Abstract
Photocaging provides a method to spatially and temporally control biological function and gene expression with high resolution. Proteins can be photochemically controlled through the site-specific installation of caging groups on amino acid side chains that are essential for protein function. The photocaging of a synthetic gene network using unnatural amino acid mutagenesis in mammalian cells was demonstrated with an engineered bacteriophage RNA polymerase. A caged T7 RNA polymerase was expressed in cells with an expanded genetic code and used in the photochemical activation of genes under control of an orthogonal T7 promoter, demonstrating tight spatial and temporal control. The synthetic gene expression system was validated with two reporter genes (luciferase and EGFP) and applied to the light-triggered transcription of short hairpin RNA constructs for the induction of RNA interference.
INTRODUCTION
Activation and deactivation of genes is precisely controlled with high spatial and temporal resolution.1-3 In order to understand and reprogram genetic circuits with the same spatio-temporal resolution, light can be used as an external control element. The installation of light-removable protecting groups on biological macromolecules is a versatile approach to the photochemical regulation of RNA, DNA, and protein activity.4-10 For the generation of caged proteins in live cells, the genetic code can be expanded allowing for the site-specific incorporation of caged amino acids into proteins.11-20 The caged amino acids are inserted in response to an amber stop codon, TAG, by suppressor tRNAs that have been misacylated by engineered tRNA synthetases.21-28
Advantages of this methodology over other approaches are that the location of the unnatural amino acid is determined through genetic mutagenesis and the light-activated proteins are produced in live cells eliminating the requirement for transfection or injection. The addition of non-mammalian RNA polymerases to the genetic circuitry of cells enables the development of orthogonal genetic expression platforms that can be manipulated to activate specific genes that will not be expressed by the endogenous cellular genetic machinery. Caged polymerases are versatile building blocks for the construction of synthetic gene networks to control gene expression with light as an input.29 Protein expression under photochemical control can be used to build synthetic biological circuits that respond to light with specific gene function outputs, thus conveying precise spatial and temporal control over these circuits. Related methods for the light-regulation of gene expression in mammalian cells, such as those based on phytochromes or cryptochromes, have been developed to produce gene outputs dependent on light-induced conformational protein changes that initiate or repress transcription through a number of pathways and protein-protein interactions.30-35 Additionally, zinc finger-based technologies have been created for photochemical regulation of transcription with spatio-temporal control.12,36 These optogenetic approaches rely on the engineering of fusion constructs between the natural photosensitive proteins and the target protein of interest in order to render it light-responsive, often in a reversible fashion. However, the photochemical regulation achieved with a single caging group modification does not encumber the native protein with fusion constructs that are several orders of magnitude larger (from 20 kDa to >120 kDa) than the caging group (0.24 kDa) and, importantly, photolysis generates the natural wild-type protein. Moreover, the use of a small caging group that responds to UV irradiation avoids interference by ambient light, allows for simultaneous application of the full palette of fluorescent protein reporters, and provides clean OFF → ON light switching. We developed a fully genetically encoded, light-activated T7 RNA polymerase that allows for the promoter-specific photochemical regulation of transcription for both coding and non-coding RNAs in a mammalian cell system.
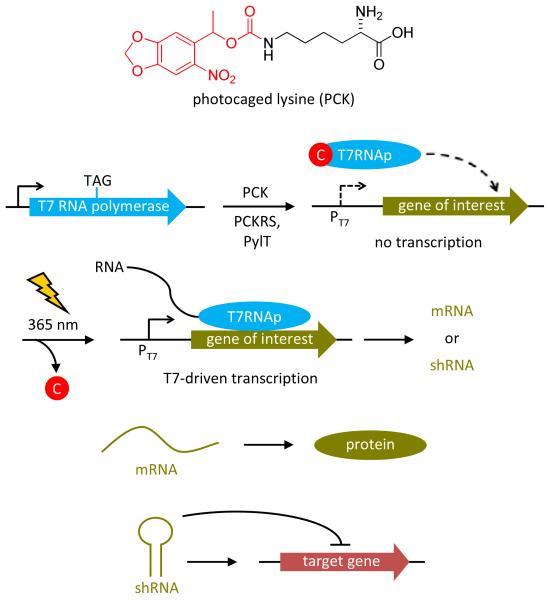
T7 RNA polymerase (T7RNAp) is a bacteriophage RNA polymerase that is related to the PolI family of DNA polymerases,37 which is especially useful for exogenous gene control in the reprogramming of genetic and network relationships, as the polymerase and its corresponding promoter are completely orthogonal to all endogenous polymerases in mammalian cells.38 Therefore, genes of interest can be specifically expressed through the light-activation of a T7RNAp temporarily rendered inactive through the incorporation of a caged amino acid in its active site. Importantly, photochemically activated transcription can be used to not only control gene expression but also to express non-coding RNA sequences of other biological function. A light-activated T7RNAp has previously been generated in our lab through the site-specific incorporation of an ortho-nitrobenzyl caged tyrosine (PCY) in E. coli cells with an expanded genetic code.39 However, its application for the photochemical control of transcription in mammalian cells was hampered since the M. janaschii tRNA synthetase/tRNA pair is not orthogonal in mammalian cells and transfection of the caged protein was required. To overcome this limitation, site-specific incorporation of a genetically encoded photocaged lysine (PCK) into T7RNAp through an engineered, fully orthogonal pyrrolysine synthetase/tRNA pair was used to photochemically control RNA polymerization in mammalian cells with (Figure 1). The photocaged lysine exhibits increased solubility in mammalian growth media, more rapid decaging due to better leaving group qualities of the released carbamate, a bathochromic shift in absorbance maximum and thus decaging wavelength, and produces a less reactive byproduct upon photolysis than the caged tyrosine.15
Figure 1. The light-activated T7RNAp is expressed in mammalian cells through site-specific incorporation of a photocaged lysine (PCK) into its active site in response to an amber stop codon (TAG) via an engineered tRNA synthetase (PCKRS) that misacylates an amber-suppressor tRNA (PylT) with PCK.
The caged T7RNAp is completely inactive until irradiation with 365 nm UV light induces decaging and activation of T7-driven transcription. Depending on the function of the transcribed RNA, light-induced protein expression from mRNA or light-induced gene silencing via RNA interference from shRNA is achieved.
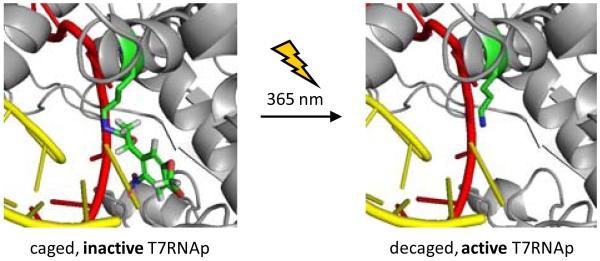
Transcription catalyzed by T7RNAp involves a number of protein conformational changes and the active site of T7RNAp, which is conserved among a number of polymerases found in nature, has been studied in detail.40 These structural and kinetic studies indicate that the active site lysine 631 (K631) is critical for T7RNAp function, as it interacts with phosphate groups of the incoming nucleotide triphosphates (NTPs) at the interface of the DNA template and RNA product strands, stabilizing the NTPs through hydrogen bonding with the 1α-phosphate residue.41-43 Mutations at the K631 position have been shown to inhibit transcription.44 Thus, we hypothesized that a sterically demanding caging group installed on K631 will block incoming NTPs from the active site and inhibit T7RNAp activity. In addition, the carbamate linker changes the electronic nature of the ε-amino group, effectively preventing protonation and removing the positive charge. The caged T7RNAp is expected to be completely inactive until a brief UV irradiation at 365 nm removes the caging group from K631 and enables transcription (Figure 2).
Figure 2. Structural representation of K631-caged (left) and wild-type (right) T7RNAp.
The K631 residue is indicated in green, while the DNA template and RNA transcript are indicated in red and yellow, respectively. The photocaged lysine is predicted to block transcription until UV irradiation removes the caging group and restores T7RNAp activity. Images were generated in PyMol from PDB 1S76.43
RESULTS AND DISCUSSION
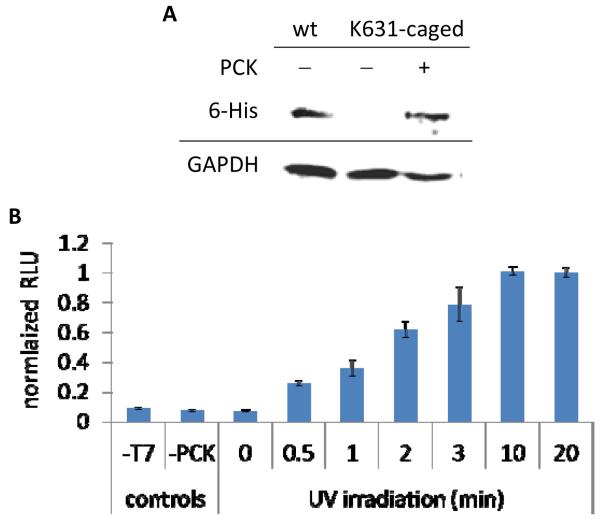
An amber stop codon TAG was introduced at K631 of T7RNAp (Supporting Table 1) and the gene was cloned into a plasmid containing the MbPylRS synthetase (PCKRS) mutant for PCK incorporation (M241F, A267S, Y271C, and L274M), derived from the M. barkeri pyrrolysine tRNA synthetase.15 Previous studies for the incorporation of unnatural amino acids have shown that there are no off-target effects on endogenous gene expression through the use of the low-frequency TAG stop codon in mammalian cells.45,46 As a positive control, wild-type (WT) T7RNAp was also cloned into the same vector. The T7RNAp expression plasmid was co-transfected with a pyrrolysine tRNACUA (PylT) expression plasmid (Supporting Figure 1A) into HEK293T cells. Western blot analysis of cells that were incubated for 24 hours in the absence or presence of PCK revealed the expression of hexahistidine-tagged T7RNAp only in the presence of PCK, indicative of the high fidelity of the engineered synthetase (Figure 3A). The activity and photochemical control of K631-caged T7RNAp was investigated using a firefly luciferase reporter gene under control of the T7 promoter (Supporting Figure 1B).39 First, the effects of UV exposure on the luciferase assay were studied, revealing that an irradiation time of 2 minutes and even up to 10-20 minutes had only small effects on T7-driven luciferase activity in cells expressing the wild-type polymerase (Supporting Figure 2). Light-activated luciferase expression was observed in cells expressing the caged T7RNAp, with a tunable linear response of gene expression to an increasing irradiation time (Figure 3B). A 3 minute irradiation exhibited a 9-fold increase compared to the non-irradiated control, and luciferase activity did not significantly increase with additional UV exposure of up to 20 minutes. Importantly, the presence of the caging group at K631 fully blocks transcriptional activity of the enzyme, since the absence of polymerase (−T7), caged T7RNAp (−PCK), and light exposure (0 min UV) all show identical, very low levels of background luminescence.
Figure 3. Expression of caged T7RNAp and photochemical activation.
A) Mammalian cells were transfected with expression plasmids for the PylT, PCKRS, and T7RNApK631TAG and grown in the presence of PCK (2 mM) for 24 hours. Western blot analysis was performed using a 6-His primary antibody with a FITC secondary antibody for detection, showing PCK dependent expression. B) HEK293T cells were transfected with plasmids expressing the PylT, PCKRS, T7RNApK631TAG, and T7-IRES-Luc were incubated with PCK overnight. UV irradiations were performed on a 365 nm transilluminator at increasing intervals. A luciferase bright glow assay was performed 24 hours after UV exposure. Luminescence units were normalized to the activation observed after a 20 minute irradiation and error bars represent the standard deviation obtained from experimental triplicates.
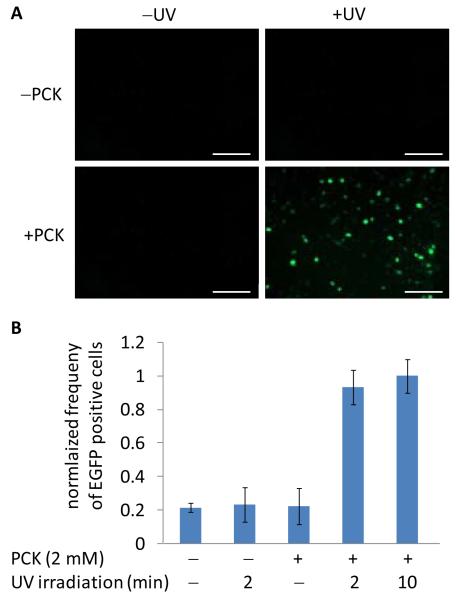
A T7-IRES-EGFP reporter plasmid was constructed for fluorescent imaging of the photochemical gene expression system (Supporting Figure 1B). The T7-driven EGFP reporter construct was validated through co-transfection with the PCKRS expression machinery, and conditions were optimized for optimal off → on switching of EGFP expression (Supporting Figures 3 and 4). Photochemical control over EGFP expression was achieved through treatment of cells with both PCK and UV irradiation (Figure 4A). No background EGFP fluorescence was observed in cells containing the caged T7RNAp construct in the absence of PCK or the absence of UV exposure. Fluorescent cell counting was performed to quantify the photochemical activation of T7RNAp-dependent expression of EGFP (Figure 4B). A five-fold increase in the number of fluorescent cells was observed after both 2 and 10 minute UV irradiations, while the absence of either PCK or UV exposure only showed basal levels of fluorescence, indicating that the caged polymerase in completely inactive and displays no background expression.
Figure 4. Light-activation of EGFP transcription in HEK293T cells.
A) Cells were co-transfected with the PCKRS, PylT, T7RNApK631TAG and T7-IRES-EGFP constructs then incubated with 2 mM PCK for 24 hours. Irradiation was then performed for 2 minutes at 365 nm, and cells were imaged 48 hours after UV exposure. Scale bars indicate 200 μm. B) Fluorescent cell counting of caged T7RNAp-driven EGFP expression of cells 48 hours after UV exposure. The frequencies of EGFP-positive cells (gated/total) were normalized to the 10 minute UV irradiation and error bars represent the standard deviation obtained from three experimental replicates.
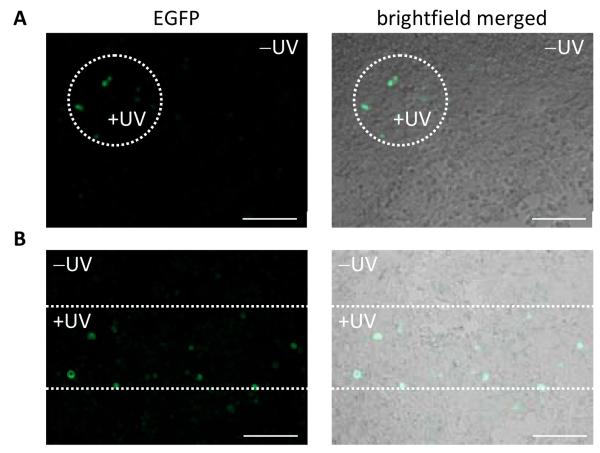
In order to demonstrate spatial control of RNA expression in mammalian tissue culture, the photochemical activation of the T7RNAp-driven EGFP reporter was performed as in Figure 4A; however, spatially restricted UV irradiations were performed through specifically shaped masks (pin hole and horizontal slit). Only areas exposed to the patterned UV light were EGFP positive due to spatially restricted decaging and activation of RNA polymerization (Figure 5). These experiments successfully demonstrate spatial and temporal control of gene function using a site-specifically caged T7RNAp expressed in mammalian cells.
Figure 5. Spatial control of photochemically activated gene expression.
Cells were transfected as previously described with the EGFP reporter for T7RNAp activity. Masks were used to irradiate a small population of cells at 365 nm for 2 minutes, and EGFP expression was observed at 48 hours. Irradiations were performed through a small pin hole (A) and a horizontal slit (B). Scale bars indicate 200 μm. Only the cells within the irradiated region express EGFP, demonstrating spatial control of K631-caged T7RNAp in mammalian cells.
To further demonstrate the applicability of the light-activated T7RNAp/promoter system, it was subsequently applied to the photochemical activation of RNA interference through T7-driven expression of short hairpin RNAs (shRNAs).47 These shRNAs are processed into siRNAs exhibiting prolonged activity for gene knockdown.48 The shRNAs can be designed to exhibit promoter-specific expression,49 and can be genetically integrated to generate stable RNAi regulators.50 The use of plasmid-based shRNA for gene suppression has been successfully applied in a variety of mammalian cell systems,51-54 and T7RNAp-driven shRNAs have been reported for the regulation of gene expression in live mammalian cells55,56 and zebrafish embryos.57 Here, we coupled our developed photochemical gene regulation system with an Eg5 shRNA component as a proof of principle study in the photochemical regulation of RNA interference. The Eg5 gene encodes a motor protein involved in mitosis58 and inhibition of Eg5 with siRNA oligonucleotides has been shown to produce binucleated cells,59,60 providing a distinct phenotypic readout for RNAi activity. The Eg5 shRNA construct was cloned into the T7-IRES-EGFP reporter plasmid with built-in T7RNAp expression components including the T7 RNA promoter and terminator sequences (Supporting Figure 1C and Table 2). This construct was designed to fluorescently track cells that were successfully transformed as well as exhibit expression and photochemical activation of T7RNAp.
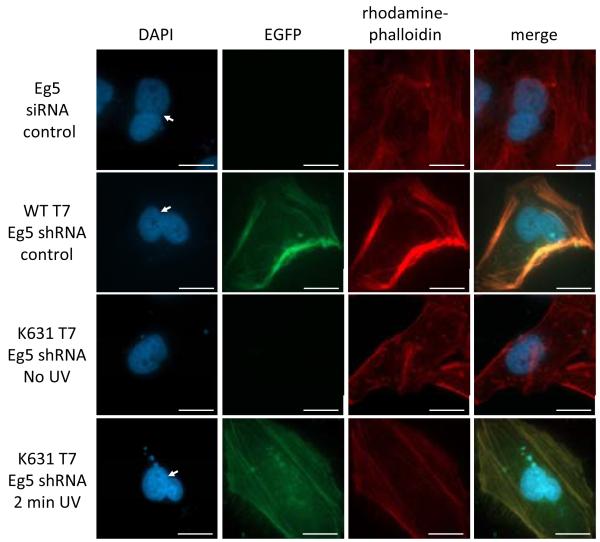
First, the shRNA expression construct was tested for activity in the presence of a WT T7RNAp gene in HeLa cells. The resulting binucleated cellular phenotype was observed, in agreement with the phenotype observed through transfection of an siRNA oligonucleotide positive control, as expected (Figure 6). The K631-caged T7RNAp expression system was then used to achieve UV activation of Eg5 shRNA expression through co-transfection of the required plasmid constructs, followed by incubation in the presence of PCK and UV irradiation (Figure 6). Light-activation of shRNA expression was verified through the observation of a binucleated cell phenotype only after UV irradiation. The relative frequency of binucleated cells observed indicates that the expression of Eg5 shRNA is photochemically regulated with K631-caged T7RNAp and is activated to levels identical to wild-type activity (Supporting Figure 5). Additionally, qRT-PCR was performed on cellular RNA extracts to quantify the relative Eg5 mRNA expression levels to further validate the photochemical silencing of an endogenous gene with shRNA expression by a light-activated polymerase. A 55% reduction of Eg5 mRNA is observed in the case of both wild-type T7RNAp-driven shRNA expression and light-activated K631-caged T7RNAp, in accordance with an Eg5 siRNA reagent as an additional positive control (Supporting Figure 6). These findings demonstrate that the caged T7RNAp-controlled genetic circuit can be applied to photochemically regulation of gene expression and the light-induction of non-coding RNAs to achieve external control over RNA interference.
Figure 6. Light-activation of Eg5 shRNA using the caged T7RNAp system, followed by RNA interference and triggering of binucleated cell phenotype.
Cells were transfected with an Eg5 siRNA (50 pmol) and the K631-caged T7RNAp expression system as previously described, followed by fixing and staining with DAPI (nucleus) and Rho-phalloidin (actin) prior to imaging. Scale bars indicate 20 μm and binucleated cells are labeled with a white arrow. A binucleated phenotype was observed with both the Eg5 siRNA and plasmid-based shRNA controls, while EGFP expression successfully tracked cells for the identification of T7RNAp-driven shRNA activity. The shRNA construct has been validated to enable light-activated RNA interference. The EGFP expression and cellular binucleation were fully dependent upon UV irradiation and activation of the caged T7RNAp.
CONCLUSION
In summary, a light-activated T7RNAp gene expression system was successfully developed in mammalian cells engineered with additional protein biosynthetic machinery for genetically-encoded protein modification with a caged lysine amino acid (PCK). Site-specific incorporation of PCK at the K631 active site residue in T7RNAP led to full inactivation of RNA polymerization. Light-irradiation at 365 nm removed the caging group from the active site and enabled the expression of transcripts that were placed under control of the T7 promoter. This was successfully demonstrated for two reporter genes, luciferase and EGFP. Temporal control of UV exposure allowed for finely tuned gene expression that gradually increased with longer durations of irradiation. In addition, precise spatial control of T7RNAp activity through localized UV irradiation was achieved, as shown by the patterned expression of an EGFP reporter in a monolayer of mammalian cells. Furthermore, photochemical control of T7RNAP activity was used to achieve precise control over the expression of non-coding RNAs, as exemplified by the light-induction of shRNA transcription and thus RNA interference. Here, a unique combination of individual parts, including an orthogonal tRNA/tRNA synthetase pair, a photocaged lysine amino acid, a caged T7RNAp, and T7 promoter-driven shRNA expression cassette were interfaced with the RNA interference pathway to construct the circuitry for light-triggered, sequence-specific gene silencing. The ability to photochemically control the expression of shRNAs allows for spatial and temporal dissection of the RNAi pathway. Overall, a genetically encoded expression platform has been developed that can photochemically regulate the expression of specific genes under control of the T7 promoter in mammalian cells. In theory, any gene under control of a T7 promoter can now be photochemically controlled in live cells. The engineered spatial and temporal control can be applied to the precise triggering of genetic networks allowing for a wide range of cellular applications.
METHODS
Genetic Constructs
An amber stop was introduced into T7 RNA polymerase through mutagenesis of the pBH161 plasmid.61 A single nucleotide was changed for the K631 AAG → TAG mutation using a QuikChange Site-Directed Mutagenesis Kit (Invitrogen) and primers shown in Supporting Table 1. The T7RNAp gene was then PCR amplified from the pBH161 plasmid with primers to introduce NheI and EcoRI restriction sites. The ~2.7 kB T7RNAp gene insert was digested, purified, and cloned between the NheI and MfeI restriction sites of the pAG31 plasmid. Ligations were performed at 1:6 vector to insert ratios with the Quick Ligation Kit (New England Biolabs). All plasmid constructs were confirmed by sequencing with promoter-specific primers (Genewiz).
Mammalian cell expression of caged T7RNAp
HEK293T cells were seeded at ~100,000 cell per well into 6-well plates (BD Falcon) and incubated overnight in DMEM growth media supplemented with 10% FBS and 2% penicillin/streptomycin at 37 °C in 5% CO2. Transfections were performed with 2 μg of each plasmid using 10 μL lipofectamine transfection reagent (Invitrogen) in 2 mL Opti-Mem media (Invitrogen) at 37 °C for 4 hours. After 4 hours the Opti-Mem transfection mixtures were removed from the cells and replaced with DMEM growth media supplemented with PCK (2 mM) for a 24 hour incubation.
Western blot
Total protein was extracted from ~1 million cells using 200 μL mammalian cell lysis buffer (Sigma-Aldrich) and was size separated on a 10% SDS-PAGE gel then transferred to a PVDF membrane. Standard western blot conditions were followed using a mouse-anti-6His primary antibody in addition to a mouse-anti-GADPH control (Santa Cruz Biotechnology). The primary antibodies were detected with a goat-anti-mouse-FITC fluorescent secondary antibody and analyzed with a Storm Phosphoimager.
Photochemical regulation of reporter genes
HEK293T cells were seeded at ~10,000 cell per well into 96-well plates (BD Falcon) and incubated overnight in DMEM growth media supplemented with 10% FBS and 2% penicillin/streptomycin at 37 °C in 5% CO2. Transfections were performed with 200 ng of each plasmid using 2 μL lipofectamine transfection reagent (Invitrogen) in 200 μL Opti-Mem media (Invitrogen) at 37 °C for 4 hours. After 4 hours the Opti-Mem transfection mixtures were removed from the cells and replaced with DMEM growth media supplemented with PCK (2 mM) for a 24 hour incubation. Branched PEI (Sigma-Aldrich) was also used for overnight transfections in PCK-supplemented media following the standard protocol. The PCK containing media was removed after the overnight incubation followed by exposure to 365 nm using a UV transilluminator (25 W). For the luciferase reporter, a bright glow assay (Promega) was performed 24 hours post UV irradiation to quantify luciferase activity. The bright glow reagent was added at 1:1 ratio to media containing cells (100 μL each) and luminescence was read from a white 96-well plate on a BioTek Synergy 4 plate reader, with the gain set at 150 and 1 sec integration per well. The luminescence was normalized to the maximum activation observed and error bars represent the standard deviation of experimental triplicates. For the EGFP reporter, fluorescent imaging was performed after 24 and 48 hour incubations (see below). Spatially distinct UV irradiations were performed through pre-cut designs (circular or linear slits) in tinfoil 96-well plate covers and irradiated at 365 nm with a transilluminator (25 W) for 2 minutes.
Fluorescence imaging of EGFP
Media was replaced with clear DMEM-high modified growth media (Thermo Scientific) for microscopy imaging. Expression of EGFP was observed using a Zeiss Observer Z1 microscope (20× objective, NA 0.8 plan-apochromat; Zeiss) and a GFP (38 HE) filter (ex: BP470/40; em: BP525/50) then processed in Zen Pro 2011 imaging software.
Fluorescent cell sorting
HEK293T cells were seeded and transfected in 96-well plates as described above. After transfection, media was replaced with DMEM growth media supplemented with PCK (2 mM) for a 24 hour incubation. The PCK-containing media was removed after the overnight incubation followed by exposure to 365 nm using a UV transilluminator (25 W) for 2 and 10 min. Following a 48 hour incubation cells were trypsinized, pooled, washed, and resuspended in PBS. Flow cytometry was performed on a FACSCalibur (Becton-Dickinson) instrument (488 nm argon laser, 530/50 nm BPF) and analyzed using Cellquest Pro Software. Cells were gated for EGFP fluorescence (above 102.5 RFUs) and analyzed until 20,000 cells had been counted for each condition tested. The frequency of EGFP positive cells (gated/total) was normalized to the UV activated EGFP expression.
Eg5 shRNA construct assembly
The Eg5 shRNA insert was prepared from the T7 RNA promoter, Eg5 shRNA sense and antisense strands connected with a loop sequence, and a T7 RNA terminator region (Supporting Table 2). These elements were ordered as single stranded DNA oligos (IDT) as shown and annealed at 1 μM, followed by purification of the DNA duplex insert, which was then ligated into the T7-IRES-EGFP plasmid at BglII and SacI restriction sites. Ligations were performed at 1:10 vector to insert ratios with the Quick Ligation Kit (New England Biolabs). All plasmid constructs were confirmed by sequencing through with promoter specific primers (Genewiz).
Eg5 shRNA expression and imaging
HeLa cells were seeded at ~50,000 cell per well into 4-well chamber slides (Lab-Tek) and incubated overnight in DMEM growth media supplemented with 10% FBS and 2% penicillin/streptomycin at 37 °C in 5% CO2. Transfections were performed with 1 μg of each plasmid using 10 μL lipofectamine transfection reagent (Invitrogen) in 1 mL Opti-Mem media (Invitrogen) at 37 °C for 4 hours. The Eg5 siRNA oligonucleotide (Supporting Table 2)62 was annealed at 1 μM and transfected at 50 pmol as a positive control with 5 uL X-tremeGENE siRNA transfection reagent (Roche) under the same conditions. After 4 hours the Opti-Mem transfection mixtures were removed from the cells and replaced with DMEM growth media supplemented with PCK (2 mM) for 24 hour incubation. The PCK containing media was removed after the overnight incubation followed by a 2 minute exposure to 365 nm using a UV transilluminator (25 W). After a 48 hour incubation cells were fixed with 3.75% formaldehyde, followed by DAPI (Invitrogen) and Rhodamine-phalloidin (Invitrogen) counter stains according to standard protocols. Imaging was performed with a Zeiss Observer Z1 microscope (63× oil objective, NA 1.4 plan-apochromat; Zeiss) using DAPI [BFP (68 HE), ex:BP377/28; em: BP464/100], EGFP [GFP (38 HE), ex: BP470/40; em: BP525/50], and Rhodamine [RFP (43 HE), ex: BP575/25; em: BP605/70] filter cubes then processed in Zen Pro 2011 imaging software. Relative frequencies of binucleated cells were determined through three counts of 50 cells for each condition and were normalized to the Eg5 siRNA positive control.
Quantification of Eg5 mRNA by qRT-PCR
HEK293T cells were seeded at ~100,000 cell per well into 6-well plates (BD Falcon) and incubated overnight in DMEM growth media supplemented with 10% FBS and 2% penicillin/streptomycin at 37 °C in 5% CO2. Transfections were performed with 2 μg of each plasmid using 10 μL lipofectamine transfection reagent (Invitrogen) in 2 mL Opti-Mem media (Invitrogen) at 37 °C for 4 hours. The Eg5 siRNA oligonucleotide (Supporting Table 2)62 was annealed at 1 μM and transfected at 200 pmol as a positive control with 5 uL X-tremeGENE siRNA transfection reagent (Roche). After 4 hours, the Opti-Mem transfection mixtures were removed from the cells and replaced with DMEM growth media supplemented with PCK (2 mM) followed by a 24 hour incubation. The PCK containing media was removed after the overnight incubation followed by a 2 minute exposure to 365 nm using a UV transilluminator (25 W). After a 48 hour incubation, total RNA was isolated from cells using the Trizol reagent (Invitrogen) and reverse transcribed with the iScript cDNA Synthesis kit (Bio-Rad). Quantitative RT-PCR was performed with the SsoFast Evagreen Supermix (Bio-Rad) using an Eg5 primer set [forward 5′ CAGCTGAAAAGGAAACAGCC, reverse 5′ATGAACAATCCACACCAGCA]59 and a GAPDH primer set [forward 5′ TGCACCACCAACTGCTTAGC, reverse 5′ GGCATGGACTGTGGTCATGAG].63 The threshold cycles (Ct) of each sample were normalized to the GAPDH control gene and the inhibition of Eg5 expression is represented relative to nontreated cells. Error bars represent the standard deviations of three replicates.
Supplementary Material
Plasmid maps, wild-type T7RNAp luciferase controls, EGFP expression and UV irradiation optimization, Eg5 shRNA cellular binucleated frequency and qRT-PCR, and sequence information. This material is also available free of charge via the internet at http://pubs.acs.org.
ACKNOWLEDGEMENT
This work was support in part by the National Science Foundation (CHE-0848398).
REFERENCES
- (1).Brevini TA, Cillo F, Antonini S, Tosetti V, Gandolfi F. Reprod Fertil Dev. 2007;19:35. doi: 10.1071/rd06119. [DOI] [PubMed] [Google Scholar]
- (2).Li E, Davidson EH. Birth Defects Res C Embryo Today. 2009;87:123. doi: 10.1002/bdrc.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chan TM, Longabaugh W, Bolouri H, Chen HL, Tseng WF, Chao CH, Jang TH, Lin YI, Hung SC, Wang HD, Yuh CH. Biochim Biophys Acta. 2009;1789:279. doi: 10.1016/j.bbagrm.2008.09.005. [DOI] [PubMed] [Google Scholar]
- (4).Young DD, Deiters A. Org Biomol Chem. 2007;5:999. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- (5).Deiters A. Curr Opin Chem Biol. 2009;13:678. doi: 10.1016/j.cbpa.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Deiters A. Chembiochem. 2010;11:47. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Mayer G, Heckel A. Angew Chem Int Ed Engl. 2006;45:4900. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- (8).Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. Angew Chem Int Ed Engl. 2012;51:8446. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- (9).Curley K, Lawrence DS. Curr Opin Chem Biol. 1999;3:84. doi: 10.1016/s1367-5931(99)80015-5. [DOI] [PubMed] [Google Scholar]
- (10).Lee HM, Larson DR, Lawrence DS. ACS Chem Biol. 2009;4:409. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gautier A, Deiters A, Chin JW. J Am Chem Soc. 2011;133:2124. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chou C, Deiters A. Angew Chem Int Ed Engl. 2011;50:6839. doi: 10.1002/anie.201101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Deiters A, Groff D, Ryu Y, Xie J, Schultz PG. Angew Chem Int Ed Engl. 2006;45:2728. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]
- (14).Edwards WF, Young DD, Deiters A. ACS Chem Biol. 2009;4:441. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]
- (15).Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW. J Am Chem Soc. 2010;132:4086. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- (16).Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Nat Chem Biol. 2007;3:769. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- (17).Arbely E, Torres-Kolbus J, Deiters A, Chin JW. J Am Chem Soc. 2012;134:11912. doi: 10.1021/ja3046958. [DOI] [PubMed] [Google Scholar]
- (18).Wu N, Deiters A, Cropp TA, King D, Schultz PG. J Am Chem Soc. 2004;126:14306. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- (19).Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR. Mol Biosyst. 2010;6:1557. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]
- (20).Groff D, Chen PR, Peters FB, Schultz PG. Chembiochem. 2010;11:1066. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Young TS, Schultz PG. J Biol Chem. 2010;285:11039. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Davis L, Chin JW. Nat Rev Mol Cell Biol. 2012;13:168. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- (23).Liu CC, Schultz PG. Annu Rev Biochem. 2010;79:413. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- (24).Hoesl MG, Budisa N. Curr Opin Biotechnol. 2012;23:751. doi: 10.1016/j.copbio.2011.12.027. [DOI] [PubMed] [Google Scholar]
- (25).Wang Q, Parrish AR, Wang L. Chem Biol. 2009;16:323. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chalker JM, Davis BG. Curr Opin Chem Biol. 2010;14:781. doi: 10.1016/j.cbpa.2010.10.007. [DOI] [PubMed] [Google Scholar]
- (27).Wang K, Schmied WH, Chin JW. Angew Chem Int Ed Engl. 2012;51:2288. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- (28).Neumann H. FEBS Lett. 2012;586:2057. doi: 10.1016/j.febslet.2012.02.002. [DOI] [PubMed] [Google Scholar]
- (29).Gardner L, Deiters A. Curr Opin Chem Biol. 2012;16:292. doi: 10.1016/j.cbpa.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Toettcher JE, Voigt CA, Weiner OD, Lim WA. Nat Methods. 2011;8:35. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Losi A, Gärtner W. Annu Rev Plant Biol. 2012;63:49. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- (32).Miesenböck G. Annu Rev Cell Dev Biol. 2011;27:731. doi: 10.1146/annurev-cellbio-100109-104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Nat Methods. 2010;7:973. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Nat Biotechnol. 2009;27:941. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- (35).Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Le Cong, Platt RJ, Scott DA, Church GM, Zhang F. Nature. 2013 doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Polstein LR, Gersbach CA. J Am Chem Soc. 2012;134:16480. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cramer P. Bioessays. 2002;24:724. doi: 10.1002/bies.10127. [DOI] [PubMed] [Google Scholar]
- (38).Moffatt BA, Dunn JJ, Studier FW. J Mol Biol. 1984;173:265. doi: 10.1016/0022-2836(84)90194-3. [DOI] [PubMed] [Google Scholar]
- (39).Chou C, Young DD, Deiters A. Chembiochem. 2010;11:972. doi: 10.1002/cbic.201000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Steitz TA. EMBO J. 2006;25:3458. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Maksimova TG, Mustayev AA, Zaychikov EF, Lyakhov DL, Tunitskaya VL, Akbarov A. K. h., Luchin SV, Rechinsky VO, Chernov BK, Kochetkov SN. Eur J Biochem. 1991;195:841. doi: 10.1111/j.1432-1033.1991.tb15773.x. [DOI] [PubMed] [Google Scholar]
- (42).Woody AY, Osumi-Davis PA, Hiremath MM, Woody RW. Biochemistry. 1998;37:15958. doi: 10.1021/bi9805801. [DOI] [PubMed] [Google Scholar]
- (43).Yin YW, Steitz TA. Cell. 2004;116:393. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- (44).Osumi-Davis PA, de Aguilera MC, Woody RW, Woody AY. J Mol Biol. 1992;226:37. doi: 10.1016/0022-2836(92)90122-z. [DOI] [PubMed] [Google Scholar]
- (45).Xie J, Schultz PG. Methods. 2005;36:227. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- (46).Wang L, Xie J, Schultz PG. Annu Rev Biophys Biomol Struct. 2006;35:225. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- (47).Sharp PA, Zamore PD. Science. 2000;287:2431. doi: 10.1126/science.287.5462.2431. [DOI] [PubMed] [Google Scholar]
- (48).Amarzguioui M, Rossi JJ, Kim D. FEBS Lett. 2005;579:5974. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- (49).Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Antisense Nucleic Acid Drug Dev. 2003;13:83. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- (50).Lambeth LS, Smith CA. Methods Mol Biol. 2013;942:205. doi: 10.1007/978-1-62703-119-6_12. [DOI] [PubMed] [Google Scholar]
- (51).Sui G, Soohoo C, Affar e. B., Gay F, Shi Y, Forrester WC. Proc Natl Acad Sci U S A. 2002;99:5515. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Genes Dev. 2002;16:948. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Paddison PJ, Caudy AA, Hannon GJ. Proc Natl Acad Sci U S A. 2002;99:1443. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- (55).Yu JY, DeRuiter SL, Turner DL. Proc Natl Acad Sci U S A. 2002;99:6047. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Peng Y, Lu JX, Shen XF. Biochem Biophys Res Commun. 2007;360:496. doi: 10.1016/j.bbrc.2007.06.100. [DOI] [PubMed] [Google Scholar]
- (57).Wang N, Sun YH, Liu J, Wu G, Su JG, Wang YP, Zhu ZY. J Biomed Sci. 2007;14:767. doi: 10.1007/s11373-007-9189-8. [DOI] [PubMed] [Google Scholar]
- (58).Ferenz NP, Gable A, Wadsworth P. Semin Cell Dev Biol. 2010;21:255. doi: 10.1016/j.semcdb.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Mol Biol Cell. 2005;16:3187. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. Mol Biol Cell. 2010;21:979. doi: 10.1091/mbc.E09-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Protein Expr Purif. 1997;9:142. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- (62).Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. J Cell Sci. 2001;114:4557. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- (63).Nielsen R, Courtoy PJ, Jacobsen C, Dom G, Lima WR, Jadot M, Willnow TE, Devuyst O, Christensen EI. Proc Natl Acad Sci U S A. 2007;104:5407. doi: 10.1073/pnas.0700330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmid maps, wild-type T7RNAp luciferase controls, EGFP expression and UV irradiation optimization, Eg5 shRNA cellular binucleated frequency and qRT-PCR, and sequence information. This material is also available free of charge via the internet at http://pubs.acs.org.