Schizophrenia is arguably the most puzzling syndrome in medicine. Why do young adults develop chronic and bizarre signs and symptoms after decades of normal or near-normal development? The public health impact of schizophrenia is massive with considerable mortality and enormous morbidity1,2 in addition to the human pain that often follows in its wake. The fruits of a century of research into its etiology were few, and false leads were regrettably prominent. However, at long last, recent genetic studies3,4 have struck gold in the form of multiple associations that meet modern standards in human genetics for robustness and replication. New but unpublished data are likely to extend the published findings importantly, and perhaps crack the genetics of schizophrenia wide open.
These studies highlight a provocative empirical finding: schizophrenia is highly polygenic.5 A parsimonious way to connect these many dots is via the hypothesis that distributed genetic variation in one (or perhaps several) pathways underlies risk for schizophrenia. The functional unit conferring risk may not be any single node in the pathway but rather the pathway itself. Thus, schizophrenia may be a cardinal example of a pathway disease.
In schizophrenia research, as in much of genetics and molecular medicine, experimental methods usually focus on single loci or molecules. Such approaches are pragmatically reductionist and often highly informative in dissecting a more fundamental biological network link by link, edge by edge. However, what if liability to schizophrenia is conferred not by genetic variants in one or several loci but rather hundreds or even thousands of loci that together confer risk? What if the genetic risk for schizophrenia is at the level of a pathway? Such a “network medicine” paradigm,6 requires a rather different conceptualization of the nature of the beast.
What is the evidence that schizophrenia might be a pathway disease? First, Mendelian sub-forms of schizophrenia have not been identified. The informal clinical surveillance network (hundreds of thousands of physicians taking family histories from millions of patients with schizophrenia worldwide for a century) has not identified unequivocal Mendelian sub-forms of schizophrenia in contrast to Mendelian forms identified for many common diseases (e.g., Alzheimer’s disease, breast cancer, autism, type 2 diabetes mellitus, etc.). Highly effortful studies of rare, childhood onset forms of schizophrenia have not revealed Mendelian sub-forms – again in contrast to many other diseases where early onset has been a critical clue. More than twenty years of ascertaining multiplex pedigrees for genetic studies has also failed to identify Mendelian sub-forms. Unpublished reports from relatively large sequencing studies of schizophrenia have not identified compelling exonic mutations. It is impossible to prove the absence of Mendelian forms – and some may eventually be found – but none have been identified despite considerable effort.
Second, several genes implicated in schizophrenia and bipolar disorder (which now seems to be a genetic cousin of schizophrenia)5,7 also have strong Mendelian variants. However, their clinical presentation does not include early onset schizophrenia but rather mental retardation and/or autism (i.e., TCF4, CACNA1C, and possibly NRXN1).3,8 Thus, strong disruption to a node in a putative underlying pathway yields a different clinical presentation rather than a more severe and early onset schizophrenia.
Third, the beginnings of a plausible network has emerged. There is now robust and replicable evidence for genetic variation near MIR137 (the gene encoding the microRNA miR-137) in schizophrenia.3 Intriguingly, four genes that had genome-wide significant associations also contain binding sites for miR-137.9 The genotypic relative risks were all 1.1–1.25, subtle effects typical for complex trait associations. Given the known roles of miR-137 in adult neurogenesis and neuronal maturation, this yields a testable hypothesis about a definable pathway that might mediate some component of risk.
Fourth, schizophrenia is highly polygenic. We now know that polygenicity is a cardinal feature of many human diseases and anthropometric traits characterized by complex inheritance of dozens to hundreds of loci (e.g., Crohn’s disease, type 2 diabetes, height, and body mass). The genetic variants involved are common with subtle effects, and some of these gene sets may have singular clinical utility. For schizophrenia, there is strong and replicated evidence that number of loci is in the thousands.5 Indeed, this polygenic component (that must include common variation rather than being a reflection of multiple rare variants) accounts for 23–33% of variance in liability to schizophrenia (on the order of half the heritability).5,10 This also suggests that so-called “missing heritability” is merely hidden and imperfectly assessed by current genotyping technologies.
A recent paper about non-syndromic autism provided a tantalizing glimpse of how genetic variation might relate to altered biological pathways.11 Typical patterns of gene expression in frontal and temporal cortex were attenuated in autism. An empirically-derived gene expression module that was under-expressed in autism was enriched for known autism susceptibility genes and genetic association signals. While not completely elucidated, these data support the notion that polygenetic variation for autism alters the expression and regulation of a transcriptional network that mediates risk for autism. A similar model could hold for schizophrenia.
For schizophrenia, the hypothesis is that polygenetic variation alters a biological pathway. Removal of any single node (e.g., via a protein-killing mutation) either has no effect (due to the emergent network property of robustness) or, for network hubs, yields a different phenotype (e.g., mental retardation or autism). There are many ways in which such a pathway could mediate liability to schizophrenia (e.g., by being insufficiently robust or overly rigid in response to environmental insult, or by coding an inappropriate developmental program).
The conceptualization of schizophrenia as a pathway disease has an immediate implication. Above all else, a priority for the field must be to complete genomic screens of a sufficient number of cases in order to define the pathway components with precision.12 If this can be accomplished, it should be possible to develop assays to monitor pathway function in living cells. Knowledge derived from this work could lead to the fulfillment of the ultimate promise of genomics – primary prevention of the development of schizophrenia in those at risk and the development of more effective therapeutics (in an era where big pharma have turned sharply away from CNV drug development).
Critically, it is possible that any such pathway is intrinsically modifiable, that people with schizophrenia are not “doomed from the womb” but rather could anticipate return to relatively normal long-term function.
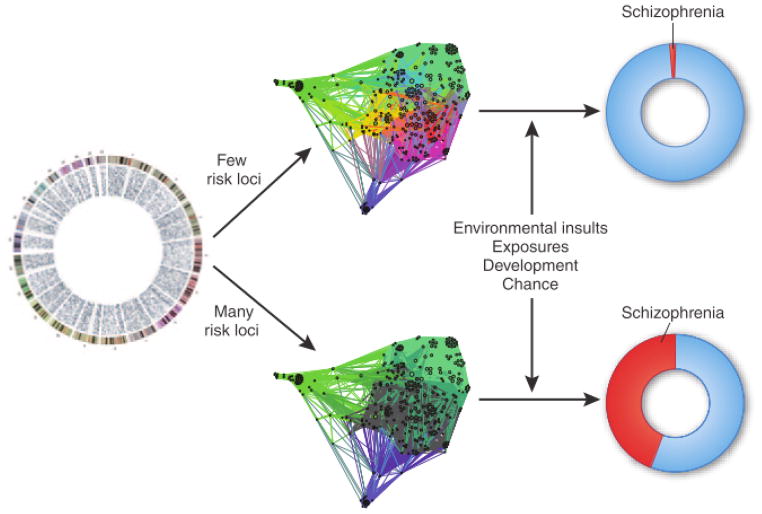
Figure.
Schematic of schizophrenia as a pathway disease. Genetic variants at many loci encode the components of a pathway or pathways. Many risk loci can be affected, resulting in a disease pathway greatly modified owing to polygenetic variation. When a few risk loci are affected, there may be limited impact on the disease pathway. This pathway itself—in conjunction with environmental risk factors or other factors—mediates risk of schizophrenia.
Acknowledgments
I thank my colleagues in the Psychiatric Genomics Consortium for years of stimulating discussion. Supported by NIH grants MH077139 and MH085520.
References
- 1.Collins PY, et al. Grand challenges in global mental health. Nature. 2011;475:27–30. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. The Global Burden of Disease: 2004 Update. WHO Press; Geneva: 2008. [Google Scholar]
- 3.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study of schizophrenia identifies five novel loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levinson DF, et al. Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews Genetics. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichtenstein P, et al. Common genetic influences for schizophrenia and bipolar disorder: A population-based study of 2 million nuclear families. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Molecular psychiatry. 2011 doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nature Genetics. doi: 10.1038/ng.1108. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan P. Don’t give up on GWAS. Molecular Psychiatry. 2011 [Google Scholar]