Abstract
Mutations in the gene encoding the single transmembrane receptor multiple epidermal growth factor-like domain 10 (MEGF10) cause an autosomal recessive congenital muscle disease in humans. Although mammalian MEGF10 is expressed in the central nervous system as well as in skeletal muscle, patients carrying mutations in MEGF10 do not show symptoms of central nervous system dysfunction. drpr is the sole Drosophila homolog of the human genes MEGF10, MEGF11, and MEGF12 (JEDI, PEAR). The functional domains of MEGF10 and drpr bear striking similarities, and residues affected by MEGF10 mutations in humans are conserved in drpr. Our analysis of drpr mutant flies revealed muscle degeneration with fiber size variability and vacuolization, as well as reduced motor performance, features that have been observed in human MEGF10 myopathy. Vacuolization was also seen in the brain. Tissue-specific RNAi experiments demonstrated that drpr deficiency in muscle, but not in the brain, leads to locomotor defects. The histological and behavioral abnormalities seen in the affected flies set the stage for further studies examining the signaling pathway modulated by MEGF10/Drpr in muscle, as well as assessing the effects of genetic and/or pharmacological manipulations on the observed muscle defects. In addition, the absence of functional redundancy for Drpr in Drosophila may help elucidate whether paralogs of MEGF10 in humans (eg, MEGF11) contribute to maintaining wild-type function in the human brain.
Muscular dystrophy is a heterogeneous group of inherited muscle diseases characterized by persistent muscle degeneration and regeneration leading to muscle wasting and loss. Recently, mutations in the gene encoding multiple epidermal growth factor–like domain protein 10 (MEGF10) were found to cause a novel autosomal recessive congenital muscle disease in humans.1–3 Patients show progressive muscle weakness with features of muscular dystrophy and congenital myopathy.1–4 MEGF10 is expressed in the central nervous system (CNS), retina, and skeletal muscle.2,5 In the brain, where it is abundantly expressed, MEGF10 is enriched in astrocytes and myelinating oligodendrocytes.6 This protein has been shown to mediate engulfment of apoptotic neurons7 as well as synapse pruning in the developing and adult CNS8 and to participate in the uptake of amyloid-β peptide.9 In addition, elegant studies in rodents have shown that MEGF10, together with MEGF11, regulates the arrangement of retinal mosaic.5 In resting muscle, MEGF10 expression is observed in myoblasts and quiescent satellite cells, where it suppresses the differentiation program.10 Many gaps remain, however, in our understanding of the signaling pathway/physiological function mediated by this receptor in muscle, as well as of the molecular consequences of the pathogenic mutations that underlie the muscle disease in humans. MEGF10 encodes a single-pass membrane protein with an N-terminus EMI domain followed by multiple extracellular EGF-like domains. These structural features, together with the intracellular noncanonical immunoreceptor tyrosine-based activation motif (ITAM) signaling motifs, have been conserved from invertebrates to humans.5,7 Previous structure–function analyses have highlighted similarities between human MEGF10 and its Drosophila (fruit fly) homolog Drpr (the corresponding gene is draper, drpr, CG2086),7,11 raising the possibility that these proteins mediate parallel functions. In addition to its well-characterized role in cell corpse engulfment12 and glial response to degenerating axons,13 Drpr mediates the remodeling of the neuromuscular junction, where it is expressed both in glia that wrap around the motor neurons as well as postsynaptically.14 Despite the phylogenetic distance between Drosophila and humans, fly models of human disease have shown potential for facilitating a better understanding of the development of the pathophysiological processes that underlie neuromuscular disorders. These include fly models of spinal muscular dystrophy, lamin-associated myopathies, actin myopathies, dystrophinopathies, and dystroglycanopathies.15–22 The identification of adult muscle precursor cells in Drosophila that share features with vertebrate satellite cells23,24 provides further rationale for using this model organism in muscle biology investigations. This study was designed to assess the extent to which drpr mutant Drosophila may be used to model skeletal muscle phenotypes with relevance to human MEGF10 myopathy. Of note, Megf10−/− mice5 do not have an obvious muscle phenotype (Peter Kang, personal communication). The characterization of drpr mutant and RNAi flies establishes a baseline on which to probe the signaling pathway modulated by MEGF10/Drpr in muscle, as well as to assess the effects of genetic and/or pharmacological manipulations on the observed muscle defects.
Materials and Methods
Drosophila Stocks and Culture
The drprΔ5 mutant fly line [genotype: w–; sp/CyOact::GFP; drprΔ5rec8 (9)/TM6, sb, Tb, e; where w–; sp/CyOact::GFP; drprΔ5rec8 (9)/drprΔ5rec8 (9) null are adult viable] and UAS-ds-drpr RNAi mutant fly line (genotype: yw; UAS-drprRNAi#7b/CyO;+/TM6, sb, Tb, e) were gifts from Marc R. Freeman (University of Massachusetts Medical School, Boston, MA).12 The UAS-ds-drpr RNAi stock transformant ID 27086 (genotype: w1118; P{GD14423}v27086, FBst0456744) was purchased from the Vienna Drosophila RNAi Center (Vienna, Austria). The genetic background strain w1118 (FBal0018186) was purchased from the Bloomington Drosophila Stock Center, as were the following Gal4 driver lines: Actin5C-Gal4 (y1 w*; P{Act5C-Gal4}25FO1/CyO, y+; FBst0004414: ubiquitous expression), how-Gal4 (w∗; P{GawB}how24B; FBst0001767: expression in mesoderm), Elav-Gal4 (P{GawB}elavC155; FBst0000458: pan-neuronal expression), and twist-Gal4 (P{GAL4-twi.G}108.4, w1; FBst0000914: expression in adult muscle precursors). The repo-Gal4 driver line (expression in glia) was a generous gift from Mary Roberts (F. Rob Jackson laboratory, Tufts University School of Medicine, Boston, MA). All strains were raised at 25°C in a 12-hour light/12-hour dark cycle on standard Drosophila media. To generate transgenic drpr RNAi flies, UAS-ds-drpr flies were crossed at 29°C with flies carrying the Gal4 transgene.
Assessment of Transcript Levels
RT-PCR was used to assess transcription levels of the drpr gene. Total RNA was isolated from pooled adult male Drosophila by RNA STAT-60 (Tel-Test, Inc., Friendswood, TX) following the manufacturer's recommendations. Complementary cDNA was generated using MuLV Reverse Transcriptase (Applied Biosystems Inc/Life Technologies Corp., Grand Island, NY). Alternatively, genomic DNA was extracted from the corresponding flies and used. PCR was performed according to standard protocols. The sequences of the drpr locus–specific primers used for the PCR reaction were as follows: drpr_2, forward, 5′-AGGACCTGGAATCCACTGC-3′; CG18171, reverse, 5′-GCGGCAAGTAATCTGAGTCC-3′; drpr_5, reverse, 5′-GCCTGAAAAGGGCTCACATA-3′; drpr_5, forward, 5′-CGGTATGTGAGCCCTTTTCA-3′; drpr_6, reverse, 5′-GCAGGTCATGCTGCAGTT-3′; CG12035_1, forward, 5′-GCTGCTTAATATCCCCAGAGG-3′; and CG12035_1, reverse, 5′-GTCGTTACTCTTGGCAATGG-3′.
Western Blot Analysis
Protein extraction and western blot analysis were performed as previously reported.25 Primary antibodies were used at the following dilutions: rabbit anti-Draper, 1:5000 (generated in the Freeman Laboratory12); and mouse anti-Discs-Large, 1:500 (clone 4F3, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA).
Lifespan Analysis
To assess lifespan, 1-day-old male adults were collected and then regularly transferred to fresh medium every 7 days. The number of dead flies was recorded at the time of each transfer. For the drpr heterozygous versus null lifespan study, raw percentage survival data were collected weekly on three populations in each group. The total numbers of flies analyzed were 83 and 70 in the heterozygous and null groups, respectively. Statistical analysis on the lifespan study was performed using a t-test in SigmaPlot version 11 software (Systat Software Inc., Chicago, IL). Data are presented as means ± SEM.
Negative Geotaxis
As an index of motor function, age-matched male adults in the control and drpr mutant groups (n = 10 to 12 flies per group) were examined using a negative geotaxis climbing assay. One-day-old flies were collected and maintained for 3 to 21 days in fresh vials before testing. Corresponding flies were transferred (without CO2 anesthesia) into a clear vial. The vial was gently tapped four times to collect flies at the bottom, thus prompting a negative geotaxis response. The number of flies crossing a 5-cm threshold during a 6-second interval was recorded. Each genotype was assessed three times consecutively. Independent groups of flies/genotype were evaluated on different days.26
Histological Analyses
Histological analyses were performed in the Department of Pathology, Tufts Medical Center. Aged-matched (30 to 40 days old) control and drpr mutant Drosophila were immersed in Telly's fixative for 7 days, embedded in paraffin, and sectioned at 5 μm using standard techniques. All sections were stained with hematoxylin and eosin, then examined using standard bright-field light microscopy.
Results
Conservation of Disease-Causing MEGF10 Mutations in Drpr
Drpr is the Drosophila homolog of human MEGF10. Sequence alignment (CLUSTAL 2.1) of the longest Drpr isoform, Drpr-PE (1042 aa, http://www.ncbi.nlm.nsih.gov/protein, accession number NP_001261276.1), and human MEGF10 protein variant a (1140 aa, http://www.ncbi.nlm.nih.gov/protein, accession number EAW62406.1) revealed a high degree of similarity within the EGF-like domains. The fly protein comprises 15 EGF-like domains12 versus 17 in the human.27 Notably, within these domains, two cysteines that are targeted by known pathogenic human gene mutations (ie, C326R and C774R)1 are conserved in Drosophila. In contrast, the elastin microfibril interface located protein (EMILIN) domain–nested arginine, which is mutated in another patient (R71W),1 was not conserved in Drosophila (an arginine is found in close proximity) (Figure 1). In addition to MEGF10, Drpr also shows homology to human MEGF11 and MEGF12 [alias platelet endothelial aggregation receptor 1 (PEAR1)/Jedi], with approximately 40% identity found in the region encompassing the EGF-like domains.
Figure 1.
Drpr is homologous to MEGF10. The highest degree of similarity is found within the EGF-like domains (gray). Residues that are mutated in disease are indicated on the right. A boxed area in the sequence denotes conservation of the wild-type residue between the fly and human counterparts. In contrast, a circled area in the sequence denotes lack of conservation.
Effects of drprΔ5 Mutant Allele on the Expression of drpr and on the Other Two Open Reading Frames Nested within the Locus
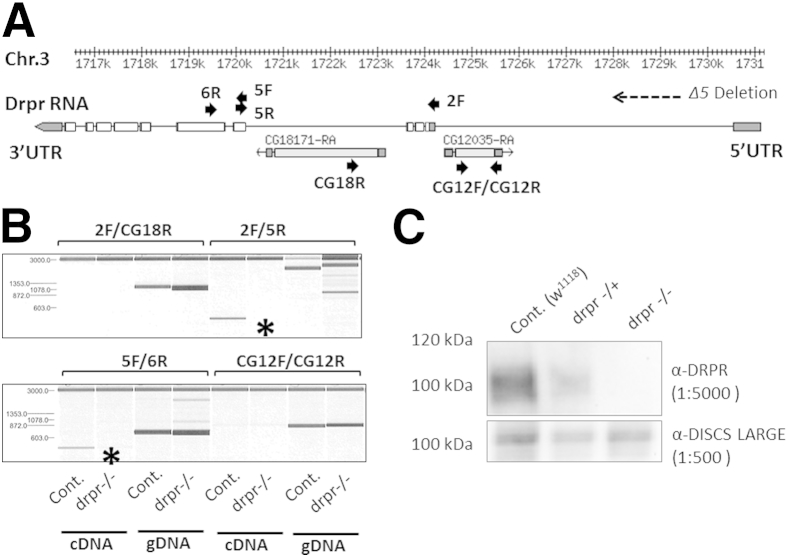
The Drosophila drpr/CG2086 gene is localized on chromosome 3 (cytolocation 3L:1715595.. 731107). Recent annotation updates of the Drosophila genome have revealed that within the drpr locus two additional genes of unknown function are found: CG18171 in cis- (ie, same strand) and CG12035 in trans- (ie, opposite strand) (Figure 2A). The Δ5 drpr mutant allele is a deletion that was generated by imprecise excision of the EP(3)522 P-element inserted in the 5′ untranslated region of the drpr gene.12 Corresponding mutant flies are commonly used to study the function of drpr. To assess whether the molecular Δ5 lesion affects the expression of the two other genes nested in the drpr locus, PCR primers were designed to amplify selected exons within this region. PCR analysis of genomic DNA as well as cDNA revealed that the drprΔ5 lesion disrupts transcription of the drpr gene but did not affect the other two genes in the locus (Figure 2B). Western blot analysis confirmed that Δ5 homozygotes did not produce Drpr, using a polyclonal antibody directed against the intracellular domain of the protein, common to all Drpr isoforms, as previously reported12,25 (Figure 2C).
Figure 2.
Molecular characterization of the drprΔ5 lesion in mutant flies. A: Schematic representation of the drpr gene locus (chromosome 3), adapted from FlyBase. Noncoding (dark gray) and coding (white/light gray) exons appear as boxes. CG18171 and CG12035 are two genes nested within the drpr locus. The arrows indicate PCR primers. B: PCR analysis of genomic DNA and cDNA reveals that the Δ5 lesion disrupts transcription of the drpr gene (asterisks). The two other genes, that is, CG12035 and CG18171, are not affected by the Δ5 deletion. C: Western blot analysis confirms that drprΔ5 homozygous mutants do not produce Drpr. Cont, control (w1118) flies; drpr−/−, drprΔ5 homozygous (null) mutants; drpr–/+, drprΔ5 heterozygous mutants.
Effects of drpr Null Mutant Display on Lifespan
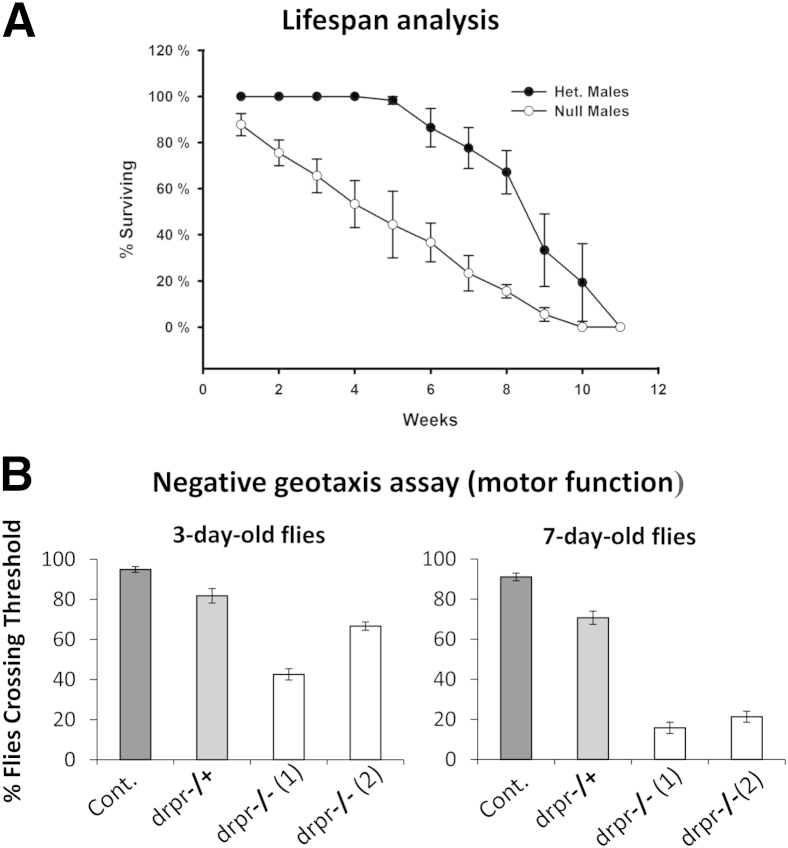
drprΔ5 homozygous null flies were viable into adulthood. However, lifespan analyses demonstrated that these mutants displayed a significantly reduced median lifespan versus corresponding heterozygotes (drprΔ5 homozygous mutants versus drprΔ5 heterozygotes, approximately 6 weeks versus 8 weeks; P = 0.014) (Figure 3).
Figure 3.
drprΔ5 Homozygous mutants display reduced lifespan, as well as a rapid and age-dependent decline in locomotor activity (versus heterozygotes). A:drprΔ5 Homozygous (null) flies display reduced lifespan compared with corresponding drprΔ5 heterozygous (het) flies. One-day-old adult male flies were collected. Every 7 days, the flies were transferred to fresh food; the number of dead flies was recorded at the time of transfer. Two independent groups of each genotype were assessed. B:drpr Null flies (drpr−/−) show reduced climbing performance compared with corresponding drprΔ5 heterozygous (drpr−/+) flies. Locomotor function in 3- or 7-day-old flies was assessed using a negative geotaxis assay. The number of flies that could reach 5-cm threshold in a 6-second period was recorded. Each genotype was assayed three times consecutively when the group reached the indicated age. The assay was repeated three times by alternating genotypes. Two groups of drpr−/− flies were assessed (the flies had emerged in different stock bottles). Data are expressed as the percentage surviving as a function of time (in weeks) (A); data are expressed as means ± SD of nine data points (B). n = 30 to 39 flies per genotype (A); n = 11 or 12 flies per genotype (B).
Regulation of the Locomotor Activity of the Fly by Drpr Expression in Muscle Versus Nervous System
To investigate whether reduced Drpr levels affect motor function, drprΔ5 null mutant flies were assessed using an established negative geotaxis assay.26 Using this approach, a significant decrease in performance of drprΔ5 mutants was demonstrated versus that in corresponding control flies (Figure 3). The impaired motor function was readily noticeable in drpr null mutants, which often display abnormal position of the legs and showed a rapid age-dependent decline in locomotor activity from 3 days to 7 to 10 days of life compared with age-matched controls (Figure 3 and Supplemental Figure S1).
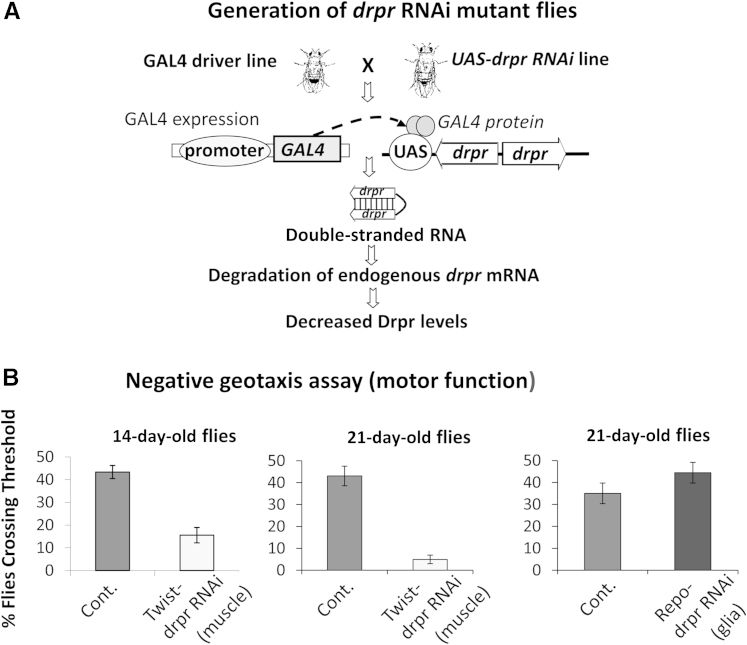
To dissect the cellular requirement that underlies the observed motor phenotype, RNA-interference flies were generated in which drpr gene expression is down-regulated in a tissue-specific manner (using the well-characterized Gal4/UAS bipartite system28). RNAi flies that down-regulate drpr in the entire organism (genotype Actin5C-Gal4; UAS-ds drpr, using the ubiquitous cytosolic actin5C-Gal4 driver line) also displayed altered locomotor function (25-day-old flies versus age-matched controls) (data not shown). The locomotor phenotype was recapitulated when drpr RNA interference was either induced in the mesoderm (how-Gal4 driver), or in the adult muscle precursor cells, which express the twist transcription factor (twist-Gal4 driver) (Figure 4). Confirming a muscle-specific effect, the locomotor phenotype was not recapitulated in flies that down-regulate drpr in neurons (pan-neuronal effect using the Elav-Gal4 driver, Elav encodes a neuronal RNA-binding protein), or in glia (using repo-Gal4 driver, repo encodes a glial-specific homeodomain protein) (Figure 4).
Figure 4.
Down-regulation of drpr in muscle cell progenitors (twist-positive cells), but not in glia (repo-positive cells), leads to reduced locomotor activity in adult Drosophila. A: Generation of drpr RNAi flies. Crossing UAS-drpr RNAi transgenic flies with flies expressing GAL4 in a tissue-specific manner results in double-transgenic RNAi flies. These flies express double-stranded (U-shaped) drpr RNA, leading to reduction/depletion of the endogenous drpr mRNA in the targeted tissue/cell type. B: Locomotor function assessment using a negative geotaxis assay. The number of flies that could reach 5-cm threshold in a 6-second period was recorded. RNAi flies that down-regulate drpr in muscle cell precursors (twist-Gal4; UAS-drpr RNAi flies) show a decline in climbing performance that progresses with age (compared with corresponding w1118/twist-Gal4 control flies). RNAi flies that down-regulate drpr in glia (repo-Gal4; UAS-drpr RNAi flies) display normal locomotor activity versus corresponding w1118; repo-Gal4 control flies. Data are expressed as means ± SD of nine data points (B). n = 8 to 13 flies per genotype (B).
Time to Abnormalities in Thoracic Muscle Morphology, Nervous System, and Retina in Adult Drpr Mutant Flies
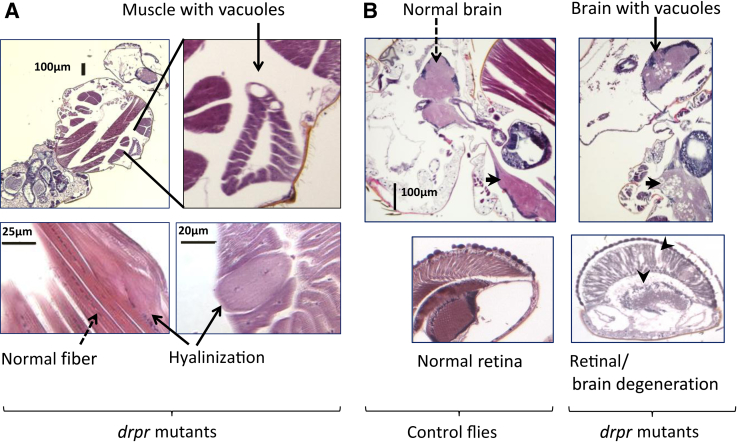
Assessment of the locomotor activity of the flies was complemented by histological examination of drprΔ5 adult male Drosophila. Flies that were 30 to 40 days old were compared to age-matched controls. Histological analyses of drpr null mutants revealed pathological changes in the skeletal muscles of the thorax. The defect, which includes hyalinization with loss of striation, variability in fiber size, as well as vacuolization, primarily affected the tergal depressor of the trochanter (jump muscle, composed of skeletal tubular fibers). In contrast, the flight muscles (which are composed of skeletal fibrillar fibers) appeared normal (Figure 5). The muscle phenotype was recapitulated in drpr RNAi flies that down-regulate drpr ubiquitously (genotype Actin5C-Gal4; UAS-ds drpr). Penetrance of the muscle defect was low to moderate in both drprΔ5 homozygotes and drpr RNAi flies (approximately 10%–20%). Parallel abnormalities were not observed in age-matched corresponding control flies (drprΔ5 heterozygotes, and w1118; Actin5C-Gal4, respectively). In addition, marked degeneration was readily observed in the nervous systems of adult drprΔ5 mutant flies as well as in drpr RNAi flies, both within the brain and in the thoracic ventral ganglion (Figure 5 and Supplemental Figure S2). Many vacuoles were seen, which clustered on the ventral part of the nervous system. Degeneration of the visual center was also evident in drpr mutant flies in which alterations were seen in the retina and within the optic ganglia (ie, lamina, medulla) (Figure 5). The brain/retinal phenotype was highly penetrant (100% adult flies were affected, with individual variability in the degree of severity).
Figure 5.
drpr Mutant Drosophila display abnormal muscle on histological examination, as well as brain and retinal degeneration. Histological sections from age-matched drpr mutant and control flies, stained with hematoxylin and eosin. A: Analysis of drpr mutants reveals foci of pathological muscular tissue affecting the thoracic muscles (solid arrows). Top panels: left, head, thorax, and upper abdomen of the organism; right, magnification of the tergal depressor of the trochanter (jump muscle) shows vacuolized fibers. Bottom panels: longitudinal section (left) and cross section (right) of affected muscles showing additional abnormalities (ie, loss of striation, loss of nuclei, enlarged fiber; solid arrows) in the vicinity of normal striated fibers (dashed arrow). B: Analysis of drpr mutants reveals extensive vacuolization in the brain, ventral nerve cord, and optic lobes. Left panels: Normal brain (top, dashed line), ventral nerve cord (top, arrow), and retina (bottom) are shown. Right panels: widespread vacuolization/degeneration is seen in the brain (solid line), the ventral nerve cord (arrow), and the optic lobe (arrowheads) of drpr mutant flies. Of note, comparable muscle and brain abnormalities were observed in drprΔ5 homozygotes (drpr null flies) and drpr RNAi flies; these defects were absent in control flies (comparison of the brain defects is shown in Supplemental Figure S2). The different groups of flies were: 30-day-old drprΔ5 homozygotes [drpr nulls: w–; sp/CyOact::GFP; drprΔ5rec8 (9)/drprΔ5rec8 (9)], 40-day-old drpr RNAi flies (w–, Act5C-Gal4/UAS-ds drpr RNAi, leading to down-regulation of drpr in the entire organism), and 40-day-old control flies (w–, Act5C-Gal4).
Discussion
The clinical and pathological features associated with the rare MEGF10 myopathy vary broadly, depending on the specific mutation. Missense mutations have been shown to result in a mild neuromuscular phenotype with prolonged preservation of ambulation,1 whereas frameshift mutations leading to nonsense-mediated mRNA decay (null mutations) cause the more severe early onset myopathy, areflexia, respiratory distress, and dysphagia (EMARDD) phenotype, which involves diaphragmatic paralysis requiring continuous respiratory assistance.2,3 One severe missense mutation impairs tyrosine phosphorylation more than does a milder missense mutation in vitro, offering a potential explanation for the phenotypic variability.29 The corresponding histological analyses of the muscles show characteristics that are common to all forms of the disease (fatty tissue replacement of the muscle), as well as features that have to date been described only in the milder form (moth-eaten fibers/minicores, occasional regenerating fibers).1,2 Muscle magnetic resonance imaging in a patient carrying the null mutation has facilitated visualization of the extensive fatty replacement of the proximal muscles.3 MEGF10 plays a role in the proliferation of Pax7-positive satellite cells and in muscle regeneration.10 Pax7-positive satellite cells were found to be absent in the muscle tissue of one EMARDD patient.2
To further understand MEGF10-mediated function in muscle, we used Drosophila as a model organism. Flies provide a unique opportunity to expand corresponding studies: i) the fly homolog of MEGF10, Drpr, shows conservation of the structurally and functionally important cysteines that are targeted by human mutations,30 ii) previously generated drpr null mutants are viable,12 enabling characterization of the knockout phenotype in fly muscles, iii) the Gal4/UAS bipartite system28 enables genetic manipulation of Drpr expression in a tissue-specific manner (eg, muscle progenitors), and iv) similar expression patterns of fly Drpr and mammalian MEGF10 have been reported in mature muscle fibers (at the postsynaptic membrane of the neuromuscular junction2,14). In addition, Drpr is the sole homolog of mammalian MEGF10, MEGF11, and MEGF12, thus eliminating, in Drosophila, possible functional redundancy of paralogs and providing insight into the role of this family of transmembrane proteins in different tissues.
drpr null mutant flies [genotype w–; sp/CyOact::GFP; drprΔ5rec8 (9)/drprΔ5rec8 (9)] display a shortened lifespan (Figure 3), and an age-dependent locomotor defect (versus heterozygotes) that is noticeable starting at approximately 2 weeks of age. The motor phenotype is readily quantified in behavioral assays and often is associated with an abnormal positioning of the legs (eg, a rigid limb that is either held upright or drags). The motor defect is also observed when drpr expression is down-regulated in a different genetic background (drpr RNAi flies). This phenotype offers a potential means to identify pharmacological modifiers of the MEGF10/Drpr depletion–induced muscle defect, using the fly model in medium throughput screens. Notably, fly models of other human muscle diseases (oculopharyngeal muscular dystrophy, myotonic dystrophy type 1) have been used successfully to establish in vivo platforms for identifying potential therapeutic compounds.31
In contrast to the human phenotype, in which only skeletal muscle alterations have been reported, histopathological examination of drpr knockout and RNAi flies reveals marked defects in the muscles as well as in the CNS (brain hemispheres, ventral cord, and retina). Vacuole formation is observed across all affected fly tissues. In muscles these lesions may be the result of fat infiltration, a feature that is widespread in human EMARDD biopsies. Well-defined vacuoles have also been observed in a limited number of other Drosophila models of muscle disease, including oculopharyngeal muscular dystrophy and myotonic dystrophy type 1.26,32–34 Further studies in this group of selected fly models may enable the dissection of the molecular processes that result in vacuolization and fat deposition in degenerating muscle. In addition, vacuolization has been described as a distinctive feature in the brain of numerous fly models of neurodegenerative disease.35
The brain lesions are reminiscent of those described in Drosophila Swiss cheese (sws) mutants. sws Is expressed in neurons and in a subset of glial cells that encode an evolutionarily conserved protein with lysophospholipase activity.35–37 In these mutants, hyperwrapping of neurons by glial sheaths precedes vacuole formation. It thus appears that in Drosophila, mutations in either drpr or sws, two genes that have a role in neuron–glia communication and the engulfment of apoptotic neurons,13,37 result in the development of brain lesions with similar histopathological features.
Sws and its mammalian homolog neuropathy target esterase possess phospholipase activity and regulate the metabolism of membrane-associated lipids, including lysophosphatidylcholine and phosphatidylcholine (PC).38,39 Mutations in sws lead to increased levels of cellular PC.37 Altered levels of PC/lysophosphatidylcholine have been associated with membrane breakdown and the development of neurodegenerative disease.40 Lysophosphatidylcholine is also a known bioactive signaling factor that acts as a find me and eat me signal when produced in apoptotic cells.41 Intriguingly, it was recently shown that another classic eat me phospholipid signal, that is, phospatidylserine, is a ligand for the Drpr receptor and regulates Drpr-mediated clearance of apoptotic neurons.8,42 Parallels in humans may be proposed, and it is possible that either phospatidylserine or PC, the levels of which increase in response to A-β1–40 accumulation,43 regulates MEGF10-mediated removal of the β-amyloid peptide.9
In muscle, in vitro studies performed in C2C12 cells have shown that selected lysophospholipids stimulated the proliferation, and inhibited the differentiation, of myoblasts.44 These effects are reminiscent of those induced by MEGF10 overexpression in the same cell type.10 More recently, studies in vivo have demonstrated a beneficial effect of lysophospholipids on muscle function; in mice, sphingosine 1 phosphate (S1P) promoted the regeneration of injured mdx muscle45 and in Drosophila S1P attenuated the development of dystrophic muscle alterations.46 Conversely, in humans, mutations in the choline kinase beta gene (CHKB) resulted in altered biosynthesis of PC, causing a form of congenital muscular dystrophy.29 The potential relevance of these bioactive/signaling factors as mediators of MEGF10/Drpr-dependent physiology in muscle remains to be explored.
Notably, the brain degeneration observed in Drpr-deficient flies (in addition to abnormalities in muscle) contrasted with the human phenotype, which affected only skeletal muscle. Myopathic patients carrying mutations in MEGF10 had normal brain imaging and did not show symptoms of CNS dysfunction.1,4 It is possible that in mammals, functional redundancy between MEG10 and its paralogs MEGF11 and MEGF12 contribute to maintaining normal brain function in MEGF10 myopathy. Consistent with this hypothesis is the recent study showing no gross phenotypic abnormalities in either the megf10−/− or megf11−/− knockout mouse; however, the corresponding double-knockout mouse had markedly disrupted retinas.5 The absence of functional redundancy for Drpr in Drosophila may help to elucidate whether MEGF11 and/or MEGF12 in humans prevents nervous system degeneration in patients bearing a MEGF10 mutation. Mammalian genes may be introduced in the fly background not only to study the in vivo function of the wild-type protein but also to investigate the effects of naturally occurring variants (splice variants, mutant isoforms, deletions).16,19,21,32,47 In the absence of other available in vivo assays, this approach is particularly helpful when investigating whether novel mutations found in a human muscular dystrophy gene (eg, LMNA) may be pathogenic.16 Drosophila thus provides a useful framework for pursuing structure–function analyses of important regulators of muscle physiology. A potential future direction for study could involve the introduction of the mammalian MEGF10, MEGF11, and MEGF12 cDNAs into the fly mutant background to examine the in vivo function of the corresponding proteins and the effects of mutations.
Acknowledgments
We thank Marc R. Freeman (University of Massachusetts Medical School, Worcester, MA) for the generous gift of the drprΔ5 mutant fly line, the UAS-ds-drpr RNAi mutant fly line, and the polyclonal anti-Drpr antibody; Ci Chen for excellent technical assistance; and Elicia Estrella for helpful discussions.
Footnotes
Supported by grant 186796 from the Muscular Dystrophy Association (P.B.K.), a Boston Children's Hospital Pilot grant (P.B.K.), and a grant from the NIH (R01 NS080929) (P.B.K. and I.D.).
Disclosures: I.D. and P.B.K. are collaborating with Medosome Biotec, LLC, a firm affiliated with the University of Florida that commercializes scientific discoveries and have submitted a grant application to screen drug candidates based on some of the work presented here.
Contributor Information
Isabelle Draper, Email: idraper@tuftsmedicalcenter.org.
Peter B. Kang, Email: pbkang@ufl.edu.
Supplemental Data
The locomotor activity of drpr null flies declines rapidly after eclosion. The locomotor function of w1118 control or drpr mutant Drosophila was assessed using a negative geotaxis assay. The number of flies that could reach 5-cm threshold in a 6-second period was recorded. Each genotype was assayed three times consecutively when the group reached the indicated age. The assay was repeated three times by alternating genotypes. The first set of control and mutant flies was aged and assessed at 3 (A), 7 (B), or 10 (C) days of age. D: The second set of flies, obtained from a different generation, was assessed at 7 days of age. Data are expressed as means ± SD of nine data points. n = 10 to 12 flies per genotype.
Drosophila with reduced drpr expression show marked brain degeneration. A and B: Comparable histological defects are readily observed in the neuropil of two drpr mutant fly lines that were generated in distinct genetic backgrounds. A: Brain (left) and ventral nerve cord (right) of drprΔ5 homozygotes [drpr nulls: w–; sp/CyOact::GFP; drprΔ5rec8 (9)/ drprΔ5rec8 (9)]. B: Ventral nerve cord of drpr RNAi flies (w–, Act5C-Gal4/UAS-ds drpr RNAi, obtained by crossing the w1118; P{GD14423}v27086 VDRC RNAi line with the ubiquitous Gal4 driver, y1 w*; P{Act5C-Gal4}25FO1/CyO, y+). The vacuoles tend to cluster on the ventral part of the nervous system (solid arrows). C: Histological examination of normal brain and ventral ganglion (dashed arrows) obtained from corresponding w–, Act5C-Gal4 control flies.
References
- 1.Boyden S.E., Mahoney L.J., Kawahara G., Myers J.A., Mitsuhashi S., Estrella E.A., Duncan A.R., Dey F., DeChene E.T., Blasko-Goehringer J.M., Bönnemann C.G., Darras B.T., Mendell J.R., Lidov H.G., Nishino I., Beggs A.H., Kunkel L.M., Kang P.B. Mutations in the satellite cell gene MEGF10 cause a recessive congenital myopathy with minicores. Neurogenetics. 2012;13:115–124. doi: 10.1007/s10048-012-0315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logan C.V., Lucke B., Pottinger C., Abdelhamed Z.A., Parry D.A., Szymanska K., Diggle C.P., van Riesen A., Morgan J.E., Markham G., Ellis I., Manzur A.Y., Markham A.F., Shires M., Helliwell T., Scoto M., Hubner C., Bonthron D.T., Taylor G.R., Sheridan E., Muntoni F., Carr I.M., Schuelke M., Johnson C.A. Mutations in megf10, a regulator of satellite cell myogenesis, cause early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD) Nat Genet. 2011;43:1189–1192. doi: 10.1038/ng.995. [DOI] [PubMed] [Google Scholar]
- 3.Pierson T.M., Markello T., Accardi J., Wolfe L., Adams D., Sincan M., Tarazi N.M., Fajardo K.F., Cherukuri P.F., Bajraktari I., Meilleur K.G., Donkervoort S., Jain M., Hu Y., Lehky T.J., Cruz P., Mullikin J.C., Bonnemann C., Gahl W.A., Boerkoel C.F., Tifft C.J. Novel SNP array analysis and exome sequencing detect a homozygous exon 7 deletion of MEGF10 causing early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD) Neuromuscul Disord. 2013;23:483–488. doi: 10.1016/j.nmd.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartley L., Kinali M., Knight R., Mercuri E., Hubner C., Bertini E., Manzur A.Y., Jimenez-Mallebrera C., Sewry C.A., Muntoni F. A congenital myopathy with diaphragmatic weakness not linked to the SMARD1 locus. Neuromuscul Disord. 2007;17:174–179. doi: 10.1016/j.nmd.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Kay J.N., Chu M.W., Sanes J.R. MEGF10 AND MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483:465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., Thompson W.J., Barres B.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheib J.L., Sullivan C.S., Carter B.D. Jedi-1 and MEGF10 signal engulfment of apoptotic neurons through the tyrosine kinase Syk. J Neurosci. 2012;32:13022–13031. doi: 10.1523/JNEUROSCI.6350-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung W.S., Clarke L.E., Wang G.X., Stafford B.K., Sher A., Chakraborty C., Joung J., Foo L.C., Thompson A., Chen C., Smith S.J., Barres B.A. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh T.D., Park S.Y., Bae J.S., Yun Y., Bae Y.C., Park R.W., Kim I.S. MEGF10 functions as a receptor for the uptake of amyloid-β. FEBS Lett. 2010;584:3936–3942. doi: 10.1016/j.febslet.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 10.Holterman C.E., Le Grand F., Kuang S., Seale P., Rudnicki M.A. Megf10 regulates the progression of the satellite cell myogenic program. J Cell Biol. 2007;179:911–922. doi: 10.1083/jcb.200709083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziegenfuss J.S., Biswas R., Avery M.A., Hong K., Sheehan A.E., Yeung Y.G., Stanley E.R., Freeman M.R. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008;453:935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman M.R., Delrow J., Kim J., Johnson E., Doe C.Q. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald J.M., Beach M.G., Porpiglia E., Sheehan A.E., Watts R.J., Freeman M.R. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 14.Fuentes-Medel Y., Logan M.A., Ashley J., Ataman B., Budnik V., Freeman M.R. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 2009;7:e1000184. doi: 10.1371/journal.pbio.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H.C., Dimlich D.N., Yokokura T., Mukherjee A., Kankel M.W., Sen A., Sridhar V., Fulga T.A., Hart A.C., Van Vactor D., Artavanis-Tsakonas S. Modeling spinal muscular atrophy in Drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dialynas G., Flannery K.M., Zirbel L.N., Nagy P.L., Mathews K.D., Moore S.A., Wallrath L.L. LMNA variants cause cytoplasmic distribution of nuclear pore proteins in Drosophila and human muscle. Hum Mol Genet. 2012;21:1544–1556. doi: 10.1093/hmg/ddr592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein J.A., Kelly S.M., LoPresti P.P., Heydemann A., Earley J.U., Ferguson E.L., Wolf M.J., McNally E.M. SMAD signaling drives heart and muscle dysfunction in a Drosophila model of muscular dystrophy. Hum Mol Genet. 2011;20:894–904. doi: 10.1093/hmg/ddq528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajendra T.K., Gonsalvez G.B., Walker M.P., Shpargel K.B., Salz H.K., Matera A.G. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevdali M., Kumar V., Peckham M., Sparrow J. Human congenital myopathy actin mutants cause myopathy and alter Z-disc structure in Drosophila flight muscle. Neuromuscul Disord. 2013;23:243–255. doi: 10.1016/j.nmd.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Shcherbata H.R., Yatsenko A.S., Patterson L., Sood V.D., Nudel U., Yaffe D., Baker D., Ruohola-Baker H. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007;26:481–493. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taghli-Lamallem O., Akasaka T., Hogg G., Nudel U., Yaffe D., Chamberlain J.S., Ocorr K., Bodmer R. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wairkar Y.P., Fradkin L.G., Noordermeer J.N., DiAntonio A. Synaptic defects in a Drosophila model of congenital muscular dystrophy. J Neurosci. 2008;28:3781–3789. doi: 10.1523/JNEUROSCI.0478-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figeac N., Jagla T., Aradhya R., Da Ponte J.P., Jagla K. Drosophila adult muscle precursors form a network of interconnected cells and are specified by the rhomboid-triggered EGF pathway. Development. 2010;137:1965–1973. doi: 10.1242/dev.049080. [DOI] [PubMed] [Google Scholar]
- 24.Maqbool T., Jagla K. Genetic control of muscle development: learning from Drosophila. J Muscle Res Cell Motil. 2007;28:397–407. doi: 10.1007/s10974-008-9133-1. [DOI] [PubMed] [Google Scholar]
- 25.McPhee C.K., Logan M.A., Freeman M.R., Baehrecke E.H. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draper I., Tabaka M.E., Jackson F.R., Salomon R.N., Kopin A.S. The evolutionarily conserved RNA binding protein SMOOTH is essential for maintaining normal muscle function. Fly (Austin) 2009;3:235–246. doi: 10.4161/fly.9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagase T., Kikuno R., Ohara O. Prediction of the coding sequences of unidentified human genes. XXII. The complete sequences of 50 new cDNA clones which code for large proteins. DNA Res. 2001;8:319–327. doi: 10.1093/dnares/8.6.319. [DOI] [PubMed] [Google Scholar]
- 28.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuhashi S., Ohkuma A., Talim B., Karahashi M., Koumura T., Aoyama C., Kurihara M., Quinlivan R., Sewry C., Mitsuhashi H., Goto K., Koksal B., Kale G., Ikeda K., Taguchi R., Noguchi S., Hayashi Y.K., Nonaka I., Sher R.B., Sugimoto H., Nakagawa Y., Cox G.A., Topaloglu H., Nishino I. A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. Am J Hum Genet. 2011;88:845–851. doi: 10.1016/j.ajhg.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsuhashi S., Mitsuhashi H., Alexander M.S., Sugimoto H., Kang P.B. Cysteine mutations cause defective tyrosine phosphorylation in MEGF10 myopathy. FEBS Lett. 2013;587:2952–2957. doi: 10.1016/j.febslet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Draper I. Model organisms offer new possibilities for discovery of therapeutics. Drug Discov Today Technol. 2013;10:e61–e64. doi: 10.1016/j.ddtec.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Chartier A., Benoit B., Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of pabpn1. EMBO J. 2006;25:2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Haro M., Al-Ramahi I., De Gouyon B., Ukani L., Rosa A., Faustino N.A., Ashizawa T., Cooper T.A., Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Lopez A., Monferrer L., Garcia-Alcover I., Vicente-Crespo M., Alvarez-Abril M.C., Artero R.D. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS One. 2008;3:e1595. doi: 10.1371/journal.pone.0001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretzschmar D. Swiss cheese et allii, some of the first neurodegenerative mutants isolated in Drosophila. J Neurogenet. 2009;23:34–41. doi: 10.1080/01677060802471635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kretzschmar D., Hasan G., Sharma S., Heisenberg M., Benzer S. The Swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mühlig-Versen M., da Cruz A.B., Tschäpe J.A., Moser M., Büttner R., Athenstaedt K., Glynn P., Kretzschmar D. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J Neurosci. 2005;25:2865–2873. doi: 10.1523/JNEUROSCI.5097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kienesberger P.C., Lass A., Preiss-Landl K., Wolinski H., Kohlwein S.D., Zimmermann R., Zechner R. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J Biol Chem. 2008;283:5908–5917. doi: 10.1074/jbc.M709598200. [DOI] [PubMed] [Google Scholar]
- 39.van Tienhoven M., Atkins J., Li Y., Glynn P. Human neuropathy target esterase catalyzes hydrolysis of membrane lipids. J Biol Chem. 2002;277:20942–20948. doi: 10.1074/jbc.M200330200. [DOI] [PubMed] [Google Scholar]
- 40.Klein J. Membrane breakdown in acute and chronic neurodegeneration: focus on choline-containing phospholipids. J Neural Transm. 2000;107:1027–1063. doi: 10.1007/s007020070051. [DOI] [PubMed] [Google Scholar]
- 41.Hochreiter-Hufford A., Ravichandran K.S. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tung T.T., Nagaosa K., Fujita Y., Kita A., Mori H., Okada R., Nonaka S., Nakanishi Y. Phosphatidylserine recognition and induction of apoptotic cell clearance by Drosophila engulfment receptor Draper. J Biochem. 2013;153:483–491. doi: 10.1093/jb/mvt014. [DOI] [PubMed] [Google Scholar]
- 43.Koudinova N.V., Koudinov A.R., Yavin E. Alzheimer's Abeta1-40 peptide modulates lipid synthesis in neuronal cultures and intact rat fetal brain under normoxic and oxidative stress conditions. Neurochem Res. 2000;25:653–660. doi: 10.1023/a:1007511120099. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida S., Fujisawa-Sehara A., Taki T., Arai K., Nabeshima Y. Lysophosphatidic acid and bfgf control different modes in proliferating myoblasts. J Cell Biol. 1996;132:181–193. doi: 10.1083/jcb.132.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ieronimakis N., Pantoja M., Hays A.L., Dosey T.L., Qi J., Fischer K.A., Hoofnagle A.N., Sadilek M., Chamberlain J.S., Ruohola-Baker H., Reyes M. Increased sphingosine-1-phosphate improves muscle regeneration in acutely injured mdx mice. Skelet Muscle. 2013;3:20. doi: 10.1186/2044-5040-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pantoja M., Fischer K.A., Ieronimakis N., Reyes M., Ruohola-Baker H. Genetic elevation of sphingosine 1-phosphate suppresses dystrophic muscle phenotypes in Drosophila. Development. 2013;140:136–146. doi: 10.1242/dev.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf M.J., Amrein H., Izatt J.A., Choma M.A., Reedy M.C., Rockman H.A. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The locomotor activity of drpr null flies declines rapidly after eclosion. The locomotor function of w1118 control or drpr mutant Drosophila was assessed using a negative geotaxis assay. The number of flies that could reach 5-cm threshold in a 6-second period was recorded. Each genotype was assayed three times consecutively when the group reached the indicated age. The assay was repeated three times by alternating genotypes. The first set of control and mutant flies was aged and assessed at 3 (A), 7 (B), or 10 (C) days of age. D: The second set of flies, obtained from a different generation, was assessed at 7 days of age. Data are expressed as means ± SD of nine data points. n = 10 to 12 flies per genotype.
Drosophila with reduced drpr expression show marked brain degeneration. A and B: Comparable histological defects are readily observed in the neuropil of two drpr mutant fly lines that were generated in distinct genetic backgrounds. A: Brain (left) and ventral nerve cord (right) of drprΔ5 homozygotes [drpr nulls: w–; sp/CyOact::GFP; drprΔ5rec8 (9)/ drprΔ5rec8 (9)]. B: Ventral nerve cord of drpr RNAi flies (w–, Act5C-Gal4/UAS-ds drpr RNAi, obtained by crossing the w1118; P{GD14423}v27086 VDRC RNAi line with the ubiquitous Gal4 driver, y1 w*; P{Act5C-Gal4}25FO1/CyO, y+). The vacuoles tend to cluster on the ventral part of the nervous system (solid arrows). C: Histological examination of normal brain and ventral ganglion (dashed arrows) obtained from corresponding w–, Act5C-Gal4 control flies.