Abstract
Sepsis is the major cause of death in the intensive care unit (ICU). Numerous biomarkers have been studied to identify the cause and severity of sepsis but these factors cannot differentiate between infectious and non-infectious inflammatory response. MicroRNAs (miRNAs) are non-coding RNA transcripts that regulate the expression of genes by repressing translation or degrading mRNA. Importantly, miRNAs can be released outside cells and easily detectable in bodily fluids such as blood, sweat, urine and breast milk. Numerous studies have explored the idea of utilizing extracellular miRNAs as biomarkers for sepsis by profiling the dysregulation of miRNAs in blood samples of sepsis patients. So far, miR-223, miR-146a and miR-150 have been identified to have promising prognostic and diagnostic value to sepsis. In addition, various intracellular miRNAs have been implicated to play critical roles in regulating the TLR-NF-κB pathway, which is a well-known inflammatory signaling pathway involved in the process of sepsis. Here, we summarize the recent progress on the role of extracellular and intracellular miRNAs in sepsis. Specifically, we discuss the possible role of circulating miRNA biomarkers for the diagnosis of sepsis and how intracellular miRNAs regulate the inflammatory responses in sepsis.
Keywords: Sepsis, miRNAs, biomarkers, inflammatory response, NF-κB
1. Introduction
Sepsis is a medical condition characterized by a severe systemic inflammatory response due to an infection [1]. At present, sepsis is the leading cause of death among patients admitted to the intensive care units (ICU) [1]. About 28.3 to 41% of all sepsis patients do not survive because of multiple organ failure [2]. Indeed, sepsis starts with an initial systemic inflammatory response (SIRS), followed by severe sepsis with multiple organ dysfunction before progressing to septic shock with persistent hypotension [3]. Moreover, sepsis might be misdiagnosed as the SIRS because the initial inflammatory response could also be caused by non-infectious substances such as burns, trauma and acute pancreatitis [4]. There is thus a critical need to develop more accurate biomarkers for the diagnosis of sepsis.
Over the past decade, numerous studies have been conducted to identify biomarkers that are sufficient to define the stage and severity of sepsis and predict the best line of treatment. The major challenge is that these studies cannot identify the cause of the systemic inflammatory response between SIRS and sepsis. Currently, biomarkers such as C-reactive protein (CRP), procalcitonin (PCT), Interlukin-6 (IL-6), sTREM-1, amongst others are being studied for their potential to detect SIRS caused by an infection. CRP, which is an acute phase protein found in the liver, is known to help in the removal of pathogens during infection [5]. PCT, a prohormone of calcitonin found in C-cells of the thyroid gland, are released in huge quantities into the blood stream during infection [6]. sTREM-1 are triggering receptors expressed on myeloid cells-1 which are released into plasma by phagocytes during bacterial or fungi infection [7]. These factors have potential to be an indicator of an infection-induced inflammatory response but experimental data have revealed they are also present during a non-infectious inflammatory response [4].
MicroRNAs are endogenous non-coding RNA molecules of about 19-22 nucleotides that play a role in post-translational gene silencing by inhibiting translation or by degrading mRNA [8]. It is generally acknowledged that a single miRNA can globally regulate the transcription of hundreds of genes to organize complex programs of gene expression which affects the overall functioning of a cell [9]. Significantly, changes in miRNA expression have been implicated in various disease states such as cardiovascular diseases, autoimmune diseases, neurodegenerative diseases, liver diseases and inflammatory diseases [10]. Lawrie et al. [11] were the first to introduce the idea of miRNAs as potential biomarkers for diseases when they compared serum miRNA levels of B-cell lymphoma patients with healthy individuals. Subsequently, many studies have been performed to characterize the expression of various miRNAs both intracellularly and extracellularly, advocating the potential role of miRNAs as biomarkers for sepsis. In this review, we will summarize such progress and discuss some potential issues.
In addition, intracellular miRNAs have been implicated as powerful endogenous factors in regulating the inflammatory signaling cascades during sepsis. Currently, Toll-like receptor (TLR)-mediated signaling pathway is well recognized to be involved in the development of septic shock [12]. In humans, ten functional types of toll-like receptors (TLR1-10) have been identified thus far. TLR1, TLR2, TLR4, TLR5, TLR6 are stimulated by various lipids and protein in microbial walls while TLR7, TLR8 and TLR9 recognize intracellular microbial nucleic acids because of their localization to the endoplasmic reticulum, endolysosomes, lysosomes and endosomes [13]. TLR4 is believed to recognize bacterial lipopolysaccharide (LPS) while lipoteichoic acid and peptidoglycans stimulate TLR2 [13]. Recently, numerous intracellular miRNAs have been defined to regulate TLR2/4-NF-ĸB signaling cascades. Therefore, in this review, we will also discuss how miRNAs affect the inflammatory signaling pathways involved in sepsis.
2. Biogenesis and Release of MicroRNAs
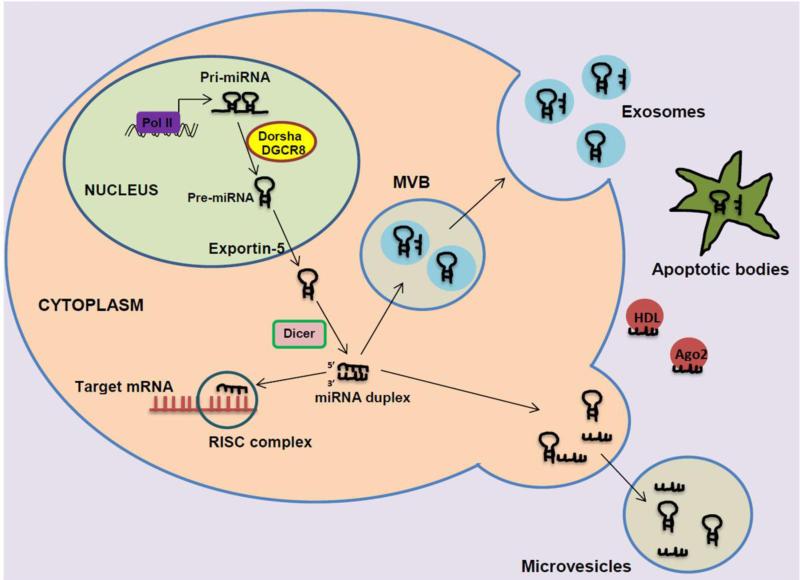
A primary miRNA transcript (pri-miRNAs) is usually formed through transcription by RNA polymerase II in the nucleus as a long capped precursor miRNA or through maturation from introns [14]. The ribonuclease (RNase) III Dorsha enzyme and the DiGeorge Syndrome Critical Region 8 (DGCR8) protein form a microprocessor complex to process the 70-100 nt premature miRNA (pre-miRNA) from the pri-miRNA [15]. With the help of nuclear export transporter Exportin 5, the pre-miRNA is exported into the cytoplasm [16]. At its terminal loop, the premiRNA interacts with RNase III endonuclease Dicer protein and the co-factor double-stranded transactivation-responsive RNA-binding protein (TRBP) to process the ~22 nt miRNA duplex [16]. The miRNA duplex is integrated into the RNA-induced silencing complex (RISC) which comprises of an argonaute (Ago) protein and a glycine-tryptophan repeat-containing protein of 182 kDa. (GW182) [17]. Previously, it was assumed that one strand at the 3’ end of the miRNA duplex (commonly called the “passenger strand”) is released and degraded, while the other strand (called “guide strand”) is remained in the RISC complex [18]. However, it has been demonstrated that both strands can be processed into mature miRNA and utilized for translational repression [19]. Mature miRNAs processed from the 3’ end are designated with a 3p suffix while miRNAs processed from the 5’end are identified with a 5p suffix [19]. The RISC complex is directed to a 3’ or 5’ untranslated region (UTR) of an mRNA where there is base pairing with 2 to 8 nucleotides (called “seed sequence”) of the mature miRNA [20].
MicroRNAs have the ability to be released outside cells into plasma and remain in circulation in a stable form [21]. Contrary to previous belief, microRNAs are highly conserved over evolutionary periods and are highly resistant to endogenous and exogenous RNase activity, acute pH and temperature conditions [22, 23]. These properties are achieved in plasma due to the fact that miRNAs are confined to membrane bound vesicles like microvesicles and exosomes. Release of miRNAs through microvesicles/exosomes is ATP and temperature dependent [23]. Microvesicles are typically about 100-1000nm in size and are generated from directly outward budding of cell membrane [23]. Conversely, exosomes are about 30-100nm in size and are produced from multivesicular bodies (MVBs) when the MVBs fuse with the cell membrane and are exocytosized out of the cell [24]. The MVBs themselves originate from vesicles in the lumen of endosomes, which collect endocytic membrane proteins within the cell. Exosomes have been found in various bodily fluids such as plasma, urine, serum, breast milk and saliva [24].
Although it happens very rarely, some miRNAs are released into plasma during tissue damage or cell apoptosis as apoptotic bodies [25]. In addition, it has also been found that argonaute 2 (Ago2) protein and high-density lipoproteins may act as carriers for miRNAs [26, 27]. Put together, the biogenesis and release of miRNAs are summarized in Figure 1.
Figure 1. Biogenesis and release of miRNAs.
After the processing by Dorsha/DGCR8 in the nucleus and followed by Dicer in the cytoplasm, miRNA is either incorporated in the RISC complex to target mRNA repression; packaged into microvesicular bodies to be released as exosomes; or exocytyzed out of the cell as microvesicles. MiRNAs are also released by apoptotic bodies during cell apoptosis and tissue damage. In addition, miRNAs may bind to some proteins such as HDL and Ago2, and released outside cells.
3. Extracellular microRNAs as potential biomarkers in sepsis
Obviously, extracellular miRNAs that exist in blood, sweat, urine and breast milk can be quantified rapidly in a clinical setting, unlike microbial cultures that are time consuming. Such property of extracellular miRNAs drives many investigators to identify possible biomarkers for the diagnosis of disease. In the field of sepsis, a number of studies have been conducted to profile plasma miRNAs of septic patients. Several miRNAs have stood out as potential diagnostic and prognostic biomarkers of sepsis and are reviewed below (Table 1).
Table 1.
Differential expression of miRNAs in various blood specimens of sepsis patients
| Specimen | Study Sample Size | Main findings | Reference |
|---|---|---|---|
| Blood leukocytes | 10 sepsis patients 12 healthy individuals |
↓ miR-150, miR-342-5p ↑ miR-486, miR-182 in sepsis patients |
[28] |
| Serum | 138 sepsis patients 85 ICU patients without sepsis 76 healthy individuals |
↑ miR-150 amongst ICU patients with or without sepsis enhances survival | [30] |
| Serum | 50 sepsis patients 30 SIRS patients 20 healthy individuals |
↓ miR-223 and miR-146a in sepsis patients | [31] |
| Serum | 117 surviving sepsis patients 97 non-surviving sepsis patients |
↑ miR-223, miR-16 ↓miR-15a, miR-122, miR-193, miR-483-5p in surviving sepsis patients |
[33] |
| Plasma | 14 non-sepsis SIRS patients 14 sepsis patients |
↑ miR-146a in non-sepsis SIRS patients | [32] |
| Blood leucocytes | 23 sepsis patients 22 SIRS patients 21 healthy people |
↑ miR-4772 family ↓ miR-150 in sepsis patients |
[35] |
| Plasma | 78 surviving sepsis patients 64 non-surviving sepsis patients |
↑ miR-574-5p ↓ miR-297 in surviving sepsis patients |
[37] |
| Serum | 17 ICU patients without sepsis 36 sepsis patients |
↓ miR-181b in sepsis patients | [38] |
| Blood leukocytes | Sepsis patients and non-sepsis ICU patients | ↓ miR-466I in non-surviving sepsis patients | [39] |
| Serum | 138 sepsis patients 85 ICU patients without sepsis 76 healthy individuals |
↑ miR-133a correlated with severity of sepsis | [40] |
3.1 miR-150
Recently, Vasilescu et al. [28] compared miRNA profiles of 12 healthy individuals and 10 sepsis patients. The authors’ analyses revealed that levels of miR-150 were mostly dysregulated in the peripheral blood leukocytes between the two groups. Significantly, the reduction in miR-150 in sepsis patients correlated well with the severity of the disease which was determined by the Sepsis-related Organ Failure (SOFA) score. Given that the SOFA score does not account for the pathology involved in sepsis [29], they further analyzed the role of miR-150 in the pathogenesis of sepsis, and observed that the decrease in miR-150 in sepsis patients correlated with the increase of pro-inflammatory cytokine, IL-18 [28].
However, with a different set of subjects, Roderburg et al. [30] analyzed serum levels of miR-150 in 223 critically ill patients (138 diagnosed with sepsis and 85 without sepsis) and 76 healthy individuals. Results of their analyses revealed that there was only a minor decrease in the levels of miR-150 in critically ill patients compared to healthy individuals. They concluded that miR-150 was not a good diagnostic tool for sepsis. Nonetheless, they observed that amongst critically ill patients, a high level of serum miR-150 correlated with survival in patients with sepsis while a lower level of miR-150 signified a high chance of organ dysfunction and mortality. They postulated that miR-150 might have a role to play as a prognostic tool for sepsis. Albeit just a minor decrease, the study by Roderburg et al.[30] was consistent with the findings of Vasilescu et al.[28] as both studies observed a reduction in miR-150 levels in sepsis patients.
3.2 miR-223 and miR-146a
miR-223 and miR-146a represent two of the most researched miRNAs in relation to their role in the pathogenesis of various diseases. In the field of sepsis, Wang et al. [31] have demonstrated that miR-223 and miR-146a are dysregulated in sepsis patients. These authors examined seven miRNAs related to inflammation and infection in 50 sepsis patients, 30 SIRS patients and 20 healthy people. The seven miRNAs they tested were miR-132, miR-146a, miR-155, miR-223, miR-15b, miR-126 and let-7i. Using quantitative PCR, they reported that serum levels of miR-146a and miR-223 were significantly reduced in patients suffering from sepsis compared to SIRS patients and the healthy individuals. Importantly, there were no significant changes in levels of miR-223 in SIRS patients compared to the healthy individuals. Subsequently, another study, using RT-PCR, examined the differential expression of miR-146a in plasma samples of 14 non-sepsis SIRS patients and 14 sepsis patients. The investigators observed higher levels of serum miR-146a in non-sepsis SIRS patients compared to sepsis patients [32]. Hence, these studies suggest that miR-146a and miR-223 may be optimal diagnostic tools for sepsis. Recently, one study profiled serum miRNAs of 214 sepsis patients (117 survivors and 97 non-survivors), and found that miR-223 levels were significantly lower in non-surviving sepsis patients compared to surviving sepsis patients [33]. Accordingly, using a global knockout mouse model, our lab has observed that the loss of pre-miR-223 aggravated sepsis-induced myocardial dysfunction, inflammatory response and mortality to a greater degree than wild-type mice [34]. Thus, miR-223 may have the potential to be used as a prognostic tool to determine the line of treatment according to the severity of the disease, and lower levels of miR-223 may contribute to septic death.
3.3 miR-4772 family
Recently, miR-4772 family [miR-4772-3p, miR-4772-5p, miR-4772-5p-iso (an isomiR of miR-4772-5p) has been shown to have prognostic and diagnostic value to sepsis. Ma et al. [35] utilized Linear Discriminant Analysis (LDA) for blood samples of 23 sepsis patients with confirmed bacterial infection, 22 SIRS patients and 21 healthy individuals. Firstly, they RNA-sequenced 4 blood samples from each group, and demonstrated that miR-4772-5p-iso was significantly increased in the sepsis group but could not distinguish between the SIRS and control groups. MiR-4772-3p and miR-4772-5p were significantly upregulated in the sepsis group compared to the healthy individuals but could not distinguish between the sepsis and SIRS group. In vitro work also revealed that miR-4772-5p-iso was upregulated in primary peripheral blood monocytes after a 24 h challenge with specific TLR ligands, providing a potential mechanistic explanation for their observed data in sepsis patients. However, further analysis in the remaining blood samples revealed that miR-150 was the only miRNA that could significantly distinguish between all three groups. Nevertheless, this study confirmed miR-150 as a potential biomarker for sepsis and showed that a combination of miR-4772 family could potentially indicate the presence of sepsis. Also, miR-4772-5p-iso has no murine homologue and it is plausible that species-specific microRNAs are likely to represent more specific biomarkers for human disease. In addition, miR-4772-5p-iso is located in an intronic region of Interleukin 18 receptor accessory protein gene (IL-18RAP), which was also increased during sepsis. More interestingly, IL-18 itself has previously been reported to be upregulated during sepsis [36] suggesting the co-regulation of IL-18RAP and IL-18.
3.4 Other miRNAs
In addition to the miRNAs outlined, the studies discussed above observed the dysregulation of other miRNAs. For example, Vasilescu et al. [28] found that levels of miR-486 and miR-182 were overexpressed while miR-342-5p levels were reduced in the peripheral blood leukocytes of sepsis patients compared to healthy individuals. Wang et al. [33] observed that levels of miR-16 were increased while levels of miR-15a, miR-122, miR-193 and miR-483-5p were decreased in the sera of surviving sepsis patients compared to the non-surviving group.
Notably, other groups have also examined the differential expression of miRNAs in varying sets of subjects. For instance, Wang et al. [37] used microarray screens to identify differentially expressed serum miRNAs by comparing samples from 12 surviving and 12 non-surviving sepsis patients. They observed that only levels of miR-297 and miR-574-5p were significantly different between survivors and non-survivors. In survivors, serum miR-297 levels were downregulated while miR-574-5p levels were upregulated. These results were further validated in additional serum samples of 66 survivors and 52 non-survivors. When miRNA expression and sepsis stages were combined with SOFA scores, they finally concluded that miR-574-5p had a better predictive value and could potentially be used for the prognosis of sepsis. Sun et al. [38] analyzed serum samples of control subjects (ICU patients without sepsis) and sepsis patients for levels of circulating miR-181b. They observed that serum miR-181b levels were reduced by about 40% in sepsis patients compared to ICU patients without sepsis. Recently, Li et al. [39] reported that levels of miR-466I in leukocytes were higher in sepsis non-survivors than in sepsis survivors. There was however no difference in serum levels of miR-466I between survivors and non-survivors.
Furthermore, Tacke et al. [40] have found that miR-133a levels were upregulated in sepsis patients and critically ill patients. In a subject pool of 223 critically ill patients (138 with sepsis and 85 without sepsis) and 76 healthy individuals, the upregulation of miR-133a correlated with sepsis severity, SOFA scores and bacterial infection markers CRP and procalcitonin. In keeping with that observation, the authors profiled serum miRNAs in septic mice induced by cecal ligation and puncture (CLP) and observed the strongest change in expression in miR-133a (16-fold) compared to miR-150, miR-155 and miR-125b. As demonstrated by Tacke et al. [40], blood samples of septic mice can be screened to determine the expression profile of microRNAs. Using septic C57BL/6 mice induced by CLP, Wu et al. [41] profiled circulating miRNAs in whole blood samples in the experimental sepsis mice. After microarray analysis, levels of miR-16, miR-17, miR-20a, miR-20b, miR-26a, miR-26b, miR-106a, miR-106b, miR-195 and miR-451 were higher in septic mice compared to controls. Further analyses of these miRNAs in TLR2-/-, TLR4-/- and NF-κB-/- mice revealed that there was no significant difference in expression levels compared to TLR2+/+, TLR4+/+ and NF-κB +/+ mice after CLP. These results signify that the expression of these ten miRNAs in blood is not dependent on TLR2, TLR4 or NF-κB-dependent pathways.
Subsequently, the validity of miRNAs as biomarkers for sepsis has been explored by Huang et al. [42] using microarray expression profiles and miRNA regulatory network analysis. The authors analyzed the expression profiles of miRNAs from various studies through validation and prediction databases such as TarBase and HOCTAR, and measured statistical significance with the student t-test. The resulting miRNAs were further examined with pathway analysis, disease oncology analysis, protein-protein interaction network (PIN) analysis and ROC curves to ascertain their prognostic or diagnostic value to sepsis. In all, miR-15a, miR-16, miR-122, miR-146a, miR-223, miR-499-5p and miR-150 were found to have diagnostic value to sepsis while miR-483-5p, miR-574-5p and miR-193b were found to have prognostic value.
As reviewed above, a number of extracellular miRNAs have been proposed to be candidate biomarkers for the diagnosis of sepsis. Given the limited sample size they analyzed and the complex nature of sepsis/SIRS, it would be necessary to employ randomized prospective large scale clinical trials in larger patient groups for evaluating the accuracy of these proposed miRNA biomarkers in the future.
4. Intracellular microRNAs as regulators of inflammatory signaling in sepsis
It is well recognized that the inflammatory response during sepsis is mediated through the activation of TLRs and the downstream NF-κB pathway in macrophages and monocytes [12]. After recognition of LPS/endotoxin, the TLR recruits the adaptor protein myeloid differentiation primary response gene 88 (MyD88) [43]. MyD88 recruits IL-1 receptor –associated kinases IRAK1, IRAK2 and IRAK4. The IRAK kinases phosphorylate the tumor necrosis factor receptor-activated factor 6 (TRAF6) proteins, an ubiquitin protein ligase, which catalyzes the production of a Lys63-linked polyubiquitin on the Iκ-B kinase subunit γ [IKK- γ, also called the NF-κB modulator (NEMO)] and on the TRAF6 protein itself [44]. This interaction causes the recruitment of the transforming growth factor-β (TGF-β)-activated kinase 1(TAK1) which phosphorylates the β subunit of IKK complex and activates IKK complex [44]. IKK complex is responsible for phosphorylating the endogenous inhibitor of NF-κB (IκBa) in order to release the inhibitory effects of IκBa on the NF-κB transcription factor [44]. The NF-κB transcription factor contains the reticuloendotheliosis (Rel) proteins p50 and p65, which form heterodimers and are transferred to the nucleus to activate the expression of inflammatory genes [45]. Alternatively, activated TLR can recruit adaptor protein TIR-domain-containing adapter-inducing interferon-B (TRIF) [46]. TRIF protein recruits receptor-interacting protein-1 (RIP1) kinase which activates TAK1. Like the myD88-dependent pathway, TAK1 phosphorylates IKK which leads to the activation of NF-κB [46]. Unlike the myD88 pathway, the TRIF pathway has not been studied extensively in sepsis.
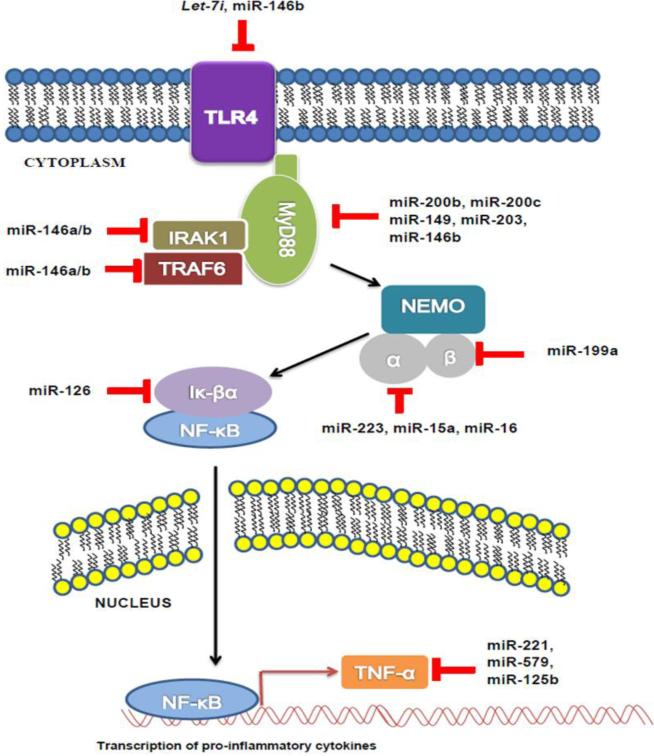
Nowadays, it is very clear that the TLR-NF-κB inflammatory response is involved in the process of sepsis. However, the use of anti-inflammatory agents such as corticosteroids, antiendotoxin antibodies, tumor necrosis factor (TNF) antagonists, interleukin-1–receptor antagonists have not been successful in treating sepsis [47]. This suggests that other factors might be associated with sepsis. Indeed, recent studies have shown that numerous intracellular miRNAs play critical roles in the regulation of TLR signaling and NF-κB-mediated inflammatory response (Figure 2), as reviewed below.
Figure 2. Negative regulation of TLR signaling by miRNA.
TLR4 is negatively regulated by let-7i and miR-146b. MyD88 is negatively regulated by miR-200b, miR-200c, miR-149 and miR-203 and miR-146b. IRAK1 and TRAF6 can be targeted by miR-146 family. IKK-α is negatively regulated by miR-223, miR-15a and miR-16. IKK-β is negatively regulated by miR-199a. IkBα is targeted by miR-126. TNFα is negatively regulated by miR-221, miR-579 and miR-125b.
4.1 Let-7i targets TLR4
The TLR4 receptor itself is predicted to be a target of the let-7i. Chen et al. [48] reported that let-7i regulates TLR4 expression via post-transcriptional suppression in cultured human biliary epithelial cells (cholangiocytes). They observed that an infection of cultured human cholangiocytes with Cryptosporidium parvum (a parasite that causes intestinal and biliary disease) results in decreased expression of primary let-7i and mature let-7i in a MyD88/NF-κB-dependent manner. Significantly, the decreased let-7i expression is associated with C. parvum-induced up-regulation of TLR4 in infected cells. Moreover, overexpression of a let-7i precursor in human cholangiocytes resulted in a reduction of TLR4 protein content. These results indicate that let-7i regulates TLR4 expression in cholangiocytes and contributes to epithelial immune responses against C. parvum infection. Furthermore, the data raises the possibility that miRNA-mediated post-transcriptional pathways may be critical to host-cell regulatory responses to microbial infection in general.
4.2 miR-200 family, miR-149 and miR-203 affect MyD88
A recent study conducted by Wendlandt et al. [49] showed that the miR-200 family (miR-200a, miR-200b, and miR-200c) could regulate TLR4 signaling and NF-κB activation. Using HEK293 cells and a luciferase reporter assay, they observed that there was a dose-dependent repression of luciferase NF-κB activity when miR-200b and miR-200c mimics were transfected while miR-200a had no effect. A luciferase assay with reporter gene for the 3‘UTR of MyD88 showed decreased activity when miR-200b and miR-200c were transfected. In contrast, when miR-200b and miR-200c were transfected in a mutated 3‘UTR of myD88, there was no change in activity. In addition, all three miRNAs in the miR-200 family had no effect on the 3‘UTR of TLR4, IRAK1 and TRAF6. Together, this study indicates that MyD88 is a bona fide target for miR-200b and miR-200c.
In addition to miR-200b and miR-200c, Xu et al. [50] showed that miR-149 could negatively regulate MyD88 protein levels. They observed that when the RAW264.7 cells were stimulated with Myobacterium bovis, there was an increase in levels of MyD88 and a decrease in miR-149 levels. When a lentiviral vector expressing miR-149 was transfected in RAW264.7 cells, there was no change in levels of MyD88 mRNA compared to controls but there was a significant decrease in myD88 protein levels. Thus, miR-149 represses the translation of MyD88 but has no effect on the transcription. Similarly, Wei et al. [51] recently demonstrated that the overexpression of miR-203 in RAW264.7 cells significantly reduced protein rather than mRNA levels of MyD88.
4.3 miR-146 family negatively regulates IRAK1 and TRAF6
miR-146 family (miR-146a and miR-146b) has been shown to negatively modulate the translation of IRAK1and TRAF6. Tagonov et al. [52] investigated the role of miR-146 family on the NF-κB-dependent immune response and observed a significant upregulation of miR-146, miR-155 and miR-132 in THP-1 cells upon stimulation with LPS. miR-146 was chosen for further analysis because it showed a rapid response to LPS stimulation. Using a luciferase construct that contained a reporter gene for the 3’-UTR of IRAK1 and TRAF6, they found that both miR-146a and miR-146b caused a significant reduction in luciferase activity when transfected in HEK293 cells.
With more focus on miR-146b, Curtale et al. [53] showed that miR-146b affects the TLR4 signaling pathway by targeting the TLR4 receptor, myD88, IRAK1 and TRAF6. In luciferase assays of HEK293 cells, there was decrease in activity of the reporter genes containing the 3’-UTR of TLR4, myD88, IRAK1 and TRAF6 when miR-146b was transfected. To confirm specificity of targets, miR-146b was transfected into reporter genes containing mutated regions of the 3’-UTR of TLR4, myD88, IRAK1 and TRAF6 which resulted in no change in activity. In addition, the authors observed an increase in miR-146b levels but not miR-146a and miR155 when human monocyte cells were stimulated with IL-10.
4.4 miR-15, miR-16, miR-223 and miR-199a for the IKK complex and miR-126 for Iĸ-Bα
The IKK complex, consisting of IKK-α, IKK-β and IKK-γ (NEMO), is essential for NF-κB activation. Using the Memorial Sloan-Kettering Cancer Center miRNA database, Li et al. [54] found that miR-15a, miR-16 and miR-223 had complementary sequences to the 3’UTR of the IKK-α gene. There was a decrease in miR-15a, miR-16 and miR-223 levels and an increase in IKK-α levels when differentiation of human monocytes was stimulated with granulocyte-macrophage colony-stimulating factor (GM-CSF). The transfection of miR-15a, miR-16 and miR-223 mimics into the human monocytes resulted in a decrease in IKK-α protein levels. In the same vein, Chen et al. [55] employed the PicTar algorithm, and found that the 3’-UTR of the IKK-β mRNA had three complementary sequences for miR-199a. The co-transfection of miR-199a with a luciferase construct containing the 3’UTR for IKK-β into EOC ovarian cancer cells resulted in a decrease in luciferase activity. A recent study by Feng et al. [56] has demonstrated that the 3’-UTR of IĸBα was complementary to the sequence of miR-126 through MicroCosm Targets, Targetscan and PicTar. The transfection of a miR-126 mimic in HT29 cells resulted in a 57% reduction in IĸBα protein levels.
4.5 miR-221, miR-579 and miR-125b target pro-inflammatory cytokine TNFα
The production of pro-inflammatory cytokines during the inflammatory response can be directly targeted by miRNAs. El Gazzar and McCall [57] utilized a computational target prediction algorithm to identify miR-221, miR-579 and miR-125b as possible candidate miRNAs with sequences complementary to the 3‘UTR of TNFα. They confirmed the repressive effect of miR-221, miR-579 and miR-125 on TNFα in LPS-tolerant THP-1 cells. Transfection of miR-221 in THP-1 cells promoted the degradation of TNFα mRNA levels while the transfection of miR-579 and miR-125b resulted in a significant reduction in TNFα protein levels. The inhibitory effects of miR-125b on TNFα were also reported by Tili et al. [58]. The authors observed a decrease in luciferase activity when miR-125b was co-transfected with the 3’UTR-TNFα construct in HEK-293 cells. Accordingly, in LPS-treated RAW264.7 macrophages, miR-125b was downregulated whereas TNFα was up-regulated.
4.6 Other miRNAs affect the inflammatory signaling
PDCD4, a pro-inflammatory protein, is known to aid in the activation of NF-κB [59]. miR-21 has been shown to regulate the expression of PDCD4 by Sheedy et al.[60]. The authors transfected RAW264.7 cells with antisense oligonucleotides for miR-21, which resulted in more PDCD4 production after stimulation with LPS. Furthermore, the transfection of a miR-21 precursor resulted in less PDCD4 and more production of IL-10 after LPS stimulation, compared to controls. Thus miR-21 may inhibit the actions of PDCD4 on NF-κB activity and promote the production of anti-inflammatory cytokines. Likewise, receptor-interacting protein 140 (RIP140) is a nuclear protein believed to associate with NF-κB to aid in the transcription of pro-inflammatory cytokines in macrophages [61]. Using an acute septic mouse model, Ho et al. [62] identified that miR-33 targeted the 3’ UTR of RIP140 mRNA. They observed that the transfection of miR-33 in RAW264.7 macrophages resulted in the decrease in RIP140 mRNA, TNFα and IL-1β mRNA levels after LPS stimulation.
Furthermore, Sweeny et al. [63] has reported that levels of Ppargc1a are dysregulated in TLR2−/− and TLR4−/− mice infected with Staphylococcus aureus. Ppargc1a, a member of PPAR-gamma coactivator (PGC) family of transcriptional co-activators, is involved in the biogenesis of mitochondria and is believed to be stimulated when there is damage to the mitochondria during sepsis [64]. The authors observed that when a miR-202-3p mimic was transfected into AML12 hepatocyte cells, levels of Ppargc1a mRNA were reduced. In silico analyses of the 3‘UTR of Ppargc1a mRNA revealed binding sites for miR-202-3p, confirming that the activity of miR-202-3p is inversely related to Ppargc1a [63].
Activated protein C (aPC), an endogenous protein in the liver, is known to decrease inflammation and reduce thrombosis [65]. The administration of activated protein C (aPC) has been shown to improve mortality in sepsis patients by modulating hepatic sinusoidal vasoregulation and restoring hepatic oxygenation [66]. Moore and colleagues [67] have demonstrated that the actions of aPC are due to changes in the miRNA expression during sepsis shock. Using the CLP model in male Sprague-Dawley rats, they reported an increase in levels of miR-182, miR-199a-5p, miR-203, miR-211, miR-222, and miR-29b in hepatic tissue. They concluded that these microRNAs may be responsible for the formation of focal adhesions in hepatic tissue through the aPC pathway. Another study by Wang et al. [68] examined the expression of miR-155 in LPS-induced sepsis in the liver of female BALB/c mice. Some mice were pretreated with Dexamethasone (DMX), an anti-inflammatory glucocorticoid, before LPS was administered intraperitoneally. Analysis of liver tissue of the two groups of mice revealed that levels of miR-155 were upregulated 70-fold in LPS-treated mice compared to those pretreated with DMX. Moreover, the DMX group also displayed a decrease in pro-inflammatory cytokines TNFα and IL-6 compared to the LPS group. Indeed, miR-155 has been found to target FADD and Ripk1 transcripts and indirectly promote translation of TNFα [58]. The effects of miRNAs on inflammatory response mediators are summarized in Table 2.
Table 2.
miRNAs regulate intracellular inflammatory response through interactions with various targets
| microRNA | Targets | Effects | Reference |
|---|---|---|---|
| miR-21 | pro-inflammatory protein PDCD4 | Inhibit the activation of NF-κB | [60] |
| miR-33 | Receptor-interacting protein 140 (RIP140) | Reduce the production of TNFα and IL-1β | [62] |
| miR-202-3p | Ppargc1a | Inhibit PGC family –mediated biogenesis of mitochondria during sepsis | [63] |
| miR-182, miR-203, miR-211, miR-222, miR-199a-5p, miR-29b | Activated Protein C (aPC) | Inhibit formation of focal adhesions in hepatic tissue | [67] |
| miR-155 | FADD and Ripk1 | Promote translation of TNFα | [58] |
6. Conclusion
Sepsis can cause multiple organ dysfunction in the liver, kidney, lungs and heart. The mechanisms underlying the clinical manifestations and pathology of sepsis are complex. However, the prospect of miRNAs being used as an indicator of the pathology of a specific condition is promising, which may provide rapid diagnosis and confirmation of infection in sepsis patients. While numerous miRNAs have been identified to regulate the inflammatory response as reviewed here, whether these intracellular miRNAs have therapeutic potential for the treatment of sepsis remains largely unexplored. In addition, the role of the miRNAs, albeit been confirmed as critical regulators of the TLR- NF-κB pathway, have not been studied extensively in sepsis models. Furthermore, all therapeutic interventions aimed at specific mediators/pathways to dampen the inflammatory response in sepsis have failed to improve the outcome [69, 70]. Thus, it will be of great significance to continue examining the role of extracellular and intracellular miRNAs in sepsis, because it will not only help us understand the complex nature of sepsis but also provide new insights for efficient therapies.
Highlights.
MicroRNAs silence genes by inhibiting translation or degrading mRNA.
Extracellular microRNAs are dysregulated in blood samples of sepsis patients.
Intracellular microRNAs regulate the TLR-NF-κB-mediated inflammatory response.
ACKNOWLEDGEMENTS
The research in Dr. Guo-Chang Fan's lab is supported by NIH grant 2R01-087861.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Critical Care Medicine. 2001;29(7):1303–10. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Levy MM, Artigas A, Phillips GS, Rhodes A, Beale R, Osborn T, Dellinger RP. Outcomes of the Surviving Sepsis Campaign in intensive care units in the USA and Europe: a prospective cohort study. The Lancet Infectious Diseases. 2012;12(12):919–24. doi: 10.1016/S1473-3099(12)70239-6. [DOI] [PubMed] [Google Scholar]
- 3.Bone RC, Sibbald WJ, Sprung CL. The ACCP-SCCM consensus conference on sepsis and organ failure. Chest. 1992;101(6):1481–3. doi: 10.1378/chest.101.6.1481. [DOI] [PubMed] [Google Scholar]
- 4.Sankar V, Webster NR. Clinical application of sepsis biomarkers. J Anesth. 2013;27(2):269–83. doi: 10.1007/s00540-012-1502-7. [DOI] [PubMed] [Google Scholar]
- 5.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. The New England Journal of Medicine. 1999;340(17):448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 6.Bloos F, Marshall JC, Dellinger RP, Vincent J-L, Gutierrez G, Rivers E, Brunkhorst FM. Multinational, observational study of procalcitonin in ICU patients with pneumonia requiring mechanical ventilation: a multicenter observational study. Critical Care. 2011;15(2):R88. doi: 10.1186/cc10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410(6832):1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna Journal of Medical Biotechnology. 2010;2(4):161–79. [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. British Journal of Haematology. 2008;141(5):672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsujimoto H, Ono S, Efron P. a, Scumpia PO, Moldawer LL, Mochizuki H. Role of Toll-like receptors in the development of sepsis. Shock. 2008;29(3):315–21. doi: 10.1097/SHK.0b013e318157ee55. [DOI] [PubMed] [Google Scholar]
- 13.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunology. 2010;11(5):373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. The EMBO Journal. 2002;21(17):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 17.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Structural & Molecular Biology. 2012;19(6):586–93. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 18.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ, Yang BB. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41(21):9688–704. doi: 10.1093/nar/gkt680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Athey BD. New class of microRNA targets containing simultaneous 5’-UTR and 3'-UTR interaction sites. Genome Research. 2009;19(7):1175–83. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Zhang C-Y. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 22.Okamura K, Phillips MD, Tyler DM, Duan H, Chou Y, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3’ UTR evolution. Nature Structural & Molecular Biology. 2008;15(4):354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature Reviews Immunology. 2009;9(8):581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran S, Palanisamy V. Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdisciplinary Reviews RNA. 2012;3(2):286–93. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Science Signaling. 2009;2(100):1–11. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 26.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Research. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Current Opinion in Lipidology. 2012;23(2):91–7. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, Calin GA. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PloS One. 2009;4(10):e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent J-L, de Mendonca A, Cantraine F, Moreno R, Takala J, Suter PM, Blecher S. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units. Critical Care Medicine. 1998;26(11):1793–800. doi: 10.1097/00003246-199811000-00016. [DOI] [PubMed] [Google Scholar]
- 30.Roderburg C, Luedde M, Vargas Cardenas D, Vucur M, Scholten D, Frey N, Luedde T. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PloS One. 2013;8(1):e54612. doi: 10.1371/journal.pone.0054612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Yu M, Yu G, Bian J, Deng X, Wan X, Zhu K. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochemical and Biophysical Research Communications. 2010;394(1):184–8. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Wang H-C, Chen C, Zeng J, Wang Q, Zheng L, Yu H-D. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Experimental and Therapeutic Medicine. 2013;5(4):1101–1104. doi: 10.3892/etm.2013.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PloS One. 2012;7(6):e38885. doi: 10.1371/journal.pone.0038885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Huang W, Yang Y, Wang Y, Peng T, Chang J, Fan G-C. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochimica et Biophysica Acta. 2014;1842(5):701–11. doi: 10.1016/j.bbadis.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Y, Vilanova D, Atalar K, Delfour O, Edgeworth J, Ostermann M, Lord GM. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PloS One. 2013;8(10):e75918. doi: 10.1371/journal.pone.0075918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Standage SW, Wong HR. Biomarkers for pediatric sepsis and septic shock. Expert Review of Anti-Infective Therapy. 2011;9(1):71–79. doi: 10.1586/eri.10.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Meng K, Chen WJ, Feng D, Jia Y, Xie L. Serum miR-574-5p: a prognostic predictor of sepsis patients. Shock. 2012;37(3):263–7. doi: 10.1097/SHK.0b013e318241baf8. [DOI] [PubMed] [Google Scholar]
- 38.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, Feinberg MW. MicroRNA-181b regulates NF-κB–mediated vascular inflammation. Journal of Clinical Investigation. 2012;122(6):1973–90. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Dalli J, Chiang N, Baron RM, Quintana C, Serhan CN. Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators. Immunity. 2013;39(5):885–98. doi: 10.1016/j.immuni.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tacke F, Roderburg C, Benz F, Cardenas DV, Luedde M, Hippe HJ, Luedde T. Levels of Circulating miR-133a Are Elevated in Sepsis and Predict Mortality in Critically Ill Patients. Critical Care Medicine. 2014;42(5):1096–104. doi: 10.1097/CCM.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 41.Wu SC, Yang JCS, Rau CS, Chen YC, Lu TH, Lin MW, Hsieh CH. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PloS One. 2013;8(10):e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Sun Z, Yan W, Zhu Y, Lin Y, Chen J, Wang J. Identification of microRNA as sepsis biomarker based on miRNAs regulatory network analysis. BioMed Research International. 2014;2014:594350. doi: 10.1155/2014/594350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145–51. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, Akira S. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nature Immunology. 2008;9(6):684–91. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. Journal of Molecular Cell Biology. 2011;3(3):159–66. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunology. 2010;11(5):373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 47.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. The New England Journal of Medicine. 2003;348(2):138–50. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 48.Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. The Journal of Biological Chemistry. 2007;282(39):28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The role of MicroRNAs miR-200b and miR-200c in TLR4 signaling and NF-κB activation. Innate Immunity. 2012;18(6):846–55. doi: 10.1177/1753425912443903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X, Huang X. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. Journal of Cellular Biochemistry. 2014;115(5):919–27. doi: 10.1002/jcb.24734. [DOI] [PubMed] [Google Scholar]
- 51.Wei J, Huang X, Zhang Z, Jia W, Zhao Z, Zhang Y, Xu G. MyD88 as a target of microRNA-203 in regulation of lipopolysaccharide or Bacille Calmette-Guerin induced inflammatory response of macrophage RAW264.7 cells. Molecular Immunology. 2013;55(3-4):303–9. doi: 10.1016/j.molimm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Taganov KD, Boldin MP, Chang K, Baltimore D. NF-κB-dependent induction of micro-RNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. PNAS. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(28):11499–504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu Z. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nature Immunology. 2010;11(9):799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Mor G. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27(34):4712–23. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng X, Wang H, Ye S, Guan J, Tan W, Cheng S, Zhou Y. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IκBα. PloS One. 2012;7(12):e52782. doi: 10.1371/journal.pone.0052782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. The Journal of Biological Chemistry. 2010;285(27):20940–51. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. Journal of Immunology. 2007;179(8):5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 59.Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20(6):669–76. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 60.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nature Immunology. 2010;11(2):141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 61.Zschiedrich I, Hardeland U, Krones-Herzig A, Berriel Diaz M, Vegiopoulos A, Müggenburg J, Herzig S. Coactivator function of RIP140 for NFkappaB/RelA-dependent cytokine gene expression. Blood. 2008;112(2):264–76. doi: 10.1182/blood-2007-11-121699. [DOI] [PubMed] [Google Scholar]
- 62.Ho PC, Chang KC, Chuang YS, Wei LN. Cholesterol regulation of receptor-interacting protein 140 via microRNA-33 in inflammatory cytokine production. FASEB Journal. 2011;25(5):1758–66. doi: 10.1096/fj.10-179267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sweeney TE, Suliman HB, Hollingsworth JW, Piantadosi CA. Differential regulation of the PGC family of genes in a mouse model of Staphylococcus aureus sepsis. PloS One. 2010;5(7):e11606. doi: 10.1371/journal.pone.0011606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haden DW, Suliman HB, Carraway MS, Welty-Wolf KE, Ali AS, Shitara H, Piantadosi CA. Mitochondrial biogenesis restores oxidative metabolism during Staphylococcus aureus sepsis. American Journal of Respiratory and Critical Care Medicine. 2007;176(8):768–77. doi: 10.1164/rccm.200701-161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher NC, Wilde JT, Roper J, Elias E. Deficiency of natural anticoagulant proteins C, S, and antithrombin in portal vein thrombosis: a secondary phenomenon? Gut. 2000;46(4):534–9. doi: 10.1136/gut.46.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, Fisher CJ., Jr. Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med. 2001;344(10):699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- 67.Moore CC, McKillop IH, Huynh T. MicroRNA expression following activated protein C treatment during septic shock. The Journal of Surgical Research. 2013;182(1):116–26. doi: 10.1016/j.jss.2012.07.063. [DOI] [PubMed] [Google Scholar]
- 68.Wang Z, Liang Y, Tang H, Chen Z, Li Z, Hu X, Ma Z. Dexamethasone down-regulates the expression of microRNA-155 in the livers of septic mice. PloS One. 2013;8(11):e80547. doi: 10.1371/journal.pone.0080547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ, Kurosawa S, Stepien D, Valentine C, Remick DG. Sepsis: multiple abnormalities, heterogeneous responses, and evolving understanding. Physiological Reviews. 2013;93(3):1247–88. doi: 10.1152/physrev.00037.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russell JA, Walley KR. Update in sepsis 2012. American Journal of Respiratory and Critical Care Medicine. 2013;187(12):1303–7. doi: 10.1164/rccm.201303-0567UP. [DOI] [PubMed] [Google Scholar]