Abstract
AIM: To develop a novel non-sequencing method for the detection of hepatitis B virus (HBV) pre-S deletion mutants in HBV carriers.
METHODS: The entire region of HBV pre-S1 and pre-S2 was amplified by polymerase chain reaction (PCR). The size of PCR products was subsequently determined by capillary gel electrophoresis (CGE). CGE were carried out in a PACE-MDQ instrument equipped with a UV detector set at 254 nm. The samples were separated in 50 μm ID eCAP Neutral Coated Capillaries using a voltage of 6 kV for 30 min. Data acquisition and analysis were performed using the 32 Karat Software. A total of 114 DNA clones containing different sizes of the HBV pre-S gene were used to determine the accuracy of the CGE method. One hundred and fifty seven hepatocellular carcinoma (HCC) and 160 non-HCC patients were recruited into the study to assess the association between HBV pre-S deletion and HCC by using the newly-established CGE method. Nine HCC cases with HBV pre-S deletion at the diagnosis year were selected to conduct a longitudinal observation using serial serum samples collected 2-9 years prior to HCC diagnosis.
RESULTS: CGE allowed the separation of PCR products differing in size > 3 bp and was able to identify 10% of the deleted DNA in a background of wild-type DNA. The accuracy rate of CGE-based analysis was 99.1% compared with the clone sequencing results. Using this assay, pre-S deletion was more frequently found in HCC patients than in non-HCC controls (47.1% vs 28.1%, P < 0.001). Interestingly, the increased risk of HCC was mainly contributed by the short deletion of pre-S. While the deletion ≤ 99 bp was associated with a 2.971-fold increased risk of HCC (95%CI: 1.723-5.122, P < 0.001), large deletion (> 99 bp) did not show any association with HCC (P = 0.918, OR = 0.966, 95%CI: 0.501-1.863). Of the 9 patients who carried pre-S deletions at the stage of HCC, 88.9% (8/9) had deletions 2-5 years prior to HCC, while only 44.4%4 (4/9) contained such deletions 6-9 years prior to HCC.
CONCLUSION: CGE is a sensitive approach for HBV pre-S deletion analysis. Pre-S deletion, especially for short DNA fragment deletion, is a useful predictive marker for HCC.
Keywords: Hepatitis B virus, Pre-S deletion, Capillary gel electrophoresis, Sequence analysis, Hepatocellular carcinoma
Core tip: Hepatitis B virus pre-S deletions are one of the risk factors associated with hepatocellular carcinoma (HCC). However, the lack of a convenient method hampers its clinical application. In this study, we develop a capillary gel electrophoresis approach that can quantitatively identify low amounts of deletions within a heterogeneous viral population. By applying this method in a case-control study, we not only confirmed previous observations that pre-S deletions increased the risk for HCC but also reported, for the first time, that only small deletions (≤ 99 bp) have a significant association with HCC, rather than large deletions.
INTRODUCTION
Hepatitis B virus (HBV) infection plays a major causative role in the development of hepatocellular carcinoma (HCC)[1]. HBV is a small enveloped 3.2 kb DNA virus with four open reading frames (ORFs). HBV envelope proteins are encoded by three different in-frame start codons. Depending on the translated initiation site among S, pre-S1, or pre-S2, three different sized proteins are produced: a small hepatitis B surface protein (small HBs), a medium hepatitis B surface protein (middle HBs), and a large hepatitis B surface protein (large HBs)[2]. The pre-S1 region mediates the attachment of the virus to hepatocytes[3]. HBV pre-S deletion, as well as C1653T, T1753V, and A1762T/G1764A in the basal core promoter /enhancer II region[4-6] and G1899A[4,7] in the pre-C gene, have been reported to increase the risk of HCC. Pre-S mutations are highly prevalent in countries where HBV infection is endemic[8]. In the past few decades, many studies have demonstrated that the presence of pre-S deletions are significantly associated with the development of HCC[5,9-15]. A meta-analysis that included a total of 16745 HBV-infected cases revealed that pre-S1 and pre-S2 deletions had a summary odds ratio (OR) of 2.94 (95%CI: 2.22-3.89) and 3.02 (95%CI: 2.03-4.50) for HCC, respectively[16].
Several hypotheses have been proposed to explain the pathogenic effects of HBV pre-S deletion mutants. Because the pre-S region contains several epitopes for T or B cells, HBV mutants carrying pre-S deletions can escape host immune surveillance and cause persistent infections[17]. Large HBs with deletions in the pre-S region are found to be retained in the endoplasmic reticulum (ER), resulting in ER stress, which in turn induces oxidative DNA damage and genomic instability[18,19]. Meanwhile, mutant large HBs can regulate the expression of many cellular genes, including cyclooxygenase-2, cyclin A[20], p27 (Kip1)[21-23], Bcl-2, Bcl-xL, and Mcl-1[24], thus promoting hepatocyte proliferation and inhibiting cell death induced by 5-fluorouracil. In transgenic mice, pre-S deletion mutants induced dysplasia of hepatocytes and led to the development of HCC, further supporting the fact that this variant has striking oncogenic properties[20].
To detect pre-S deletions, the most common method used previously was direct sequencing of polymerase chain reaction (PCR) products[9-15,25,26]. However, most pre-S deletion mutants co-exist with wild-type viruses in the blood of HBV carriers[27,28]. Therefore, to reveal the minority mutants in the viral population, a cloning procedure is required before sequence analysis. This methodology makes the detection of pre-S deletions both tedious and expensive, which is not suitable for large-scale screening tests. Other assays available to identify HBV pre-S deletions include polyacrylamide gel electrophoresis (PAGE)[27] and pre-S gene chip[28]. However, these techniques are relatively laborious and yield low-resolution results[29,30]. Therefore, there is an urgent need for developing a convenient and reliable method for detecting pre-S deletions.
Capillary gel electrophoresis (CGE) is a separation method based on using soluble polymers to create a replaceable molecular sieve that allows for size-based separation. This method is characterized by high sensitivity, a short analysis time, and a small sample size requirement. These features have led to many applications of CGE in analytical chemistry, pharmaceutical analysis, endocrinology, and immunology[31-34]. This method has also been successfully applied to the analysis of nucleic acids, including rapid separation of DNA restriction fragments[35] and DNA sequencing[36,37]. In this study, we established a rapid method based on CGE to identify pre-S deletion mutations in HBV-infected patients. This approach has proved to be a useful tool in the molecular epidemiological study of HBV pre-S deletions and the risk of HCC.
MATERIALS AND METHODS
Samples
The HCC cases and HBV-infected non-HCC controls were obtained from a prospective cohort started in 1992 that included 807 hepatitis B surface antigen (HBsAg)-positive and 761 HBsAg-negative individuals from 7 towns in Qidong[38]. Until the end of 2012, a total of 181 HCC were diagnosed, of which 169 were from the HBsAg-positive group and 12 from HBsAg-negative group. The 157 HCC patients who showed positive PCR results for pre-S gene amplification were selected as the case group. The inclusion criteria for the non-HCC control group were: (1) HBV-infected individual without HCC; (2) positive in HBV pre-S gene amplification; and (3) age, sex, and living location matched with the subjects in the HCC group. According to analysis of the phylogenetic tree constructed by pre-C/C sequence (nt. 1824-2275), genotype C1 infection was found in 96.8% (5/157) of HCC cases and 96.2% (6/160) of the controls. The diagnosis of HCC was based on one of the following criteria: positive histological findings or positive imaging on either computerized tomography or ultrasonography along with α-fetoprotein (AFP) serum levels ≥ 400 ng/mL. The diagnosis of chronic hepatitis B (CHB) was based on the Guideline on Prevention and Treatment of Chronic Hepatitis B in China[39]. The study was approved by the local ethical committee at the Qidong Liver Cancer Institute and Shanghai Cancer Institute and conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from each participant.
Extraction of viral DNA and amplification of the pre-S region
The QIAamp MinElute Virus Spin Kit (QIAGEN, Hilden, Germany) was used to extract HBV DNA from 100 μL plasma according to the manufacturer’s protocol. The HBV pre-S gene from nt. 2815 to 175 was amplified by semi-nested PCR using pre-SF1 [5’-GGTCACCATATTCTTGGGAAC-3’ (nt. 2815-2835), forward] and pre-SR1 [5’-CTGTAACACGAGCAGGGGT-3’ (nt. 184-202), reverse] as the first-round primers, and pre-SF1 and pre-SR2 [5’-CTGATGTTGTGTTCTCCATG-3’ (nt. 155-175), reverse] as the second-round primers. PrimeSTAR Max polymerase (Takara, Dalian, China) was used for PCR under the following conditions: 94 °C for 3 min, followed by 35 cycles of 94 °C for 25 s, 60 °C for 25 s, and 72 °C for 1 min, and a final extension at 72 °C for 7 min. The PCR products were purified using AxyPrep™ PCR Cleanup kits (Axygen Scientific, Union City, CA, United States).
Detection of pre-S deletions by CGE
Analyses of PCR samples were carried out in a PACE-MDQ instrument (Beckman Instruments, Fullerton, CA, United States) equipped with a UV detector set at 254 nm. Separations were performed in 50 μm ID eCAP Neutral Coated Capillaries (Beckman P/N 477441). The separation buffer (Beckman Coulter eCAP dsDNA 1000 kit) was injected into the capillary using 50 psi of pressure for 1 min. The samples were then injected using a voltage of 4 kV for 50 s and separated using a voltage of 6 kV for 30 min. Data acquisition and analysis were performed using the 32 Karat software (Beckman Instruments).
Detection of pre-S deletions by DNA clone sequencing
The HBV pre-S gene from nt. 2815-175 was amplified from 19 serum samples by PCR. The PCR products were cloned into the pUCm-T plasmid (Shanghai Shenergy Biocolor BioScience and Technology Co.). The recombinant plasmids were introduced into the Escherichia coli DH5α strain. A total of 114 clones were selected for sequencing analysis. BioEdit software (version 5.0.9; available at http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was used to align the validated sequences against the wild-type genotype C HBV DNA reference sequence (GenBank accession no. GU434374.1).
Statistical analysis
Statistical analyses were performed using the SPSS software, version 12.0 (SPSS Inc., Chicago, IL, United States). The ORs and 95%CIs were calculated using a two-by-two frequency table. The comparison between categorical variables was tested using the χ2 test. All of the tests were two-tailed, and a P value of < 0.05 was considered to be statistically significant.
RESULTS
Establishment of a rapid method based on CGE to detect the HBV pre-S deletions
According to previous results from our group[27] and others[40-45], pre-S deletions can occur at any position within the pre-S1 and pre-S2 genes, with deletion sizes ranging from 3 bp to 321 bp. Thus, in this study, a DNA fragment from nt. 2815-175, which covers the entire pre-S1 and pre-S2 region, was PCR amplified and subsequently analyzed using CGE. Theoretically, the length of the PCR products amplified from the HBV wild-type and mutants with the maximum deletion in the pre-S region could be 576 bp and 255 bp, respectively. Therefore, a DNA marker containing molecular weights of 246 bp, 325 bp, 388 bp, 435 bp, 545 bp, and 576 bp was chosen to construct a standard curve. CGE showed a linear detection range between 246 bp and 576 bp, with a regression coefficient (R2) of 0.999 (Figure 1A). Figure 1B demonstrates the profiles of CGE separation for the DNA marker and the 5 PCR products of the pre-S gene amplified from the serum samples of Qidong HCC patients. The size of the PCR fragment calculated according to the CGE migration time was exactly the same as that obtained from DNA sequencing. The CGE method could clearly identify PCR products differing in size by 6 bp (sample b). More importantly, CGE could quantitatively characterize the mixed HBV population by calculating the different peak areas. In sample d, wild-type HBV co-existed with 40.1% deletion mutants, whereas in sample e, the wild-type virus was mixed with 14.7% type I mutants (-21 bp) and 34.1% type II mutants (-54 bp).
Figure 1.
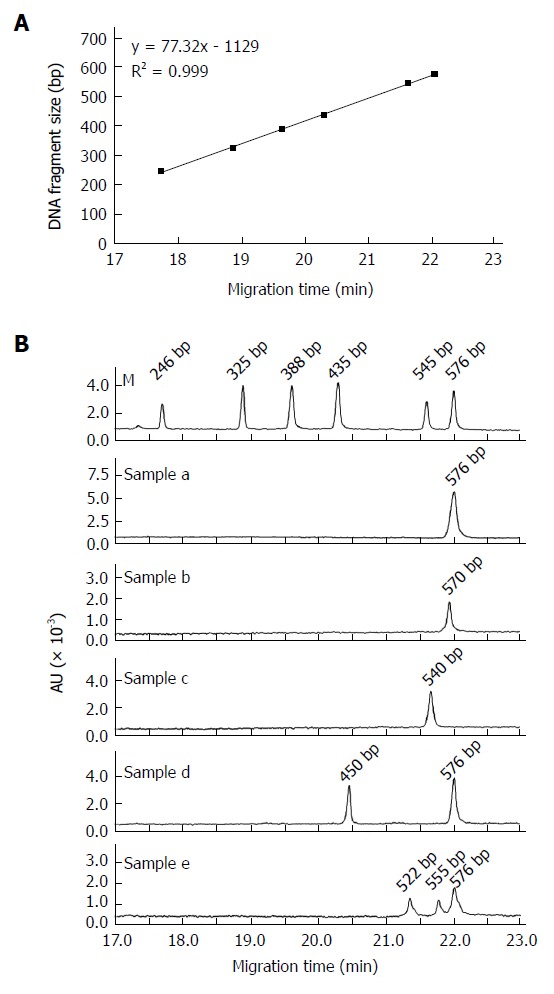
Establishment of a rapid method based on capillary gel electrophoresis to detect HBV pre-S deletions. A: A DNA marker was applied to construct the standard curve. CGE was used to detect the size of DNA fragments with a linear range of 246 bp to 576 bp and a regression coefficient (R2) of 0.999; B: Representative CGE profiles of the pre-S DNA amplification products from the serum samples of 5 HBV-infected subjects. Sample a: wild-type HBV; Sample b: 6 bp deletion mutant alone; Sample c: 36 bp deletion mutant alone; Sample d: mixture of wild-type virus and a 126 bp deletion mutant; Sample e: mixture of wild-type virus and 21 bp and 54 bp deletion mutants. M: Maker (246 bp, 325 bp, 388 bp, 435 bp, 545 bp, and 576 bp); AU: Absorbance units.
Evaluation of the accuracy of CGE in the detection of HBV pre-S deletions
A total of 114 DNA clones containing different sizes of the HBV pre-S gene were used to test the accuracy of the CGE method. The characteristics of the 19 cases used for clone construction are presented in Table 1. After PCR amplification, DNA fragments from HBV nt. 2815 to nt. 175 were subjected to CGE analysis. Among the 114 pre-S clone inserts, 77 were wild-type sequences, while the remaining 37 were truncated sequences with deletions ranging in size from 3 bp to 294 bp. For these 114 samples, the CGE method was able to correctly characterize 113 DNA samples. It only failed to detect one sample with a 3 bp deletion (Table 2). The overall accuracy of the CGE approach was 99.1% (113/114). These data indicate that CGE is a sensitive method for DNA size prediction and that it can generate comparable results to DNA sequencing.
Table 1.
Characteristics of the 19 cases used for clone sequencing
| Cases | Sex | Age | Clinical diagnosis | Clone No. | Pre-S deletion size (bp) | Pre-S deletion site (nt) |
| 486 | Male | 33 | HCC | 486-1, 486-2, 486-3, 486-4, 486-5, 486-6 | 0 | / |
| 6-200 | Male | 60 | HCC | 6-200-1,6-200-2,6-200-3,6-200-4,6-200-5,6-200-6 | 0 | / |
| 440 | Male | 52 | HCC | 440-1, 440-2, 440-3, 440-4, 440-5, 440-6 | 6 | 6-11 |
| 221 | Male | 38 | HCC | 221-1 | 0 | / |
| 221-2, 221-3, 221-4, 221-5, 221-6 | 78 | 3107-3184 | ||||
| 644 | Male | 53 | HCC | 644-1, 644-2, 644-3, 644-5, 644-6 | 0 | / |
| 644-4 | 33 | 18-50 | ||||
| 606 | Male | 41 | HCC | 606-1, 606-2, 606-3, 606-4, 606-6 | 0 | / |
| 606-5 | 3 | 3199-3201 | ||||
| 6-133 | Male | 50 | HCC | 6-133-1, 6-133-3, 6-133-4, 6-133-5 | 0 | / |
| 6-133-2, 6-133-6 | 42 | 25-66 | ||||
| 4-066 | Female | 58 | HCC | 4-066-1, 4-066-2, 4-066-4, 4-066-5, 4-066-6 | 0 | / |
| 4-066-3 | 33 | 34-66 | ||||
| 140 | Male | 64 | CH | 140-1, 140-2, 140-3, 140-4, 140-5, 140-6 | 0 | / |
| 79 | Female | 45 | CH | 79-1, 79-2, 79-3, 79-4, 79-5, 79-6 | 0 | / |
| 96 | Male | 43 | CH | 96-1, 96-2, 96-3, 96-4, 96-5, 96-6 | 0 | / |
| 141 | Male | 32 | CH | 141-1, 141-2, 141-3, 141-4, 141-5, 141-6 | 0 | / |
| 207 | Male | 60 | CH | 207-1, 207-2, 207-3, 207-4, 207-5, 207-6 | 36 | 62-97 |
| 259 | Male | 70 | CH | 259-1, 259-2, 259-3, 259-4, 259-5, 259-6 | 183 | 3015-3197 |
| 70 | Male | 49 | CH | 70-1, 70-5 | 126 | 2970-3095 |
| 70-2, 70-3, 70-4, 70-6 | 0 | / | ||||
| 296 | Male | 56 | CH | 296-1, 296-4, 296-5, 296-6 | 0 | / |
| 296-2, 296-3 | 123 | 2922-3044 | ||||
| 356 | Male | 38 | CH | 356-1, 356-4, 356-5, 356-6 | 0 | / |
| 356-2 | 21 | 33-53 | ||||
| 356-3 | 54 | 2906-2959 | ||||
| 229 | Male | 67 | CH | 229-1, 229-6 | 15 | 42-56 |
| 229-2, 229-3, 229-4, 229-5 | 0 | / | ||||
| 322 | Male | 58 | CH | 322-1. 322-2, 322-3, 322-4, 322-6 | 0 | / |
| 322-5 | 24 | 45-68 |
HCC: Hepatocellular carcinoma.
Table 2.
Comparison of cloning-based sequencing and capillary gel electrophoresis analysis of HBV pre-S deletions
| Size of deletion (bp) | Cloning-based sequencing (No.) | Capillary gel electrophoresis1 (No.) |
| Wild type | 77 | 77 |
| Pre-S deletion | ||
| 0-51 | 21 | 20 |
| 54-99 | 6 | 6 |
| 102-294 | 10 | 10 |
| Total | 114 | 113 |
The number of clones for which the result was in agreement with that of the sequencing analysis.
Analysis of the pre-S deletions in HCC and non-HCC patients by CGE
We used this CGE-based method to assess the association between HBV pre-S deletions and HCC in serum samples from 157 HCC patients and 160 non-HCC controls. The pre-S deletion was more frequently found in HCC patients than in non-HCC controls (47.1% vs 28.1%, respectively, P < 0.001, Table 3). The presence of the pre-S deletion was associated with a 2.278-fold higher risk of HCC (95%CI: 1.430-3.630). It is worth pointing out that among the 119 patients who had pre-S deletion mutants, 86 (72.2%) were co-infected with wild-type viruses.
Table 3.
Analysis of pre-S deletions in hepatocellular carcinoma and non- hepatocellular carcinoma patients by capillary gel electrophoresis n (%)
| Characteristics | HCC n = 157 | Non-HCC n = 160 | P value | OR (95%CI) |
| Age range, yr | ||||
| ≤ 48 | 78 (49.7) | 88 (55.0) | 0.343 | |
| > 48 | 79 (50.3) | 72 (45.0) | / | |
| Gender | ||||
| Male | 136 (86.6) | 133 (83.1) | 0.385 | / |
| Female | 21 (13.4) | 27 (16.9) | / | |
| Mutation pattern | ||||
| Pre-S deletion alone | 22 (14.0) | 11 (6.9) | 0.037 | 2.207 (1.032-4.722) |
| Wild type + deletion | 52 (33.1) | 34 (21.2) | 0.017 | 1.835 (1.109-3.038) |
| Total deletions | 74 (47.1) | 45 (28.1) | < 0.001 | 2.278 (1.430-3.630) |
HCC: Hepatocellular carcinoma.
The relationship between HBV pre-S deletion size and HCC was analyzed for the first time in this study. Interestingly, while small deletions (≤ 99 bp) occurred more frequently in the HCC group than in the non-HCC group (34.4% vs 15.0%, respectively, P < 0.001), deletions larger than 99 bp showed no distribution difference between the two groups (12.7% vs 13.1%, respectively, P = 0.918, Table 4). This result highlights the importance of deletion size in HCC risk prediction.
Table 4.
Analysis of the correlation between the size of pre-S deletion and hepatocellular carcinoma n (%)
| Characteristics | HCC n = 157 | Non-HCC n = 160 | P value | OR (95%CI) |
| Wild type | 83 (52.9%) | 115 (71.9%) | ||
| Pre-S deletion | ||||
| Deletion ≤ 99 bp | 54 (34.4%) | 24 (15.0%) | < 0.001 | 2.971 (1.723-5.122) |
| Deletion > 99 bp | 20 (12.7%) | 21 (13.1%) | 0.918 | 0.966 (0.501-1.863) |
Longitudinal observation of pre-S deletions during the development of HCC
To determine the temporal relationship between HBV pre-S deletions and HCC occurrence, we examined pre-S deletions from serial serum samples collected 2-9 years prior to HCC diagnosis. Figure 2 shows the evolution of pre-S deletion mutants during the development of HCC. Of the 9 patients who carried pre-S deletions at the stage of HCC, 88.9% (8/9) had shown deletions 2-5 years prior to HCC, suggesting it was an early predictive marker for HCC. However, for the samples collected 6-9 years prior to HCC, only 4 patients (44.4%) contained HBV pre-S deletions, indicating that these deletions were not acquired from the beginning of the infection but arose de novo during the progression of liver disease. We did not find the reverse mutation in any of the 9 cases (Figure 3). It is noteworthy that, for patient 372, the wild-type HBV co-existed with approximately 10% deletion mutants 6 years before the onset of HCC, indicating that CGE can quantitatively identify a small number of deletions in a heterogeneous viral population.
Figure 2.
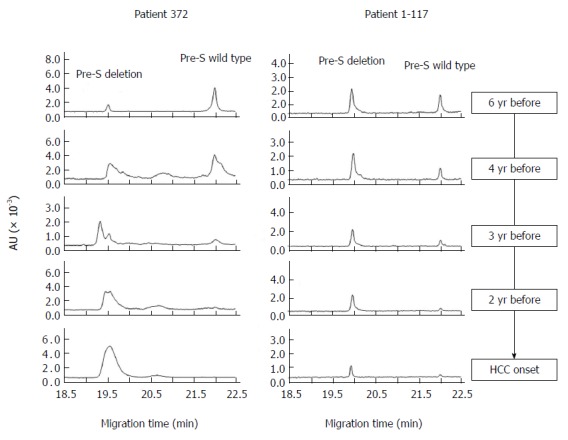
Longitudinal observation of hepatocellular carcinoma-related pre-S deletions during the development of hepatocellular carcinoma. HCC: Hepatocellular carcinoma.
Figure 3.

Longitudinal observation of hepatocellular carcinoma-related pre-S deletions in Qidong hepatocellular carcinoma patients. Black: Pre-S deletion; Grey: Wild type; HCC: Hepatocellular carcinoma.
DISCUSSION
HBV pre-S deletions have been consistently reported as a risk factor for HCC[5,9-15]. However, the lack of an effective detection system hampers its clinical application. In this study, we developed a CGE-based approach that can quantitatively identify a small number of deletions in a heterogeneous viral population. It is rapid, convenient, and can generate a result comparable to DNA sequencing. It has potential usage in the large-scale screening of pre-S deletions in chronic hepatitis B carriers.
Most of the previous studies used PCR direct sequencing methods to reveal pre-S deletions[9-15,25,26]. However, PCR direct sequencing is only suitable for samples containing single viral populations. Indeed, most pre-S mutant-positive cases simultaneously contain wild-type viruses, and the prevalence of mixed infections could be up to 90.9%[27,28]. In this study, we found the prevalence rates of mixed infections in patients with HCC and chronic hepatitis to be 70.3% and 75.6%, respectively. Currently, for patients infected with mixed strains of HBV, a time-consuming clone sequencing method must be performed. It is of great importance to develop an easy system that can concurrently identify two or more viral strains existing in one sample. To achieve this, two non-sequencing approaches have been reported previously: (1) PAGE followed by silver staining[27]; and (2) an oligonucleotide chip hybridization system[28]. Compared with these methods, CGE has a higher resolution and simpler detection procedure; it can automatically examine 96 samples without any manual treatment and quantitatively analyze the distribution of different types of HBV mutants. The overall accuracy of CGE in the measurement of pre-S deletions was 99.1%. In a total of 114 sequence-known DNA clones, only one sample with a deletion of 3 bp failed to be identified by CGE. Because a 3 bp deletion of pre-S occurred in a very small number of patients, this methodological limitation should not drastically influence the overall specificity.
We applied this method to assess the HBV pre-S deletion rate in HCC and non-HCC patients. Consistent with previous studies, we found there was a significantly higher rate of pre-S deletions in HCC patients than in non-HCC patients, confirming that pre-S deletions can be used as a predictive marker for HCC (OR = 2.278; 95%CI: 1.430-3.630). Interestingly, we observed that the short stretch pre-S deletions, not the large ones, were significantly associated with HCC. While pre-S deletions ≤ 99 bp had an OR of 2.971 (95%CI: 1.723-5.122) for HCC risk, deletions > 99 bp lost their association with HCC (OR = 0.918, 95%CI: 0.501-1.863). To our knowledge, this phenomenon has never before been reported. In one recent study, Yeh et al[46] found that, in liver tissues, short pre-S deletions located between codon 107 and 141 (nt. 3166-52) were strongly associated with a worse postoperative prognosis. As for blood samples, several previous papers have suggested that the location of the pre-S deletions play a role in the outcome of liver disease. Pre-S deletions can occur in pre-S1, pre-S2, or in the region spanning these genes. Yeung et al[44] found that pre-S2 deletions, but not pre-S1, were significantly associated with the development of HCC (P = 0.020). Pre-S deletions generally occur in the 5’ half of the pre-S2 region[43,47], and it is speculated that the small deletions in the pre-S2 region have a more significant impact on HCC development than the large deletions occurring in the pre-S1 or pre-S1+ pre-S2 region. To confirm this hypothesis, we carried out a pool analysis of the pre-S deletions in HCC and non-HCC controls by using the sequence information provided by previous studies[40-45]. The results indicated that small deletions (≤ 99 bp) have a significantly closer association with HCC than large deletions (data not shown). Thus, the CGE-based method, which only measures the size of the deletion, might have a similarly predictive value for HCC compared to the sequencing method.
Consistent with the previous reports from our group[48] and others[14], genotype C is highly prevalent in the Qidong area. In this study, only 5 (3.2%) in the HCC group and 6 (3.8%) in the control group were found to be infected with the genotype B virus. Therefore, it is unclear from this study whether HBV pre-S deletion can be served as a predictive marker for HCC in patients infected with other viral genotypes. Pre-S deletion has been reported to occur more frequently in HBV carriers with genotype C than in those with genotype B[9,11], but very few studies have focused on the relationship between pre-S deletion and HCC in genotype B-infected patients. This intriguing issue awaits further investigation in the future.
In addition to the cross-sectional study, we also applied CGE technology to our longitudinal study. This allowed us to clearly observe the appearance and evolution of the pre-S deletion mutants during liver disease progression. The mutants seemed to have a higher survival advantage than the wild-type virus in vivo, as their population increased constantly until they eventually dominated the viral population. We observed no cases in which the pre-S deletion mutants were replaced by wild-type viruses. This observation strengthens the importance of early detection of pre-S deletions. Because CGE can detect the presence of pre-S deletions representing as little as 10% of the viral population, its value in HCC prediction will surpass that of the sequencing of PCR products.
Taken together, our results provided evidence to support the idea that pre-S deletions, especially short DNA fragment deletions, are a useful predictive marker for HCC. CGE is a rapid and sensitive method that can be used to detect low numbers of pre-S mutants among a mixed viral population. It will thus be a useful tool in the screening of chronic hepatitis patients who are at an increased risk for HCC.
ACKNOWLEDGMENTS
The authors wish to thank all contributors for their assistance in the Qidong prospective cohort.
COMMENTS
Background
HBV pre-S deletions have been consistently reported as a risk factor for HCC. However, the lack of an effective detection system hampers its clinical application. Most previous studies used PCR direct sequencing methods to reveal pre-S deletions. However, PCR direct sequencing is only suitable for samples containing single viral populations. Indeed, most pre-S mutant-positive cases simultaneously contained wild-type viruses. For patients infected with mixed strains of HBV, a time-consuming clone sequencing method must be performed. Thus, it is necessary to develop a sensitive, simple, and accurate method to detect HBV pre-S deletions.
Research frontiers
Various techniques have been used for the detection of HBV pre-S deletions. The capillary gel electrophoresis (CGE) method has allowed for the sensitive detection of low amounts of HBV pre-S deletion among a mixed viral population.
Innovations and breakthroughs
This is the first study aiming to develop a CGE-based approach that can quantitatively identify a small numbers of deletions within a heterogeneous viral population. By applying this method in a case-control study, we not only confirmed previous observations that pre-S deletions increased the risk of HCC, but also reported, for the first time, that only small deletions (≤ 99 bp) have a significant association with HCC, rather than large deletions.
Applications
The CGE method developed in this study is a sensitive approach for the detection of low amounts of HBV pre-S deletion. It will be a useful tool in the screening of chronic hepatitis patients who are at an increased risk for HCC.
Terminology
CGE is a separation method based on using soluble polymers to create a replaceable molecular sieve that allows for size-based separation.
Peer review
The rapid PCR-CGE method in this study is able to accurately determine pre-S deletions associated with HCC and has greater utility than sequencing based approaches. The paper is well written.
Footnotes
Supported by Chinese state key project specialized for infectious diseases, No. 2012ZX10002-008-002
P- Reviewer: McGarvey MJ, Netter HJ, Pazienza V S- Editor: Qi Y L- Editor: Rutherford A E- Editor: Liu XM
References
- 1.Chen CJ, Yang HI, Iloeje UH. Hepatitis B virus DNA levels and outcomes in chronic hepatitis B. Hepatology. 2009;49:S72–S84. doi: 10.1002/hep.22884. [DOI] [PubMed] [Google Scholar]
- 2.Schmitt S, Glebe D, Alving K, Tolle TK, Linder M, Geyer H, Linder D, Peter-Katalinic J, Gerlich WH, Geyer R. Analysis of the pre-S2 N- and O-linked glycans of the M surface protein from human hepatitis B virus. J Biol Chem. 1999;274:11945–11957. doi: 10.1074/jbc.274.17.11945. [DOI] [PubMed] [Google Scholar]
- 3.Chai N, Gudima S, Chang J, Taylor J. Immunoadhesins containing pre-S domains of hepatitis B virus large envelope protein are secreted and inhibit virus infection. J Virol. 2007;81:4912–4918. doi: 10.1128/JVI.02865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jang JW, Chun JY, Park YM, Shin SK, Yoo W, Kim SO, Hong SP. Mutational complex genotype of the hepatitis B virus X /precore regions as a novel predictive marker for hepatocellular carcinoma. Cancer Sci. 2012;103:296–304. doi: 10.1111/j.1349-7006.2011.02170.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsukawa M, Takaki A, Shiraha H, Koike K, Iwasaki Y, Kobashi H, Fujioka S, Sakaguchi K, Yamamoto K. Hepatitis B virus core promoter mutations G1613A and C1653T are significantly associated with hepatocellular carcinoma in genotype C HBV-infected patients. BMC Cancer. 2011;11:458. doi: 10.1186/1471-2407-11-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CH, Changchien CS, Lee CM, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis. 2008;198:1634–1642. doi: 10.1086/592990. [DOI] [PubMed] [Google Scholar]
- 8.Huy TT, Ushijima H, Win KM, Luengrojanakul P, Shrestha PK, Zhong ZH, Smirnov AV, Taltavull TC, Sata T, Abe K. High prevalence of hepatitis B virus pre-s mutant in countries where it is endemic and its relationship with genotype and chronicity. J Clin Microbiol. 2003;41:5449–5455. doi: 10.1128/JCM.41.12.5449-5455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007;133:1466–1474. doi: 10.1053/j.gastro.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Qu LS, Liu TT, Jin F, Guo YM, Chen TY, Ni ZP, Shen XZ. Combined pre-S deletion and core promoter mutations related to hepatocellular carcinoma: A nested case-control study in China. Hepatol Res. 2011;41:54–63. doi: 10.1111/j.1872-034X.2010.00732.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153–1168. doi: 10.1053/j.gastro.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Lin CL, Liu CH, Chen W, Huang WL, Chen PJ, Lai MY, Chen DS, Kao JH. Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22:1098–1103. doi: 10.1111/j.1440-1746.2006.04515.x. [DOI] [PubMed] [Google Scholar]
- 13.Sugauchi F, Ohno T, Orito E, Sakugawa H, Ichida T, Komatsu M, Kuramitsu T, Ueda R, Miyakawa Y, Mizokami M. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003;70:537–544. doi: 10.1002/jmv.10428. [DOI] [PubMed] [Google Scholar]
- 14.Qu L, Kuai X, Liu T, Chen T, Ni Z, Shen X. Pre-S deletion and complex mutations of hepatitis B virus related to young age hepatocellular carcinoma in Qidong, China. PLoS One. 2013;8:e59583. doi: 10.1371/journal.pone.0059583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Qin Y, Li W, Shi Q, Xue Y, Li J, Liu C, Hollinger FB, Shen Q. Molecular analysis of the hepatitis B virus presurface and surface gene in patients from eastern China with occult hepatitis B. J Med Virol. 2013;85:979–986. doi: 10.1002/jmv.23556. [DOI] [PubMed] [Google Scholar]
- 16.Liao Y, Hu X, Chen J, Cai B, Tang J, Ying B, Wang H, Wang L. Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One. 2012;7:e38394. doi: 10.1371/journal.pone.0038394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 18.Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683–688. doi: 10.1111/j.1349-7006.2006.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Cao Y, Wang T, Xiang G, Lu J, Zhang J, Hou P. The N-Glycosylation Modification of LHBs (Large Surface Proteins of HBV) Effects on Endoplasmic Reticulum Stress, Cell Proliferation and its Secretion. Hepat Mon. 2013;13:e12280. doi: 10.5812/hepatmon.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HC, Chang WT, Chang WW, Wu HC, Huang W, Lei HY, Lai MD, Fausto N, Su IJ. Hepatitis B virus pre-S2 mutant upregulates cyclin A expression and induces nodular proliferation of hepatocytes. Hepatology. 2005;41:761–770. doi: 10.1002/hep.20615. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh YH, Su IJ, Yen CJ, Tsai TF, Tsai HW, Tsai HN, Huang YJ, Chen YY, Ai YL, Kao LY, et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid suppresses the pro-oncogenic effects induced by hepatitis B virus pre-S2 mutant oncoprotein and represents a potential chemopreventive agent in high-risk chronic HBV patients. Carcinogenesis. 2013;34:475–485. doi: 10.1093/carcin/bgs365. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh YH, Su IJ, Wang HC, Tsai JH, Huang YJ, Chang WW, Lai MD, Lei HY, Huang W. Hepatitis B virus pre-S2 mutant surface antigen induces degradation of cyclin-dependent kinase inhibitor p27Kip1 through c-Jun activation domain-binding protein 1. Mol Cancer Res. 2007;5:1063–1072. doi: 10.1158/1541-7786. [DOI] [PubMed] [Google Scholar]
- 23.Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N, Kato JY. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277:2302–2310. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- 24.Hung JH, Teng YN, Wang LH, Su IJ, Wang CC, Huang W, Lee KH, Lu KY, Wang LH. Induction of Bcl-2 expression by hepatitis B virus pre-S2 mutant large surface protein resistance to 5-fluorouracil treatment in Huh-7 cells. PLoS One. 2011;6:e28977. doi: 10.1371/journal.pone.0028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajiya Y, Hamasaki K, Nakata K, Nakagawa Y, Miyazoe S, Takeda Y, Ohkubo K, Ichikawa T, Nakao K, Kato Y, et al. Full-length sequence and functional analysis of hepatitis B virus genome in a virus carrier: a case report suggesting the impact of pre-S and core promoter mutations on the progression of the disease. J Viral Hepat. 2002;9:149–156. doi: 10.1046/j.1365-2893.2002.00335.x. [DOI] [PubMed] [Google Scholar]
- 26.Mu SC, Lin YM, Jow GM, Chen BF. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J Hepatol. 2009;50:264–272. doi: 10.1016/j.jhep.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Cao Z, Bai X, Guo X, Jin Y, Qian G, Tu H. High prevalence of hepatitis B virus pre-S mutation and its association with hepatocellular carcinoma in Qidong, China. Arch Virol. 2008;153:1807–1812. doi: 10.1007/s00705-008-0176-9. [DOI] [PubMed] [Google Scholar]
- 28.Shen FC, Su IJ, Wu HC, Hsieh YH, Yao WJ, Young KC, Chang TC, Hsieh HC, Tsai HN, Huang W. A pre-S gene chip to detect pre-S deletions in hepatitis B virus large surface antigen as a predictive marker for hepatoma risk in chronic hepatitis B virus carriers. J Biomed Sci. 2009;16:84. doi: 10.1186/1423-0127-16-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaufils N, Ben Lassoued A, Essaydi A, Dales JP, Formisano-Tréziny C, Bonnet N, Grob JJ, Gabert J. Analysis of T-cell receptor-γ gene rearrangements using heteroduplex analysis by high-resolution microcapillary electrophoresis. Leuk Res. 2012;36:1119–1123. doi: 10.1016/j.leukres.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal S, Sarwai S, Nigam N, Singhal P. Rapid detection of alpha + thalassaemia deletion & amp; alpha-globin gene triplication by Gap-PCR in Indian subjects. Indian J Med Res. 2002;116:155–161. [PubMed] [Google Scholar]
- 31.Bortolotti F, De Paoli G, Pascali JP, Tagliaro F. Fully automated analysis of Carbohydrate-Deficient Transferrin (CDT) by using a multicapillary electrophoresis system. Clin Chim Acta. 2007;380:4–7. doi: 10.1016/j.cca.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 32.Hurtado-Fernández E, Contreras-Gutiérrez PK, Cuadros-Rodríguez L, Carrasco-Pancorbo A, Fernández-Gutiérrez A. Merging a sensitive capillary electrophoresis-ultraviolet detection method with chemometric exploratory data analysis for the determination of phenolic acids and subsequent characterization of avocado fruit. Food Chem. 2013;141:3492–3503. doi: 10.1016/j.foodchem.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 33.García-Cañas V, González R, Cifuentes A. Detection of genetically modified maize by the polymerase chain reaction and capillary gel electrophoresis with UV detection and laser-induced fluorescence. J Agric Food Chem. 2002;50:1016–1021. doi: 10.1021/jf011033g. [DOI] [PubMed] [Google Scholar]
- 34.Luo S, Feng J, Pang HM. High-throughput protein analysis by multiplexed sodium dodecyl sulfate capillary gel electrophoresis with UV absorption detection. J Chromatogr A. 2004;1051:131–134. [PubMed] [Google Scholar]
- 35.Cohen AS, Najarian D, Smith JA, Karger BL. Rapid separation of DNA restriction fragments using capillary electrophoresis. J Chromatogr. 1988;458:323–333. doi: 10.1016/s0021-9673(00)90576-4. [DOI] [PubMed] [Google Scholar]
- 36.Froim D, Hopkins CE, Belenky A, Cohen AS. Method for phosphorothioate antisense DNA sequencing by capillary electrophoresis with UV detection. Nucleic Acids Res. 1997;25:4219–4223. doi: 10.1093/nar/25.21.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong W, Yeung ES. Multiplexed capillary electrophoresis for DNA sequencing with ultra violet absorption detection. J Chromatogr A. 2002;960:229–239. doi: 10.1016/s0021-9673(01)01393-0. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Chen TY, Lu PX, Wang JB, Wu Y, Zhang QN, Jin Y, Qian GS, Tu H. A seventeen-year prospective cohort study on association of hepatitis B virus e antigen with the risk of hepatocellular carcinoma. Tumor. 2011;31:841–845. [Google Scholar]
- 39.Chinese Society of Hepatology, Chinese Medical Association, Chinese Society of Infectious Diseases, Chinese Medical Association. Guideline on prevention and treatment of chronic hepatitis B in China (2005) Chin Med J (Engl) 2007;120:2159–2173. [PubMed] [Google Scholar]
- 40.Fang ZL, Sabin CA, Dong BQ, Wei SC, Chen QY, Fang KX, Yang JY, Huang J, Wang XY, Harrison TJ. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol. 2008;89:2882–2890. doi: 10.1099/vir.0.2008/002824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeung P, Wong DK, Lai CL, Fung J, Seto WK, Yuen MF. Profile of pre-S deletions in the natural history of chronic hepatitis B infection. J Med Virol. 2010;82:1843–1849. doi: 10.1002/jmv.21901. [DOI] [PubMed] [Google Scholar]
- 42.Utama A, Siburian MD, Fanany I, Intan MD, Dhenni R, Kurniasih TS, Lelosutan SA, Achwan WA, Arnelis B, Yusuf I, et al. Low prevalence of hepatitis B virus pre-S deletion mutation in Indonesia. J Med Virol. 2011;83:1717–1726. doi: 10.1002/jmv.22172. [DOI] [PubMed] [Google Scholar]
- 43.Abe K, Thung SN, Wu HC, Tran TT, Le Hoang P, Truong KD, Inui A, Jang JJ, Su IJ. Pre-S2 deletion mutants of hepatitis B virus could have an important role in hepatocarcinogenesis in Asian children. Cancer Sci. 2009;100:2249–2254. doi: 10.1111/j.1349-7006.2009.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung P, Wong DK, Lai CL, Fung J, Seto WK, Yuen MF. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in chronic hepatitis B. J Infect Dis. 2011;203:646–654. doi: 10.1093/infdis/jiq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang HP, Hsu HY, Chen CL, Ni YH, Wang HY, Tsuei DJ, Chiang CL, Tsai YC, Chen HL, Chang MH. Pre-S2 deletions of hepatitis B virus and hepatocellular carcinoma in children. Pediatr Res. 2010;67:90–94. doi: 10.1203/PDR.0b013e3181c1b0b7. [DOI] [PubMed] [Google Scholar]
- 46.Yeh CT, So M, Ng J, Yang HW, Chang ML, Lai MW, Chen TC, Lin CY, Yeh TS, Lee WC. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology. 2010;52:1922–1933. doi: 10.1002/hep.23898. [DOI] [PubMed] [Google Scholar]
- 47.Kao JH, Liu CJ, Jow GM, Chen PJ, Chen DS, Chen BF. Fine mapping of hepatitis B virus pre-S deletion and its association with hepatocellular carcinoma. Liver Int. 2012;32:1373–1381. doi: 10.1111/j.1478-3231.2012.02826.x. [DOI] [PubMed] [Google Scholar]
- 48.Jin Y, Zhu Y, Chen TY, Guo X, Zhang J, Wang JB, Chen WZ, Qian GS, Tu H. [The association of hepatitis B virus genotype and the basal core promoter mutation in Qidong, China] Zhonghua Ganzangbing Zazhi. 2010;18:511–515. doi: 10.3760/cma.j.issn.1007-3418.2010.07.010. [DOI] [PubMed] [Google Scholar]


