Abstract
Background
B cell infiltrates are common in rejected kidney allografts, yet their composition is still unclear. The aim of our study was to characterize the clonal composition of B cell infiltrates of rejected human kidney grafts.
Methods
We used a molecular approach to characterize the partial B cell repertoires of 5 failed human kidney grafts with detectable B cell infiltrates. A comparison between the intragraft and blood repertoire was also conducted for one case.
Results
Redundant sequences were observed in both blood and graft, although the level of clonal amplification was significantly higher for the graft. Somatic hypermutations (SHM) were also more frequent in sequences found in the graft compared to the blood. The rate of non-silent mutations was significantly higher in complementarity determining regions (CDR) compared to framework regions in blood sequences as well as in graft sequences found at low frequency. In contrast, this preferential distribution was lost in sequences found at high frequency in the graft, suggesting a lack of affinity maturation in situ. Lastly, follicular dendritic cells were undetectable in CD20+ infiltrates in all samples examined.
Conclusions
We provide here evidence that B cell clones expand and undergo SHM in situ. However, the even distribution of non-silent SHM in high frequency graft sequences together with the absence of FDC do not support the view that infiltrating B cells are part of functional germinal centers.
Keywords: Infiltrating B cells, graft rejection, kidney allografts
Introduction
It is now well established that large aggregates of B cells tend to develop in allograft tissue during rejection (1-4). Studies have associated such infiltrates with local inflammation, scar tissue and interstitial fibrosis as well as steroid refractory rejection (5-8). While some of these studies report a correlation between the presence of these B cell infiltrates and poor graft outcome (9-12), others do not appear to reach the same conclusions (13-15). This discrepancy may be explained by the heterogeneity of the infiltrates examined in these studies. Perhaps, the clearest indication that infiltrating B cells play an active role in the rejection process, however, comes from clinical responses to anti-CD20 therapy in patients with active rejection (16-19). Some of these responses were associated with the elimination of intragraft B cells while circulating donor specific antibodies (DSA) were unchanged, underscoring the role of B cells, rather than antibodies in the rejection process (20).
In recent years, a number of groups have investigated the composition of B cell infiltrates in kidney grafts and attempted to unravel their biological significance. Cheng et al. showed the predominance of uniquely rearranged immunoglobulin gene sequences in biopsy samples collected from patients with chronic allograft nephropathy, supporting the view of B cell clonal expansion in situ (21). Other studies used immunohistochemistry (IHC) to reveal the presence of CD20+ B cells at different stages of differentiation in kidney biopsies, suggesting an ongoing B cell maturation process (22). This phenomenon has been attributed to the development of ectopic germinal centers (eGC) directly in the graft tissue with properties comparable to those found in lymph nodes (2, 21, 23-25). These eGC would act as tertiary lymphoid organs (TLOs) where recruited B cells would undergo differentiation, somatic hypermutations (SHM) and affinity maturation.
Here, we further investigated the clonal composition of B cell infiltrates from failed kidney grafts by analyzing the repertoire of rearranged immunoglobulin heavy chain variable gene (IGHV) sequences in comparison to peripheral blood B cells. We particularly examined the presence and distribution of somatic mutations in the rearranged IGHV sequences as a molecular footprint of this clonal history. Lastly, we looked for the presence of markers associated with affinity maturation in B cell clusters to evaluate their resemblance to traditional GCs.
Results
Tissue samples from a total of 21 human kidney allograft recipients were included in this study, 11 males, ranging in age from 6 to 59 years. All patients experienced graft failure resulting in transplant nephrectomy. Approximately 75% of the explanted grafts exhibited evidence of chronic rejection (Supplementary Table S1). Nearly 50% of the graft tissue samples stained positive for CD20+ B cell infiltrates, and most of the negative cases could be explained by recent B cell specific therapy. Blinded analyses of the tissues differentiated between diffuse infiltrates (Figure S1A) and dense infiltrate aggregates (Figure S1B-1F). Samples exhibiting dense clustered lymphoid infiltrates (Patients 2, 5, 10, 17 and 19) were further analyzed.
IGHV repertoire analysis in graft infiltrates and peripheral blood
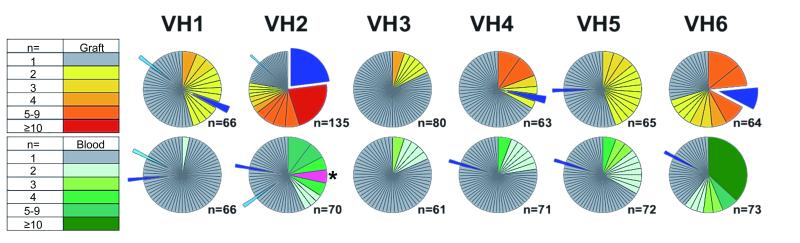
We used a PCR-based strategy to analyze IGHV sequences from graft infiltrates and peripheral blood B cells collected at time of nephrectomy in patient 2. This analysis examines >60 sequences in each IGHV family (IGHV1-6) for both blood and graft. A comparison of the 2 B cell repertoires reveals that the composition of intragraft B cell infiltrates is largely distinct from that of peripheral B cell populations, with minimal overlap observed between the two compartments (Figure 1). Moreover, we observed a higher level of redundancy among graft sequences when compared to peripheral blood sequences, indicating B cell clonal expansion in situ (Figure 1). Such amplification was more apparent in the IGHV2 subfamily, in which two clones accounted for nearly 50% of the entire IGHV2 repertoire. Comparable evidence of clonal expansion in the IGHV2 family was observed for all additional patients assessed using the same approach (Figure S2).
Figure 1. IGHV comparative repertoire analysis in blood and graft.
Pie chart representation of the frequency of clonotypic CDR3 sequences for each the 6 IGHV families (VH1-6) in the graft tissue (top row) and the peripheral blood (bottom row) of patient 2. Each pie chart section represents a distinct clonotypic CDR3. The size of each section corresponds to the frequency of the clonotypic sequences among all analyzed sequences. The number of clonotypic sequences observed is also indicated by a color code (left panel). The total number of sequences analyzed for each IGHV family is indicated below each pie. Pie sections filled in light or dark blue indicate clonotypic sequences found both in the patient blood and graft tissue. One clonotypic IGHV2 sequence highlighted in pink and indicated with an asterisk, corresponds to a polyreactive B cell clone expanded in the blood and further characterized in previous studies (29).
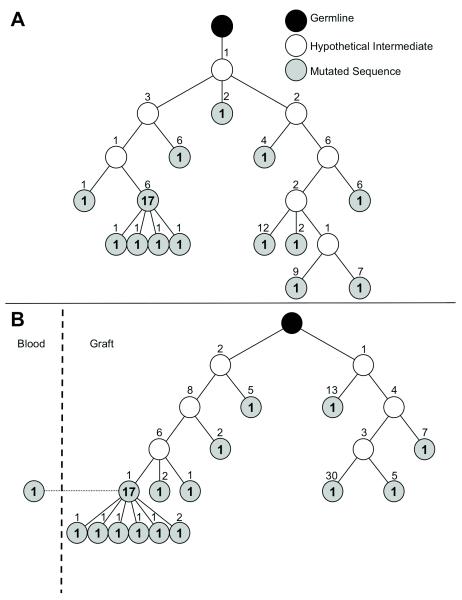
Virtually all redundant sequences exhibited evidence of SHM, resulting in divergence from germline sequences. Multiple sequences sharing identical CDR3 also showed distinct SHM indicating ongoing mutations of the corresponding B cell clones. Retrospective reconstruction of the phylogeny of the 2 most expanded graft infiltrating clones in IGHV2 using a statistical maximum-likelihood method revealed a branched multi-generational tree, which culminated in a predominant consensus sequence (Figure 2). One such consensus sequence was also detected in the periphery (Figure 2B), suggesting that the corresponding B cell clone trafficked between the graft and the peripheral blood.
Figure 2. Phylogeny of expanded graft-infiltrating B cell clones.
Phylogenies were constructed using a Maximum-Likelihood method for the 2 most abundant graft infiltrating B cell clones in the IGHV2 family corresponding to the red and blue sections in Figure 1 VH2 graft pie chart. All sequences within each tree (A: n=30; B: n=31 in the graft and n=1 in the blood) shared the same identical CDR3 sequence, but differed by the presence of somatic mutations elsewhere. Each circle represents a distinct sequence that differs from the upstream neighboring sequence by the number of amino acid indicated above the circle. Bolded numbers in the circles indicate the number of occurrences of the corresponding sequence.
Comparative analysis of IGHV somatic hypermutations
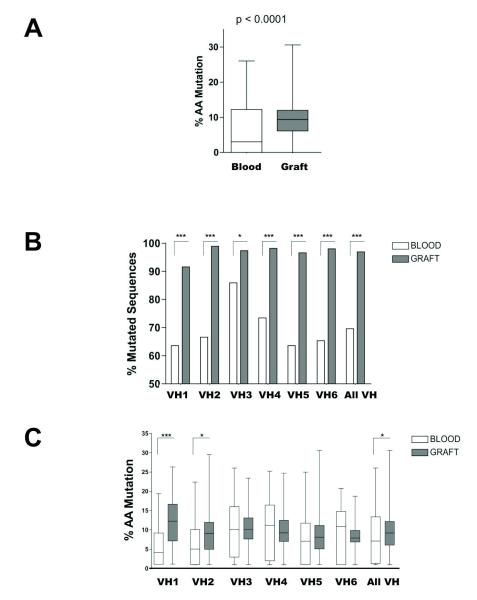
All IGHV sequences were compiled to further analyze SHM in graft infiltrates as well as peripheral blood B cells. As illustrated in Figure 3A (amino acid level) and figure S3A (nucleic acid level), graft IGHV sequences showed significantly more mutations than sequences cloned from peripheral blood B cells (p<0.0001). This difference resulted primarily from a higher percentage of mutated sequences among all unique IGHV sequences cloned from the graft tissue compared to the sequences cloned from the blood (Figure 3B and Figure S3B). Note that over 90% of unique graft sequences were mutated, suggesting an intensive SHM activity in situ. However, only marginal differences were observed when comparing the number of SHM within each individual sequences cloned from the graft or the blood (Figure 3C and Figure S3C).
Figure 3. Comparative analysis of blood and intragraft B cell IGHV mutation rates.
A) Overall percentage of mutated residues within all sequences analyzed in the blood (N=413) and the graft (N=473) in patient 2. B) Percentage of mutated sequences at the amino acid level for the 6 IGHV families among all unique sequences found in the blood (N=362) and the graft (N=398) of patient 2. All sequences sharing the same clonotypic CDR3 motifs were counted once regardless of the number of occurrence). C) Percentage of mutated amino acids for the 6 IGHV families within all unique sequences found in the blood (N=252) and the graft (N=386) of patient 2. D) Percentage and distribution of silent and non-silent mutations for all unique sequences from the peripheral blood of patient 2 (N=252). E) Percentage and distribution of silent and non-silent mutations for all mutated sequences from the graft of patient 2 (N=386). [* p<0.05; *** p<0.0001]
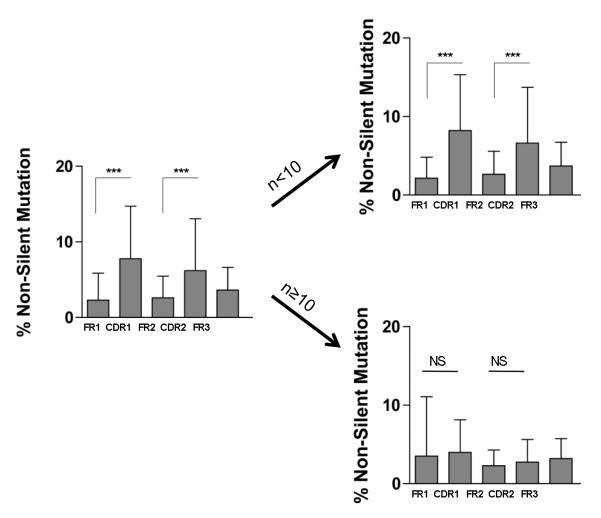
We next investigated the type (silent or non-silent) and distribution of SHM in all unique mutated IGHV sequences cloned from the graft tissue and contemporary blood sample. As depicted in Figure 3D (Blood) and 3E (Graft), silent SHM were distributed evenly between framework regions (FR1-3) and complementarity determining regions (CDR1-2) in both graft and blood. In contrast, the rate of non-silent mutation in CDR1 and CDR2 was significantly higher (p<0.0001) than in FR1 and FR2, respectively for the 2 compartments, consistent with the effect of selective pressure towards clones of higher affinity. This initial data suggested that the overall mutation profile of graft infiltrating B cells was indistinguishable from that of peripheral blood B cells. Mutations observed in graft IGHV sequences could have originated from in situ SHM or could have already been present in B cells at the time of their recruitment in the graft. To discriminate between the two possibilities, we compared the distribution of non-silent mutations in IGHV sequences detected at high frequency in the graft (N≥10), and therefore more likely to have resulted from in situ SHM, to lower frequency sequences (N<10). Whereas non-silent mutations were more frequent in CDR1-2 compared to FR1-3 in low frequency IGHV sequences, this uneven distribution was no longer observed in high frequency sequences (Figure 4). These findings suggest that B cell clones that underwent SHM in situ were not subjected to a comparable selection as B cells detected in the blood.
Figure 4. Comparative analysis of intragraft B cell IGHV non-silent mutations by VH segment.
Percentage and distribution of non-silent mutations for all analyzed mutated sequences (N=529) from the graft of patient 2 (VH1-VH6) and patients 5, 10, 17 and 19 (VH2). Sequences were analyzed together (left bar graph) or separately depending on their low or high frequencies in the IGHV repertoire (number of occurrences, n<10 or n≥10 respectively). [NS, non significant; *** p<0.0001]
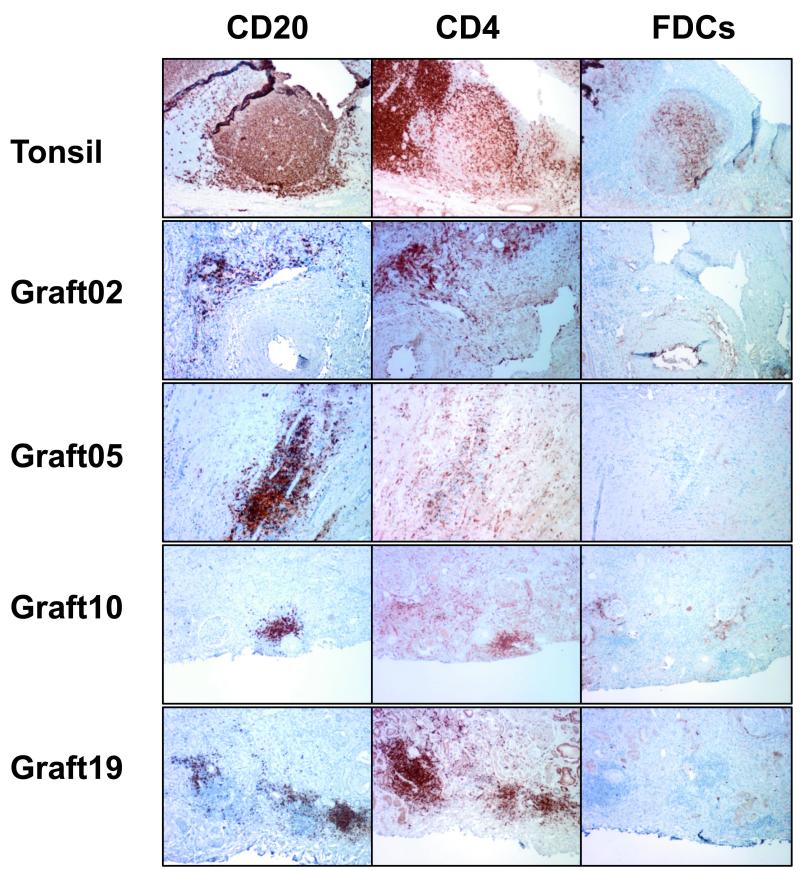
Intragraft lymphoid structures lack follicular dendritic cell networks
Follicular dendritic cells (FDC) are essential cells located in GC in secondary lymphoid organs and involved in the selection of B cell clones that underwent SHM. We examined whether FDC were present in intragraft lymphoid structures as a marker of ectopic GC. Using a specific antibody for FDC (CNA.42), we could not detect FDC in the vicinity of CD20+ B cells and CD4+ T cell clusters in any of the graft tissue assessed (Figure 5). In contrast, GC from tonsils revealed intricate FDC networks using the same antibody. Moreover, the architecture of intragraft lymphoid structures we observed in all the samples we examined was also significantly different from that of conventional GC (Figure 5).
Figure 5. Immunohistochemical staining of intragraft lymphoid aggregate structures.
Paraffin-embedded tissue sections from rejected human renal allografts showing representative lymphoid structures were stained for GC markers. Sections from human tonsil including GC were used as controls.
Discussion
B cells infiltrates in kidney allografts during rejection have been described for more than 30 years (26). In 2003, gene expression profiling studies sparked a renewed interest in these cells by revealing an association between dense intra-graft B cell infiltrates and steroid refractory rejection (5, 6). More recent gene expression analyses associated B cell infiltrates, inflammation, scarring tissue, interstitial fibrosis and poor graft survival (7, 8). In this study, we provided evidence of highly expanded B cell clones in explanted kidney graft specimens. These findings appear to corroborate results from a previous study that also reported B cell clonal expansion in human kidney allografts (21). We also showed the serial accumulation of mutations throughout redundant clonal IGHV sequences, indicating that the corresponding infiltrating B cell clones undergo active SHM in situ. Such expansion and mutations indicate the recruitment and activation of selective B cell clones in situ, presumably in response to specific antigens. It is often assumed that the antigens driving these responses are the same as the ones targeted by the humoral immunity, i.e. allo-HLA or self-antigens. However, there is no convincing evidence to support a link between serological responses and intragraft B cell infiltrates. On the contrary, several studies before ours have reported a lack of correlation between intragraft B cell clusters and circulating DSA(5, 9, 12, 22). This important observation strongly suggests that infiltrating B cells are distinct from B cells producing allospecific antibodies. Moreover, clinical responses following rituximab treatment appear to be associated with the elimination of graft infiltrating B cells but not any modification of serum antibodies, suggesting that these 2 components of B cell immunity are distinct (20). To date, the only indication on the specificity of kidney allograft infiltrating B cells come from Lerner and Salomon’s group (27). Their studies showed the reactivity of antibodies cloned from graft infiltrates to lipopolysaccharide (LPS), suggesting a role for bacterial wall products in the initiation of B cell immunity associated with antibody mediated rejection.
Infitrating B cells have been reported to be part of neatly organized lymphoid structures in explanted graft specimens (24). Some of these structures appeared to exhibit a number of markers also found in lymph node GC and were therefore termed ectopic GC (eGC). It was proposed that eGC support the differentiation and affinity maturation of B cell clones in a manner comparable to conventional GC (28). Our findings appear to contrast with this view. In particular, two elements seem to indicate that intragraft lymphoid structures differ from functional GC. Firstly, the even distribution of non-silent SHM along the different segments of the high frequency rearranged IGHV genes without any preferential accumulation in CDR suggests a lack of clonal selection in situ. Secondly, we could not detect the presence of FDC in or around B cell infiltrates in any sample we studied whereas a clear FDC network was observed in control tonsil specimens. As FDCs are primary participants in the affinity maturation process, their absence pointed to a different kind of organized lymphocytic infiltrates, which lack the capacity to select B cell clones. While our studies do not exclude the possibility of fully operational GC in rejected kidney grafts, these lymphoid structures do not appear to be prevalent.
Although no FDC could be detected in the graft specimens we analyzed, we constantly observed CD4+ T cell infiltrates in direct contact to B cells, confirming previous reports. There again, the patterns of the mixed infiltrates were not consistent with that of conventional GC. Nevertheless this frequent association evoked a functional interaction between the two cell types. Remarkably, intragraft B cell infiltrates have been found in cases of cellular rejection in the absence of any indication of humoral rejection (12, 22). Collectively, these data suggest that B cells play a supportive role to the T cell response in situ without necessarily generating a concomitant humoral response. This view would support the use of anti-B cell therapy to treat cases of ACR with documented B cell infiltrate and regardless of the presence of circulating DSA.
In conclusion, our results further support that B cell immunity develops in situ in a majority of rejected kidney allografts. These responses were associated with the expansion and somatic hypermutation of discrete clonal B cell populations and involved a substantial CD4+ T cell component. In contrast to previous reports, we did not find convincing signs of functional eGC as a prominent structural element in these infiltrates. Rather, the jumbled architecture of the lymphoid structures we observed, together with the absence of FDC and lack of affinity maturation, suggest that they result from local B cell-T cell activation and expansion without a definite organized structure. The unequivocal evidence of somatic mutations in the absence of eGC also suggests that infiltrating B cells may be distinct from conventional follicular B cells. Additional studies will be required to verify if these B cells contribute to the severity of allograft rejection and should be targeted as such.
Materials and methods
Patients and biological samples
All kidney transplant recipients included in this study underwent nephrectomy due to graft failure as detailed in supplementary Table S1. The collection and use of the specimens was approved by the Massachusetts General Hospital (MGH) and Brigham and Women’s Hospital (BWH) internal review boards. Upon receipt, the majority of graft tissue was flash frozen in liquid nitrogen and stored at −80°C. Some tissue was immediately subjected to total mRNA extraction as described below. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by density gradient medium (Ficoll-Paque Plus, GE Healthcare, Waukesha, WI) and cryopreserved in liquid nitrogen.
IGHV Repertoire analysis
RNA was extracted from graft tissue and PBMC using TRIzol Reagent (Ambion, Foster City, CA) and a gentleMACS dissociator (Miltenyi, Auburn, CA) for tissue homogenization. Complementary DNA (cDNA) was synthesized using the extracted RNA as template and the SuperScript III First Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. PCR was carried out using the cDNA to amplify the different families of IGHV genes. Six forward primers specific to the different families of IGHV genes (VH1-VH6) were used together with a consensus IGHJ reverse primer as previously described (29). Primers for GAPDH were also used as reaction controls.
PCR reactions were carried out using Platinum PCR SuperMix High Fidelity (Invitrogen) in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA) as follows: 94°C/5′, (94°C/30″; 56°C/30″; 68°C/30″) X 35 cycles; 68°C/7′. PCR products were ligated into the pCR8/GW/TOPO expression vector (Invitrogen) and used to transform chemically competent TOP10 E coli (Invitrogen). Transformed bacteria were grown on LB-Agar (Becton-Dickinson, Franklin Lakes, NJ) plates with 100ng/ml spectinomycin (Sigma-Aldrich). Single colonies of transformed bacteria were selected and expanded in LB (Becton-Dickinson) supplemented with 100ng/ml spectinomycin overnight at 37°C with shaking at 225rpm. Populations were harvested the following day and plasmid DNA was purified from cell pellets using Miniprep Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. Purified plasmid DNA was submitted to the DNA core facility at MGH and sequenced using the GW1 forward sequencing primer.
IGHV sequences were analyzed using the Human Ig Reference Directory Set compiled by the International Immunogenetics Information System (www.imgt.org) to construct partial B cell repertoires for the given IGHV gene families. Using this tool, the different framework (FR1-FR3) domains and complementarity determining regions (CDR1-CDR3) were identified for each sequence. All sequences including identical CDR3 amino acid (AA) segments were considered originating from the same clone and grouped together to constitute partial IGHV repertoires. The reproducibility of our technique was assessed by carrying out 2 separate sequence analyses using tissue fragments from the same graft (Figure S4).
Clone phylogenies were determined for larger clonal families (N≥10 sequences). Phylogenic trees were constructed following a Maximum-Likelihood method using Molecular Evolutionary Genetics Analysis (MEGA) software (30). MEGA was also used to generate alignments between clonal sequences sharing identical CDR3 and the nearest IGHV germline sequence. These alignments were used to identify somatic mutations in framework or complementarity determining regions as well as determine their silent or non-silent nature. Rates of mutation away from germline were then determined at both the NT and AA levels. These values were calculated for the sequence as a whole or on a segment by segment basis.
Immunohistochemistry
Previously frozen graft tissue samples were fixed in 10% phosphate buffered formalin (Thermo Scientific) and embedded in paraffin blocks from which successive sections were cut. Successive sections were then stained to assess for the expression of the following markers: CD20 (Clone L26, Dako, Carpinteria, CA), CD4 (Clone BC/1F6, BioCare, Concord, CA) and Follicular Dendritic Cells (Clone CNA.42, eBioscience, San Diego, CA).
Statistical analysis
SHM were all determined and classified using the IMGT database tool. Statistical significance of observed differences was assessed using an unpaired Students’ t- test. Clone phylogenies were determined using the Maximum-likelihood method encoded in the MEGA software. Three types of analysis were conducted depending on the IGHV sequences examined. 1) All sequences: Every sequence generated was considered and weighted equally; 2) All mutated sequences: Only sequences with a percent identity to germline <100% were considered. Each sequence was weighted equally, regardless of its clonal representation; 3) All unique mutated sequences: Only sequences with a percent identity to germline <100% were considered. All redundant sequences were considered only once.
Supplementary Material
Table 1.
Patient characteristics.
| Patient | Gender | Age at Tx (yrs) |
Original Disease | Time to Graft Failure* (mos) |
DSA at Return to Dialysis |
Time from Failure to Nephrectomy (mos) |
DSA at Time of Nephrectomy |
Cause(s) of Graft Loss | B cell infiltrate pattern | B cell Specific Therapy (mos. before nephrectomy) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 59 | Diabetes, Type II | 2.5 | − | 1 | − | ACRIII, Chronic Rejection | − | − |
| 2 | M | 41 | FSGS | 17 | − | 2.5 | − | ACRIII, Chronic Rejection, Recurrent FSGS | Perivascular Clusters | − |
| 3 | F | 34 | Diabetes, Type I | 120 | − | 12 | + | Chronic Humoral and Cellular Rejection, CIT | − | Rituximab (<1); Alemtuzumab (12) |
| 4 | F | 44 | Diabetes, Type I | 60 | − | 1 | − | ACRIII, Acute and Chronic Humoral Rejection | − | − |
| 5 | F | 25 | Zonal Displasia and FSGS | 33 | − | 6.5 | + | Acute and Chronic Humoral and Cellular Rejection | Non-Perivascular Clusters | − |
| 6 | F | 17 | Bilateral Displasia | 37 | + | 0.5 | + | Acute and Chronic Cellular Rejection | − | Rituximab (11) |
| 7 | F | 55 | FSGS | 24 | − | 3.5 | − | ACRIII, Chronic Allograft Vasculopathy, Thrombosis, Infarction, Recurrent FSGS | Diffuse | †TPE × >20 |
| 8 | M | 43 | Lupus Nephritis | 84 | + | 5.5 | + | Chronic Humoral and Cellular Rejection | − | − |
| 9 | F | 57 | Presumed HTN and chronic GN | 115 | − | 5 | − | ACRIII, Chronic rejection | Diffuse with small Non-perivascular clusters | |
| 10 | M | 23 | Unknown | 132 | − | 4 | − | ACRIII, Chronic Rejection | Perivascular and Non-Perivascular Clusters | |
| 11 | M | 37 | Reflux Nephropathy | 6 | + | 1 | + | Chronic Cellular Rejection II, Chronic Humoral Rejection | − | Rituximab (4); Bortezomib (4), Splenectomy (6), TPE × >40 |
| 12 | M | 29 | Infantile Nephrotic Syndrome | 48 | 84 | + | ACRIII, Acute and Chronic Pyelonephritis | Diffuse Subcapsular | Rituximab (18) | |
| 13 | F | 32 | Diabetes, Type I | 116 | − | 10 | + | Renal Vein Thrombosis, Infarction | − | − |
| 14 | M | 6 | Nephronophthisis | 32 | − | 2 | + | ACRII, Acute AMR, Chronic Arteriopathy | − | − |
| 15 | F | 48 | Polycystic Kidney Disease | 123 | − | 0.5 | − | Acute and Chronic Cellular Rejection III | Diffuse | − |
| 16 | M | 56 | Diabetes, Type I | 50 | − | 1 | − | Acute and Chronic Cellular Rejection, Interstitial Hemorrhage | ||
| 17 | M | 44 | Hemolytic-Uremic Syndrome | 6 | + | 3 | + | Acute Humoral Rejection, Acute and Chronic Cellular Rejection | Non-Perivascular Clusters | Bortezomib (6) |
| 18 | F | 41 | Malignant Hypertension | 30 | + | 4 | + | Infarction | − | Rituximab (3) |
| 19 | M | 16 | Nephronophthisis and FSGS | 102 | − | 0.5 | − | Thrombotic Microangiopathy | Perivascular Clusters | − |
| 20 | M | 57 | FSGS | 0 | − | immediate | − | Acute Humoral Rejection, Thrombotic Microangiopathy | − | Rituximab (<1) |
| 21 | F | 37 | SLE | 65 | immedate | ACRIII and Chronic Rejection, Chronic Pyelonephritis | Pericalyceal and Non-Perivascular Clusters | IVIg (65), Rituximab (65) |
Defined as return to dialysis
Abbreviations: FSGS- Focal Segmental Glomerulosclerosis; HTN - Hypertension; GN - Glomerulonephropathy; SLE - Systemic Lupus Erythematosus; ACRIII - Acute Cellular Rejection, Type III; CIT - Calcineurin Inhibitor Toxicity; TPE - Therapeutic Plasma Exchange
TPE treatments are listed as number of rounds of therapy over the life of the graft NOT months prior to nephrectomy
Acknowledgements
This work was supported by the Fahd and Nadia Alireza’s Research Fund, the Roche Organ Transplantation Research Foundation (ROTRF) and the National Institute of Health, National Institute of Diabetes, Digestive and Kidney Diseases Grant DK083352 and National Institute of Allergy and Infectious Diseases Grant AI94524.
Abbreviations
- IHC
Immunohistochemistry
- FDC
Follicular dendritic cells
- GG
Germinal centers
- IGHV
immunoglobulin heavy chain variable gene
- SHM
Somatic hypermutation
- CDR
Complementarity determining region
- DSA
Donor specific antibodies
Footnotes
EZ was the principal investigator and takes primary responsibility for the paper.
JF, FP, BG and CM performed the laboratory work for this study.
JDV, SD and EF participated in laboratory work or data analysis.
MVT, NE, TK, SKM, SGT and WW recruited the patients and provided clinical data.
EZ coordinated the research.
JF and EZ wrote the paper.
Disclosure The authors of this manuscript have no conflicts of interest to disclose.
References
- 1.Segerer S, Schlondorff D. B cells and tertiary lymphoid organs in renal inflammation. Kidney Int. 2008;73(5):533–537. doi: 10.1038/sj.ki.5002734. [DOI] [PubMed] [Google Scholar]
- 2.Thaunat O, Field AC, Dai J, Louedec L, Patey N, Bloch MF, et al. Lymphoid neogenesis in chronic rejection: evidence for a local humoral alloimmune response. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(41):14723–14728. doi: 10.1073/pnas.0507223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thaunat O, Patey N, Gautreau C, Lechaton S, Fremeaux-Bacchi V, Dieu-Nosjean MC, et al. B cell survival in intragraft tertiary lymphoid organs after rituximab therapy. Transplantation. 2008;85(11):1648–1653. doi: 10.1097/TP.0b013e3181735723. [DOI] [PubMed] [Google Scholar]
- 4.Wehner J, Morrell CN, Reynolds T, Rodriguez ER, Baldwin WM., 3rd Antibody and complement in transplant vasculopathy. Circulation research. 2007;100(2):191–203. doi: 10.1161/01.RES.0000255032.33661.88. [DOI] [PubMed] [Google Scholar]
- 5.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349(2):125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 6.Zarkhin V, Li L, Sarwal M. “To B or not to B?” B-cells and graft rejection. Transplantation. 2008;85(12):1705–1714. doi: 10.1097/TP.0b013e318177793e. [DOI] [PubMed] [Google Scholar]
- 7.Einecke G, Reeve J, Mengel M, Sis B, Bunnag S, Mueller TF, et al. Expression of B cell and immunoglobulin transcripts is a feature of inflammation in late allografts. Am J Transplant. 2008;8(7):1434–1443. doi: 10.1111/j.1600-6143.2008.02232.x. [DOI] [PubMed] [Google Scholar]
- 8.Mengel M, Reeve J, Bunnag S, Einecke G, Sis B, Mueller T, et al. Molecular correlates of scarring in kidney transplants: the emergence of mast cell transcripts. Am J Transplant. 2009;9(1):169–178. doi: 10.1111/j.1600-6143.2008.02462.x. [DOI] [PubMed] [Google Scholar]
- 9.Hippen BE, DeMattos A, Cook WJ, Kew CE, 2nd, Gaston RS. Association of CD20+ infiltrates with poorer clinical outcomes in acute cellular rejection of renal allografts. Am J Transplant. 2005;5(9):2248–2252. doi: 10.1111/j.1600-6143.2005.01009.x. [DOI] [PubMed] [Google Scholar]
- 10.Martins HL, Silva C, Martini D, Noronha IL. Detection of B lymphocytes (CD20+) in renal allograft biopsy specimens. Transplantation proceedings. 2007;39(2):432–434. doi: 10.1016/j.transproceed.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 11.Moreso F, Seron D, O’Valle F, Ibernon M, Goma M, Hueso M, et al. Immunephenotype of glomerular and interstitial infiltrating cells in protocol renal allograft biopsies and histological diagnosis. Am J Transplant. 2007;7(12):2739–2747. doi: 10.1111/j.1600-6143.2007.02013.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsai EW, Rianthavorn P, Gjertson DW, Wallace WD, Reed EF, Ettenger RB. CD20+ lymphocytes in renal allografts are associated with poor graft survival in pediatric patients. Transplantation. 2006;82(12):1769–1773. doi: 10.1097/01.tp.0000250572.46679.45. [DOI] [PubMed] [Google Scholar]
- 13.Doria C, di Francesco F, Ramirez CB, Frank A, Iaria M, Francos G, et al. The presence of B-cell nodules does not necessarily portend a less favorable outcome to therapy in patients with acute cellular rejection of a renal allograft. Transplantation proceedings. 2006;38(10):3441–3444. doi: 10.1016/j.transproceed.2006.10.173. [DOI] [PubMed] [Google Scholar]
- 14.Kayler LK, Lakkis FG, Morgan C, Basu A, Blisard D, Tan HP, et al. Acute cellular rejection with CD20-positive lymphoid clusters in kidney transplant patients following lymphocyte depletion. Am J Transplant. 2007;7(4):949–954. doi: 10.1111/j.1600-6143.2007.01737.x. [DOI] [PubMed] [Google Scholar]
- 15.Scheepstra C, Bemelman FJ, van der Loos C, Rowshani AT, van Donselaar-Van der Pant KA, Idu MM, et al. B cells in cluster or in a scattered pattern do not correlate with clinical outcome of renal allograft rejection. Transplantation. 2008;86(6):772–778. doi: 10.1097/TP.0b013e3181860a74. [DOI] [PubMed] [Google Scholar]
- 16.Becker YT, Becker BN, Pirsch JD, Sollinger HW. Rituximab as treatment for refractory kidney transplant rejection. Am J Transplant. 2004;4(6):996–1001. doi: 10.1111/j.1600-6143.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 17.Genberg H, Hansson A, Wernerson A, Wennberg L, Tyden G. Pharmacodynamics of rituximab in kidney allotransplantation. Am J Transplant. 2006;6(10):2418–2428. doi: 10.1111/j.1600-6143.2006.01497.x. [DOI] [PubMed] [Google Scholar]
- 18.Lehnhardt A, Mengel M, Pape L, Ehrich JH, Offner G, Strehlau J. Nodular B-cell aggregates associated with treatment refractory renal transplant rejection resolved by rituximab. Am J Transplant. 2006;6(4):847–851. doi: 10.1111/j.1600-6143.2006.01246.x. [DOI] [PubMed] [Google Scholar]
- 19.Steinmetz OM, Lange-Husken F, Turner JE, Vernauer A, Helmchen U, Stahl RA, et al. Rituximab removes intrarenal B cell clusters in patients with renal vascular allograft rejection. Transplantation. 2007;84(7):842–850. doi: 10.1097/01.tp.0000282786.58754.2b. [DOI] [PubMed] [Google Scholar]
- 20.Zarkhin V, Li L, Kambham N, Sigdel T, Salvatierra O, Sarwal MM. A randomized, prospective trial of rituximab for acute rejection in pediatric renal transplantation. Am J Transplant. 2008;8(12):2607–2617. doi: 10.1111/j.1600-6143.2008.02411.x. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J, Torkamani A, Grover RK, Jones TM, Ruiz DI, Schork NJ, et al. Ectopic B-cell clusters that infiltrate transplanted human kidneys are clonal. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(14):5560–5565. doi: 10.1073/pnas.1101148108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarkhin V, Kambham N, Li L, Kwok S, Hsieh SC, Salvatierra O, et al. Characterization of intra-graft B cells during renal allograft rejection. Kidney Int. 2008 doi: 10.1038/ki.2008.249. [DOI] [PubMed] [Google Scholar]
- 23.Thaunat O, Nicoletti A. Comment on “Activation-induced cytidine deaminase expression in follicular dendritic cell networks and interfollicular large B cells supports functionality of ectopic lymphoid neogenesis in autoimmune sialoadenitis and MALT lymphoma in Sjogren’s syndrome”. Journal of immunology. 2008;180(4):2007–2008. doi: 10.4049/jimmunol.180.4.2007-b. author reply 2008-2009. [DOI] [PubMed] [Google Scholar]
- 24.Thaunat O, Patey N, Caligiuri G, Gautreau C, Mamani-Matsuda M, Mekki Y, et al. Chronic rejection triggers the development of an aggressive intragraft immune response through recapitulation of lymphoid organogenesis. J Immunol. 2010;185(1):717–728. doi: 10.4049/jimmunol.0903589. [DOI] [PubMed] [Google Scholar]
- 25.Thaunat O, Patey N, Morelon E, Michel JB, Nicoletti A. Lymphoid neogenesis in chronic rejection: the murderer is in the house. Curr Opin Immunol. 2006;18(5):576–579. doi: 10.1016/j.coi.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Strom TB, Tilney NL, Carpenter CB, Busch GJ. Identity and cytotoxic capacity of cells infiltrating renal allografts. N Engl J Med. 1975;292(24):1257–1263. doi: 10.1056/NEJM197506122922402. [DOI] [PubMed] [Google Scholar]
- 27.Grover RK, Cheng J, Peng Y, Jones TM, Ruiz DI, Ulevitch RJ, et al. The costimulatory immunogen LPS induces the B-Cell clones that infiltrate transplanted human kidneys. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(16):6036–6041. doi: 10.1073/pnas.1202214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaunat O. Pathophysiologic significance of B-cell clusters in chronically rejected grafts. Transplantation. 2011;92(2):121–126. doi: 10.1097/TP.0b013e31821f74fe. [DOI] [PubMed] [Google Scholar]
- 29.Porcheray F, Devito J, Helou Y, Dargon I, Fraser JW, Nobecourt P, et al. Expansion of Polyreactive B Cells Cross-Reactive to HLA and Self in the Blood of a Patient With Kidney Graft Rejection. Am J Transplant. 2012;12(8):2088–2097. doi: 10.1111/j.1600-6143.2012.04053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.