Abstract
The assembly of two covalently linked monomers into dimeric complexes is a prerequisite for metabotropic glutamate receptor 1 (mGluR1) function. The former concept of a strictly homodimeric subunit contribution in metabotropic glutamate receptor complexes has recently been brought into question. Alternative splicing of the GRM1 gene results in expression of variants that vary within their intracellular C-termini. Here we bring evidence that the short mGluR1b variant is found preferentially in a complex with the long mGluR1a variant in the rodent brain. The mGluR1a and mGluR1b variants distribution overlaps in Purkinje cells and the two variants colocalize in their spines. However mGluR1a and mGluR1b show distinct sub-cellular localization when expressed alone in neurons. We discovered that trafficking of mGluR1b to distal dendrites is reliant on its association with mGluR1a and that the long C-terminus of mGluR1a within the mGluR1a/b dimer is necessary for trafficking of the complex.
Keywords: G-protein coupled receptors (GPCR), metabotropic glutamate receptors, alternative splicing, protein-protein interactions, trafficking
1. Introduction
Alternative splicing is tightly regulated spatially and temporally and governs the properties of many synthesized proteins, including several neurotransmitter receptors, both metabotropic and ionotropic. The distinct protein variants often have unique and functionally relevant characteristics that are being recognized.
Alternative splicing of GRM1 gene transcripts results in the expression of at least three mGluR1 variants (a,b,d) in rodents, however the expression of variants other than mGluR1a and mGluR1b in the human brain is uncertain [1]. The mGluR1 variants are identical within N-terminal domains including the ligand binding domain and cysteine residues responsible for covalent bonds between the two subunits within the dimeric receptor complexes, followed by a cysteine-rich region and seven transmembrane domains up to short adjacent intracellular sequences including a basic RRKK motif. After alanine 887 the C-terminal sequences of the variants diversify. The long variant, mGluR1a, is characterized by a unique sequence of 313 amino acids while the short variant, mGluR1b, has only 20 exclusive residues. In the mGluR1b variant, the RRKK motif is responsible for endoplasmic reticulum retention when this subunit is expressed alone [2]. An undetermined sequence within the extensive C-terminus of mGluR1a masks its own retention signal.
Previously, we reported that in transfected HEK293 cells, mGluR1a and mGluR1b readily combine in mGluR1a-mGluR1b (mGluR1a/b) complexes. In these mGluR1a/b dimers, the single long C-tail of mGluR1a neutralizes the RRKK motif from its own poplypetide chain, and also that of the adjacent mGluR1b. Thus the mGluR1a/b complexes are readily trafficked to the cell membranes [2]. However, the situation within neurons is more complex due to compartment specific distribution of the receptors and requirement of trafficking these to distal dendrites.
There is accumulating evidence for association of the two splice variants in vivo. The indications include the overlapping pattern of mGluR1a and mGluR1b distribution in the neurons of the central nervous system (CNS) [3] and also the observation that mGluR1b distribution might be dependent on association with the mGluR1a variant [4]. However, direct evidence has not yet been shown. This prompted our interest to understand the targeting of the mGluR1b variant in tandem with mGluR1a in neurons.
Targeting of receptors and signaling molecules to the correct subcellular compartment is essential for their specific functions in inter-neuronal communication. Several motifs within the mGluR1a C-terminus are required for protein interactions with trafficking partners that are involved in sorting of the newly synthesized molecules to dendrites and their clustering at postsynaptic portions of dendritic spines [5]. Our aim was to determine whether the C-tail of the mGluR1a would also administer the distribution of the mGluR1a/b complexes in neurons. In addition to biochemical and immunohistochemical studies of native tissue we transfected cortical neurons to study trafficking of the variants alone and in tandem.
2. Material and methods
Chemicals were obtained from Sigma-Aldrich (Prague, Czech Republic), serum, culture media and other solutions used for cell culture were from Gibco BRL (Prague, Czech Republic), unless indicated otherwise. For the histological studies and for primary neuronal cultures preparations animals were treated in accordance to local applicable laws and all efforts were made to minimise animal suffering, to reduce the number of animals used, and to utilise alternatives to in vivo techniques, if available.
2.1. Antibodies used
For mGluR1a variant detection we used the following antibodies: monoclonal mouse anti-mGluR1a (mono a-1a) (BD Transduction Laboratories) and guinea pig polyclonal anti-mGluR1a C7 (GP a-1a); the homemade antibodies were raised against synthetic peptide corresponding to epitope derived from the extreme mGluR1a carboxyl-terminus (CTPPNVTYASVILRDYKQSSSTL-COOH) linked to thyroglobulin. The short mGluR1b subunit was detected using antibodies generated against a peptide corresponding to the extreme C-terminal mGluR1b sequence (KRQPEFSPSSQCPS AHAQL-COOH) linked to thyroglobulin. These were homemade mouse monoclonal anti-mGluR1b 1D7.F11.D10 (mono a-1b), guinea pig polyclonal anti-mGluR1b O13 (GP a-1b) and rabbit polyclonal anti-mGluR1b D4 (RB a-1b) antibodies [2]. Mice monoclonal antibodies PAN-mGluR1 2E2.0.G4 (mono a-1PAN) that stain both variants equally were raised against common sequence derived from N-terminal portion of mGluR1 (ASSQRSVARMDGDVII) linked to thyroglobulin. The antibodies were employed based on their performance: GP a-1a and GP a-1b for immunoprecipitations; mono a-1a, mono a-1b and mono a-1PAN for the protein detection on immunoblots; mono a-1a in combination with RB a-1b were used for immunohistochemistry co-localization studies and electron microscopy analysis as shown in Figs. 2 and 3. Other primary antibodies used in these studies were: polyclonal rabbit anti-HA (Sigma-Aldrich), monoclonal mouse anti-cMyc (Covance) and chicken anti-MAP2 (Millipore). Primary antibodies were used either as supernatants in case of monoclonal antibodies, crude sera, or where indicated purified on ProSep-vA High Capacity columns (Millipore). Antibodies that were used in this study for the first time were characterized on immunoblots using HEK293 cells transfected with plasmids coding for mGuR1a and mGluR1b variants to verify their specificity and to exclude cross-reactivity towards the variants.
Figure 2. Immunohistochemistry of cerebellar cortex.
a) 4 μm sagittal sections of Rat cerebella stained with anti-mGluR1a in the red channel (left panel) and anti-mGluR1b in the green channel (middle panel). Right panel shows co-localization as a merger (Yellow). b) Purkinje cells details enlarged from panel merge of anti-mGluR1a and anti-mGluR1b stain together with DAPI stain from the selected areas as indicated by asterisks. ML-molecular layer, GC-granular layer, PC-Purkinje cells. Scale bar is 20μm.
Figure 3. Subcellular localization of mGluR1a and mGluR1b in Purkinje cell spine synapses.
Immunogold localization of 5 nm gold (arrows) anti-mGluR1b antibody and 15 nm gold (arrowheads) for anti-mGluR1a antibody at parallel fiber/Purkinje cell spine synapses in the cerebellar molecular layer from two P37 male rats. Labelling can be seen commonly in the perisynaptic region as well as on the postsynaptic membrane of the synapse. Colocalization of 5 and 15 nm gold is seen in the five right micrographs. Pre-presynaptic terminal. Asterisks indicate the approximate centers of the postsynaptic densities. Scale bar is 100 nm.
2.2. Detergent solubilization and co-immunoprecipitation
20 μl of Protein A/G beads (Thermo Scientific) slurry was incubated with 1 μl GP a-1a or GP a-1b sera overnight. Next day rat cerebella or transfected HEK293 cells were homogenized at 5 μg/μl total protein in buffer A (100 mM NaCl, 20 mM Tris; pH 7.4; with protease inhibitor cocktail tablet (Roche)). Sodium dodecyl-sulfate (SDS) was added to a final concentration of 1%, kept at 37°C for 1hour and centrifuged at 100,000x g for 1 hr. A sample of the pellet was kept as a control and the supernatant diluted 1:1000 with buffer A with 0.1% SDS, 0.1% sodium deoxycholate (DOC) and 0.1% Triton X-100. 1 ml of the diluted sample was mixed with the A/G beads with bound antibody and incubated at 4°C for 3-4 hrs while rotating. After brief centrifugation at 5,000x g, the supernatant (unbound fraction) was transferred to 4x volume of cold acetone and incubated at −20°C overnight. The precipitate was harvested by centrifugation at 15,000x g for 20 min. The beads with bound fraction were washed 3 times in buffer A including 0.1% SDS, 0.1% DOC and 0.1% Triton X-100; the samples were then incubated with 100 μl of SDS-PAGE treatment buffer (0.25 M Tris-Cl, 8% SDS, 20% glycerol, 0.02% bromophenol blue, 0.04 M DTT, pH 6.8) for 10 min at 60°C.
2.3. SDS-PAGE and Immunoblot analysis
Samples of the supernatants or pellets corresponding to 2 μg of protein from the total brain homogenate samples were loaded onto a SDS polyacrylamide tris-glycine gradient gel (4-20%) and proteins were separated electrophoretically. Resolved proteins were either stained in gel using Coomasie blue, or transferred to a nitrocellulose membrane. Following overnight blocking in 5% blotting-grade powdered milk (Roth), membranes were washed 3× 10 min in phosphate buffered saline-tween (136.9 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4, 8.1 mM Na2HPO4, 0.1% Tween 20) (PBST). Membranes were then incubated with mono a-1a 1:5000, mono a-1b 1:1000 or mono a-1PAN 1:1000 for 2 hrs at 4°C while agitating. After 3× 10 min washes in PBST, membranes were incubated with goat anti-mouse IgG-HRP (Santa Cruz Biotechnology), 1:5000 for 2 hrs at 4°C. Blots were then washed 3×10 min in PBST and visualized using SuperSignal™ West PICO chemiluminescent substrate system (ThermoScientific) and detected either on LAS-300 system (Fujifilm) or by X-ray film detection (Medical X-ray Film 13× 18 cm, Kodak).
2.4. Subcellular fractionation
Subcellular fractionation was done according to [6] and synaptic membrane preparation was adapted from [7]. Briefly, all samples, solutions and glassware were kept on ice or at 4°C during centrifugation. Samples from each fraction were stored at −80°C until processed. Deep-frozen rat brains (−80°C) were thawed on ice in 10 ml of 0.32 M sucrose in buffer A and homogenized with a chilled glass homogenizer. Brain homogenate was spun for 15 minutes at 1,000x g resulting in P1 pellets and S1 supernatant. The supernatant (S1) was further centrifuged for 20 minutes at 10,000x g at 4°C. Supernatant (S2) was centrifuged for 30 minutes at 12,000x g to obtain P3 pellets and supernatant S3. The supernatant was further centrifuged for 2 hrs at 140,000x g resulting in supernatant S4 and pellet P4. P4 was resuspended in 0.32 M sucrose and used in a discontinuous gradient of 0.8 M, 1 M, 1.3 M and 2 M, and centrifuged for 16 hours at 97,000x g. Then each of the interface microsomal fractions /2/, /3/ and /4/ were collected. The pellets (P2) were washed 3 × 0.5 ml of 0.32 M sucrose in buffer A and spun for 20 minutes at 10,000x g. Washed pellets (P2) were dissolved in 1 ml of buffer A and homogenized. This P2 homogenate was mixed with concentrated sucrose to a resulting concentration of 1.3 M and a discontinuous gradient was made by an overlay of 0.8 M sucrose and thin 0.32 M sucrose layer and spun for 20 minutes at 60,000x g. The synaptosomal fraction /5/ was collected from the interface between the 0.8 M and 1.3 M fractions.
2.5. Immunohistochemistry
Adult male Wistar rats were anesthetized using a mixture of Narketan (Vetoquinol) and Rometar (Spofa). Animals were then trans-cardially perfused with 0.9% saline solution followed by 4% Paraformaldehyde (PFA). Following fixation, the brains were removed and placed in 4% PFA and further fixed overnight, cut along the sagittal plane, dehydrated, embedded into paraffin blocks and stored at 4°C until required. Sagittal sections (4 μm) were made and placed directly onto Superfrost™ Ultra plus adhesion slides (Thermo Scientific). Following drying overnight, paraffin was removed and samples were rehydrated in descending alcohol concentrations. Antigen retrieval was achieved by boiling slides in 10 mM sodium citrate for 25 min in a pressure cooker. Slides were then blocked for 1 hour in the following solution: 2% goat serum, 1% bovine serum albumin (BSA), 0.1% Triton X-100, 0.05% Tween 20 and 0.1% gelatin in 1x Tris-buffered saline pH 7.4 (TBS). Incubation of slices in primary antibodies was done at 4°C overnight. Concentrations of primary antibodies used for immunohistochemistry were: 1 mg/ml of mono a-1a and 5 mg/ml purified RB a-1b. The secondary antibodies Alexa Fluor 488 goat anti-mouse IgG (H+L; Molecular Probes), and Cy3-conjugated AffiniPure donkey anti-rabbit (H+L; Jackson ImmunoResearch) were used at a dilution of 1:500 and incubated for 1 hour at room temperature in the dark. Slides were sealed using Fluoroshield + DAPI. All sections were imaged using the Leica DM 6000 upright microscope and the Leica Application Suite Advanced Fluorescence software.
2.6. Electron microscopy
Postembedding immunogold labeling of the cerebellum was performed using established materials and methods [8-10]. Fixed tissue sections from two P37 Sprague-Dawley rats were cryoprotected and frozen in a Leica CPC using liquid propane, then embedded in Lowicryl HM-20 resin in a Leica AFS freeze-substitution instrument. After blocking with normal goat serum, sections were incubated with the rabbit RB a-1b and mono a-1a antibodies. Following incubation with 5 and 15 nm gold-conjugated secondary antibodies, sections were stained with uranyl acetate and lead citrate.
2.7. Neuronal cell culture, transfection and immunofluorescence labelling
Rat embryonic cortical neurons E18 were isolated from Wistar rats and cultured as described [11]. The cell isolation was done in buffer HBSS 1x supplemented with 10 mM HEPES (pH 7.0) and 1x Trypsin using Pasteur pipettes for tissue disruption. Cells were settled on poly-L-lysine coated cover slips (18 mm diameter) in a density of approximately 750 cells/mm2. Young cortical neurons up to 7 days in vitro (DIV) were cultured in the presence of 125 μM glutamate and 0.5 mM L-glutamine in Neurobasal medium supplemented with 1x B27 (Invitrogen), and antibiotic mix (Gibco) and older without glutamate. At 5 DIV cultures were transfected using the calcium-phosphate method [12]. Transfection medium NMEM-B27 consisted of MEM (Life Technologies/Invitrogen) supplemented with 1 mM sodium pyruvate, 15 mM HEPES (Invitrogen), 2 mM L-glutamine, 1x B27 supplement (Invitrogen) and 10 mM glucose (pH 7.76). The transfection buffer was 2x BES-buffered saline (2x BBS) consisting of 50 mM BES, 1.5 mM Na2HPO4, 280 mM NaCl with optimal pH 7.0. The HBSS wash buffer (pH 7.3) included: 135 mM NaCl, 20 mM HEPES, 4 mM KCl, 1 mM Na2HPO4, 2 mM CaCl2, 1 mM MgCl2, and 10 mM glucose. All media (except 2x BBS and 2.5 M CaCl2) were pre-incubated at 37°C at least 1hr before transfection. Cover slips with neuronal cultures were transferred to new culture dishes with pre-warmed NMEM-B27 and undergone transfection procedure as described [12]. After that each cover slip was returned back to the culture dish with cultivation medium and incubated in 37°C for 3-4 days.
Neurons were fixed usually before 9 DIV by incubation in fresh 3% paraformaldehyde solution for 13 minutes. They were then permeabilized using a 0.01 M Tris, 0.025% Triton X-100, 0.025% Tween 20 and 1% BSA (pH=7,4) solution for 10 minutes. Primary and secondary antibodies were diluted in the same buffer: rabbit anti-HA (Sigma-Aldrich) 1:200, mouse anti-c-Myc (Covance) 1:500 and chicken anti-MAP2 (Millipore) 1:500. Secondary antibodies Alexa Fluor 488 goat anti-rabbit IgG (H+L; Molecular Probes), Cy3-conjugated AffiniPure donkey anti-rabbit and anti-mouse IgG (H+L; Jackson ImmunoResearch), and Alexa Fluor 633 goat anti-chicken IgG (H+L; Molecular Probes) were used according to manufacturers’ recommendations. FluoromountG (Fisher Scientific) was used to mount the slides.
2.8. Image acquisition and processing
Microscope slides were scanned on an inverted Leica confocal microscope (Leica DMI6000 with confocal superfast scanner Leica TCS SP5 AOBS TANDEM) using a 20x objective (numerical aperture 0.7). Deconvolution was performed using Huygens Software. Point spread function and chromatic aberration of objective was determined by measurement of 100 nm beads.
3. Results
3.1 Antibody characterization
Antibodies that were newly developed for these studies (GP a-1a, mono a-1b and mono a-1PAN) were characterized on immunoblots of extracts from HEK293 cells transfected with mGluR1a or mGluR1b variants separated on SDS-PAGE (Fig. 1a). Antibodies GP a-1b, RB a-1b mono and mono a-1a were used and characterized previously [2, 13].
Figure 1. Immunochemical analysis of association of mGluR1 splice variants.

a) Antibody characterization. Transfected HEK293 cells with either mGluR1a (lane A) or mGluR1b (lane B) coding expression vectors were resolved on SDS-PAGE and stained using the indicated antibodies. b) The GP anti-mGluR1a variant-specific antibodies were used for immunoprecipitation of proteins from detergent soluble fraction of cells co-expressing both subunits (line AB) or extracts from cells expressing the variants in separate single transfections and mixed prior to solubilization (line A+B). The immunoblot was visualized using mono-PAN antibodies. c) Co-immunoprecipitation of the mGluR1a and mGluR1b variants from the SDS soluble fraction from rat brain homogenates. d) Subcellular fractionation. Proteins from fractions of gradient centrifugation as described in methods. 1= S1 fraction, homogenate without nuclear pellet, 2,3,4= microsomal fractions, 5= synaptic membrane fraction.
STAIN: primary antibodies used for visualization of proteins; IP: Antibodies used for immunoprecipitation and staining: 1a= anti-mGluR1a, 1b= anti-mGluR1b. B= bound to the antibodies, U= unbound fraction. Arrows mark expected position of mGluR1a homodimer (a/a); mGluR1a/b heterodimer (a/b); and monomers after partial disruption of the dimers in the treatment buffer upon DTT mediated reduction: mGluR1a (a) and mGluR1b (b). Molecular weight markers, from bottom: 100,000; 180,000, 250,000 and 300,000 Da.
We also tested performance and specificity of the novel GP a-mGluR1a antibodies in immunoprecipitation. The HEK293 cell were transfected either with both variants together (AB sample), or they were expressing each variant alone and mixed prior to solubilization (A+B sample) (Fig. 1b). The precipitate from the SDS-soluble fraction from AB samples contained the mGluR1b subunit arising from the mGluR1a/b dimer, while in the precipitate from A+B sample we detected only mGluR1a subunit. Previously, the GP a-mGluR1b sera were characterized in the same way [2].
3.2. Immunoprecipitation of SDS soluble receptors from brain
Next we aimed at immunoprecipitation of the mGluR1 receptor complexes with the splice-variant specific antibodies, either anti-mGluR1a or anti-mGluR1b from rat brain SDS soluble fraction. Detergent solubilization of membrane proteins is often incomplete when mild conditions are required for the preservation of structure or function. In this study our aim was the opposite, to break any non-covalent interactions between the polypeptides of interest, because subunit composition of metabotropic glutamate receptor complexes that are formed by dimers made of covalently linked subunits was analyzed. In line with this notion SDS was employed for mGluR1 solubilization from rat brain homogenate. On top of excellent yields, this strong ionic detergent is known to disrupt non-covalent protein-protein interactions. The detergent-soluble fraction was next diluted and neutralized by addition of detergents compatible with immunoprecipitation.
The resolved precipitate was detected on immunoblot with complementary antibodies or antibodies directed against the same polypeptide, but made in different species (Fig. 1c). Clearly, the anti-mGluR1a did pull-down majority of the mGluR1b subunit. Thus, major portion of the mGluR1b protein was associated with the mGluR1a presumably within the mGluR1a/b complexes and only a relatively low fraction of mGluR1b remained in the unbound supernatant when the anti-mGluR1a antibodies were employed for the immunoprecipitation. In the opposite situation, the anti-mGluR1b antibodies did pull-down not only the mGluR1b, but also mGluR1a. Visible band migrating in velocity corresponding to that estimated for mGluR1a/b complexes (from dimers that did not disassemble on the gel upon reducing conditions within the SDS-PAGE gel loading buffer) was detected with both, anti-mGluR1a and anti-mGluR1b antibodies on immunoblots in the anti-mGluR1b precipitate. On the other hand, no band corresponding to mGluR1a homodimers was detected when anti-mGluR1b antibodies were employed for immunoprecipitation and anti-mGluR1a antibodies for detection on immunoblots. These data suggest that the mGluR1b protein is found preferentially in complexes with mGluR1a in the CNS.
3.3. Subcellular fractionation
Rat brain homogenate was used to further separate distinct sub-cellular compartments into fractions with a high content of microsomes (comprising mostly Golgi and endoplasmic reticulum (ER) membranes) and into synaptosomal fraction enriched with synaptic membranes (Fig. 1d). The homodimers made of mGluR1a and mGluR1a/b heterodimers were detected in all fractions, with relatively higher levels detected in the synaptic membranes. In the fractions that have enrichment of the early trafficking membranes and corresponding sub-cellular particles, we were able to detect mGluR1b homodimers; these were almost absent from the synaptosomal fraction. It is possible, that a portion of mGluR1b does form homodimers in the early proteo-synthetic pathways and these are retained within the ER and destined for proteolysis and not trafficked to the synapses.
3.4. Co-localization of mGluR1a and mGluR1b in cerebellum
Using the immunohistochemistry technique we analyzed the co-localization of mGluR1a and mGluR1b. In order to shed more light on the association of the two variants in vivo, we aimed at selecting cells with relatively high and comparable levels of expression of the studied polypeptides in anatomically well distinguishable cells. We chose to study the mGluR1 receptor splice variant relations in the Purkinje cells of the cerebellum because they express both mGluR1a and mGluR1b splice variants and also the function of mGluR1 has been extensively studied in these neurons. Moreover, immunohistochemical studies involving co-labelling with both mGluR1a and mGluR1b variants have not been presented so far in these cells. As shown in Fig. 2, we observed labelling of the cerebellum with our antibodies directed against mGluR1a similar as was shown previously [13]. Importantly, mGluR1b had a comparable distribution as mGluR1a. The mGluR1a and mGluR1b proteins were found to overlap in both the cell bodies and dendrites of the Purkinje cells as well as a population of interneurons. This result supports the probable co-trafficking of the two splice variants; however whether they co-localize at the post-synaptic membrane could only be determined using the electron microscopy technique.
3.5. Co-localization of mGluR1a and mGluR1b on Purkinje cell spines
We performed electron microscopy immunogold co-localization studies of mGluR1a and mGluR1b, using double labelling with 15 nm and 5 nm gold secondary antibodies, respectively (Fig. 3). The distribution of mGluR1a at Purkinje spine synapses was identical to that described previously [9, 13]. Basically, labelling of the spine synapses was found on the postsynaptic membrane within the synaptic cleft, within the perisynaptic region comprising a ring of about 100 nm around the active zone, as well as on other areas of the extrasynaptic membrane of the spine. However, labelling tended to concentrate in the perisynaptic region and the lateral parts of the postsynaptic membrane [14]. Labelling for mGluR1b was similar to that seen for mGluR1a, including labelling on the postsynaptic, perisynaptic and extrasynaptic areas of the spine, and again being particularly prevalent in the perisynaptic region [3]. In some cases, co-localization of gold labelling of mGluR1a and mGluR1b was seen on the same spine, with 15 and 5 nm gold particles occurring very close to each other.
3.6. Trafficking of mGluR1 variants in neurons
Primary cultures of cortical prefrontal neurons were chosen for transfection of mGluR1 variants in this study for two reasons. Firstly they have low endogenous expression of mGluR1 that was found not to interfere with our experiments, a situation that may not be probable in case of neurons with higher levels of endogenous receptors e.g. Purkinje neurons (see above and Fig. 2) [13]. Thus, we chose cortical prefrontal neurons because the endogenous levels of mGluR1 are low, but not absent, and therefore we expect that regulatory, trafficking and scaffolding molecules involved in the mGluR1 processing are present in these cells.
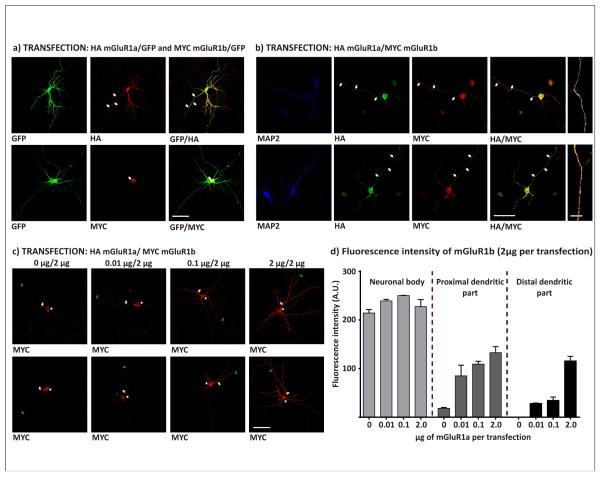
The differently tagged splice variants mGluR1a and mGluR1b were transfected into these neurons either alone, or in tandem (Fig. 4a and 4b; respectively). Green Fluorescent protein (GFP) was employed as a cell volume marker. The mGluR1a variant (labelled with HA tag) alone was readily trafficked into proximal and distal dendrites. In contrast, when expressed alone, MYC-mGluR1b was restricted mainly to the somatic compartment, and in certain cases, with low staining in proximal dendrites (Fig. 4a). However, mGluR1b distribution changed dramatically when co-transfected with mGluR1a. When both mGluR1a and mGluR1b were expressed in single neuron, the variants distribution was clearly overlapping not only within cell bodies but also in dendrites, including their distal portions. The mGluR1b delivery to distal dendrites is therefore dependent on co-trafficking with the mGluR1a (Fig. 4b).
Figure 4. Distribution of mGluR1a or mGluR1b in transfected cortical neurons.
Cells from primary prefrontal cortical neurons were transfected a) with GFP and corresponding single tagged subunits and stained 72 hours later against HA and MYC epitopes or visualized using GFP fluorescence. UPER ROW: Left panel: GFP; middle panel: HA-mGluR1a detected using anti-HA antibodies; right panel-merge. LOWER ROW: left panel: GFP; middle panel MYC-mGluR1b detected using anti-MYC antibodies, right panel: merge; b) Two primary cortical neurons co-transfected with HA-mGluR1a (Green) and MYC-mGluR1b (Red); right panel: merge; dendritic marker MAP2 shown in left panel in blue. Co-localization HA-mGluR1a and MYC-mGluR1b in MAP2 positive distal dendrites is shown in extreme right in detail as merge for green and red channels (HA-mGluR1a and MYC-mGluR1b, respectively). c) Neurons were transfected with stable amounts of MYC-mGluR1b coding plasmid (2μg per transfection) and increasing amounts of HA-mGluR1a plasmid (from left to right: 0 μg; 0.01 μg; 0.1 μg and 2 μg). MYC-mGluR1b was detected using anti-MYC antibodies. Asterisk, yellow and green arrows show regions where fluorescence was measured. Scale bars are 20 μm. d) Fluorescence of selected areas from 3 neurons for each transfection within neuronal cell bodies and areas from proximal and distal dendrites. (A.U.: Arbitrary Units)
To further explore reliance of mGluR1b trafficking to dendrites on association with mGluR1a we expressed the variants in different ratios between mGluR1a and mGluR1b (0:1, 0.02:1, 0.2:1 and 1:1). Clearly, trafficking efficiency of mGluR1b from the cell soma to the dendrites improved with increasing amounts of the mGluR1a protein expressed. This observation was possible to quantify as fluorescence of the MYC-mGluR1b signal in the soma, and proximal and distal dendrites of the transfected neurons (Fig. 4c, d). The cells expressing low amounts of mGluR1a had only a little of the mGluR1b subunit delivered to their dendrites in contrast to cells in which mGluR1a has been expressed in higher levels.
4. Discussion
Dimerization is a prerequisite for Class C GPCR function [15-18]. Heterodimerization is well documented and recognized as a receptor formation requirement for GABAb and sweet and umami taste receptors [19]. In the case of mGluRs the concept (until recently) was that dimeric complexes are assembled as homodimers. Our initial studies revealed the possible formation of receptor complexes containing short and long mGluR1 splice variants in heterologous expression systems [2]. Later it was shown that distinct mGluRs may combine in receptor complexes and that dimer formation between Group I mGluRs, mGluR1 and mGluR5, or association between subunits that fall into Groups II and III is possible in heterologous systems [20]. Heterodimerization between the mGluR2 and mGluR4 in vivo in complexes with novel pharmacological properties was reported [21]. This finding was confirmed recently using novel allosteric modulators acting on the mGluR2/4 heterodimers [22]. Here we bring evidence that endogenously expressed mGluR1a and mGluR1b variants assemble into dimeric receptor complexes mGluR1a/b and that this association affects the trafficking of the variants in neurons, namely on the mGluR1b delivery to the dendrites.
First we used the SDS soluble fraction from rat brain tissue homogenates for immunoprecipitation. This detergent solubilization protocol disrupts non-covalent protein-protein interactions but does not disturb covalently linked mGluR1 subunits associated within a single receptor complexes. Therefore, only the mGluR1a covalently linked to mGluR1b would co-precipitate in further manipulation of the SDS-detergent soluble fraction. Indeed, we detected co-precipitation of mGluR1a with mGluR1b and also mGluR1b with mGluR1a. The use of strong ionic detergent prior to immunoprecipitation minimizes the possibility of pulling down mGluR1a dimers with mGluR1b dimers. Not only would oligomeric formation of non-covalently interacting complexes be disrupted by our detergent solubilization procedure, also, formation of higher oligomeric complexes is unlikely in the case of the mGluRs [20].
Subcellular fractionation revealed that mGluR1b homodimers are detected in relatively low amounts, so that a rather minor portion of mGluR1b was incorporated into homodimers when compared with the amounts integrated into mGluR1a/b dimers. These marginal mGluR1b homodimers are almost entirely excluded from the synaptosomal fraction. The mGluR1a/b heterodimer was the major form of mGluR1b detected, including in synaptic membrane fractions, where it was enriched over the relative amounts found in the microsomal fractions. Our explanation for this is that mGluR1b homodimers are probably not trafficked in the same way as mGluR1a/b dimers, but are rather destined for degradation after retention within the ER.
For several ionotropic and metabotropic glutamate receptors splicing alters the targeting properties by introducing, masking or unmasking the endoplasmic reticulum (ER) retention sequence, which in most instances is a stretch of basic amino acid residues [2, 23-25]. Retention of the nascent polypeptide within the ER and its release by an interacting subunit assures quality control of proper formation of the oligomeric receptor complexes. Our results suggest that mGluR1b trafficking from the ER within the mGluR1a/b dimer is reminiscent of that for GB1 subunit dimerization with GB2 during GABAb receptor assembly [25]. The short mGluR1b C-tail basic amino acid residue motif, RRKK, most likely mediates interaction with the coat protein I complex (COPI). ER retention then may be an important checkpoint for mGluR1a/b heterodimer formation. Within the mGluR1a/b heterodimer, the single long mGluR1a tail is dominant, masks the effect of RRKK motifs on both C-tails and governs trafficking of the mGluR1a/b dimer to distal dendrites.
Immunohistochemical staining using antibodies specific for mGluR1a and mGluR1b show that the two splice variants have overlapping distributions, especially in Purkinje cells in the cerebellum. This option was already discussed beforehand by authors of a careful study that revealed an mGluR1b subcellular distribution pattern reminiscent of that of mGluR1a [3]. In our studies, electron microscopy with a double labelling strategy for the first time allowed the side by side detection of the two variants in discrete perisynaptic membrane regions of dendritic spines.
As our results indicate that mGluR1b distribution overlaps with that of mGluR1a in vivo, next we addressed the question of the assembly of the mGluR1a/b dimers in neurons and its consequence on targeting of such heterodimer. When we transfected primary cortical neurons with the mGluR1a subunit, it was readily trafficked to distal dendrites in accordance with the anticipated distribution [13]. The striking difference between the distribution of the mGluR1b variant in neurons when expressed alone, and of that with co-transfected mGluR1a is apparent. The mGluR1b expressed without the mGluR1a was retained mainly in the soma and only a small portion was found in dendrites. However, when the mGluR1b subunit was co-expressed with mGluR1a, they co-localize, even in distal dendrites. The pattern of distribution of both variants was reminiscent of that of mGluR1a in neurons when this subunit was expressed alone. The rate of mGluR1b trafficking to distal dendrites did correlate with the expression levels of the mGluR1a protein. Higher levels of mGluR1a co-expression resulted in elevated amounts of mGluR1b transported from neuronal cell bodies to dendrites. Thus mGluR1b is co-transported to distal dendrites within heterodimeric mGluR1a/b complexes and these heterodimers are most likely formed already in the neuronal soma.
Distribution of mGluR1 variants was studied previously in transfected neurons [24, 26]. The authors used primary neuronal cultures with either mGluR1a or mGluR1b variants transfection, but never in tandem, possibly assuming that only homodimers form functional receptors. In a study that employed retinal neurons, the transfected mGluR1a was observed in cell bodies and in dendrites. The mGluR1b variant expressed alone showed a different distribution compared to mGluR1a, as it was excluded from dendrites and directed to axons [24]. In other studies, (including ours) the axonal targeting of mGluR1b variant in distinct neuronal cultures was not observed [26]. However, in certain neuronal populations, notably in striatal neurons, mGluR1a was observed also in the presynaptic compartments [27]. Therefore, when describing targeting properties of molecules, the specific pattern in distinct cell types has to be taken into account.
The mGluR1a/b dimers might have functional significance, as the two distinct intracellular C-termini combine in one receptor complex. The posttranslational modifications will certainly be different for the two variants C-termini. Also, the mGluR1b C-tail was reported to have distinct interacting partners from the mGluR1a [28]. On the other hand, in the mGluR1a/b dimer compared to the mGluR1a homodimer, there will be a numerical loss of motifs that confer important interactions, such as those seen with Homer proteins or Tamalin. It remains to be elucidated whether two binding motifs within a dimer are required for some of these functionally relevant interactions, e.g. the Homer proteins modulation of the mGluR1a activity [29]. It might well be that in the case of mGluR1a/b heterodimers the impact of association with such interacting molecules would be altered.
Therefore, the mGluR1a/b heterodimers described herein are expected to have some unique functional properties compared to the mGluR1a homodimers, but their relevance remains to be clarified.
Highlights.
metabotropic glutamate receptor 1 (mGluR1) is expressed in several splice variants
alternative splicing of mGluR1 results in complexes in which variants may combine
mGluR1b variant is found mainly in association with mGluR1a in mGluR1a/b complexes
long mGluR1a is required for proper mGluR1b trafficking within the mGluR1a/b complex
Acknowledgments
We would like to thank Jean-Philippe Pin from Montpellier for constant support and inspiring critical discussions and Ondrej Horvath for the help with fluorescent microscopy. These studies were supported by the Czech Science Foundation (GACR P303/12/2408) and RVO:68378050 to JB. RSP and YXW were supported by the intramural research program of NIDCD/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferraguti F, Crepaldi L, Nicoletti F. Metabotropic glutamate 1 receptor: current concepts and perspectives. Pharmacol Rev. 2008;60(4):536–81. doi: 10.1124/pr.108.000166. [DOI] [PubMed] [Google Scholar]
- 2.Kumpost J, et al. Surface expression of metabotropic glutamate receptor variants mGluR1a and mGluR1b in transfected HEK293 cells. Neuropharmacology. 2008;55(4):409–18. doi: 10.1016/j.neuropharm.2008.06.073. [DOI] [PubMed] [Google Scholar]
- 3.Mateos JM, et al. Immunolocalization of the mGluR1b splice variant of the metabotropic glutamate receptor 1 at parallel fiber-Purkinje cell synapses in the rat cerebellar cortex. J Neurochem. 2000;74(3):1301–9. doi: 10.1046/j.1471-4159.2000.741301.x. [DOI] [PubMed] [Google Scholar]
- 4.Ohtani Y, et al. The Synaptic Targeting of mGluR1 by Its Carboxyl-Terminal Domain Is Crucial for Cerebellar Function. J Neurosci. 2014;34(7):2702–12. doi: 10.1523/JNEUROSCI.3542-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock C, et al. Assembly-dependent surface targeting of the heterodimeric GABAB Receptor is controlled by COPI but not 14-3-3. Mol Biol Cell. 2005;16(12):5572–8. doi: 10.1091/mbc.E05-05-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurd JW, et al. Isolation and partial characterization of rat brain synaptic plasma membranes. J Neurochem. 1974;22(2):281–90. doi: 10.1111/j.1471-4159.1974.tb11591.x. [DOI] [PubMed] [Google Scholar]
- 7.Carlin RK, et al. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86(3):831–45. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petralia RS, et al. Subcellular distribution of patched and smoothened in the cerebellar neurons. Cerebellum. 2012;11(4):972–81. doi: 10.1007/s12311-012-0374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petralia RS, et al. Glutamate receptor targeting in the postsynaptic spine involves mechanisms that are independent of myosin Va. Eur J Neurosci. 2001;13(9):1722–32. doi: 10.1046/j.0953-816x.2001.01553.x. [DOI] [PubMed] [Google Scholar]
- 10.Petralia RS, Wenthold RJ. Immunocytochemistry of NMDA receptors. Methods Mol Biol. 1999;128:73–92. doi: 10.1385/1-59259-683-5:73. [DOI] [PubMed] [Google Scholar]
- 11.Brewer GJ, et al. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35(5):567–76. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 12.Goetze B, et al. Chemically controlled formation of a DNA/calcium phosphate coprecipitate: application for transfection of mature hippocampal neurons. J Neurobiol. 2004;60(4):517–25. doi: 10.1002/neu.20073. [DOI] [PubMed] [Google Scholar]
- 13.Petralia RS, et al. A monoclonal antibody shows discrete cellular and subcellular localizations of mGluR1 alpha metabotropic glutamate receptors. J Chem Neuroanat. 1997;13(2):77–93. doi: 10.1016/s0891-0618(97)00023-9. [DOI] [PubMed] [Google Scholar]
- 14.Baude A, et al. The metabotropic glutamate receptor (mGluR1 alpha) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron. 1993;11(4):771–87. doi: 10.1016/0896-6273(93)90086-7. [DOI] [PubMed] [Google Scholar]
- 15.Hlavackova V, et al. Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci Signal. 2012;5(237):ra59. doi: 10.1126/scisignal.2002720. [DOI] [PubMed] [Google Scholar]
- 16.Hlavackova V, et al. Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 2005;24(3):499–509. doi: 10.1038/sj.emboj.7600557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goudet C, et al. Asymmetric functioning of dimeric metabotropic glutamate receptors disclosed by positive allosteric modulators. J Biol Chem. 2005;280(26):24380–5. doi: 10.1074/jbc.M502642200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tateyama M, et al. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1alpha. Nat Struct Mol Biol. 2004;11(7):637–42. doi: 10.1038/nsmb770. [DOI] [PubMed] [Google Scholar]
- 19.Kniazeff J, et al. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol Ther. 2011;130(1):9–25. doi: 10.1016/j.pharmthera.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Doumazane E, et al. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 2011;25(1):66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- 21.Kammermeier PJ. Functional and pharmacological characteristics of metabotropic glutamate receptors 2/4 heterodimers. Mol Pharmacol. 2012;82(3):438–47. doi: 10.1124/mol.112.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin S, et al. Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J Neurosci. 2014;34(1):79–94. doi: 10.1523/JNEUROSCI.1129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petralia RS, Al-Hallaq RA, Wenthold RJ. Trafficking and Targeting of NMDA Receptors. In: Van Dongen AM, editor. Biology of the NMDA Receptor. Boca Raton (FL): 2009. [PubMed] [Google Scholar]
- 24.Francesconi A, Duvoisin RM. Alternative splicing unmasks dendritic and axonal targeting signals in metabotropic glutamate receptor 1. J Neurosci. 2002;22(6):2196–205. doi: 10.1523/JNEUROSCI.22-06-02196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagano A, et al. C-terminal interaction is essential for surface trafficking but not for heteromeric assembly of GABA(b) receptors. J Neurosci. 2001;21(4):1189–202. doi: 10.1523/JNEUROSCI.21-04-01189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das SS, Banker GA. The role of protein interaction motifs in regulating the polarity and clustering of the metabotropic glutamate receptor mGluR1a. J Neurosci. 2006;26(31):8115–25. doi: 10.1523/JNEUROSCI.1015-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paquet M, Smith Y. Group I metabotropic glutamate receptors in the monkey striatum: subsynaptic association with glutamatergic and dopaminergic afferents. J Neurosci. 2003;23(20):7659–69. doi: 10.1523/JNEUROSCI.23-20-07659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francesconi A, Kumari R, Zukin RS. Proteomic analysis reveals novel binding partners of metabotropic glutamate receptor 1. J Neurochem. 2009;108(6):1515–25. doi: 10.1111/j.1471-4159.2009.05913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ango F, et al. Agonist-independent activation of metabotropic glutamate receptors by the intracellular protein Homer. Nature. 2001;411(6840):962–5. doi: 10.1038/35082096. [DOI] [PubMed] [Google Scholar]