Abstract
Recent advances in genomic medicine have opened up the possibility of tailored medicine that may eventually replace traditional “one-size-fits all” approaches to the treatment of inflammatory bowel disease (IBD). In addition to exploring the interactions between hosts and microbes, referred to as the microbiome, a variety of strategies that can be tailored to an individual in the coming era of personalized medicine in the treatment of IBD are being investigated. These include prompt genomic screening of patients at risk of developing IBD, the utility of molecular discrimination of IBD subtypes among patients diagnosed with IBD, and the discovery of proteome biomarkers to diagnose or predict cancer risks. Host genetic factors influence the etiology of IBD, as do microbial ecosystems in the human bowel, which are not uniform, but instead represent many different microhabitats that can be influenced by diet and might affect processes essential to bowel metabolism. Further advances in basic research regarding intestinal inflammation may reveal new insights into the role of inflammatory mediators, referred to as the inflammasome, and the macromolecular complex of metabolites formed by intestinal bacteria. Collectively, knowledge of the inflammasome and metagenomics will lead to the development of biomarkers for IBD that target specific pathogenic mechanisms involved in the spontaneous progress of IBD. In this review article, our recent results regarding the discovery of potential proteomic biomarkers using a label-free quantification technique are introduced and on-going projects contributing to either the discrimination of IBD subtypes or to the prediction of cancer risks are accompanied by updated information from IBD biomarker research.
Keywords: Inflammatory bowel disease, Biomarker, Proteomics, Tailored medicine, Colitic cancer
Core tip: Our recent achievements in discovering biomarkers to predict cancer risk are introduced. Ultimately, models based on combinations of genotype and gene expression data referenced with clinical, biochemical, and serological data may permit the development of tools for individualized risk stratification and efficient treatment selection, as well as complete rescue from complications, including colitis-associated cancer, in the near future.
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic inflammatory disease that causes injury to the gastrointestinal (GI) tract and is accompanied by clinical characteristics of remission and relapse. The two common types of IBD are ulcerative colitis (UC) and Crohn’s disease (CD). Although many molecular methods for the investigation of protein and gene sequences have contributed to diagnostic methodologies, the diagnosis of IBD is primarily based on clinical, endoscopic, radiological, and histological criteria. Unfortunately, there has been little to no change in this traditional approach to diagnosis, despite modern advances in genomics and proteomics. However, progress in treatment strategies involving the incorporation of marketed biologicals and molecular targeted therapeutics has lead to the development of the concept of “deep remission” or “mucosal healing” in the treatment of IBD[1,2]. Furthermore, there has been major innovation in diagnostic methods, including the development of more complicated endoscopic and non-invasive imaging methods. These techniques are used to improve quality of life (QOL) of patients, predict complications, and contribute to the prevention or surveillance of cancer associated with IBD such as colitis-associated cancer (CAC)[3].
Recent developments in the molecular pathogenesis of IBD have highlighted three aspects. First, IBD is caused by complex disorders influenced by susceptibility genes, and is characterized by disturbed epithelial barrier function, and abnormal innate and adaptive immunity. Second, the compositions of gut microflora are altered or the epithelial barrier function is disorganized, which leads to a response from the immune system. Third, a murine model has been very helpful in unraveling the pathogenesis/mucosal immunopathology of IBD[4] by suggesting that the abnormal immune reaction to normal microbiota results from dysregulation of the mucosal immune system[5]. For example, the composition of microbes in the gastrointestinal tract may impair the patient’s lifestyle in developed countries and a pathogenic infection in the gastrointestinal tract has a significant function in modulating the immune system. These data may explain why developing and some Asian countries are confronting steep increases in the incidence of IBD. Developments in gene-sequencing technologies, such as next generation sequencing, as well as the emergence of several bioinformatic tools have led to novel insights into the microbe balance in the human gastrointestinal tract and the effect of microbes on human physiology and pathology[6].
Additional innovative technologies, such as mass spectrometry (MS)-based proteomics, also referred to as next generation proteomics, have discovered new classes of proteomic biomarkers that can be used to explore the accurate and comprehensive molecular characterization of IBD genes and proteomes. These advances are expected to lead to more reliable identification of IBD diagnostic- or progression-specific targets and enable molecular diagnosis, as well as provide guidance regarding the selection of treatment options and the risk of cancer development in cases with longstanding remission and relapse[7]. A more robust molecular definition of IBD subtypes is likely to be based on specific molecular pathways that determine not only disease susceptibility, but also disease characteristics, such as location, natural history and therapeutic response. Furthermore, such advances could be applicable in defining “deep remission”, which has not been feasible with currently-used scoring systems or endoscopic evaluation. Discovering biomarkers for IBD may allow objective measurements of disease activity and severity while also serving as prognostic indicators for therapeutic outcomes[8]. Furthermore, the discovery of one or more biomarkers predictive of the risk of IBD-associated cancers, such as CAC, combined with advanced therapeutics, may lead to tremendous improvements in patient QOL in the near future.
In this review article, our recent achievements in discovering biomarkers to predict cancer risk are introduced. Ultimately, models based on combinations of genotype and gene expression data referenced with clinical, biochemical, and serological data may permit the development of tools for individualized risk stratification and efficient treatment selection, as well as complete rescue from complications, including CAC, in the near future[9].
Molecular pathogenesis of colorectal cancer and CAC
Chronic inflammatory diseases are associated with cancer incidence, which is dependent on the duration and severity of the diseases. For example, Barrett’s esophagus is relevant to esophageal cancer, chronic Helicobacter pylori-associated chronic atrophic gastritis is relevant to gastric cancer, and UC or CD are relevant to CAC. These are well-acknowledged examples that support a connection between gut inflammation and cancer. In fact, it has been reported that patients with IBD are at enhanced risk of colorectal cancer (CRC): approximately 15% of CRC patients have related IBD etiology[10]. Although general carcinogenesis is a multi-factorial process that combines accumulation of genetic mutations, post-translational modification, and cell-matrix reciprocal action, inflammation-prone carcinogenesis is somewhat different[11]. The utility of biomarkers for CAC can be extended to permit earlier detection of dysplasia; therefore, the targeted manipulation of biomarkers might lead to advances in cancer therapies and cancer preventions, and may prove to be effective in reducing the development of CAC, with clinical interventions such as blocking agents or endoscopic treatments. Regarding molecular aspects, the mechanism of CAC in IBD differs from CRC ,which is a well-known adenoma-to-carcinoma sequence. CAC appears to take place from either flat dysplastic tissue or dysplasia-associated lesions or masses[12]. The major pathways of sporadic CRC and CAC comprise chromosomal instability, hypermethylation and microsatellite instability; however, CAC shows inflamed colonic mucosa before histological changes of dysplasia or cancer. Patients with IBD have a high risk of CAC following diagnosis, and patients have common symptom, such as colitis[13]. The risk of CAC is increased in younger patients, those with more extensive colitis, those with concomitant primary sclerosing cholangitis, and a family history of CRC[14]. Most cancers show no high risk related to proctitis; however, increased in pancolitis, i.e., left-sided colitis, carries an intermediate cancer risk[15]. Patients with CD and UD have the same risk of CRC and the prevalence in the US is greater than 200 cases per 100000, representing a total of between 1 and 1.5 million patients with IBD. Fortunately, the incidence of CAC is lower than CRC in the United States and other western countries[16]. A biological background for the high risk of CRC in IBD gives a one-sided interpretation, patients with high levels of inflammatory mediators production may progress to CAC. The key signal of IBD-induced carcinogenesis is inflammatory cytokines induced by mucosal and immune cells in the gut. The key molecules of inflammation, including nuclear factor kappaB (NF-κB) and cyclooxygenase-2 (COX-2), are important links between inflammation and cancer. Recently, other factors, such as tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) -induced signaling, have been proven to induce cancer development in animal models of CAC[17]. Based on these aspects of molecular carcinogenesis, we have tried to apply biologicals such as infliximab to neutralize TNF-α, a proton pump inhibitor based on mechanisms including the inhibition of NF-κB as well as attenuation of oxidative stress, and aspirin/celecoxib to inhibit cancer-prone COX enzymes. As expected, all of these efforts to block inflammation-promoted carcinogenesis efficiently prevented CAC in our mouse model experiments[18,19]. The recent descriptions of epigenetic alterations, in particular alterations in DNA methylation, that have been observed during inflammation and inflammation-associated carcinogenesis, led us to explore nutritional interventions as a means of targeting and correcting epigenetic oncogenic abnormalities, as a form of CAC prevention[20].
Prediction of CAC
Patients with long-standing UC and CD are at increased risk of developing CRC, and patients with small intestinal CD have a high risk for developing small bowel adenocarcinoma. Unlike the sporadic CRC that can develop in those with IBD, CAC development is intimately associated with IBD. In those with IBD, CAC results from a process that is believed to begin with mutagenic benign inflammation that develops into indeterminate, low-grade and high-grade dysplasia, and eventually to carcinoma. Regarding the risk factors predisposing to carcinoma in IBD, the risk is increased depending on duration, severity of colitis, presence of sclerosing cholangitis, degree of inflammation and family history of CRC. Evidence-based medicine advises that patients with colitis should be kept under surveillance colonoscopy after diagnosis for 8 to 10 years. As surveillance guidelines for early detection of CAC, the general approach of periodic endoscopic examinations and systematic random biopsies of involved mucosa is generally recommended[21]. Recently, advanced colonoscopic techniques, including narrow band imaging, chromoendoscopy and confocal microendoscopy, have been used to identity abnormal areas in targeted, but not random, biopsies, and biomarkers could be adopted for high resolution endoscopy[22]. Although medications such as aminosalicylates, folic acid and ursodeoxycholic acid seem to be chemopreventive, potent preventive therapeutics, as well as surveillance of high risk patients through the use of potential biomarkers, would seem to be ideal[23].
Current Status Of Biological Markers For IBD
Serological markers for IBD are rapidly developing. However, the most studied antibodies, anti-Saccharomyces cerevisiae antibodies (ASCA) and atypical perinuclear antineutrophil cytoplasmic antibodies (P-ANCA), have limited sensitivity. Thus, the relationship between serological markers of disease pattern and phenotype may be of greater value than the use of serological markers as diagnostic tools. For example, patients with CD who have high titers of various serological makers have more serious small intestine disease than those with low titers of antibodies[24]. On the genetic level, application of a genome wide association study design in CD has provided new insights into the immunopathogenesis of CD, identifying links to genes of the innate and adaptive immune system[25]. One patient had CD associated with gene mutations of NOD2 and the ATG16L1 autophagy gene, both of which affect the intracellular processing of bacterial components. In addition, genetic variation of the IL-23 receptor, STAT3 and NKX2-3 genes, were associated with CD and UC in Asian patients. Although comparative analyses of gene associations between CD and UD can identify unique mechanisms of immunopathogenesis of IBD, such results have limited applicability in real-world clinical settings because of ethnic, racial, and environmental differences in the samples studied. Since the advent of the concept of proteomics, a plethora of proteomic technologies have been developed to study proteomes. In IBD, several studies have used proteomics to better understand the disease and discover molecules that could serve as therapeutic targets. The advance of proteomic technologies will have an important effect on the development of new biomarkers for IBD[26]. Further advances in proteomic technologies have allowed us to use label-free quantification to detect biomarkers in various IBD patients for the first time. The results are expected to provide additional insights in to the molecular biomarkers of IBD that may be used in predicting responses to treatment[27].
Classic serological and fecal markers in IBD
Currently, diagnosis of IBD from the blood and stool of patients represent reliable and quantitative tools to clinicians[28]. The C-reactive protein (CRP) and fecal-based leukocyte markers, calprotectin (Cal) or lactoferin (Lf), can help clinicians to assess disease activity and to distinguish IBD from non-inflammatory diarrhea and simple colitis. Serological tests including both ASCA and P-ANCA can be used to determine the current status and risk of IBD[29]. The progression of IBD and inflammatory processes are assessed by tests for CRP and the erythrocyte sedimentation rate (ESR). In addition, clinicians might measure the levels of drug metabolites and antibodies against therapeutic agents that aim to determine why patients do not respond to treatment and to select alternative therapy. The advantages of using the fecal markers including, Cal or Lf, are their ease of detection and use of an inexpensive ELISA technique, as well as their long-term stability in feces[30]. However, several limitations have been associated with these classical serological and fecal markers in IBD. The ESR technique is simple to perform, widely available, and inexpensive; however, it has several disadvantages, such as a concentration that depends on age, several confounders, and the use of certain drugs[31]. Several factors can affect the utility of CRP, including long half-life and prolonged latency period after changes in chronic IBD. Determination of fecal Cal or Lf markers is very helpful into diagnosis chronic IBD, while other GI diseases, ischemic colitis, and non-steroidal anti-inflammatory drug-associated intestinal damage show greater leukocyte elimination in feces[32]. In spite of 80%-100% diagnostic accuracy levels, fecal markers are not specific for IBD and may be elevated in a range of organic conditions. To compensate for these limitations, Langhorst et al[33] evaluated fecal levels of PMN-elastase (PMN-e) in addition to the aforementioned markers and concluded the IBD and IBS can be discriminated by the fecal markers Cal, Lf, and PMN-e. However, these fecal markers have shown a similar capacity to indicate endoscopic pathology, and are more efficient for diagnosis than CRP. Therefore, the combined diagnosis using fecal markers and CRP with disease-specific activity index will be very useful when assessing endoscopic inflammation in UC. It should be noted that these fecal markers were proven to be very efficient in diagnosing IBD as well as in predicting impending clinical relapse in pediatric patients with IBD[34]. Despite the benefits that can be derived from these serum and fecal biomarkers, there remains considerable room for improvement regarding disease prediction and prognosis assessment, as none of them can be applied to predict future risk of CAC development in IBD.
Proteomic biomarkers in IBD
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) and surface-enhanced laser desorption/ionization (SELDI)-TOF MS, have become popular methods recently for the analysis of macromolecules of biological origin such as tissues, serum, or plasma[35]. MALDI-TOF MS is used in clinical medicine to identify disease markers in combination with classic protein gel analysis (1-D) and two dimensional gel electrophoresis separation, accomplished through either peptide mass fingerprinting or peptide sequence tag (2-D) analysis followed by a data base search using proteome blot analysis software[36]. Using these applications, evaluation of samples by MALDI-TOF MS can give novel data regarding peptides present at high molecular mass and may therefore be valuable to assess potential disease markers of IBD. For example, Nanni et al[37] determined serum proteins of 22 healthy subjects and 41 patients with IBD (15 CD, 26 UC) extracted with reversed-phase (C18) and subsequently performed MALDI-TOF MS . The results of serum protein profiles showed the highest overall prognosis capability (96.9%) of identifiable protein biomarkers involved in IBD discrimination . Similarly, in a study by Liu et al[38], serum proteins from 74 CRC samples compared with 48 healthy samples were applied to SELDI-TOF MS using a ProteinChip reader. The diagnostic pattern could distinguish samples according to status of CRC from normal samples with sensitivity and specificity of 95% by independent analysis of the samples. These two studies demonstrated the high potential for biomarker discovery in patients with IBD or CRC in clinical settings, and further clinical validation in large patient cohorts is expected to promote the use of novel biomarkers in clinical practice[39]. In addition to protein identification, 2-D difference gel electrophoresis (2D DIGE) is good method protein quantification; however, isobaric tags for relative and absolute quantification (iTRAQ) and stable isotope labeling by amino acids (SILAC) are the best methods developed recently. Sample preparation is important to the success or failure of such analysis and subcellular fractionation can be used to give more specific protein localization analysis than total cellular proteins[40-42]. In our recent study[43], we applied advanced technologies such as iTRAQ and SILAC for label-free quantification in samples obtained from a mouse model of UC and CAC to identify potential biomarkers for cancer-prone inflammation in IBD and to evaluate empirical therapeutics.
PROTEOMIC BIOMARKER DISCOVERY FOR CAC: A CLASSICAL PROTEOMIC APPROACH
Multiple chemical and biological systems, including intestinal tissue, its associated immune system, the gut microbiota, xenobiotics, and their metabolites, meet and interact to form a tightly regulated state of tissue homeostasis. Disturbance to this state of homeostasis can cause IBD as well as CAC through intercalated multi-factorial mechanisms. Many strong pathological and mechanistic correlates exist between mouse models of CAC and the clinically relevant situation in humans, allowing for the use of systems biology approaches[44]. Furthermore, the close proximity of colonic tumors to the myriad of intestinal microbes, as well as the instrumental implication of microbiota in IBD, introduces microbes as new factors capable of triggering inflammation and possibly promoting CAC, necessitating high throughput metabolomic approaches[45]. Additionally, a detailed understanding of these interactions may also provide a means of preventing CAC[46]. In a small study, Watanabe et al[47] performed a low density array analysis of 149 genes implicated in CAC and identified 20 genes showing differential expression between UC- and non-UC-associated CAC, including cancer-related genes such as CYP27B1, runt-related transcription factor 3, sterile alpha motif domain-containing protein 1, EGF-like repeats and discoidin I-like domain 3, nucleolar protein 3, CXCL9, integrin beta2, and LYN. Colliver et al[48] demonstrated that 392 transcripts showed differential expression during progression from UC to CAC. Both dysplasia and CAC showed 224 transcripts in common and it was concluded that some genes showed same modification both in dysplasia and CAC, signifying that they might be related to tumor initiation and progression. Following these studies, a host of potential biomarkers have been reported in the literature, including cytokeratin 7/20[49], a-methylacyl-CoA-racemase[50], transgelin, a frame shift mutation in the TGF-β type II receptor[51,52], HSP47[53], methylation of the estrogen receptor[54], association with certain HLA class II alleles[55], DNA methyltransferase-1[56], 8-nitroguanine or 8-oxo-deoxyguanine[57-59], CCL20[60] and activation-induced cytidine deaminase[61]. We used our experimental animal model for colitic cancer, which was provoked with repeated bouts of UC, and an additional proteomic method based on 2-D electrophoresis and MALDI-TOF MS to analyze proteins related in CAC. In detail, 38 proteins were differentially expressed between CAC and healthy samples, using comparative 2-D electrophoresis analysis. Through validation studies, 27 proteins, including enolase, GRP94, HSC70 prohibitin and transgelin, were identified. Among these identified proteins, the downregulation of transgelin in mouse colitic cancer was supported by western blotting and immunohistochemistry. Moreover, transgelin was significantly decreased in colon tumors compared with non-tumorous regions in humans, implying that reduced levels of transgelin could be a good biomarker for CAC[62].
BIOMARKER DISCOVERY FOR CAC: THE NEXT GENERATION OF PROTEOMICS
Currently, knowledge of proteomics is important and provides researches with complicated label and label-free techniques. This next generation of proteomics may provide effective perspectives in the diagnosis and treatment of gastrointestinal disease and allow for translation of proteomics from the bench to the bedside. Currently, proteomic studies focus not only in the identification of proteins in a sample, but also the quantification of them. Various protein expression profiles are examined because they provide useful information, especially in clinical proteomics, involving molecular targets related to specific diseases. The technology for proteomics study is continuously developing, and both of labeled and label-free methods have their several advantages. Currently, label-free and isotope-label techniques are used to study proteins quantitatively. The label-free approach, combining spectral and signal quantitation, for identifying amino acids provides accurate relative protein expression data and is an easy way to determine quantitative information. However, this strategy is subject errors from variation in protein preparation that may be reduced when various stable isotopes are inserted in the specimens to make protein isotopomers, which have different spectra according to their different masses. Therefore, several metabolic labeling strategies that apply stable isotopes to minimize error have been developed recently and have been applied in animal models.
Biomarkers to predict CAC risk discovered via label-free quantification analyses
A comparative label-free quantification analysis was conducted in eight patients with UC, eight patients with CD and eight patients with irritable bowel syndrome (IBS). Colonic tissue biopsies were obtained during colonoscopy after written consent and stored in a deep freeze until the assay. Using an Agilent HPLC-Chip 6520 Q-TOF MS system and a label-free quantitative technique (IDEAL-Q v1.0.6.3), signal pathway analysis associated with carcinogenesis was conducted to discover potential biomarkers in CAC (Figure 1). The analysis was conducted according to the degree of intestinal inflammation, type of IBD, and extent of inflammation. To compare against the analysis from IBS samples, the proteins implying CAC risk were isolated (Figure 2A and B). As seen in Figure 2A, 22 significant proteins were found to be potential biomarkers predicting CAC risk in patients with UC . Figure 2B shows the 19 proteins found to be potential biomarkers for CAC risk in patients with CD. Further analysis yielded four important protein biomarkers: proteoglycan 2 (PRG2), S100A6 (calcyclin), ribosomal protein L18 (RPL18), UDP-glucose dehydrogenase (UGDH) as potential target proteins and predictive biomarkers for CAC risk in IBD (Figure 3). PRG is a major component of the animal extracellular matrix and has been shown to be involved in the differentiation process across the epithelial-mesenchymal axis. It is a potential biomarker inferred principally through its ability to bind growth factors and modulate their downstream signaling; malignant tumors have their individual characteristic PRG profiles closely associated with their differentiation and biological behavior. PRG2 has further been implicated as a biomarker for neuropathic pain attributable to advanced pancreatic cancer[63] in an animal model of CAC[27], and as an inflammation related gene of several cancers, including prostate, lung, CRC[64] and pancreatic cancer[65]. The expression of S100 calcium binding protein A6 (S100A6) is upregulated in proliferating and differentiating cells[66], and has been reported to be a possible biomarker for hepatocellular carcinoma[67,68], pancreatic cancer[69], acute lymphoblastic leukemia[70], CRC[71] and breast cancer[72]. RPL18 is a 60S ribosomal protein expressed in stem cells[73] and during adipogenesis[74] UGDH has been suggested as biomarker for cancer metabolism. The oxidation of UDP-glucose is catalyzed by UGDH to generate UDP-glucuronic acid (UDP-GlcA), a precursor of glycosaminoglycans (GAGs). Wang et al[75] showed decreases in the expressions of UGDH, UDP-GlcA and GAG expression after treatment with a UGDH-siRNA in HCT-8 colon cancer cells and concluded that UGDH could be a new target for CRC clinical treatment[76]. Additionally, UGDH has been identified as a potential biomarker for prostate cancer[77,78], hepatocellular carcinoma[79] and breast cancer[80].
Figure 1.
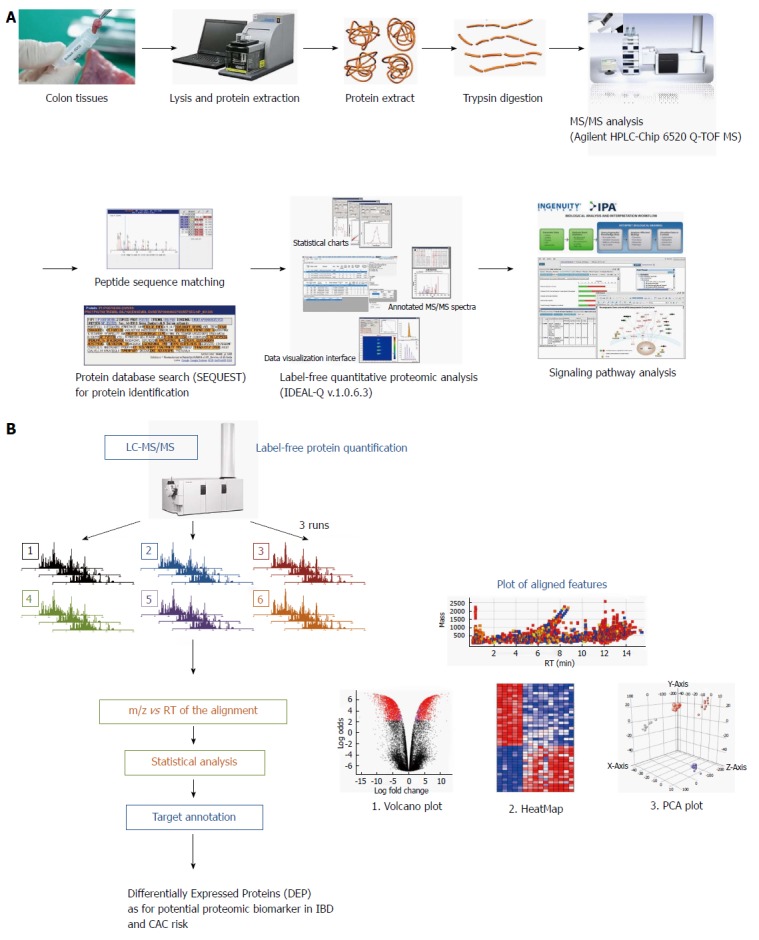
Schematic presentation showing proteome analysis to discover potential biomarkers and label-free quantification analysis in inflammatory bowel disease. A: Applying the label-free quantification method to discover proteomic biomarkers in patients with different types and different stage of inflammatory bowel disease (IBD). Comparative analysis was done in eight patients with ulcerative colitis (UC), eight patients with Crohn’s disease (CD) and eight patients with irritable bowel syndrome (IBS). Biopsied colon tissues were obtained during colonoscopy after written consent, and stored in a deep freeze until assayed. Using Agilent HPLC-Chip 6520 Q-time-of-flight mass spectrometry (TOF-MS) and label-free quantitative proteome analysis (IDEAL-Q v1.0.6.3), significant signal pathway analysis was done. In the current review, the analysis done according to the degree of intestinal inflammation, type of IBD, and extent of inflammation from 24 patients, eight from non-IBD normal patients; i.e., IBS patients, eight from patients with UC, and from patients with CD; B: Label-free protein quantification scheme for potential biomarker for colitis-associated cancer (CAC) risk in 16 patients with IBD.
Figure 2.
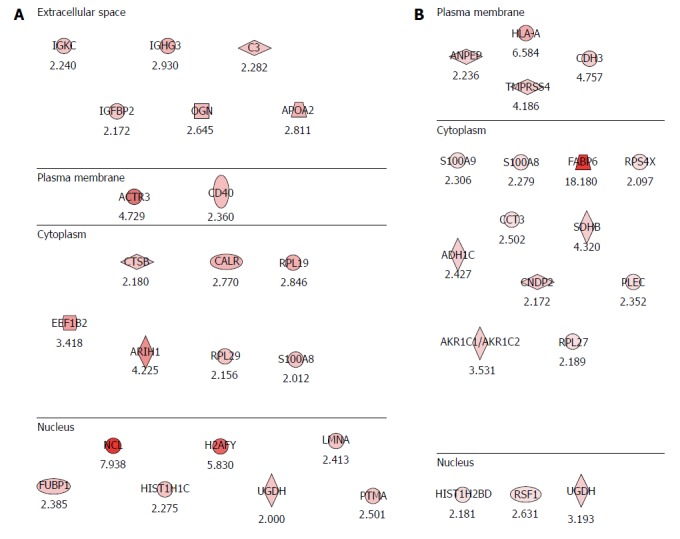
Potential proteomic markers signifying colitis-associated cancer risks in inflammatory bowel disease. A: Proteomic markers for colitis-associated cancer (CAC) risk in patients with ulcerative colitis (UC). The analysis was performed according to the degree of intestinal inflammation, type of inflammatory bowel disease and extent of inflammation. Compared with the analysis of CAC, 22 significant potential biomarkers of CAC risk were obtained from patients as p with UC, six biomarkers existing in the extracellular space, two biomarkers at the plasma membrane, seven are from the cytoplasm, and seven are nuclear proteins; B: Proteomic markers for CAC risk in patients with Crohn’s disease (CD). Eighteen potential biomarkers for the risk of CAC were identified in patients with CD. Four were from the plasma membrane, 11 from the cytoplasm, and three from the nucleus.
Figure 3.
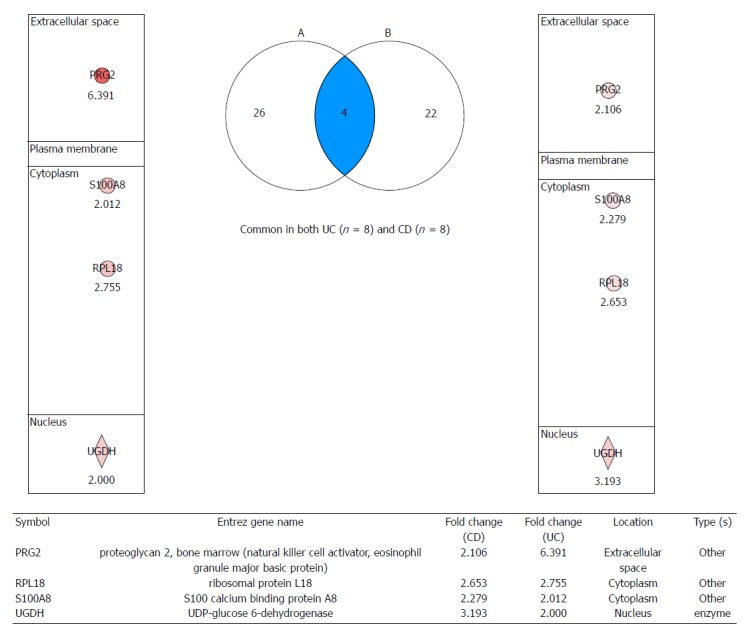
Proteomic markers for colitis-associated cancer risk in patients with both ulcerative colitis and Crohn’s disease. After analyzing signaling pathways from label-free quantitative analysis, four important proteome biomarkers were identified: proteoglycan 2 (PRG2), S100 calcium binding protein A8 (S100A8), ribosomal protein L18 (RPL18), and UDP-glucose dehydrogenase (UGDH), all of which showed fold changes. Validation is ongoing to investigate these biomarkers for predicting colitis-associated cancer risk in patients with inflammatory bowel disease. UC: Ulcerative colitis; CD: Crohn’s disease.
Biomarkers to predict CAC risk discovered via label-based protein quantification analyses
Proteomic techniques with blood and biopsy provide reliable and accurate tools that provide support to clinicians in the diagnosis and treatment of IBD. For example, clinically meaningful biomarkers may be used in the differential diagnosis of CD and UC, or as predictors of treatment responses. Tandem mass spectrometry (MS/MS) is commonly used proteomic analysis. However, various workflows are possible for peptide analysis before MS/MS, as well as bioinformatics, to identify peptides, for which 2-D electrophoresis and subsequent MS, liquid chromatography-MS, 2D DIGE, and iTRAQ are under development. In our previous publication[43], the present status and perspectives regarding these developed proteomic methods were discussed, with descriptions of examples of new biomarkers for the diagnosis, treatment and prognosis of IBD and CAC in mouse models and in humans. In detail, we showed new concepts and technologies of proteomics, such as protein identification and proteome coverage, as determined by iTRAQ with different shotgun proteomic methods in samples from an animal model of CAC that used repeated oral administration of dextran sulfate sodium (DSS) to induce CAC. As previously reported, iTRAQ protein quantification analysis identified fibrinogen beta, prohibitin, transgelin, Hsc-70-interacting protein, suppression of tumorigenicity 13 (ST13), a TKL kinase of the MLK family, dual leucine zipper kinase (MKL1), actin, beta, protein-coding gene (ACTB), ubiquitin carboxyl-term, esterase L3 (UCHL3), coronin, actin binding protein, 1A (CORO1A), hypoxanthin-guanine phosphoribosyltransferase (HPRT), glutathione peroxidase 1 (GPX1), estrogen receptor 1 (ESR1), transcriptional repressor protein 1 (YY1), transcription activator1, ATP-dependent helicase SMARCA4 (BRG1), brahma gene (BRM) and ornithine aminotransferase (OAT) as potential biomarkers for CAC in the DSS-induced colitic cancer model[43].
CONCLUSION
MALDI imaging mass spectrometry (IMS) is a new technology to analyze small peptides in various samples and is a novel tool for studying molecular mechanisms in biological tissues. Recent studies have demonstrated considerable diagnostic and prognostic value, which should be applicable to clinical settings in the near future[81,82]. However, one challenge associated with the use of MALDI-IMS in the identification of potential biomarkers involves the systematic identification of peptides introduced in the MALDI matrix with association of top-down and bottom-up analysis[83]. IMS will be investigated without target-specific reagents to identify new markers for the diagnosis, treatment, and prognosis of CAC, as well as the determination of effective therapies. In the near future, an era of tailored medicine will provide for diagnostic algorithms that include molecular parameters for the detection of early disease and treatment algorithms guided by predicting the individual course of the disease. However, more trials focused on discovering proteomic biomarkers will be necessary to guide the treatment of IBD with more advanced levels of biologicals or molecular targeted therapeutics for inflammation. Using label-free quantification methods on biopsied tissue from patients with IBD, four potential biomarkers, PRG2, S100A6 (calcyclin), RPL18, and UGDH, have been discovered. Further validation of these potential biomarkers will be necessary to ascertain their clinical value.
Footnotes
Supported by Grant from the Ministry of Education and Science Technology, 2010-0002052, Korea and JSPS Asian CORE Program, Japan
P- Reviewer: Prasad KK, van Langenberg DR, Vynios D, Voortman J S- Editor: Gou SX L- Editor: Stewart G E- Editor: Wang CH
References
- 1.Vucelic B. Inflammatory bowel diseases: controversies in the use of diagnostic procedures. Dig Dis. 2009;27:269–277. doi: 10.1159/000228560. [DOI] [PubMed] [Google Scholar]
- 2.Nikolaus S, Schreiber S. Diagnostics of inflammatory bowel disease. Gastroenterology. 2007;133:1670–1689. doi: 10.1053/j.gastro.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Graham DB, Xavier RJ. From genetics of inflammatory bowel disease towards mechanistic insights. Trends Immunol. 2013;34:371–378. doi: 10.1016/j.it.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 5.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 7.Fassan M, Baffa R, Kiss A. Advanced precancerous lesions within the GI tract: the molecular background. Best Pract Res Clin Gastroenterol. 2013;27:159–169. doi: 10.1016/j.bpg.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Mendoza JL, Abreu MT. Biological markers in inflammatory bowel disease: practical consideration for clinicians. Gastroenterol Clin Biol. 2009;33 Suppl 3:S158–S173. doi: 10.1016/S0399-8320(09)73151-3. [DOI] [PubMed] [Google Scholar]
- 9.Gerich ME, McGovern DP. Towards personalized care in IBD. Nat Rev Gastroenterol Hepatol. 2014;11:287–299. doi: 10.1038/nrgastro.2013.242. [DOI] [PubMed] [Google Scholar]
- 10.Goel GA, Kandiel A, Achkar JP, Lashner B. Molecular pathways underlying IBD-associated colorectal neoplasia: therapeutic implications. Am J Gastroenterol. 2011;106:719–730. doi: 10.1038/ajg.2011.51. [DOI] [PubMed] [Google Scholar]
- 11.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 12.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 13.Herszenyi L, Miheller P, Tulassay Z. Carcinogenesis in inflammatory bowel disease. Dig Dis. 2007;25:267–269. doi: 10.1159/000103898. [DOI] [PubMed] [Google Scholar]
- 14.Viennot S, Deleporte A, Moussata D, Nancey S, Flourié B, Reimund JM. Colon cancer in inflammatory bowel disease: recent trends, questions and answers. Gastroenterol Clin Biol. 2009;33 Suppl 3:S190–S201. doi: 10.1016/S0399-8320(09)73154-9. [DOI] [PubMed] [Google Scholar]
- 15.Jawad N, Direkze N, Leedham SJ. Inflammatory bowel disease and colon cancer. Recent Results Cancer Res. 2011;185:99–115. doi: 10.1007/978-3-642-03503-6_6. [DOI] [PubMed] [Google Scholar]
- 16.Rubin DC, Shaker A, Levin MS. Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front Immunol. 2012;3:107. doi: 10.3389/fimmu.2012.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraus S, Arber N. Inflammation and colorectal cancer. Curr Opin Pharmacol. 2009;9:405–410. doi: 10.1016/j.coph.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Kim YJ, Hong KS, Chung JW, Kim JH, Hahm KB. Prevention of colitis-associated carcinogenesis with infliximab. Cancer Prev Res (Phila) 2010;3:1314–1333. doi: 10.1158/1940-6207.CAPR-09-0272. [DOI] [PubMed] [Google Scholar]
- 19.Kim YJ, Lee JS, Hong KS, Chung JW, Kim JH, Hahm KB. Novel application of proton pump inhibitor for the prevention of colitis-induced colorectal carcinogenesis beyond acid suppression. Cancer Prev Res (Phila) 2010;3:963–974. doi: 10.1158/1940-6207.CAPR-10-0033. [DOI] [PubMed] [Google Scholar]
- 20.Hartnett L, Egan LJ. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis. 2012;33:723–731. doi: 10.1093/carcin/bgs006. [DOI] [PubMed] [Google Scholar]
- 21.Rubin DT, Ulitsky A, Poston J, Day R, Huo D. What is the most effective way to communicate results after endoscopy? Gastrointest Endosc. 2007;66:108–112. doi: 10.1016/j.gie.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 22.Dyson JK, Rutter MD. Colorectal cancer in inflammatory bowel disease: what is the real magnitude of the risk? World J Gastroenterol. 2012;18:3839–3848. doi: 10.3748/wjg.v18.i29.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmadi A, Polyak S, Draganov PV. Colorectal cancer surveillance in inflammatory bowel disease: the search continues. World J Gastroenterol. 2009;15:61–66. doi: 10.3748/wjg.15.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papp M, Norman GL, Altorjay I, Lakatos PL. Utility of serological markers in inflammatory bowel diseases: gadget or magic? World J Gastroenterol. 2007;13:2028–2036. doi: 10.3748/wjg.v13.i14.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 26.Alex P, Zachos NC, Nguyen T, Gonzales L, Chen TE, Conklin LS, Centola M, Li X. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. 2009;15:341–352. doi: 10.1002/ibd.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han NY, Choi W, Park JM, Kim EH, Lee H, Hahm KB. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J Dig Dis. 2013;14:166–174. doi: 10.1111/1751-2980.12035. [DOI] [PubMed] [Google Scholar]
- 28.Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826.e2. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iskandar HN, Ciorba MA. Biomarkers in inflammatory bowel disease: current practices and recent advances. Transl Res. 2012;159:313–325. doi: 10.1016/j.trsl.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham BP, Kane S. Fecal markers: calprotectin and lactoferrin. Gastroenterol Clin North Am. 2012;41:483–495. doi: 10.1016/j.gtc.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Gisbert JP, González-Lama Y, Maté J. [Role of biological markers in inflammatory bowel disease] Gastroenterol Hepatol. 2007;30:117–129. doi: 10.1157/13100073. [DOI] [PubMed] [Google Scholar]
- 32.Langhorst J, Boone J. Fecal lactoferrin as a noninvasive biomarker in inflammatory bowel diseases. Drugs Today (Barc) 2012;48:149–161. doi: 10.1358/dot.2012.48.2.1732555. [DOI] [PubMed] [Google Scholar]
- 33.Langhorst J, Elsenbruch S, Koelzer J, Rueffer A, Michalsen A, Dobos GJ. Noninvasive markers in the assessment of intestinal inflammation in inflammatory bowel diseases: performance of fecal lactoferrin, calprotectin, and PMN-elastase, CRP, and clinical indices. Am J Gastroenterol. 2008;103:162–169. doi: 10.1111/j.1572-0241.2007.01556.x. [DOI] [PubMed] [Google Scholar]
- 34.Walkiewicz D, Werlin SL, Fish D, Scanlon M, Hanaway P, Kugathasan S. Fecal calprotectin is useful in predicting disease relapse in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:669–673. doi: 10.1002/ibd.20376. [DOI] [PubMed] [Google Scholar]
- 35.Marvin LF, Roberts MA, Fay LB. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin Chim Acta. 2003;337:11–21. doi: 10.1016/j.cccn.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Hortin GL. The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clin Chem. 2006;52:1223–1237. doi: 10.1373/clinchem.2006.069252. [DOI] [PubMed] [Google Scholar]
- 37.Nanni P, Parisi D, Roda G, Casale M, Belluzzi A, Roda E, Mayer L, Roda A. Serum protein profiling in patients with inflammatory bowel diseases using selective solid-phase bulk extraction, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and chemometric data analysis. Rapid Commun Mass Spectrom. 2007;21:4142–4148. doi: 10.1002/rcm.3323. [DOI] [PubMed] [Google Scholar]
- 38.Liu XP, Shen J, Li ZF, Yan L, Gu J. A serum proteomic pattern for the detection of colorectal adenocarcinoma using surface enhanced laser desorption and ionization mass spectrometry. Cancer Invest. 2006;24:747–753. doi: 10.1080/07357900601063873. [DOI] [PubMed] [Google Scholar]
- 39.Gemoll T, Roblick UJ, Auer G, Jörnvall H, Habermann JK. SELDI-TOF serum proteomics and colorectal cancer: a current overview. Arch Physiol Biochem. 2010;116:188–196. doi: 10.3109/13813455.2010.495130. [DOI] [PubMed] [Google Scholar]
- 40.Alex P, Gucek M, Li X. Applications of proteomics in the study of inflammatory bowel diseases: Current status and future directions with available technologies. Inflamm Bowel Dis. 2009;15:616–629. doi: 10.1002/ibd.20652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brewis IA, Brennan P. Proteomics technologies for the global identification and quantification of proteins. Adv Protein Chem Struct Biol. 2010;80:1–44. doi: 10.1016/B978-0-12-381264-3.00001-1. [DOI] [PubMed] [Google Scholar]
- 42.Vaiopoulou A, Gazouli M, Theodoropoulos G, Zografos G. Current advantages in the application of proteomics in inflammatory bowel disease. Dig Dis Sci. 2012;57:2755–2764. doi: 10.1007/s10620-012-2291-4. [DOI] [PubMed] [Google Scholar]
- 43.Han NY, Kim EH, Choi J, Lee H, Hahm KB. Quantitative proteomic approaches in biomarker discovery of inflammatory bowel disease. J Dig Dis. 2012;13:497–503. doi: 10.1111/j.1751-2980.2012.00625.x. [DOI] [PubMed] [Google Scholar]
- 44.Mangerich A, Dedon PC, Fox JG, Tannenbaum SR, Wogan GN. Chemistry meets biology in colitis-associated carcinogenesis. Free Radic Res. 2013;47:958–986. doi: 10.3109/10715762.2013.832239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feagins LA, Souza RF, Spechler SJ. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat Rev Gastroenterol Hepatol. 2009;6:297–305. doi: 10.1038/nrgastro.2009.44. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe T, Kobunai T, Yamamoto Y, Ikeuchi H, Matsuda K, Ishihara S, Nozawa K, Iinuma H, Kanazawa T, Tanaka T, et al. Predicting ulcerative colitis-associated colorectal cancer using reverse-transcription polymerase chain reaction analysis. Clin Colorectal Cancer. 2011;10:134–141. doi: 10.1016/j.clcc.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Colliver DW, Crawford NP, Eichenberger MR, Zacharius W, Petras RE, Stromberg AJ, Galandiuk S. Molecular profiling of ulcerative colitis-associated neoplastic progression. Exp Mol Pathol. 2006;80:1–10. doi: 10.1016/j.yexmp.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Tatsumi N, Kushima R, Vieth M, Mukaisho K, Kakinoki R, Okabe H, Borchard F, Stolte M, Okanoue T, Hattori T. Cytokeratin 7/20 and mucin core protein expression in ulcerative colitis-associated colorectal neoplasms. Virchows Arch. 2006;448:756–762. doi: 10.1007/s00428-006-0188-3. [DOI] [PubMed] [Google Scholar]
- 50.Thorsteinsdottir S, Gudjonsson T, Nielsen OH, Vainer B, Seidelin JB. Pathogenesis and biomarkers of carcinogenesis in ulcerative colitis. Nat Rev Gastroenterol Hepatol. 2011;8:395–404. doi: 10.1038/nrgastro.2011.96. [DOI] [PubMed] [Google Scholar]
- 51.Yoo SH, Jung HS, Sohn WS, Kim BH, Ku BH, Kim YS, Park SW, Hahm KB. Volatile sulfur compounds as a predictor for esophagogastroduodenal mucosal injury. Gut Liver. 2008;2:113–118. doi: 10.5009/gnl.2008.2.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujiwara I, Yashiro M, Kubo N, Maeda K, Hirakawa K. Ulcerative colitis-associated colorectal cancer is frequently associated with the microsatellite instability pathway. Dis Colon Rectum. 2008;51:1387–1394. doi: 10.1007/s10350-008-9212-9. [DOI] [PubMed] [Google Scholar]
- 53.Araki K, Mikami T, Yoshida T, Kikuchi M, Sato Y, Oh-ishi M, Kodera Y, Maeda T, Okayasu I. High expression of HSP47 in ulcerative colitis-associated carcinomas: proteomic approach. Br J Cancer. 2009;101:492–497. doi: 10.1038/sj.bjc.6605163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujii S, Tominaga K, Kitajima K, Takeda J, Kusaka T, Fujita M, Ichikawa K, Tomita S, Ohkura Y, Ono Y, et al. Methylation of the oestrogen receptor gene in non-neoplastic epithelium as a marker of colorectal neoplasia risk in longstanding and extensive ulcerative colitis. Gut. 2005;54:1287–1292. doi: 10.1136/gut.2004.062059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrity-Park MM, Loftus EV, Sandborn WJ, Bryant SC, Smyrk TC. MHC Class II alleles in ulcerative colitis-associated colorectal cancer. Gut. 2009;58:1226–1233. doi: 10.1136/gut.2008.166686. [DOI] [PubMed] [Google Scholar]
- 56.Fujii S, Katake Y, Tanaka H. Increased expression of DNA methyltransferase-1 in non-neoplastic epithelium helps predict colorectal neoplasia risk in ulcerative colitis. Digestion. 2010;82:179–186. doi: 10.1159/000311064. [DOI] [PubMed] [Google Scholar]
- 57.Saigusa S, Araki T, Tanaka K, Hashimoto K, Okita Y, Fujikawa H, Okugawa Y, Toiyama Y, Inoue Y, Uchida K, et al. Identification of patients with developing ulcerative colitis-associated neoplasia by nitrative DNA damage marker 8-nitroguanin expression in rectal mucosa. J Clin Gastroenterol. 2013;47:e80–e86. doi: 10.1097/MCG.0b013e31828f51e1. [DOI] [PubMed] [Google Scholar]
- 58.Ock CY, Kim EH, Choi DJ, Lee HJ, Hahm KB, Chung MH. 8-Hydroxydeoxyguanosine: not mere biomarker for oxidative stress, but remedy for oxidative stress-implicated gastrointestinal diseases. World J Gastroenterol. 2012;18:302–308. doi: 10.3748/wjg.v18.i4.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gushima M, Hirahashi M, Matsumoto T, Fujita K, Fujisawa R, Mizumoto K, Nakabeppu Y, Iida M, Yao T, Tsuneyoshi M. Altered expression of MUTYH and an increase in 8-hydroxydeoxyguanosine are early events in ulcerative colitis-associated carcinogenesis. J Pathol. 2009;219:77–86. doi: 10.1002/path.2570. [DOI] [PubMed] [Google Scholar]
- 60.Hashimoto K, Saigusa S, Araki T, Tanaka K, Okita Y, Fujikawa H, Kawamura M, Okugawa Y, Toiyama Y, Inoue Y, et al. Correlation of CCL20 expression in rectal mucosa with the development of ulcerative colitis-associated neoplasia. Oncol Lett. 2013;6:1271–1276. doi: 10.3892/ol.2013.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gushima M, Hirahashi M, Matsumoto T, Fujita K, Ohuchida K, Oda Y, Yao T, Iida M, Tsuneyoshi M. Expression of activation-induced cytidine deaminase in ulcerative colitis-associated carcinogenesis. Histopathology. 2011;59:460–469. doi: 10.1111/j.1365-2559.2011.03965.x. [DOI] [PubMed] [Google Scholar]
- 62.Yeo M, Kim DK, Park HJ, Oh TY, Kim JH, Cho SW, Paik YK, Hahm KB. Loss of transgelin in repeated bouts of ulcerative colitis-induced colon carcinogenesis. Proteomics. 2006;6:1158–1165. doi: 10.1002/pmic.200500390. [DOI] [PubMed] [Google Scholar]
- 63.Demir IE, Schorn S, Schremmer-Danninger E, Wang K, Kehl T, Giese NA, Algül H, Friess H, Ceyhan GO. Perineural mast cells are specifically enriched in pancreatic neuritis and neuropathic pain in pancreatic cancer and chronic pancreatitis. PLoS One. 2013;8:e60529. doi: 10.1371/journal.pone.0060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer TE, Chu LW, Li Q, Yu K, Rosenberg PS, Menashe I, Chokkalingam AP, Quraishi SM, Huang WY, Weiss JM, et al. The association between inflammation-related genes and serum androgen levels in men: the prostate, lung, colorectal, and ovarian study. Prostate. 2012;72:65–71. doi: 10.1002/pros.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hagihara A, Miyamoto K, Furuta J, Hiraoka N, Wakazono K, Seki S, Fukushima S, Tsao MS, Sugimura T, Ushijima T. Identification of 27 5’ CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene. 2004;23:8705–8710. doi: 10.1038/sj.onc.1207783. [DOI] [PubMed] [Google Scholar]
- 66.Nowotny M, Bhattacharya S, Filipek A, Krezel AM, Chazin W, Kuznicki J. Characterization of the interaction of calcyclin (S100A6) and calcyclin-binding protein. J Biol Chem. 2000;275:31178–31182. doi: 10.1074/jbc.M001622200. [DOI] [PubMed] [Google Scholar]
- 67.Li Z, Tang M, Ling B, Liu S, Zheng Y, Nie C, Yuan Z, Zhou L, Guo G, Tong A, et al. Increased expression of S100A6 promotes cell proliferation and migration in human hepatocellular carcinoma. J Mol Med (Berl) 2014;92:291–303. doi: 10.1007/s00109-013-1104-3. [DOI] [PubMed] [Google Scholar]
- 68.Hua Z, Chen J, Sun B, Zhao G, Zhang Y, Fong Y, Jia Z, Yao L. Specific expression of osteopontin and S100A6 in hepatocellular carcinoma. Surgery. 2011;149:783–791. doi: 10.1016/j.surg.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Zihao G, Jie Z, Yan L, Jing Z, Jing C, Xue L, Jing Z, Heng LW, Ru G, Jianyu H. Analyzing S100A6 expression in endoscopic ultrasonography-guided fine-needle aspiration specimens: a promising diagnostic method of pancreatic cancer. J Clin Gastroenterol. 2013;47:69–75. doi: 10.1097/MCG.0b013e3182601752. [DOI] [PubMed] [Google Scholar]
- 70.Tamai H, Miyake K, Yamaguchi H, Takatori M, Dan K, Inokuchi K, Shimada T. Resistance of MLL-AFF1-positive acute lymphoblastic leukemia to tumor necrosis factor-alpha is mediated by S100A6 upregulation. Blood Cancer J. 2011;1:e38. doi: 10.1038/bcj.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kilańczyk E, Graczyk A, Ostrowska H, Kasacka I, Leśniak W, Filipek A. S100A6 is transcriptionally regulated by β-catenin and interacts with a novel target, lamin A/C, in colorectal cancer cells. Cell Calcium. 2012;51:470–477. doi: 10.1016/j.ceca.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 72.McKiernan E, McDermott EW, Evoy D, Crown J, Duffy MJ. The role of S100 genes in breast cancer progression. Tumour Biol. 2011;32:441–450. doi: 10.1007/s13277-010-0137-2. [DOI] [PubMed] [Google Scholar]
- 73.Chooi WH, Zhou R, Yeo SS, Zhang F, Wang DA. Determination and validation of reference gene stability for qPCR analysis in polysaccharide hydrogel-based 3D chondrocytes and mesenchymal stem cell cultural models. Mol Biotechnol. 2013;54:623–633. doi: 10.1007/s12033-012-9604-x. [DOI] [PubMed] [Google Scholar]
- 74.Fromm-Dornieden C, von der Heyde S, Lytovchenko O, Salinas-Riester G, Brenig B, Beissbarth T, Baumgartner BG. Novel polysome messages and changes in translational activity appear after induction of adipogenesis in 3T3-L1 cells. BMC Mol Biol. 2012;13:9. doi: 10.1186/1471-2199-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang TP, Pan YR, Fu CY, Chang HY. Down-regulation of UDP-glucose dehydrogenase affects glycosaminoglycans synthesis and motility in HCT-8 colorectal carcinoma cells. Exp Cell Res. 2010;316:2893–2902. doi: 10.1016/j.yexcr.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 76.Egger S, Chaikuad A, Kavanagh KL, Oppermann U, Nidetzky B. UDP-glucose dehydrogenase: structure and function of a potential drug target. Biochem Soc Trans. 2010;38:1378–1385. doi: 10.1042/BST0381378. [DOI] [PubMed] [Google Scholar]
- 77.Huang D, Casale GP, Tian J, Lele SM, Pisarev VM, Simpson MA, Hemstreet GP. Udp-glucose dehydrogenase as a novel field-specific candidate biomarker of prostate cancer. Int J Cancer. 2010;126:315–327. doi: 10.1002/ijc.24820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei Q, Galbenus R, Raza A, Cerny RL, Simpson MA. Androgen-stimulated UDP-glucose dehydrogenase expression limits prostate androgen availability without impacting hyaluronan levels. Cancer Res. 2009;69:2332–2339. doi: 10.1158/0008-5472.CAN-08-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan HZ, Liu H, Zhang C, Gao DM, Xue Q, Chen J, Sun RX, Liu YK, Yang PY. Comparative proteomics and molecular mechanical analysis in CDA-II induced therapy of LCI-D20 hepatocellular carcinoma model. J Cancer Res Clin Oncol. 2009;135:591–602. doi: 10.1007/s00432-008-0493-0. [DOI] [PubMed] [Google Scholar]
- 80.Huh JW, Choi MM, Yang SJ, Yoon SY, Choi SY, Cho SW. Inhibition of human UDP-glucose dehydrogenase expression using siRNA expression vector in breast cancer cells. Biotechnol Lett. 2005;27:1229–1232. doi: 10.1007/s10529-005-0022-z. [DOI] [PubMed] [Google Scholar]
- 81.Maier SK, Hahne H, Gholami AM, Balluff B, Meding S, Schoene C, Walch AK, Kuster B. Comprehensive identification of proteins from MALDI imaging. Mol Cell Proteomics. 2013;12:2901–2910. doi: 10.1074/mcp.M113.027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ko KH, Kwon CI, Park SH, Han NY, Lee HK, Kim EH, Hahm KB. Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Mass Spectrometry (MALDI-TOF IMS) for Premalignant Gastrointestinal Lesions. Clin Endosc. 2013;46:611–619. doi: 10.5946/ce.2013.46.6.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry--looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]


