some European ancestry populations also differ from one another in their mean EBV copy number: Toscani from Italy (TSI) differ from CEU (European ancestry in Utah, USA) (difference = −1.63, 95% CI −2.52–−0.74, p = 2×10−7).
Table 1. Samples used in this study.
| Population code | Population description* | N (samples) | N (trios) | N (duos) |
| ASW | People with African Ancestry in the Southwest United States | 89 | 13 | 21 |
| CEU | Utah residents with ancestry from Northern and Western Europe | 84 | 27 | |
| CHB | Han Chinese in Beijing, China | 44 | ||
| CHD | Han Chinese in Denver, Colorado | 55 | ||
| GIH | Gujarati Indians in Houston, Texas | 90 | 1 | |
| IBS | Iberian Populations in Spain | 99 | 1 | |
| JPT | Japanese in Tokyo, Japan | 44 | ||
| LWK | Luhya in Webuye, Kenya | 5 | ||
| MKK | Maasai in Kinyawa, Kenya | 140 | 21 | 5 |
| MXL | People with Mexican Ancestry in Los Angeles, California | 92 | 24 | |
| TSI | Toscani in Italia | 67 | ||
| YRI | Yoruba in Ibadan, Nigeria | 90 | 30 |
Figure 1. Distribution of relative EBV copy number in LCLs from 12 populations.
A. Relative EBV copy number in 915 LCLs from 12 populations, including 97 parent-offspring trios. Range 16.07–29.02, mean 23.36 (SD±2.03). The data are not normally distributed (Shapiro-Wilk: W = 0.977, p-value = 2.086−11). B. Box and whisker plots showing the range (dotted lines), interquartile range (coloured boxes) and mean relative EBV copy number (central vertical lines) for the 12 populations included in this analysis.
Association testing
Using the entire sample set we performed a genome wide association study for common host genetic variants associated with LCL EBV copy number. To determine if the EBV that immortalised the LCLs was the laboratory strain B95.8 rather than outgrowth of spontaneous LCLs with wild type EBV we assembled the EBV genomes from 77 CEU and YRI LCLs. We looked for the presence of the deletion specific for the B95.8 genome, which was present in every LCL studied. The frequency of spontaneous LCLs containing wild type EBV, and/or LCLs immortalised with B95.8 and co-infected with wild type EBV, is therefore less than 1 in 77.
After sample-level quality control, genome-wide sequence and genotype data were available for 899 samples from the 1000 Genomes Phase I and HapMap Phase III consensus releases. Mixed-effects modelling [25] was used to test each variant individually for association with EBV copy number in LCLs. Samples without full human genome sequence data were imputed using 1000 Genomes information [26]. Variants were taken forward to association analysis if they were observed to vary in all 12 populations studied with a frequency of >1% and a minimum imputation quality of 0.9. This created a set of 1.6×106 SNPs in common between the samples. The sample size of phenotyped LCLs with genotype or whole genome sequence information available is 899 individuals, including 798 unrelated individuals, once offspring of 101 trios were removed. The statistical power of the study to detect a variant explaining a given proportion of the total trait variance was calculated using GWApower [27]. Our study had an 80% power to detect a variant explaining 4.7% of the total variance in relative EBV copy number. The overall distribution of P values showed little evidence of genomic inflation (λ = 0.98), consistent with the null hypothesis and suggesting that mixed-effects modelling was able to correct for high familial relatedness and population structure (Figure 2 A and B).
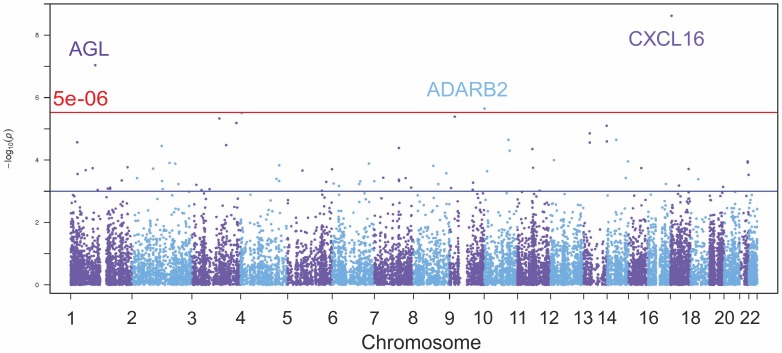
Figure 2. Manhattan plots of the association between EBV copy number and human genetic variants in LCLs.

A. Results of the stage 2 GWAS of relative EBV copy number in 899 LCLs, derived from 12 populations. In total, ∼1.6 M SNPs, polymorphic in every population studied, were analysed using FaST-LMM. Each point represents a SNP. There were no SNPs with genome-wide significant p-values after correction for multiple testing. B. QQ plot showing the distribution of observed test statistics plotted against the expected (null) distribution (red line). The SNPs which fall below the red line suggest that the GWAS may be losing power to detect SNPs associated with EBV copy number. C. The strongest signal is located within an intron of protein tyrosine phosphatase receptor delta (PTPRD), SNP rs10959089 (p = 1.17×10-6). Plot C made with LocusZoom [56].
We estimated the proportion of variance in EBV copy number within the dataset explained by the set of ∼1.6 million common genetic variants using GCTA, in 677 unrelated individuals to be 0.65 (SE ±0.38; p = 0.04). We also calculated the heritability of EBV copy number in 101 trios present within the 1000 Genomes dataset, where EBV copy number was available for all trio members. The parental mid-point EBV copy number was regressed against the offspring EBV copy number, giving a heritability estimate of 34% (SE ±11, p = 0.0028).
No variants passed the genome-wide significance threshold (P<5×10−8, Figure 2 A), although 98 SNPs achieved a genome wide significance of <1×10−5 and are suggestive of a possible association (Table S2 in File S1). Of these the top SNP, rs10959089 (P = 1.17×10−6, beta = 0.52 for the minor C allele), was located in the first intron of the gene PTPRD (protein tyrosine phosphatase receptor delta) (Figure 2 C).
Association testing of variants implicated in EBV infection, immune response and disease by previous studies
48 SNPs and small structural variants have been previously reported to influence EBV traits such as acquisition risk, antibody response, or EBV-positive disease risk. 28 of these SNPs were included in our association study (Table 2). Two SNPs had P values of nominal significance (rs2516049, p = 0.01; rs1052536, p = 0.03). It is therefore not possible to link these variants to the phenotype of relative EBV copy number in LCLs.
Table 2. Results for 27 SNPs previously reported to be associated with EBV infection, immune response or EBV-positive disease.
| SNP | Gene | Minor allele | Chr | Position | P value | Beta | Reference |
| rs17102086 | RAD54L | C | 1 | 46722939 | 0.69 | 1.05 | [49] |
| rs1801274 | FCGR2A | G | 1 | 161479745 | 0.59 | 1.05 | [50] |
| rs1800587 | IL1A | A | 2 | 113542960 | 0.47 | 1.08 | [51] |
| rs2306597 | RFC1 | A | 4 | 39297200 | 0.56 | 1.07 | [49] |
| rs10947261 | BTNL2 | T | 6 | 32373232 | 0.55 | 1.08 | [38] |
| rs10947262 | BTNL2 | T | 6 | 32373312 | 0.60 | 1.07 | [38] |
| rs1800629 | TNF | A | 6 | 31543031 | 0.47 | 1.14 | [52] |
| rs204999 | G | 6 | 32109979 | 0.75 | 1.04 | [38] | |
| rs2213585 | HLA-DRA | G | 6 | 32413150 | 0.37 | 1.09 | [38] |
| rs2213586 | HLA-DRA | A | 6 | 32413094 | 0.37 | 1.09 | [38] |
| rs2227139 | G | 6 | 32413459 | 0.37 | 1.09 | [38] | |
| rs2239803 | HLA-DRA | C | 6 | 32411833 | 0.44 | 1.08 | [38] |
| rs2294882 | BTNL2 | C | 6 | 32367515 | 0.41 | 1.11 | [38] |
| rs2294884 | BTNL2 | G | 6 | 32367259 | 0.54 | 1.08 | [38] |
| rs2516049 | LOC100507709 | C | 6 | 32570400 | 0.01 | 1.32 | [38] |
| rs28362683 | BTNL2 | A | 6 | 32372963 | 0.84 | 1.03 | [38] |
| rs3130048 | BAG6 | C | 6 | 31613739 | 0.62 | 1.06 | [38] |
| rs4248166 | BTNL2 | C | 6 | 32366421 | 0.75 | 1.04 | [38] |
| rs477515 | LOC100507709 | A | 6 | 32569691 | 0.04 | 1.25 | [38] |
| rs652888 | EHMT2 | G | 6 | 31851234 | 0.15 | 1.19 | [38] |
| rs6904029 | HCG9 | A | 6 | 29943067 | 0.86 | 1.02 | [53] |
| rs7192 | HLA-DRA | T | 6 | 32411646 | 0.37 | 1.09 | [38] |
| rs7195 | HLA-DRA | A | 6 | 32412539 | 0.37 | 1.09 | [38] |
| rs9264942 | HLA-C | C | 6 | 31274380 | 0.17 | 1.14 | [54] |
| rs9268832 | T | 6 | 32427789 | 0.37 | 1.09 | [38] | |
| rs1800450 | MBL2 | T | 10 | 54531235 | 0.85 | 1.03 | [55] |
| rs1052536 | LIG3 | T | 17 | 33331575 | 0.03 | 1.28 | [49] |
Epstein-Barr virus gene copy number, host gene expression and eQTL analysis in LCLs
Microarray gene expression data was available for 466 unrelated individuals from 8 populations [24]. A linear regression was performed for these individuals between 21,800 gene transcripts and EBV genome copy number. A statistically significant positive correlation was found between EBV relative copy number and the expression levels of two genes: CXCL16 (chemokine (C-X-C motif) ligand 16) and AGL (amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase), and a statistically significant negative correlation between EBV relative copy number and ADARB2 (adenosine deaminase, RNA-specific, B2) expression (Figure 3; Table 3). Transcripts with suggestive P values (P>5×10−3) are included in Table S3 in File S1. Evidence for the effect of EBV genome copy number on eQTL results was not observed for any of these genes; the correlation did not occur in a genotype-dependent manner. QTL mapping using EBV as a phenotype did not reveal any statistically significant SNPs located in or near these genes.
Figure 3. Association between EBV copy number and human gene expression in LCLs.
Manhattan plot showing the correlation between expression levels of 21,800 individual gene transcripts from microarray data and EBV copy number.
Table 3. Gene transcripts associated with relative EBV copy number with P<3×10−5.
| Gene | Array_tag | Chr | Beta | SE | P value |
| CXCL16 | ILMN_1672278 | 17 | 0.114 | 0.019 | 2.4×10−9 |
| AGL | ILMN_1680343 | 1 | 0.039 | 0.007 | 9.3×10−8 |
| ADARB2 | ILMN_1749493 | 10 | −0.042 | 0.009 | 2.3×10−6 |
Discussion
This is the largest study to investigate the impact of host genetic factors on Epstein-Barr virus genome copy number in lymphoblastoid cell lines. As inhibitors of the EBV lytic cycle effectively reduce EBV copy number in LCLs this suggests the higher copy numbers are the result of lytic replication induction [8]. Our study therefore also represents a proxy phenotype for the spontaneous switch from latent to lytic EBV replication in LCLs. We identified the EBV strain infecting a sample of the LCLs, quantified relative EBV genome copy number in 915 LCLs from the HapMap and 1000 Genomes study, performed a genome-wide association study of EBV genome copy number, estimated the heritability of EBV genome copy number in parent-offspring trios, and examined the relationship between EBV genome copy number and human gene expression in a subset of LCLs.
Using high-coverage sequence data from the 1000 Genomes Pilot project for 77 CEU and YRI LCLs, we estimate that the rate of LCLs containing wild-type or non-B-95.8-strain EBV is less than 1 in 77. This is consistent with the findings of Santpere and colleagues [25], who detected unambiguous evidence of EBV co-infection or wild-type infection in 10 out of 929 LCLs from the 1000 Genomes Project dataset. These 10 LCLs were not included in our analysis, therefore competition between viral strains was unlikely to be a factor influencing copy number in our study.
Our results, of relative EBV copy number varying between individuals, and within and between populations are consistent with previous studies [6], [8] of LCLs of unreported ancestry, where EBV genome copy number was a trait that varied more significantly between LCLs than between different passages or sub-cultures of the same LCL. Davies et al. [8] suggest that the variation in EBV copy number is controlled by cell-intrinsic factors. In our analysis, it remained unclear whether genetic or cell-intrinsic factors caused the differences in mean EBV copy number between the Denver Han Chinese (CHD) and Beijing Han Chinese (CHB) populations. While it has been noted that the CHD LCLs grow more slowly than the Asian (ASN) LCLs (which include the CHB LCLs) [28], another study [29] found no statistically significant relationship between LCL growth rate and EBV copy number. We conclude that although cell intrinsic factors are still the most plausible explanation, multiple genetic variants, each with a small effect of EBV copy number, also likely play a role.
The strongest association signal in the genome-wide association study was an intronic SNP, located in gene PTPRD. PTPRD functions in cellular signalling, is located on the cell surface, and has been identified as a tumour suppressor. Mutations within PTPRD have been associated with several cancers: glioblastoma multiforme and head and neck squamous cell carcinomas [30], and clear cell renal carcinoma [31]. The paralogous gene PTPRK (protein tyrosine phosphatase receptor kappa) interacts with EBV. When PTPRK is over-expressed in EBV-infected Hodgkin lymphoma cells, survival of these cells decreases; when PTPRK expression is knocked down by RNAi, survival increases; suggesting a role for PTPRK in tumour suppression. The EBV gene EBNA1 targets Smad2, a protein that regulates PTPRK expression. By decreasing the half-life of Smad2, PTPRK is down-regulated in turn [32]. PTPRC (protein tyrosine phosphatase receptor C) has recently been associated with herpes simplex encephalitis susceptibility in mice [33]. Therefore although not genome wide significant, polymorphisms in PTPRD are in a biologically plausible gene.
Changes in the relative EBV copy number of LCLs have an impact on the gene expression profiles of those LCLs in a genotype-independent manner. Our analysis of genes differentially expressed between LCLs identified three genes whose expression positively (CXCL16 and AGL) or negatively (ADARB2) correlated with relative EBV copy number. CXCL16 is regulated by the microRNA CMVmiR-M23-2 of another human herpesvirus, CMV [34]. Reptar [35] predicts that CXCL16 is targeted by seven EBV microRNAs (ebv-miR-BART17-5p, ebv-miR-BART21-3p, ebv-miR-BART21-5p, ebv-miR-BART22, ebv-miR-BART3*, ebv-miR-BART5* and ebv-miR-BART7). It is also interesting to note that CXCL16 is a chemokine which has recently been associated with disease activity in multiple sclerosis [36] and mouse models of experimental autoimmune encephalomyelitis [37]. EBV microRNAs are also predicted to interact with AGL and ADARB2 [35]. It is likely that in a larger sample of LCLs, further genes would show significant correlations with relative EBV copy number. It is therefore important to control for the effects of EBV copy number in gene expression studies utilising large samples of LCLs. Other studies of the impact of EBV on the gene expression patterns of LCLs include a study by Choy et al., which reported that ∼15% of genes to have at least 5% of their variance correlated with EBV copy number [7].
In a study of semi-quantitative antibody response to EBV gene EBNA1, Rubicz et al. [38] were able to identify 15 SNPs which were associated with EBV antibody response at genome-wide significant level. In contrast, Kuparinen et al. [17] performed a GWAS for CMV antibody response and did not identify any genome-wide significant SNPs. It is possible that a different genetic architecture underlies the host response to control of EBV copy number in LCLs than that of EBV antibody response. Additionally all of the genes associated with the EBV antibody response were within the HLA system, a region of the genome this study was not well-powered to interrogate given the differences in HLA allele frequencies between populations and our criteria that a variant must be present in all 12 populations studied for SNP inclusion. Our finding that the 1.6 M SNPs studied in this GWAS could collectively explain 65% (se = 38%) of the variance in relative EBV copy number, while no single SNP reached genome-wide significance, suggests that many variants of small effect play a collective role within LCLs. Similarly, GWAS of genetic resistance to HIV-1 infection [15] could not find any common variants (with the exception of CCR5-delta 32) which were protective, while GWAS of host control of HIV-1 viral load found a number of genome-wide significant loci associated with lower HIV-1 viral load set points and slower progression to AIDS [12], [39]. The power to identify host genetic variation of virus infection traits may greatly depend on the trait under study and the sample sizes.
We estimated the heritability of relative EBV copy number, based on data from 101 parent-child trios to be 34% (se ±11%, p = 0.003). Other studies have found EBV anti-EBNA1 antibody response to be 68% heritable when considered as a discrete trait (seropositive versus seronegative) [38] and discrete anti-VCA IgG antibody response to be 32–48% heritable [40]. Infectious mononucleosis concordance rates in twins were estimated to be 12% between monozygotic twins and 6% between dizygotic twins [41]. Therefore, host genetic factors appear to play a variable but significant role in symptomatic response to primary EBV infection, the adaptive immune response to EBV latency, and the cell-intrinsic control of EBV latency although none of the variants previously identified were significantly associated with EBV copy number in our analysis.
This study has established that relative EBV copy number within LCLs is very much a complex trait, which has a significant heritable component. Our results suggest that many genetic variants of effect size less than 4.7% of variance in relative EBV copy number exist, but which this study was not statistically powered to detect. This study also focused on 1.6 million single nucleotide polymorphisms which were common to every population studied. Because SNPs which did not vary in all 12 populations studied were excluded from this analysis due to potential population stratification, we cannot rule out the effect on relative EBV copy number of SNPs which were only variable in a subset of the populations studied. We also cannot exclude the effects of rare variants of large effect on relative EBV copy number in LCLs, as these were not studied here. It is also possible that structural variants which are poorly tagged by common SNPs may play a role. To identify the genetic factors that underpin EBV copy number, a significant increase in sample size is necessary that will become possible within on-going large-scale sequencing and genotyping projects. However, we do find that even in a relatively modest sample size, EBV copy number is correlated with LCL host-gene expression patterns, in a host genotype-independent manner. Studies using larger samples of LCLs to study host gene expression profiles may find EBV-associated changes in LCLs generate false-positive results, unless EBV copy number is controlled for.
Methods
Ethics
No primary human tissue was used in this study. Details of this project were sent to the Coriell Cell Repository to be passed on the relevant Community Advisory Groups for HapMap participants.
Samples
Separated from peripheral blood as part of the HapMap and 1000 Genomes Projects, LCLs and LCL-derived DNA were provided to Wellcome Trust Sanger Institute by the Coriell Cell Repository. DNA from 915 HapMap lymphoblastoid cell lines was obtained from Coriell Institute for Medical Research, representing 12 populations. A summary of the composition of the sample group that provided the LCLs is provided in Table 1. Cell line BCBL-1 was used as a calibrator for quantitative PCR. It is a Kaposi Sarcoma Herpesvirus-positive, EBV-negative body cavity-derived primary effusion lymphoma cell line, of B cell origin [42].
Quantification of relative EBV copy number per cell
Quantification of relative EBV copy number was performed using quantitative PCR (qPCR). An artificial gene (GeneArt, Life Technologies) was designed based on the EBV BALF5 sequence for use as a positive control in qPCR, but containing an artificially inserted sequence to distinguish it from wild-type BALF5, with the sequence: CCCTGTTTATCCGATGGAATGACGGCGCATTTCTCGTGCGTGTACACCGTCTCGAGTATGACTGGTTCCAATTGACAAGCTGGGTCGTAGACATGGAAGTCCAGAGGGCTTCCG. Quantitative PCR was performed on an Agilent MxPro 3005 machine using the QuantiTect Multiplex PCR NoROX Kit (Qiagen). The PCR reagents were: 2.5 µl nuclease-free water (Qiagen), 12.5 µl QuantiTect Multiplex PCR No ROX mastermix (Qiagen), 2 µl BALF5 primer mix (10 pmol/µl of each primer and 0.75 pmol/µl probe [43]), 2 µl GAPDH primer mix (2.5 pmol/µl of each primer and 2 pmol/µl probe [44]), 1 µl ROX (diluted 1∶10 in 10 mM Tris-HCL (Life Technologies)), and 5 µl template DNA or positive control. EBV qPCR primer and probe sequences were from Kimura, 1999 [43]. GAPDH primer and probe sequences were from Pardieu, 2010 [44]. PCR conditions were as follows: 95°C for 15 minutes, followed by 45 cycles of 94°C for 60 seconds, 57°C for 30 seconds and 72°C for 30 seconds. Fluorescence data was collected during the annealing step.
The 2−ΔΔCT method [45] was used for relative quantification of target gene abundance (target gene BALF5, endogenous control gene GAPDH). Gene copy numbers in LCLs were normalised against the BCBL-1 cell line [46]. Data analysis was performed using MXPro v4.10 qPCR software (Agilent Technologies).
Heritability analysis
Within the individuals with relative EBV copy number data available, there were 101 trios with EBV copy number information available for mother, father and offspring. The family structure was taken from the 1000 Genomes pedigree files. They were drawn from four populations - CEU, IBS, MXL and YRI. The mid-parental average for the phenotype (relative EBV copy number) was calculated and the child's phenotype regressed against the mid-parental phenotype. The regression gives an estimate of narrow sense heritability of the trait and its associated P value.
Genotyping
899 samples have been sequenced or genotyped using three different platforms. For 355 samples, sequencing data was available from the 1000 Genomes Project Phase 1 release (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/integrated_call_sets/); for 313 samples, genotypes was available from the 1000 Genomes Project Illumina HumanOmni2.5–Quad v1-0 B_SNP data release (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20120131_omni_genotypes_and_intensities/); and for 231 samples, genotypes were available from the HapMap Phase III consensus release (ftp://ftp.ncbi.nlm.nih.gov/hapmap/genotypes/2009-01_phaseIII/plink_format/). Where multiple sources of genotyping or sequencing data were available for a sample, 1000 Genomes Phase I sequence data was used in preference to other sources, followed by Illumina 2.5 M Omni genotypes, and finally HapMap Phase III genotypes. Briefly, quality control for SNP and sample inclusion was as follows: Hardy-Weinberg equilibrium P value of >1×10−6; minor allele frequency of >1%; SNP call rate of >95%; and a sample call rate of >95%. Monomorphic SNPs were excluded. All quality control was performed using PLINK [47].
Imputation and association testing
The statistical power to identify a genetic variant was calculated using GWApower [27]. Imputation of genotyped samples to 1000 Genomes Phase 1 was performed using IMPUTE2 [26]. This increased the number of SNPs in common between the samples from 600,000 to 37 million. These imputed variants were then subjected to quality control in PLINK [47], namely removing: SNPs where Hardy-Weinburg equilibrium was P<1×10−5 in at least one of the 1000 Genomes populations (n = 8,754,733); SNPs which were monomorphic among phenotyped samples (n = 8,689,910); SNP with a missingness >0.01 (n = 9,808,778) and all SNPs with MAF <0.01 (n = 16,881,983). SNPs which were called discordantly in samples where sequencing and genotyping information were both available were excluded from further analysis. As EBV copy number varies significantly between different populations, we performed additional QC in order to account for potential stratification in the association analysis: removing any SNP if HWE P<0.01 in two or more populations, and removing any SNP that was monomorphic in at least one of the populations. This left a final set of 1,595,489 SNPs which were polymorphic in every population studied. Association analysis was performed using a linear mixed model implemented in FaST-LMM-Select [25].
Transcriptional profiles
Microarray expression data from Stranger et al. [24] (“REDUCED” dataset) was obtained for 466 unrelated individuals with relative EBV copy numbers. Expression data is available on http://www.ebi.ac.uk/arrayexpress/(Series Accession Number E-MTAB-264 and E-MTAB-198.processed.1). For each individual, correlation between expression levels of individual gene transcripts and EBV copy number was determined via linear regression. P-values of correlation with EBV copy number were obtained for each gene transcript (21,800 in total). Transcripts with p-value lower than 5×10−6 were considered significantly correlated with EBV copy number. Transcripts were mapped back to the genes they correspond to according to the array design file (“A-MEXP-930.adf.txt”) available on http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-264/
eQTL analysis
Effect of EBV relative copy number on the identification of expression quantitative loci (eQTLs) was tested in PLINK via linear association model with or without EBV copy number used as a covariate. Expression data included normalized log2 quantitative gene expression measurements for the 21,800 probes (18,226 unique autosomal genes) from 466 unrelated individuals of HapMap Phase III assayed on the Illumina Sentrix Human-6 Expression BeadChip [24]. SNP genotypes were as described for the QTL analysis. SNPs within a 2 Mb window around a gene locus were defined as eQTLs. Correction for population structure was performed using principal component analysis (PCA). Difference in effect sizes of eQTLs identified in the two association studies (with or without EBV copy number as a covariate) was determined via paired t-test across all tested SNPs (1.6 million). Resulting p-values of the difference in effect sizes were plotted for each transcript across all 22 non-sex chromosomes.
Supporting Information
Standard curve of BALF5 quantitative PCR primer set dilution series, which amplifies the EBV polymerase gene BALF5. The efficiency of the BALF5 qPCR assay was 99.4% (compared to GAPDH), r2 = 0.998.
(DOCX)
Supplementary tables. Table S1 in File S1 shows the results of a Tukey honest significant difference test of mean EBV copy number between populations. Table S2 in File S1 summarises the results of the EBV copy number GWAS, including all SNPs associated with EBV copy number with P<5×10−5. Table S3 in File S1 shows changes in gene expression (microarray) correlated with EBV copy number with suggestive P values (P >5×10−3).
(DOCX)
Spreadsheet of EBV copy number data for HapMap and 1000 Genomes LCLs included in this study.
(XLSX)
Acknowledgments
The authors would like to thank Sample Logistics (Wellcome Trust Sanger Institute) for supplying many of the HapMap and 1000 Genomes DNA samples used in this study, and the Cancer Genomes Project for providing DNA samples for assay optimisation.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by the Wellcome Trust (grant 098051), providing funding to CJH, VP, JZL, CAA, AG and PK. CJH is also supported by MRC Doctoral Training Grant G0900209 and ERC FP7 grant 261213. The funders had no role in study design, data collection and analysis, preparation of the manuscript, or in the decision to publish.
References
- 1. Miyashita EM, Yang B, Lam KM, Crawford DH, Thorley-Lawson DA (1995) A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80: 593–601. [DOI] [PubMed] [Google Scholar]
- 2. Babcock GJ, Hochberg D, Thorley-Lawson AD (2000) The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13: 497–506. [DOI] [PubMed] [Google Scholar]
- 3. Laichalk LL, Thorley-Lawson DA (2005) Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J Virol 79: 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hadinoto V, Shapiro M, Sun CC, Thorley-Lawson DA (2009) The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog 5: e1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fafi-Kremer S, Morand P, Brion JP, Pavese P, Baccard M, et al. (2005) Long-term shedding of infectious epstein-barr virus after infectious mononucleosis. J Infect Dis 191: 985–989. [DOI] [PubMed] [Google Scholar]
- 6. Caliskan M, Cusanovich DA, Ober C, Gilad Y (2011) The effects of EBV transformation on gene expression levels and methylation profiles. Hum Mol Genet 20: 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, et al. (2008) Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet 4: e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies ML, Xu S, Lyons-Weiler J, Rosendorff A, Webber SA, et al. (2010) Cellular factors associated with latency and spontaneous Epstein-Barr virus reactivation in B-lymphoblastoid cell lines. Virology 400: 53–67. [DOI] [PubMed] [Google Scholar]
- 9. Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, et al. (2012) An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 12: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uphoff CC, Denkmann SA, Steube KG, Drexler HG (2010) Detection of EBV, HBV, HCV, HIV-1, HTLV-I and -II, and SMRV in human and other primate cell lines. J Biomed Biotechnol 2010: 904767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khor CC, Chau TN, Pang J, Davila S, Long HT, et al. (2011) Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet 43: 1139–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, et al. (2010) The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330: 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomas R, Thio CL, Apps R, Qi Y, Gao X, et al. (2012) A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol 86: 6979–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim AY, Kuntzen T, Timm J, Nolan BE, Baca MA, et al. (2011) Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology 140: 686–696 e681. [DOI] [PMC free article] [PubMed]
- 15. McLaren PJ, Coulonges C, Ripke S, van den Berg L, Buchbinder S, et al. (2013) Association Study of Common Genetic Variants and HIV-1 Acquisition in 6,300 Infected Cases and 7,200 Controls. PLoS Pathog 9: e1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rubicz R, Yolken R, Drigalenko E, Carless MA, Dyer TD, et al. A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1). PLoS Genet 9: e1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuparinen T, Seppala I, Jylhava J, Marttila S, Aittoniemi J, et al. (2012) Genome-wide association study does not reveal major genetic determinants for anti-cytomegalovirus antibody response. Genes Immun 13: 184–190. [DOI] [PubMed] [Google Scholar]
- 18. Hobbs MR, Jones BB, Otterud BE, Leppert M, Kriesel JD (2008) Identification of a herpes simplex labialis susceptibility region on human chromosome 21. J Infect Dis 197: 340–346. [DOI] [PubMed] [Google Scholar]
- 19. Kriesel JD, Jones BB, Matsunami N, Patel MK, St Pierre CA, et al. (2011) C21orf91 genotypes correlate with herpes simplex labialis (cold sore) frequency: description of a cold sore susceptibility gene. J Infect Dis 204: 1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li DF, Lian L, Qu LJ, Chen YM, Liu WB, et al. (2012) A genome-wide SNP scan reveals two loci associated with the chicken resistance to Marek's disease. Anim Genet. [DOI] [PubMed]
- 21. Wolc A, Arango J, Jankowski T, Settar P, Fulton JE, et al. (2013) Genome-wide association study for Marek's disease mortality in layer chickens. Avian Dis 57: 395–400. [DOI] [PubMed] [Google Scholar]
- 22. The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 23. Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, et al. (2012) Patterns of cis regulatory variation in diverse human populations. PLoS Genet 8: e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Listgarten J, Lippert C, Heckerman D (2013) FaST-LMM-Select for addressing confounding from spatial structure and rare variants. Nat Genet 45: 470–471. [DOI] [PubMed] [Google Scholar]
- 26. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng S, Wang S, Chen CC, Lan L (2011) GWAPower: a statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genet 12: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stark AL, Zhang W, Zhou T, O'Donnell PH, Beiswanger CM, et al. (2010) Population differences in the rate of proliferation of international HapMap cell lines. Am J Hum Genet 87: 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Im HK, Gamazon ER, Stark AL, Huang RS, Cox NJ, et al. (2012) Mixed effects modeling of proliferation rates in cell-based models: consequence for pharmacogenomics and cancer. PLoS Genet 8: e1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, et al. (2009) The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A 106: 9435–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du Y, Su T, Tan X, Li X, Xie J, et al. (2013) Polymorphism in protein tyrosine phosphatase receptor delta is associated with the risk of clear cell renal cell carcinoma. Gene 512: 64–69. [DOI] [PubMed] [Google Scholar]
- 32. Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, et al. (2008) Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood 111: 292–301. [DOI] [PubMed] [Google Scholar]
- 33. Caignard G, Leiva-Torres GA, Leney-Greene M, Charbonneau B, Dumaine A, et al. (2013) Genome-Wide Mouse Mutagenesis Reveals CD45-Mediated T Cell Function as Critical in Protective Immunity to HSV-1. PLoS Pathog 9: e1003637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dolken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, et al. (2010) Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog 6: e1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elefant N, Berger A, Shein H, Hofree M, Margalit H, et al. (2011) RepTar: a database of predicted cellular targets of host and viral miRNAs. Nucleic Acids Res 39: D188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holmoy T, Loken-Amsrud KI, Bakke SJ, Beiske AG, Bjerve KS, et al. (2013) Inflammation markers in multiple sclerosis: CXCL16 reflects and may also predict disease activity. PLoS One 8: e75021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wojkowska DW, Szpakowski P, Ksiazek-Winiarek D, Leszczynski M, Glabinski A (2014) Interactions between Neutrophils, Th17 Cells, and Chemokines during the Initiation of Experimental Model of Multiple Sclerosis. Mediators Inflamm 2014: 590409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rubicz R, Yolken R, Drigalenko E, Carless MA, Dyer TD, et al. (2013) A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1). PLoS Genet 9: e1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Apps R, Qi Y, Carlson JM, Chen H, Gao X, et al. (2013) Influence of HLA-C Expression Level on HIV Control. Science 340: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Besson C, Amiel C, Le-Pendeven C, Plancoulaine S, Bonnardel C, et al. (2009) Strong correlations of anti-viral capsid antigen antibody levels in first-degree relatives from families with Epstein-Barr virus-related lymphomas. J Infect Dis 199: 1121–1127. [DOI] [PubMed] [Google Scholar]
- 41. Hwang AE, Hamilton AS, Cockburn MG, Ambinder R, Zadnick J, et al. (2011) Evidence of genetic susceptibility to infectious mononucleosis: a twin study. Epidemiol Infect 140: 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arguello M, Sgarbanti M, Hernandez E, Mamane Y, Sharma S, et al. (2003) Disruption of the B-cell specific transcriptional program in HHV-8 associated primary effusion lymphoma cell lines. Oncogene 22: 964–973. [DOI] [PubMed] [Google Scholar]
- 43. Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, et al. (1999) Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol 37: 132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pardieu C, Vigan R, Wilson SJ, Calvi A, Zang T, et al. (2010) The RING-CH ligase K5 antagonizes restriction of KSHV and HIV-1 particle release by mediating ubiquitin-dependent endosomal degradation of tetherin. PLoS Pathog 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 46. Renne R, Zhong W, Herndier B, McGrath M, Abbey N, et al. (1996) Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2: 342–346. [DOI] [PubMed] [Google Scholar]
- 47. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen GP, Pan QH, Hong MH, Qin HD, Xu YF, et al. (2011) Human genetic variants of homologous recombination repair genes first found to be associated with Epstein-Barr virus antibody titers in healthy Cantonese. Int J Cancer 129: 1459–1466. [DOI] [PubMed] [Google Scholar]
- 50.Ghesquieres H, Dogan A, Link BK, Maurer MJ, Cunningham JM, et al. (2013) FCGR2A and FCGR3A polymorphisms in classical Hodgkin lymphoma by EBV status. Leuk Lymphoma. [DOI] [PMC free article] [PubMed]
- 51. Hatta K, Morimoto A, Ishii E, Kimura H, Ueda I, et al. (2007) Association of transforming growth factor-beta1 gene polymorphism in the development of Epstein-Barr virus-related hematologic diseases. Haematologica 92: 1470–1474. [DOI] [PubMed] [Google Scholar]
- 52.Ghesquieres H, Maurer MJ, Casasnovas O, Ansell SM, Larrabee BR, et al. (2013) Cytokine gene polymorphisms and progression-free survival in classical Hodgkin lymphoma by EBV status: Results from two independent cohorts. Cytokine. [DOI] [PMC free article] [PubMed]
- 53. Urayama KY, Jarrett RF, Hjalgrim H, Diepstra A, Kamatani Y, et al. (2012) Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status-defined subgroups. J Natl Cancer Inst 104: 240–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durovic B, Gasser O, Gubser P, Sigle J, Hirsch HH, et al. (2013) EBV negativity among individuals older than 60 years is associated with HLA-C and HLA-Bw4 variants, and tonsillectomy. J Virol. [DOI] [PMC free article] [PubMed]
- 55. Friborg JT, Jarrett RF, Koch A, Garred P, Freeland JM, et al. (2010) Mannose-binding lectin genotypes and susceptibility to epstein-barr virus infection in infancy. Clin Vaccine Immunol 17: 1484–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Standard curve of BALF5 quantitative PCR primer set dilution series, which amplifies the EBV polymerase gene BALF5. The efficiency of the BALF5 qPCR assay was 99.4% (compared to GAPDH), r2 = 0.998.
(DOCX)
Supplementary tables. Table S1 in File S1 shows the results of a Tukey honest significant difference test of mean EBV copy number between populations. Table S2 in File S1 summarises the results of the EBV copy number GWAS, including all SNPs associated with EBV copy number with P<5×10−5. Table S3 in File S1 shows changes in gene expression (microarray) correlated with EBV copy number with suggestive P values (P >5×10−3).
(DOCX)
Spreadsheet of EBV copy number data for HapMap and 1000 Genomes LCLs included in this study.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.