Abstract
Genetic efficiency in higher organisms depends on mechanisms to create multiple functions from single genes. To investigate this question for an enzyme family, we chose aminoacyl tRNA synthetases (AARSs). They are exceptional in their progressive and accretive proliferation of noncatalytic domains as the Tree of Life is ascended. Here we report discovery of a large number of natural catalytic nulls (CNs) for each human AARS. Splicing events retain noncatalytic domains while ablating the catalytic domain (CD) to create CNs with diverse functions. Each synthetase is converted into several new signaling proteins with biological activities ‘orthogonal’ to that of the catalytic parent. We suggest that splice variants with non-enzymatic functions may be more general, as evidenced by recent findings of other catalytically inactive splice-variant enzymes.
Aminoacyl tRNA synthetases (AARSs) establish the genetic code by esterifying specific amino acids to the 3’-ends of their cognate tRNAs (1–5), and have adaptations of this reaction for specific physiological responses (6). A few literature examples show natural proteolysis or alternative splicing of AARS can reveal novel AARS proteins (7, 8) with new functions (9–11). With this in mind, we investigated potential mechanisms for achieving genetic efficiency through functional expansions. The enzymes are divided into two classes of 10 proteins each, with each class being defined by the architecture of the highly conserved catalytic domain that is retained through evolution (12–14). As the Tree of Life is ascended, 13 new domains, which have no obvious association with aminoacylation or editing, have collectively been added to AARSs and maintained over the course of evolution, with no significant benefit or detriment to primary function (15–17). The extent of these domain additions appears to be particular to AARSs (15). Some of these new domains are appended to each of several synthetases, while others are specific to a single synthetase. Interestingly, these novel domain additions are accretive and progressive; and while their persistence provides no major benefit to aminoacylation, the strong evolutionary pressure for their retention suggests they are not random function-less stochastic fusions, but may be conserved for a specific biological purpose, perhaps distinct from the canonical enzymatic function.
We made a comprehensive search for alternative splice variants of AARSs to understand how splicing changes the domain organization and underlying architecture of each synthetase. We selectively targeted the AARS family of genes by enriching the AARS transcriptome in 6 distinct human samples (human fetal and adult brain, primary human leukocytes, and three cultured leukocyte cell-types Raji B-cells, Jurkat T-cells and THP1 monocytes). A PCR-based gene-capture and enrichment method was integrated with high-throughput deep sequencing to increase sequencing depth for each AARS transcript (Materials and Methods, Fig. 1A). This methodology allowed for high enrichment of AARS mRNAs, and mainly targeted exon-exon junctions for discovery of exon-skipping events. We defined the AARS transcriptome as the transcripts of 37 AARS genes, including those for 17 cytoplasmic synthetases, 17 mitochondrial synthetases, and for 3 that encode both cytoplasmic and mitochondrial forms. For efficient capture, transcripts were amplified by multiplex PCR using AARS gene-specific primers and optimized PCR conditions (see Materials and Methods). Sensitive detection of low-abundance splice variants was achieved with an optimized multiplex PCR that amplified gene regions close to exon-exon junctions of AARS transcripts and produced short PCR fragments (Fig. 1A). Fragments were assembled into cDNA libraries and sequenced by high-throughput deep sequencing (18, 19).
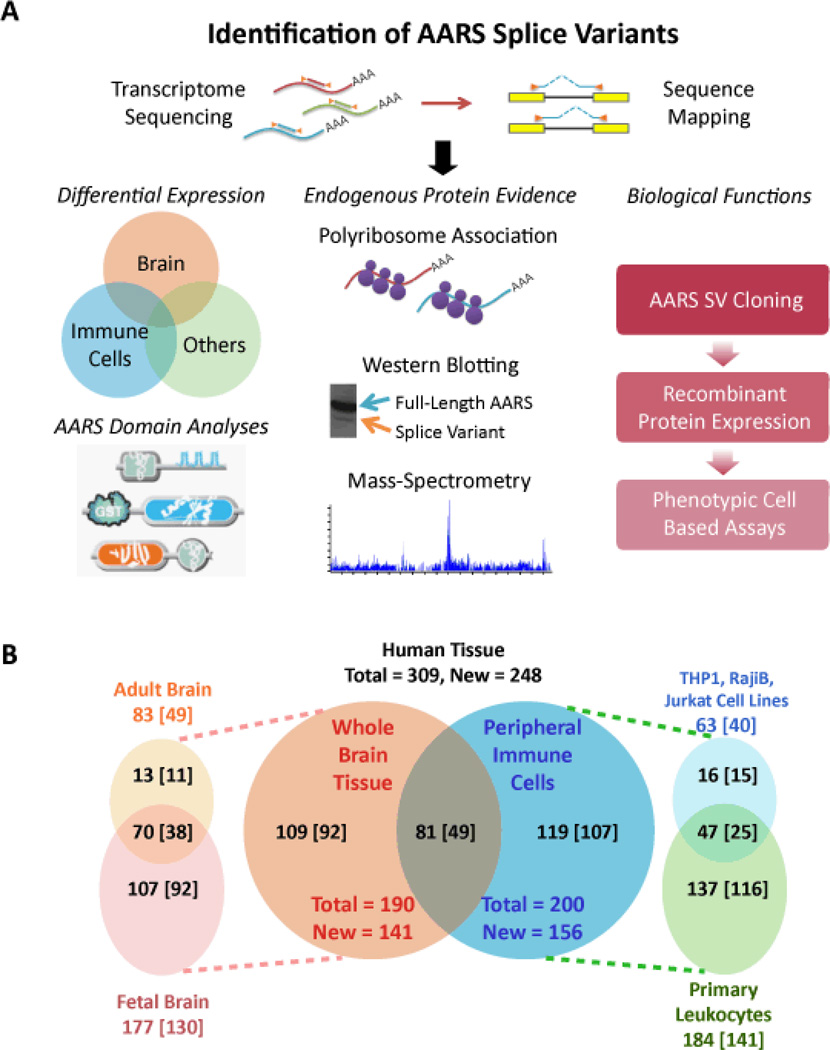
Figure 1. Identification of AARS splice variants and assays for their functions.
(A) AARS Transcriptome Discovery and Analysis Funnel. The AARS transcriptome was amplified by gene-specific multiplex PCR and sequenced by high-throughput deep sequencing. Sequencing reads were then mapped to the AARS transcriptome for identification of exon-skipping events and alternative splice sites. Products of this effort were submitted to various analyses. Regulation and functions of the identified AARS splice variants were studied in the context of their distribution across various human tissues, through identification of the endogenously expressed protein products, and the effects of expressed protein products on cells in a diverse set of in-vitro cell-based assays. (B) The Venn diagram in the center shows the total number of exon-skipping junctions that were identified by RNAseq and by ‘exon-flanking’ PCR in fetal and adult brain tissues (orange circle) and in immune cells that include primary total leukocytes and three different cultured leukocytic cell-types (blue circle). In the diagram on the left, the light pink circle annotates the number of exon-skipping junctions identified in fetal brain, while the light orange circle shows those found in adult brain. The diagram on the right gives the number of identified exon-skipping junctions in the three cultured leukocytic cell-types (Raji B-cells, Jurkat T-cells and THP1 monocytes) (light blue) and in primary leukocytes (pale green). Overlapping areas give the number of splice junctions that were common between tissues or cells. The numbers in square brackets give the total new alternative splice junctions identified in each respective tissue.
Approximately 42 million 50-base reads were obtained and analyzed, using established methods (19). About 70% (30.4 million) mapped to the 37 AARS genes and about two-thirds of the AARS-specific reads (21.4 million) covered AARS exon-exon junctions. When compared to previously published whole transcriptome studies (20, 21), the AARS transcriptome enrichment method employed here successfully improved sequencing depth so that we could detect all of the 61 previously reported exon-exon junctions for AARS transcripts, as well as identify 248 previously unreported junctions (Fig. 1B and table S1). These new splice forms allowed for the ablation of specific coding regions and simultaneous creation of new exon-exon junctions.
In addition, the tissue origin and the overlap of AARS splice variants in different tissues were examined. Although there was obvious tissue specificity for certain transcripts, many of the same splice variant transcripts were found across distinct tissue pools (Fig. 1B). Surprisingly, the majority of the splice variants of both class I and class II family members abrogated the catalytic domain (Fig. 2A and fig. S1). These included both truncations of N- or C-terminal coding regions as well as in-frame internal deletions (Fig. 2B). For instance, 79% of the 66 discovered in-frame splice variants (Fig. 2C and table S2) had a disrupted or ablated canonical catalytic domain (CD), and thereby created a catalytic null (CN) (Fig. 2B and fig. S1). Because 3-D structures are available for many human AARSs and their orthologs, events that removed entire specific exons could be diagrammatically portrayed as linear arrangements of domain structure elements (fig. S1). These virtual structures suggest that the new domain-domain interactions created by internal deletions might engender new structural conformations (cf. 22) and thereby might lead to new interactions.
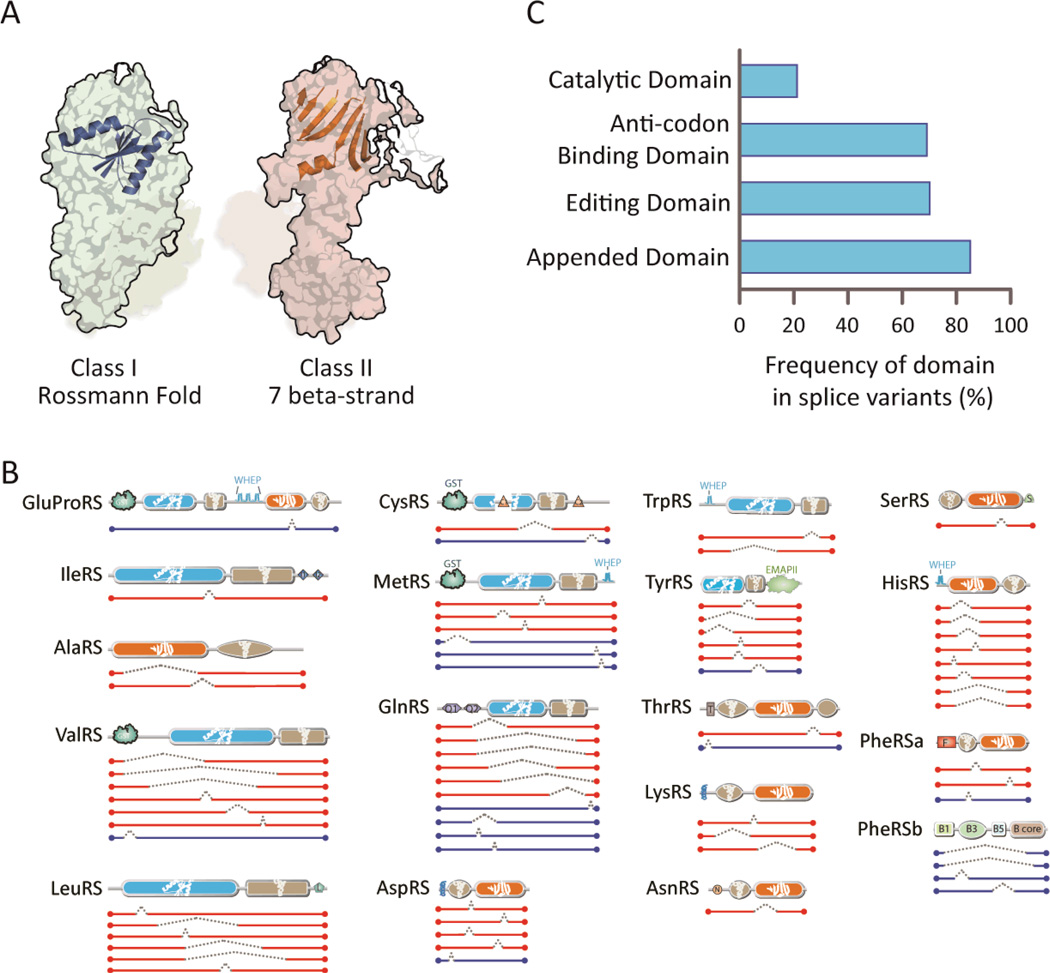
Figure 2. Majority of AARS splice variants are catalytic nulls.
(A) Architectures of the catalytic domains for aminoacylation - Class I versus Class II. The conserved core Rossmann fold is represented on the structure of MetRS (pdb2CT8) (ref: Pubmed 16155581) in Class I, and the conserved core 7 beta-strand with motif-3 helix is represented on the structure of LysRS (pdb4DPG) (ref: Pubmed23159739) in Class II. (B) In-frame splice variants of cytoplasmic AARS are illustrated. Splice variants with catalytic domains deleted (catalytic nulls) are highlighted in red while those with catalytic domains retained are represented in blue. (C) The catalytic domain is abrogated in most AARS splice variants. In contrast, domains that have functions distinct from aminoacylation are predominantly retained. Of note, the UNE domains (such as UNE-S, and UNE-L (abbreviated as “S” and “L”, respectively, in panel B), which comprise part of the “Appended Domain” category and are idiosyncratic to specific synthetases, are retained in the AARS splice variants identified here.
As specific examples, all 8 in-frame splice variants of HisRS showed an ablated CD and only one of the 6 in-frame splice variants of TyrRS retained the CD (Fig. 2B and table S2). In contrast to the consistent abrogation of the canonical CD, 60 of the 70 in-frame splice variants (85%) are CNs that retain at least one of the 13 added domains appearing in the AARSs of higher eukaryotes (Fig. 2B and table S2). Of particular interest are the UNE domains, which are specific to AARSs and have, like the other appended domains, no significant aminoacylation function. The UNE domains are almost universally retained in the CNs (Fig. 2B and fig. S1). An interesting case, suggesting a non-canonical role for the CNs, is the retention of the UNE-S domain of SerRS. Recent work established a nuclear activity for SerRS that is dependent on the UNE-S domain and showed that the addition of UNE-S to SerRS was essential for development of the closed circulatory systems of vertebrates (23). Motifs found in other proteins of higher eukaryotes, such as the GST-, single-helix-, WHEP-, and EMAPII-like-domain, also remain intact in many of the AARS CN splice variants (fig. S1 and table S2).
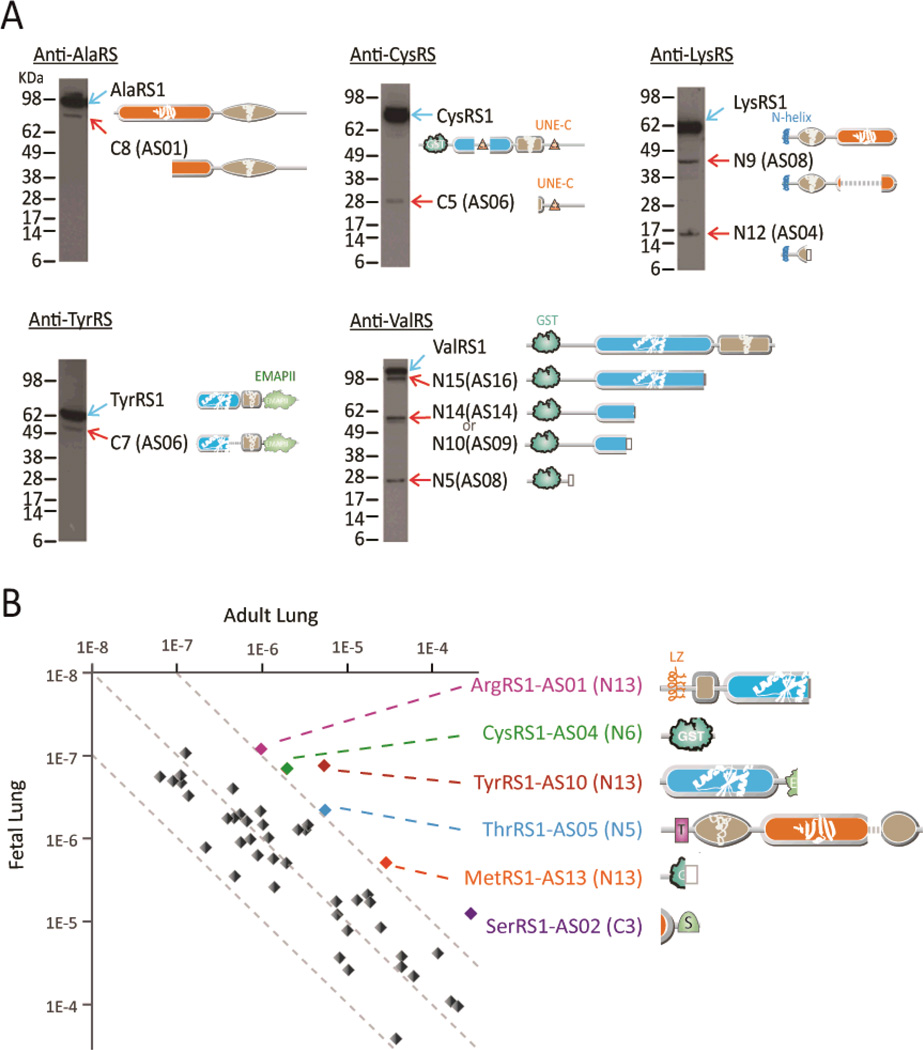
The tissue specific association of transcripts suggested that AARS mRNA splice variants encode endogenously expressed proteins. To explore this possibility, polysome-association of the splice variant-encoding mRNAs was examined [Materials and Methods as described in (24)]. Of the 48 CN mRNAs tested, all were associated with polysomes in naïve Jurkat cells (fig. S2 and listed in table S3). AARS specific antibodies probed the same Jurkat cell lysates that were used for detecting polysome-association of the mRNAs. To detect endogenous translation products of the CN splice variant transcripts, western blot analysis was done using antibodies specific for AlaRS, CysRS, LysRS, TyrRS and ValRS (Fig. 3A and table S4). These synthetase fragments were chosen based on the availability of suitable antibodies for immunoprecipitation. By western blot analysis, we detected the expected endogenous AARS splice forms that lacked CDs but retained appended domains (Fig. 3A). Mass spectrometry identified specific GlnRS, ValRS and TyrRS CN-sized fragments, and multiple peptides were identified for all of these CNs (fig. S3A). In addition to finding representative peptides from these CNs, we found no support for the possibility of proteolytic cleavage of full-length TyrRS giving rise to its assigned CN-peptides (legend to fig. S3A). In a separate vein, we identified an approximately 23kd protein as HisRS1-C9 in the public PROTOMAP MS database (25). We aligned MS-scored peptides on both sides of the sequence encompassing the splice junction reported here for HisRS1-C9 (fig. S3B). Finally, in vitro translation of a copy of the mRNA encoding an endogenously expressed TyrRS1-C7 splice variant (identified by western blot analysis of whole cell lysates as shown in Fig. 3) confirmed that the transcripts could be stably translated into proteins (fig. S3C). MS confirmed peptides on both sides of the internal splice junction.
Figure 3. Detection of endogenous AARS splice variants.
(A) Western blot detection of AARS splice variants in Jurkat cell lysates. Detailed information for these splice variants is shown in table S4. (B) Tissue specific expression of selected AARS splice variants in adult and fetal lung tissues. Gene expression of the target gene was normalized by the gene expression of house-keeping genes (see Materials and Methods) in the same sample.
We observed tissue specific expression of specific CNs. Across 19 human adult tissues or cells, 38 of 48 CN transcripts (79%) were differentially expressed with gene upregulation (by 5 times or more of median) in at least one of the tissues, while the full-length parent AARS genes were evenly distributed (table S3). We also found that some CN transcripts expressed differentially in one developmental stage over another. As an example, six specific CNs were highly expressed (by 10-fold or more) in adult versus fetal lung tissue. These included ArgRS1-AS01, CysRS1-AS04, MetRS1-AS13, SerRS1-AS02, ThrRS1-AS05, and TyrRS1-AS10 (Fig. 3B and table S3).
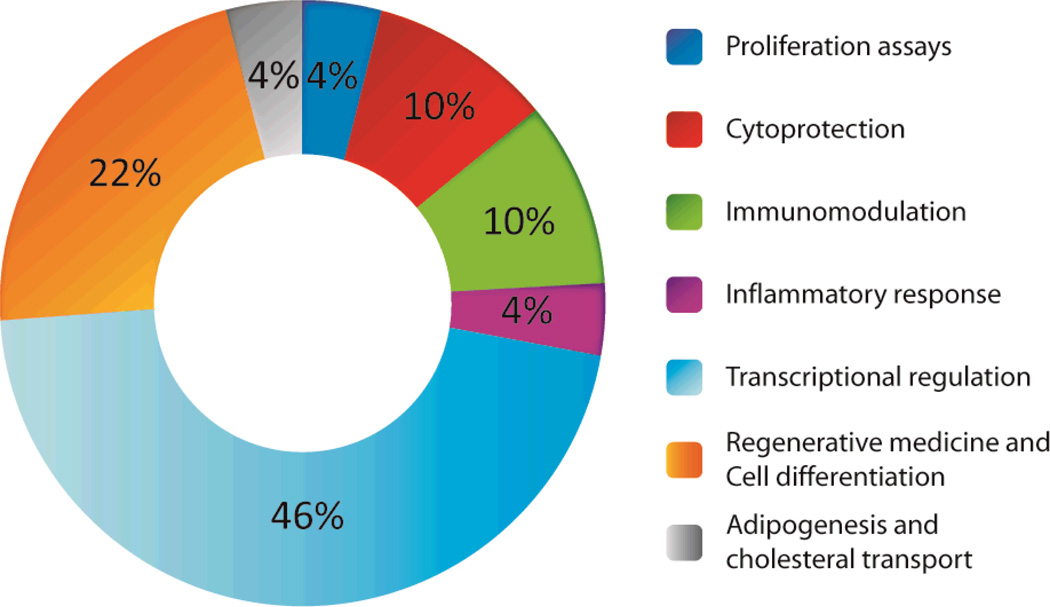
Because the splice-variant mRNAs prominently ablate the CD-encoding portion, we were interested to investigate the potential for these fragments to exert biological activities distinct from the canonical aminoacylation function. To this end, recombinant human AARS fragments, including CNs, were expressed as soluble proteins and purified to >95% homogeneity. Phenotypic cell-based assays were performed largely in primary human cells to monitor potential biological activities (fig. S4 and table S5). The assays types were clustered into assay groups (Fig. 4) including proliferation (different cell types were profiled for effects of splice variants on proliferation or cell death), cytoprotection, immunomodulation, acute inflammatory response, transcriptional regulation (4 assays in two cell types at two distinct time points across a set of 88 genes), ‘regenerative responses’, cell differentiation in primary human cell types and, finally, cholesterol transport. All assays were run at minimum in duplicate for each protein, and many proteins were run in multiple batches, and at a range of concentrations, to confirm activity. All proteins were generated as His-tagged recombinant forms, with either the N or C termini, or both, having the tag (table S6). Full length forms of AspRS, TyrRS, HisRS and AsnRS synthetases were expressed in parallel and run in assays as controls for the expressed synthetase fragments. In all cases, the full length parental form was either inactive across all assays or had a single activity that was not the same as any of its splice variants.
Figure 4. Recombinant AARS variants have specific biological activities across a spectrum of cell-based assays.
Proteins were expressed in E. coli and purified for use in cell-based assays. The majority of the proteins were soluble and highly expressed. The AAR variants (table S6) were tested in a variety of different cell based assays (fig. S4) spanning a range of biological activities, largely using primary human cells (table S5).
More than 100,000 data points were evaluated across the cell based assay panel (fig. S4). Of the 94 AARS-derived proteins interrogated here, 88% tested positive for one or more biological activities. The cell-based activities associated with each recombinant protein were specific and idiosyncratic to the variant. This observation provided a system-wide ‘internal control’, largely ruling out the potential for non-specific readouts of cell signaling by the various proteins. MetRS1-C5 is presented as a specific example. This CN strongly stimulated skeletal muscle fiber formation in vitro (fig. S5). Following exposure to the recombinant MetRS1-C5 for 2 days, quantitative PCR assessment of primary human skeletal myoblasts showed upregulation of key genes for muscle cell differentiation and metabolism, including insulin growth factor (IGF-1) and lipoprotein lipase (LPL) (fig. S6).
While deliberately ablating the canonical catalytic function, alternative splicing of the AARS family of genes has created a large ensemble of CNs that specifically retain the domain expansions. The successful expression of over 100 recombinant forms as soluble proteins suggests that splice-site selection has been tailored to create stable folded structures. The canonical function and structure of the ancient aminoacyl tRNA synthetase catalytic domain is strongly preserved throughout all taxa, which makes the ablation of this essential (for aminoacylation) domain especially provocative. The paradox of strongly conserved non-catalytic domains progressively added to AARSs protein structure over the course of evolution appears to be at least in part an evolutionary reshaping of tRNA synthetases for other functions.
While splice variants of other proteins also exist, it is the extent of these novel domain additions specifically to AARSs, and their retention by the CNs, which make the AARSs splice variants distinct. Possibly, functional expansion of AARSs was to link translation at the first step of protein synthesis to a variety of cell signaling pathways. Recent studies have demonstrated roles for specific AARSs in pathways associated with angiogenesis (9, 26–28), inflammation (29,30), the immune response, mTor signaling, apoptosis, tumorigenesis, and IFN-γ and p53 signaling (15). The work detailed here suggests the universe of AARS-derived entities, which are active for non-translational functions, may be far greater than anticipated. The mechanism of erasing the canonical function, while adding non-catalytic domains, engenders a clear implementation of orthogonal functions. Members of other enzyme families, though perhaps to a lesser extent, likely also gain new functions through splice variants. The recently reported catalytically impaired natural splice variants of several oncogenic kinases (31) and of the SIRT2 histone deacetylase (32) suggest that other enzyme families have undergone similar, though perhaps less extensive variation.
Supplementary Material
Acknowledgements
This work was supported by the Innovation and Technology Fund from the Hong Kong Government (UIM181, UIM192, and UIM199), by a fellowship from the National Foundation for Cancer Research and by NIH grants R01CA92577, R01GM088278, R01NS085092, R01HG005717 and R01GM100136. We also thank Kristi Piehl and Johnny Li (aTyr Pharma) for help with splice variant cloning and protein expression. We thank Andrew Cubitt (aTyr Pharma) for assistance on the transcriptional profiling and Vy Trinh and Dr. Ji Zhao (both formerly of aTyr Pharma) for assistance with cell profiling of recombinant tRNA synthetase fragments. Some of the authors have a financial interest in and/or compensation from aTyr Pharma.
References
- 1.Boniecki MT, Vu MT, Betha AK, Martinis SA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19223. doi: 10.1073/pnas.0809336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yadavalli SS, Musier-Forsyth K, Ibba M. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19031. doi: 10.1073/pnas.0810781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giegé R. Nat. Struct. Biol. 2003;10:414. doi: 10.1038/nsb0603-414. [DOI] [PubMed] [Google Scholar]
- 4.Carter CW., Jr Annu. Rev. Biochem. 1993;62:715. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 5.Ibba M, Söll D. Genes Dev. 2004;18:731. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 6.Netzer N, et al. Nature. 2009;462:522. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwenzer H, et al. Biochimie. 2014;100:18. doi: 10.1016/j.biochi.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 8.Miyanokoshi M, Tanaka T, Tamai M, Tagawa Y, Wakasugi K. Sci. Rep. 2013;3:3477. doi: 10.1038/srep03477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakasugi K, et al. Proc. Natl. Acad. Sci. U. S. A. 2002;99:173. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakasugi K, Schimmel P. Science. 1999;284:147. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 11.Lareau LF, Green RE, Bhatnagar RS, Brenner SE. Curr. Opin. Struct. Biol. 2004;14:273. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Ludmerer SW, Schimmel P. J. Biol. Chem. 1987;262:10807. [PubMed] [Google Scholar]
- 13.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature. 1990;347:203. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 14.Cusack S. Curr. Opin. Struct. Biol. 1997;7:881. doi: 10.1016/s0959-440x(97)80161-3. [DOI] [PubMed] [Google Scholar]
- 15.Guo M, Schimmel P. Nat. Chem. Biol. 2013;9:145. doi: 10.1038/nchembio.1158. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo M, Yang XL, Schimmel P. Nat. Rev. Mol. Cell Biol. 2010;11:668. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia J, Arif A, Ray PS, Fox PL. Mol. Cell. 2008;29:679. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeda J, et al. Nucleic Acids Res. 2006;34:3917. doi: 10.1093/nar/gkl507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang H, Wong WH. Bioinformatics. 2009;25:1026. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toung JM, Morley M, Li M, Cheung VG. Genome Res. 2011;21:991. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thierry-Mieg D, Thierry-Mieg J. Genome Biol. 2006;7(Suppl 1):S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, et al. Structure. 2012;20:1470. doi: 10.1016/j.str.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, et al. Nat. Commun. 2012;3:681. doi: 10.1038/ncomms1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, et al. J. Biol. Chem. 2013;288:29223. doi: 10.1074/jbc.C113.490599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dix MM, et al. Cell. 2008;134:679. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzima E, et al. J. Biol. Chem. 2005;280:2405. doi: 10.1074/jbc.C400431200. [DOI] [PubMed] [Google Scholar]
- 27.Kawahara A, Stainier DY. Trends Cardiovasc. Med. 2009;19:179. doi: 10.1016/j.tcm.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q, et al. Nat. Struct. Mol. Biol. 2010;17:57. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SG, et al. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6356. doi: 10.1073/pnas.0500226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arif A, Jia J, Moodt RA, DiCorleto PE, Fox PL. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1415. doi: 10.1073/pnas.1011275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anamika K, Garnier N, Srinivasan N. BMC. Genomics. 2009;10:622. doi: 10.1186/1471-2164-10-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rack JG, Vanlinden MR, Lutter T, Aasland R, Ziegler M. J. Mol. Biol. 2014;426:1677. doi: 10.1016/j.jmb.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 33.Vandesompele J, et al. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.