Abstract
We report a novel strategy for studying synaptic pathology by concurrently measuring levels of four SNARE complex proteins from individual brain tissue samples. This method combines affinity purification and mass spectrometry and can be applied directly for studies of SNARE complex proteins in multiple species or modified to target other key elements in neuronal function. We use the technique to demonstrate altered levels of presynaptic proteins in Alzheimer disease patients and prion-infected mice.
One prominent pathological feature of neuropsychiatric disorders such as Alzheimer disease (AD)1 is severe synaptic loss (1–3). Previous reports of AD patients have shown that presynaptic dysfunction might occur early in the disease process (1, 4). Cortical synapse pathology has also been shown to correlate to the severity of dementia more closely than other pathological hallmarks of AD such as plaques and neurofibrillary tangles (5, 6). The SNARE proteins are essential components for the regulation of neurotransmitter exocytosis at the presynaptic site (7). Animal models suggest that changed expression or modification of SNARE complex proteins (synaptosomal-associated protein 25 (SNAP-25), syntaxin-1, and vesicle-associated membrane protein (VAMP)) alters synaptic function and is an interesting target for the development of therapeutics for neuropsychiatric illness (8, 9). The constituents of the SNARE complex are either localized in synaptic vesicles (VAMPs) or anchored at the presynaptic plasma membrane (SNAP-25 and syntaxin). The SNARE proteins are tightly assembled, and subsequent neurotransmitter release of the complex is quickly dissociated by N-ethylmaleimide-sensitive factor (7, 10–12). Because they are both strongly associated into complexes and membrane associated, the SNARE proteins are difficult to analyze via mass spectrometry, which is incompatible with most detergents necessary for the solubilization of proteins. Each SNARE complex protein exists in several isoforms that are differently distributed within the central nervous system (13–18). Post-translational modifications and truncated variants of the SNARE proteins make investigation of the protein expression even more complicated.
In this study we developed an approach for the characterization and concurrent quantification of SNARE complex proteins that combines affinity purification by immunoprecipitation and mass spectrometry (IP-MS). We used precipitation with monoclonal antibodies against SNAP-25 to target the SNARE complex proteins and nanoflow LC–tandem mass spectrometry (LC-MS/MS) to characterize the co-immunoprecipitated interaction partners. Selected reaction monitoring (SRM) on a triple quadrupole mass spectrometer coupled to a microflow LC system was used for quantification of the SNARE proteins. To demonstrate the usability of the IP-MS method, we performed a comparison of SNARE complex protein levels in brain tissue from AD patients and age-matched controls, as well as a study of SNARE complex protein levels in brain tissue from prion-infected mice.
EXPERIMENTAL PROCEDURES
Human Brain Tissue Samples
This study involved autopsy-confirmed patients with AD (n = 15) and age-matched controls (n = 15). Brain tissues from the superior parietal gyrus were analyzed. All brain tissues were obtained from the Netherlands Brain Bank. Braak and Braak criteria, which are based on the distribution of neurofibrillary tangles, were used to categorize the stage of AD (19). All AD patients were classified as Braak stage 5 or 6, and the controls were Braak stage 0 or 1. Supplemental Table S1 shows the clinical and demographic characteristics of the groups.
Mouse Brain Tissue Samples
All animal work conformed to UK regulations and institutional guidelines and was performed under Home Office guidelines. tg37 (20) were inoculated with 1% brain homogenate of Chandler/Rocky Mountain Laboratory prions or with normal brain homogenate aged 3 to 4 weeks, as described in Ref. 21. Hippocampi were processed at 6, 7, 8, 9, and 10 weeks post-infection (w.p.i.) and stored at −80 °C prior to homogenization. For all analyses, n = 3 mice unless otherwise stated.
Homogenization of Brain Tissue
The brain extraction procedure was performed as described by Öhrfelt et al., with minor modifications (22). Briefly, 100 ± 10 mg of brain tissue was homogenized on ice in 1 ml of Tris-buffer (10 mm Tris-HCl, pH 6.8) containing complete protease inhibitor (Roche Diagnostics GmBH). Centrifugation of the homogenate occurred at 31,000g for 1 h at +4 °C, and then the supernatant was collected (Tris). One milliliter of Tris-buffer containing 0.5% Triton X-100 (Union Carbide Corporation, Danbury, CT) containing complete protease inhibitor was added to the pellet, which was then homogenized on ice and sonicated using a micro-probe sonicator. The centrifugation step was repeated, and the supernatant was collected (0.5% Triton (i.e. membrane-bound fraction)). The same procedure was repeated with the addition of Tris-buffer containing 2% Triton and complete protease inhibitor and with the addition of Tris-buffer containing 0.5% SDS and complete protease inhibitor for a final centrifugation at +12 °C (SDS fraction (i.e. membrane-raft associated fraction)). All supernatants were aliquoted and stored at −80 °C pending analysis. For protein quantitation, Protein DC assay (Bio-Rad Laboratories) reagent was used. This reagent is a reducing agent and is detergent compatible.
Antibodies and Recombinant Protein of SNAP-25
The following antibodies were used: mouse monoclonal antibody SP12 recognizing SNAP-25 (23), mouse monoclonal antibody SMI81 (Covance, Princeton, NJ) against SNAP-25 (24), and a polyclonal anti-SNAP-25 antibody raised in rabbit according to Ref. 25. Recombinant standard protein of SNAP-25 was purchased from Origene (Rockville, MD).
Immunoprecipitation
The immunoprecipitation of brain tissue extracts was performed according to Ref. 22, with minor modifications. Briefly, an aliquot (1 μg) of the mouse monoclonal antibody SP12 (1 g/l), the mouse monoclonal antibody SMI81 (1 g/l), or IgG from murine serum (1 g/l, Sigma-Aldrich) (a negative control) was separately added to 100 μl of magnetic Dynabead M-280 Sheep anti-mouse IgG (Invitrogen) and incubated for 1 h on a rocking platform at room temperature. The beads were washed three times with 1 ml of PBS (10 mm sodium phosphate, 0.15 m NaCl, pH 7.4). The antibodies were cross-linked using 20 mm dimethyl pimelimidate dihydrochloride (Sigma-Aldrich) and 0.2 m triethanolamine (pH 8.2; Sigma-Aldrich) according to the manufacturer's product description. The cross-linked beads were washed two times in PBS and blocked with Roti-Block (Carl Roth, GmbH Karlsruhe, Germany)) for 1 h on a rocking platform at room temperature. Each brain tissue extract (0.5% Triton and SDS) was adjusted with 20% Triton and PBS to a final concentration of 0.2% Triton and a final concentration of 26 μg of total protein. Samples and magnetic beads were incubated overnight on a rocking platform at +4 °C. The magnetic bead–sample solution was transferred to a KingFisher magnetic particle processor (Thermo Fisher Scientific) (tube 1). The following three wash steps (tubes 2–4) were conducted for 10 s in 1 ml of each washing buffer: tube 2, 0.025% Tween 20 in PBS; tube 3, PBS; and tube 4, 50 mm ammonium hydrogen carbonate (NH4HCO3, pH 8.0). Then, SNAP-25 and closely interacting proteins (e.g. SNARE complex proteins) were eluted from the beads with 100 μl of 0.5% formic acid (FA) (tube 5) for 4 min. The eluted fractions were transferred to 0.5-ml Protein LoBind tubes (Eppendorf AG, Hamburg, Germany) and dried in a vacuum centrifuge.
Protein Digestion
The dried samples were dissolved in 10 μl of 0.1% RapiGest SF Surfactant (Waters, Milford, MA) in 50 mm NH4HCO3 (pH 8.0) for 1 h at room temperature. Disulfide bonds were reduced by the addition of 10 μl of 10 mm DTT (Sigma-Aldrich, St. Louis, MO) in 50 mm NH4HCO3 and incubation for 3 min at +90 °C. After cooling to room temperature, 5 μl of 10 mm iodoacetamide (Sigma-Aldrich) in 50 mm NH4HCO3 was added, and the samples were incubated in the dark at room temperature for 30 min. Digestion was carried out by the addition of 5 μl of trypsin solution (20 μg of Sequencing Grade Modified Trypsin (Promega, Madison, WI) dissolved in 0.01% aqueous HCl (0.1g/l) and diluted 1 to 20 in 50 mm NH4HCO3) and overnight incubation at +37 °C. To reduce the amount of RapiGest SF Surfactant in the samples, 2 μl of 10% aqueous TFA was added (resulting in pH < 2). The samples were incubated for 45 min at +37 °C and then centrifuged (16,900g, 10 min, +4 °C). Twenty-five microliters of the resulting supernatants of each sample was carefully transferred to 0.5-ml Protein LoBind tubes (Eppendorf AG).
Addition of Heavy-isotope-labeled Peptide Standards
C-terminal isotopically labeled peptides containing U-13C6, U-15N4-arginine or U-13C6, U-15N2-lysine were supplied by Sigma-Aldrich at over 95% peptide purity as determined via reverse phase HPLC. Three labeled peptides were chosen as reference standards for SNAP-25 (amino acids 18–30, ADQLADESLEST[R], common for both SNAP-25A and SNAP-25B), syntaxin-1A (amino acids 95–108, SIEQSIEQEEGLN[R]), and syntaxin-1B (amino acids 94–107, AIEQSIEQEEGLN[R]). Heavy-isotope-labeled AQUA™ Peptide standard for VAMP-2/3 (AQ0256, AQUA™ Peptide ADALQAGASQFETSAA[K]) was also purchased (Sigma-Aldrich). The three custom synthesized peptides were dissolved and diluted in 0.1% aqueous FA and mixed to a final concentration of 5 fmol/μl (SNAP-25, syntaxin-1B) or 10 fmol/μl (syntaxin-1A). One vial of AQ0256 was dissolved in 100 μl of 20% acetonitrile in 0.1% aqueous FA, diluted in 0.1% aqueous FA, and mixed with the rest of the heavy-isotope-labeled peptides to a final concentration of 5 fmol/μl. A 25-μl aliquot of the reference peptide mixture was added to each immunoprecipitated brain homogenate sample after digestion and centrifugation.
LC-MS/MS Analysis of Digested SNARE Complex Proteins
Aliquots (25 μl) of the 1:1 mixtures of stable-isotope-labeled standard peptides and immunoprecipitated SNARE proteins were transferred to LC vials (Waters) and analyzed via LC-MS/MS. The LC-MS/MS spectra were acquired with a hybrid linear quadrupole ion trap Fourier transform ion cyclotron resonance mass spectrometer equipped with a 7 T magnet (LTQ FT Ultra, Thermo Fisher Scientific, Bremen, Germany) coupled to a multidimensional nanoflow chromatography system (Ettan MDLC, GE Healthcare). A Zorbax 300 SB-C18 trap column (length, 5 mm; inner diameter, 0.3 mm; particle size, 5 μm; Agilent Technologies, Palo Alto, CA) was used for on-line desalting, and a reversed phase Zorbax 300 SBC18 column (length, 150 mm; inner diameter, 0.075 mm; particle size, 3.5 μm; Agilent Technologies) was used for high-resolution separation. Mobile phases were 0.1% aqueous FA (v/v) (A) and 0.1% FA in 84% acetonitrile in water (v/v) (B). The separation was performed at a flow rate of ∼250 nL/min by applying a linear gradient of 0% to 60% B for 50 min. The LTQ FT Ultra was set to acquire positive ions and operated in the data-dependent mode, where a scan cycle consisted of one full scan mass spectrum (m/z 350–1500) acquired in the Fourier transform ion cyclotron resonance mode (resolution 25,000) followed by 10 tandem mass spectrometry (MS/MS) scans acquired in linear quadrupole ion trap mode using collision-induced dissociation and wideband activation. Dynamic exclusion was enabled with a repeat count of 2 and an exclusion duration of 120 s. The MS/MS isolation width was set to 3 m/z units, and the normalized collision energy to 35. Each scan consisted of three microscans.
Database Search and Bioinformatic Analysis
Database searches were submitted to the in-house database server by using Mascot Deamon 2.3.0 (Matrix Science, London, UK). Database search parameters were database (UniProtKB Human 131030, 88,266 sequences, 35,040,462 residues), taxonomy (Homo sapiens), enzyme (trypsin), variable modifications (acetyl (N-terminal), oxidation (M), Label:13C(6)15N(2) (K), and Label:13C(6)15N(4) (R)), fixed modifications (carbamidomethyl (C)), mass values (monoisotopic), peptide mass tolerance (5 ppm), fragment mass tolerance (0.5 Da), maximum number of missed cleavages (two), and instrument type (electrospray ionization trap). The Mascot result files were exported as XML files with ion score cutoff (1), threshold type (identity), protein scoring (MudPIT), same-set protein hits (additional proteins that span the same set of peptides) included, and bold red required. The Mascot XML files were imported into the ProteinCenter software package (version 3.8.2014, Thermo Fisher Scientific). In ProteinCenter, the datasets from each sample were filtered by removing first peptides with individual ion scores less than or equal to 31 (where a score >31 indicated identity or extensive homology (p < 0.05)) and subsequently proteins with less than two unique peptides. All datasets from the same biochemical fraction were then compared and clustered on indistinguishable proteins (possible peptide assignments not included). To generate the list of proteins specific to immunoprecipitation with a specific antibody, all proteins that were present in the negative controls were removed. To represent each cluster of indistinguishable proteins, the protein was chosen that was present in most datasets or, if that was the same, that had the strongest evidence of existence stated in UniProtKB. In the case that that was the same, all proteins were stated on the same row.
Quantification of High-mass-accuracy Precursor Ions
Peak detection and integration were performed using DeCyder 2.0 (GE Healthcare) using processing parameters as described in Ref. 26. In-house-developed software (Sequence and PeakExtractor) was used for in silico digestion and automatic peak mass matching.
SRM-MS
Aliquots (25 μl) of the 1:1 mixtures of stable-isotope-labeled standard peptides and immunoprecipitated SNARE proteins were transferred to LC vials (SUN-SRi) and analyzed via SRM-MS using an Accela 1250 pump (Thermo Fisher Scientific) coupled to a triple quadrupole mass spectrometer (TSQ Vantage, Thermo Fisher Scientific) with an IonMax source and HESI-II electrospray probe equipped with a high-flow metal needle (Thermo Fisher Scientific). Mobile phases were 0.1% aqueous FA (v/v) (A) and 0.1% FA in 84% acetonitrile in water (v/v) (B). Samples were loaded directly onto a Hypersil Gold-C18 column (length, 50 mm; inner diameter, 2.1 mm; particle size, 5 μm; Thermo Fisher Scientific) with 0.1% aqueous FA at 200 μl/min. After 2 min of loading, the peptides were eluted off the column using the following linear gradient steps: 4 min 17% B; 12 min 23% B; 15 min 100% B. The following global MS parameters were used: positive ion mode; spray voltage, 3.5 kV; vaporizer temperature, 350 °C; sheath gas pressure, 40 psi; auxiliary gas pressure, 25 (arbitrary units); capillary temperature, 350 °C; collision gas pressure, 1.9 mTorr. Pinpoint software version 1.1 (Thermo Fisher Scientific) was used for method optimization and data processing.
Sandwich ELISA Methods for SNAP-25
A combination of mouse monoclonal antibody SP12 or mouse monoclonal antibody SMI81, as a capturing antibody, and a polyclonal anti-SNAP-25 antibody, as a detection antibody, resulted in the most sensitive detection method for SNAP-25 in human brain tissue extract. Maxisorp plates (Nunc) were coated with 100 μl of mouse monoclonal antibody SP12 (1 g/l) or mouse monoclonal antibody SMI81 (1 g/l), diluted 1:1000 in 50 mm carbonate buffer, pH 9.6, and incubated overnight at +4 °C. The plates were washed four times with 350 μl of PBS-T (0.05% v/v Tween-20 in PBS) and blocked with 200 μl of Roti-Block (Carl Roth) (diluted with PBS-T) for 1 h at room temperature. The plates were washed four times with 350 μl of PBS-T. The washing procedure was repeated between all following incubation steps. All samples were analyzed in duplicate. Brain tissue extracts were diluted 1:500 (SP12 and SMI81, Triton) and 1:1500 (SP12 and SMI81, SDS) in 0.1× Rotiblock, 0.01% Triton X-100 in PBS-T (diluent buffer). The standard of SNAP-25 was diluted in diluent buffer to a concentration range of 31.3–8000 mg/l. Samples (100 μl) of diluted brain tissue extract and standard were incubated for 1 h on a plate shaker at room temperature. Then, 100 μl of polyclonal anti-SNAP-25 antibody was diluted in diluent buffer to a final concentration of 0.5 mg/l, and the plates were incubated for 1 h on a plate shaker at room temperature. Thereafter, 100 μl of anti-rabbit IgG-biotin antibody (Sigma-Aldrich) was diluted 1:100,000 in diluent buffer and incubated for 1 h on a plate shaker at room temperature. NeutrAvidin Horseradish Peroxidase conjugate (1 g/l) (Thermo Fisher Scientific) diluted 1:10,000 in dilution buffer was incubated for 1 h on a plate shaker at room temperature, and development occurred with the addition of 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate (TMB Peroxidase EIA Substrate Kit, Bio-Rad Laboratories). The reaction was quenched with 100 μl of H2SO4 (2 m). The absorbance was measured at 450 nm. The concentration of SNAP-25 in samples of brain tissue extracts was calculated from the standard curve. Samples below the limit of detection were conservatively set to this limit. For each sample, a ratio was calculated where the SNAP-25 level was divided by the total protein concentration.
Statistical Analysis
SPSS 20.0 was employed for the statistical analyses for human samples. Because the distributions of most investigated proteins were not normal (Shapiro–Wilk test, p < 0.05), non-parametric statistics were used for the analysis of human brain tissue samples. Data are given as median (interquartile range). The Mann–Whitney U test was used to investigate group differences. The correlation coefficients (ρ) were calculated using Spearman's two-tailed correlation test. Data from the mice study are given as mean ± S.E. Student's unpaired t test was applied using GraphPad Prism 5.
Ethics
The ethical principles adhered to by Netherlands Brain Bank can be found online.
RESULTS
In this study we used a hybrid approach that combined affinity purification through immunoprecipitation with a monoclonal antibody and quantitation and characterization with mass spectrometry analysis (22). Brain tissue from age-matched patients with AD (n = 15) and controls (n = 15) (supplemental Table S1) were biochemically fractionated according to solubility using non-ionic and ionic detergents at different concentrations. Immunoprecipitation with the monoclonal antibody SP12 (derived through immunization with a human synaptic immunoprecipitate (16, 23)) against the SNARE complex protein SNAP-25 was utilized to selectively purify proteins from the fractions containing membrane-bound proteins and membrane-raft-associated proteins. The affinity-purified proteins were digested with trypsin, and four stable-isotope-labeled peptide standards, representing the SNARE complex proteins (SNAP-25, syntaxin-1A, syntaxin-1B, and VAMP-2/3), were added.
Initially, high-resolution LC-MS/MS analysis was used to identify the SNARE proteins that were selectively immunoprecipitated from the different brain homogenate fractions. We found that all four SNARE complex proteins (SNAP-25, syntaxin-1A, syntaxin-1B, and VAMP-2) were present in the majority of the samples (Table I, supplemental Tables S2 and S3). SNAP-25 exists in two isoforms in the brain, SNAP-25A and SNAP-25B (27). We identified SNAP-25B-specific peptides in all samples, but specific SNAP-25A peptides only in a few. This indicates that SNAP-25B levels were much higher than SNAP-25A levels, consistent with earlier findings that SNAP-25B is the predominant species in adult brain (18, 28). Overall, the identified tryptic peptides of SNAP-25B covered the whole protein (amino acids 2–198, supplemental Fig. S1, supplemental Table S3). We also found SNAP-25 to be modified by methionine excision and acetylation, which is in agreement with a previous study (24). Syntaxin-1 also exists in two isoforms (1A and 1B) (14, 29). Peptides specific for both forms were identified via MS/MS in all samples. Tryptic peptides covering most of the proteins were also detected (supplemental Fig. S1, supplemental Table S3). Concerning the VAMP/synaptobrevin isoforms, peptides specific for VAMP-2 and VAMP-1 were detected in most samples, and for VAMP-3 in ∼20% of the samples (supplemental Fig. S1, supplemental Table S3). All detected peptides originated from the v-SNARE coiled-coil homology domains in the respective VAMP isoforms (supplemental Fig. S1, supplemental Table S3) (30). Apart from the SNARE complex proteins, several other proteins that are known or suspected to interact with the SNARE complex (e.g. complexin-1, complexin-2, syntaxin-binding protein-1, and synaptotagmin-1) were co-immunoprecipitated with SNAP-25 (supplemental Table S2) (31–33).
Table I. SNARE proteins identified with LC-MS/MS in immunoprecipitate (monoclonal antibody SP12) from membrane-bound and membrane-raft-associated protein fractions of superior parietal gyrus from controls and patients with Alzheimer disease (AD).
| Protein | Fraction of samples in which the protein was found (%) |
Unique peptide sequences (average) |
Sequence coverage (%) (average) |
|||
|---|---|---|---|---|---|---|
| Membrane-bound | Membrane-raft-associated | Membrane-bound | Membrane-raft-associated | Membrane-bound | Membrane-raft-associated | |
| Control/AD | Control/AD | Control/AD | Control/AD | Control/AD | Control/AD | |
| SNAP-25B | 100/100 | 100/100 | 19/15 | 19/16 | 67/63 | 68/63 |
| SNAP-25A | 13/0 | 7/7 | Too few | Too few | Too few | Too few |
| Syntaxin-1A | 100/100 | 100/100 | 9/6 | 9/7 | 24/18 | 28/21 |
| Syntaxin-1B | 100/100 | 100/100 | 16/15 | 17/15 | 47/45 | 47/44 |
| VAMP-2 | 93/100 | 100/93 | 5/5 | 4/4 | 45/44 | 38/39 |
| VAMP-1 | 73/57 | 87/50 | 4/4 | 3/3 | 30/32 | 24/26 |
| VAMP-3 | 20/7 | 53/7 | 5/too few | 4/too few | 48/too few | 39/too few |
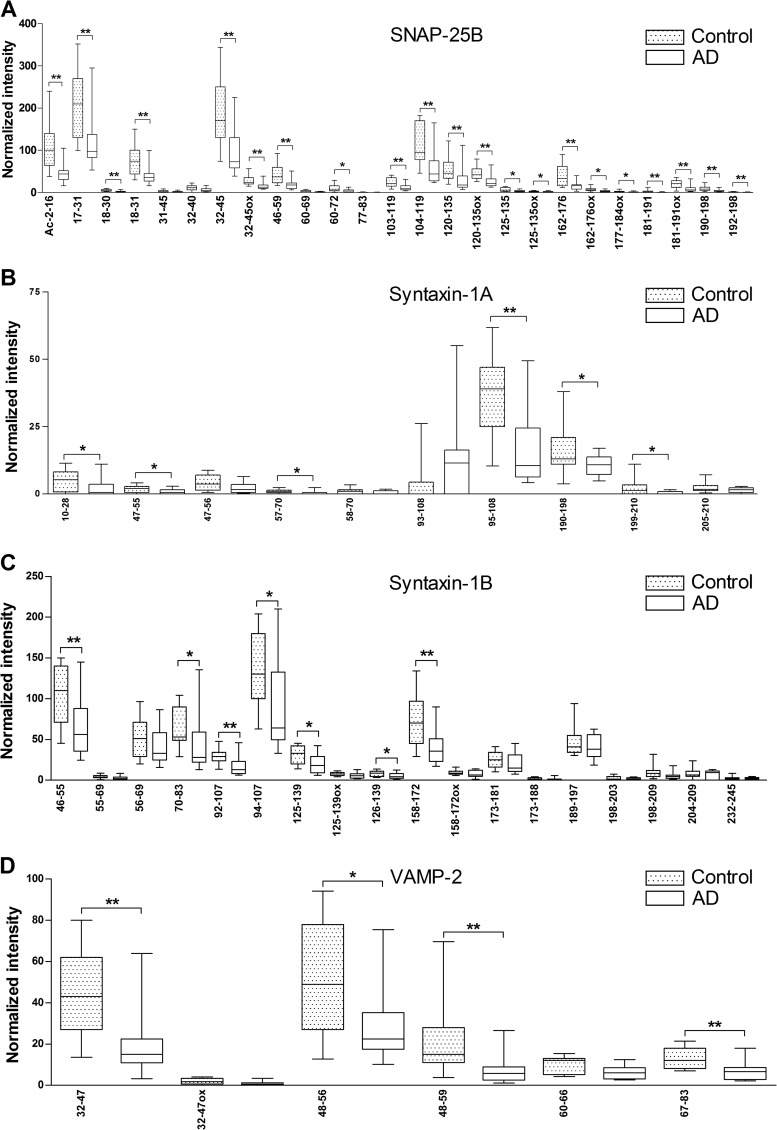
To estimate the SNARE protein levels, we extracted the acquired LC-MS ion chromatograms of all peptides belonging to a SNARE complex protein and compared the signal intensities between AD and control groups (Figs. 1A–1D, supplemental Figs. S2A–S2D, supplemental Table S4). We found that the intensities of most of the investigated tryptic peptides belonging to SNAP-25B, syntaxin-1B, syntaxin-1A, or VAMP-2 were significantly lower in the AD group than in the control group (Figs. 1A–1D, supplemental Figs. S2A–S2D, supplemental Table S4).
Fig. 1.
LC-MS signal intensities of individual tryptic peptides from immunoprecipitated SNARE proteins SNAP-25B (A), syntaxin-1A (B), syntaxin-1B (C), and VAMP-2 (D) affinity-purified with the monoclonal antibody SP12 from membrane-bound fractions of superior parietal gyrus from controls and patients with Alzheimer disease (AD). Normalized intensities of tryptic peptides measured with high-resolution LC-MS, confirmed with MS/MS, and present in at least 10 controls (dotted bars) and 10 patients with AD (white bars). The lower, upper, and middle portions of the bars correspond to the 25th percentile, 75th percentile, and median, respectively. The whiskers on the bottom extend from the 10th percentile to the top 90th percentile. p values less than 0.05 and 0.01 are denoted by * and ** (Mann–Whitney U test).
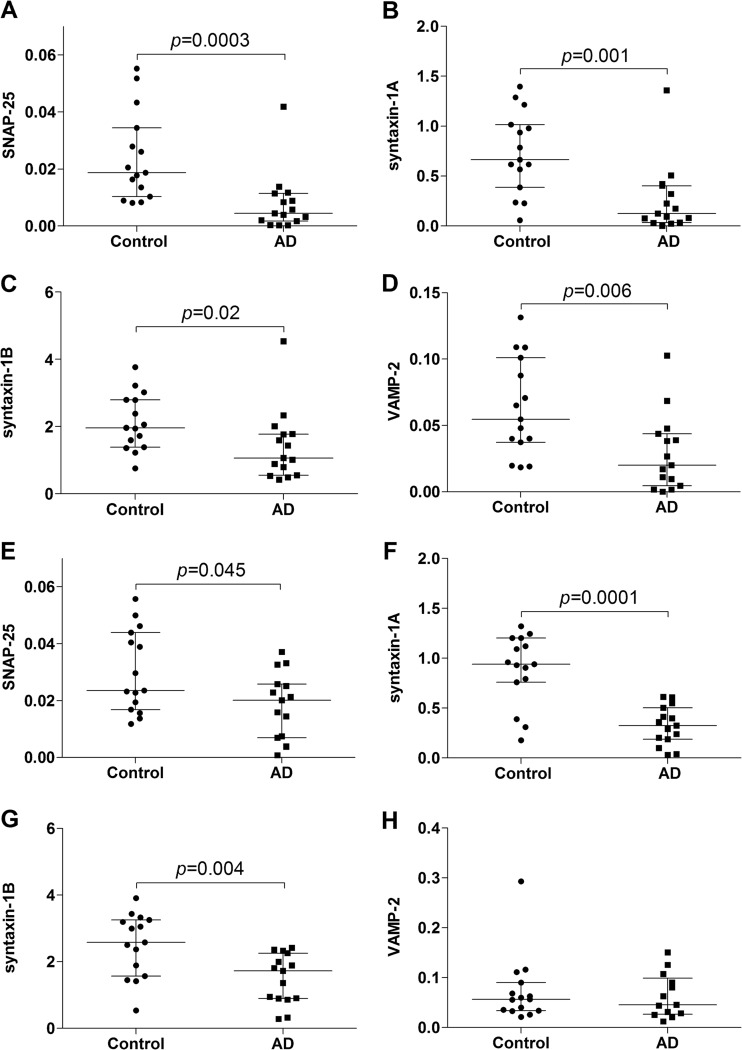
To get a more accurate quantification of the SNARE complex proteins, aliquots of immunoprecipitated (SP12) brain protein fractions from patients with AD (n = 15) and controls (n = 15) were quantified using SRM-MS. Membrane-bound fraction levels of all SNARE proteins were significantly lower in the AD group (Figs. 2A–2D, supplemental Table S5). SNAP-25 and syntaxin-1A/B levels were also significantly lower in the AD group in the membrane-raft-associated fractions (Figs. 2E–2G, supplemental Table S5). The finding of reduced SNARE protein levels in AD patients was consistent with previous studies of AD using different immunoassays (34–37). The results also demonstrate that the IP-MS method is sufficiently sensitive to detect pathological changes in SNARE protein levels in brain tissue samples from individual patients. The IP-MS levels of SNAP-25 were strongly correlated with the levels of syntaxin-1A/B and VAMP-2 (membrane-bound (ρ = 0.886, p < 0.001 for syntaxin-1A; ρ = 0.806, p < 0.001 for syntaxin-1B; and ρ = 0.779, p < 0.001 for VAMP-2) and membrane-raft-associated (ρ = 0.624, p < 0.001 for syntaxin-1A; ρ = 0.645, p < 0.001 for syntaxin-1B; and ρ = 0.513, p = 0.005 for VAMP-2)). This indicates that the membrane-bound fractions of SNARE proteins are tightly associated, supporting previous work showing that the SNARE complex is stable and even resistant to the detergent SDS (10).
Fig. 2.
Targeted mass spectrometry analyses of the SNARE complex in human brain. Individual values for the SRM-MS measured ratios (endogenous peptide/labeled peptide standard) of immunoprecipitated (SP12 antibody) SNAP-25, syntaxin-1A, syntaxin-1B, and VAMP-2/3 in biochemically fractionated membrane-bound (A–D) and membrane-raft-associated (E–H) fractions of superior parietal gyrus from controls (n = 15) and patients with Alzheimer disease (AD) (n = 15). The lower, upper, and middle lines of the error bars correspond to the 25th and 75th percentiles and medians, respectively. The Mann–Whitney U test was used to investigate group differences.
Potential disadvantages of the IP-MS method relative to ELISA may include lower sensitivity and reproducibility. However, when using an in-house-developed sandwich ELISA for SNAP-25 with SMI81 (recognizes an epitope containing the N-terminally acetylated first 11 amino acids of brain SNAP-25 (24)) or SP12 as capture antibodies, we observed results similar to those of the IP-MS assay (supplemental Fig. S3). The SNAP-25 levels measured with SMI81 and SP12 ELISA were highly correlated both in the membrane-bound fractions (ρ = 0.984, p < 0.001) and in the membrane-raft-associated fractions (ρ = 0.943, p < 0.001). Furthermore, the IP-MS-measured levels of SNAP-25 were correlated with the SNAP-25 ELISA-measured levels (membrane-bound fractions: ρ = 0.865, p < 0.001 for SP12 coating and ρ = 0.839, p < 0.001 for SMI81 coating), indicating that the two methods are comparably reproducible and sensitive. For membrane-raft-associated fractions, there was a moderate correlation between the IP-MS- and ELISA-measured levels of SNAP-25 (ρ = 0.389, p < 0.05 for SP12 coating and ρ = 0.478, p = 0.008 for SMI81 coating).
A major advantage of the IP-MS method is that because the SNARE complex proteins are highly conserved, the method can be applied directly, without modification of brain tissue samples from other species. To demonstrate this, we measured levels of SNARE proteins in mice with prion disease in which synaptic degeneration is known to occur. We used tg37 mice in which the time course of synapse loss and clinicopathological course are well mapped, with onset at around 7 weeks post-inoculation or 9 w.p.i., progressing to neuronal loss at 10 w.p.i. and death from prion disease at 12 w.p.i. when inoculated with Rocky Mountain Laboratory prions or normal brain homogenate at ∼4 weeks of age as described in Refs. 20 and 21. We sampled brains from prion-infected tg37 mice and controls 6, 7, 8, 9, and 10 w.p.i. and fractionated these according to solubility. Levels of proteins immunoprecipitated with SMI81 were quantified with SRM-MS. Similar to results from human brain tissue, all SNARE complex proteins were observed in the membrane-bound and membrane-raft-associated protein fractions. SNAP-25 levels did not show any significant change with time after prion infection in any of the protein fractions. However, levels of the SNAP-25 interacting proteins syntaxin-1A and syntaxin-1B were significantly decreased 10 weeks after prion infection (Figs. 3B, 3C, 3F, and 3G; supplemental Table S6) relative to uninfected controls. For VAMP-2, the measured levels were significantly decreased 10 weeks after prion infection in the membrane-bound protein fractions, but not in the membrane-raft-associated fractions (Figs. 3D and 3H, supplemental Table S6).
Fig. 3.
Targeted mass spectrometry analyses of the SNARE complex in prion-infected mice. Individual values for the SRM-MS measured ratios (endogenous peptide/labeled peptide standard) of immunoprecipitated (SMI81 antibody) SNAP-25, syntaxin-1A, syntaxin-1B, and VAMP-2 in biochemically fractionated membrane-bound (A–D) and membrane-raft-associated (E–H) fractions of prion-infected mice at various weeks post-infection. Three mice are represented at each time point. All data show mean ± S.E. (Student's unpaired t test).
DISCUSSION
In this paper we describe an approach for the concurrent measurement of levels of all SNARE complex proteins from individual human and mouse brain tissue samples. The main advantage of the IP-MS method relative to immunocytochemistry, Western blotting, and ELISA methods that have been used to study pathological changes in SNARE complex protein expression is its versatility. With the IP-MS method, all SNARE proteins, including modified forms, can be quantified simultaneously, which is an advantage over existing methods. The protein target can be altered or expanded by changing antibodies, adding stable-isotope-labeled standards of modified protein forms or protein complex interaction partners, or changing the fractionation method to study different subtypes of cells or specific cell compartments (13). This is of particular interest when studying the SNARE complex proteins, as it has been shown that different combinations of SNARE protein isoforms interact with dissimilar affinities and are differentially distributed in defined areas of the central nervous system, suggesting specialized functions of these isoforms (29). Other SNARE protein isoforms such as SNAP-25A and -25B have very different expression during embryonic and early postnatal development: SNAP-25A is the predominant species during embryonic and early postnatal development, whereas SNAP-25B transcripts increase dramatically after the first postnatal week to become the majority of SNAP-25 mRNA in adult brain (38, 39). Other studies have shown that different SNARE protein isoforms could have specialized roles in the neurosecretory process (40).
Recently, Varjosalo et al. demonstrated the technical feasibility of obtaining high-quality maps of human protein interactomes using immunopurification combined with mass spectrometry (41). In their study they showed that very high within-laboratory and interlaboratory IP-MS reproducibility can be achieved. Our study focused on four SNARE proteins, including two isoforms of syntaxin-1 (A and B), but there are several very interesting interaction partners that could be added to the method without major changes. Interestingly, our results with mice corroborate earlier data indicating SNARE complex destabilization in prion-infected mice (21). These data support the notion that the described IP-MS method can be utilized to investigate the stability of the SNARE complex and the strength of association between the different SNARE protein forms or other interaction partners. The study involving mouse brain tissue also confirmed another important aspect of the presented IP-MS method. The mechanism and structure of SNARE proteins are widely evolutionarily conserved (42, 43). Thus, it is possible to apply the method that we have developed for human brain tissue directly to brain tissue samples from several other species often used as animal models (e.g. rat, mouse, dog, pig, and zebrafish) without major modifications.
In summary, we have demonstrated that it is possible to use IP-MS to measure levels of four SNARE complex proteins in parallel. The applicability of the IP-MS method was demonstrated in a comparison of SNARE complex protein levels in brain tissue from AD patients and age-matched controls, as well as in a study of SNARE complex protein levels in brain tissue from prion-infected mice. Furthermore, the high correlation between the IP-MS-measured levels of SNAP-25 and the SNAP-25 levels measured with sandwich ELISA indicates that the two methods are comparably reproducible and sensitive. The presented method could be used to study the synaptic pathology in neuropsychiatric or neurodegenerative diseases and evaluate drugs aimed at restoring synaptic function in various animal models, as well as in human postmortem tissue and possibly biological fluids.
Supplementary Material
Acknowledgments
We are grateful to Rita Persson and Dzemila Secic for their technical assistance. We are also grateful to Professor Lennart Brodin and Dr. Peter Löw for providing the polyclonal SNAP-25 antibodies.
Footnotes
Author contributions: A.B., H.Z., K.B., and A.O. designed research; A.B. and A.O. performed research; A.B., G.B., W.G.H., J.A.M., J.J., G.R.M., and A.O. contributed new reagents or analytic tools; A.B., G.B., W.G.H., and A.O. analyzed data; A.B., G.B., W.G.H., J.A.M., J.J., G.R.M., H.Z., K.B., and A.O. wrote the paper.
* This work was supported by grants from the Swedish Brain Power Consortium, Swedish Alzheimer Foundation, Demensfonden, Eivind och Elsa K:son Sylvans stiftelse, Märtha och Gustaf Ågrens stiftelse, Gun och Bertil Stohnes Foundation, Stiftelsen Gamla Tjänarinnor and Svenska Läkaresällskapet, the Canadian Institutes of Health Research (MOP-14037 and CBG-101827), and Åhlén-stiftelsen.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- AD
- Alzheimer disease
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SNAP-25
- synaptosomal-associated protein 25
- VAMP
- vesicle-associated membrane protein
- IP-MS
- immunoprecipitation–mass spectrometry
- SRM
- selected reaction monitoring
- w.p.i.
- weeks post-infection
- FA
- formic acid.
REFERENCES
- 1. Davies C. A., Mann D. M., Sumpter P. Q., Yates P. O. (1987) A quantitative morphometric analysis of the neuronal and synaptic content of the frontal and temporal cortex in patients with Alzheimer's disease. J. Neurol. Sci. 78, 151–164 [DOI] [PubMed] [Google Scholar]
- 2. Overk C. R., Masliah E. (2014) Pathogenesis of synaptic degeneration in Alzheimer's disease and Lewy body disease. Biochem. Pharmacol. 88, 508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terry R. D., Masliah E., Salmon D. P., Butters N., DeTeresa R., Hill R., Hansen L. A., Katzman R. (1991) Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 30, 572–580 [DOI] [PubMed] [Google Scholar]
- 4. Masliah E., Mallory M., Alford M., DeTeresa R., Hansen L. A., McKeel D. W., Jr., Morris J. C. (2001) Altered expression of synaptic proteins occurs early during progression of Alzheimer's disease. Neurology 56, 127–129 [DOI] [PubMed] [Google Scholar]
- 5. Blennow K., Bogdanovic N., Alafuzoff I., Ekman R., Davidsson P. (1996) Synaptic pathology in Alzheimer's disease: relation to severity of dementia, but not to senile plaques, neurofibrillary tangles, or the ApoE4 allele. J. Neural Transm. 103, 603–618 [DOI] [PubMed] [Google Scholar]
- 6. DeKosky S. T., Scheff S. W. (1990) Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann. Neurol. 27, 457–464 [DOI] [PubMed] [Google Scholar]
- 7. Sollner T., Whiteheart S. W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J. E. (1993) SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324 [DOI] [PubMed] [Google Scholar]
- 8. Barakauskas V. E., Beasley C. L., Barr A. M., Ypsilanti A. R., Li H. Y., Thornton A. E., Wong H., Rosokilja G., Mann J. J., Mancevski B., Jakovski Z., Davceva N., Ilievski B., Dwork A. J., Falkai P., Honer W. G. (2010) A novel mechanism and treatment target for presynaptic abnormalities in specific striatal regions in schizophrenia. Neuropsychopharmacology 35, 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeans A. F., Oliver P. L., Johnson R., Capogna M., Vikman J., Molnar Z., Babbs A., Partridge C. J., Salehi A., Bengtsson M., Eliasson L., Rorsman P., Davies K. E. (2007) A dominant mutation in Snap25 causes impaired vesicle trafficking, sensorimotor gating, and ataxia in the blind-drunk mouse. Proc. Natl. Acad. Sci. U.S.A. 104, 2431–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayashi T., McMahon H., Yamasaki S., Binz T., Hata Y., Sudhof T. C., Niemann H. (1994) Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 13, 5051–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sudhof T. C. (2004) The synaptic vesicle cycle. Annu. Rev. Neurosci. 27, 509–547 [DOI] [PubMed] [Google Scholar]
- 12. Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 13. Barakauskas V. E., Moradian A., Cheng G. S. W., Dwork A. J., Honer W. G., Morin G. (2011) MRM-based quantitation of SNAP-25 protein isoforms in cells and human brain samples. 59th ASMS Conference on Mass Spectrometry Allied Topics, Denver, CO June 5–9 2011 [Google Scholar]
- 14. Bennett M. K., Garcia-Arraras J. E., Elferink L. A., Peterson K., Fleming A. M., Hazuka C. D., Scheller R. H. (1993) The syntaxin family of vesicular transport receptors. Cell 74, 863–873 [DOI] [PubMed] [Google Scholar]
- 15. Garbelli R., Inverardi F., Medici V., Amadeo A., Verderio C., Matteoli M., Frassoni C. (2008) Heterogeneous expression of SNAP-25 in rat and human brain. J. Comp. Neurol. 506, 373–386 [DOI] [PubMed] [Google Scholar]
- 16. Oyler G. A., Higgins G. A., Hart R. A., Battenberg E., Billingsley M., Bloom F. E., Wilson M. C. (1989) The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J. Cell Biol. 109, 3039–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trimble W. S., Gray T. S., Elferink L. A., Wilson M. C., Scheller R. H. (1990) Distinct patterns of expression of two VAMP genes within the rat brain. J. Neurosci. 10, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamamori S., Itakura M., Sugaya D., Katsumata O., Sakagami H., Takahashi M. (2011) Differential expression of SNAP-25 family proteins in the mouse brain. J. Comp. Neurol. 519, 916–932 [DOI] [PubMed] [Google Scholar]
- 19. Braak H., Braak E. (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]
- 20. Mallucci G. R., Ratte S., Asante E. A., Linehan J., Gowland I., Jefferys J. G., Collinge J. (2002) Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 21, 202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moreno J. A., Radford H., Peretti D., Steinert J. R., Verity N., Martin M. G., Halliday M., Morgan J., Dinsdale D., Ortori C. A., Barrett D. A., Tsaytler P., Bertolotti A., Willis A. E., Bushell M., Mallucci G. R. (2012) Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature 485, 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Öhrfelt A., Zetterberg H., Andersson K., Persson R., Secic D., Brinkmalm G., Wallin A., Mulugeta E., Francis P. T., Vanmechelen E., Aarsland D., Ballard C., Blennow K., Westman-Brinkmalm A. (2011) Identification of novel alpha-synuclein isoforms in human brain tissue by using an online nanoLC-ESI-FTICR-MS method. Neurochem. Res. 36, 2029–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Honer W. G., Falkai P., Young C., Wang T., Xie J., Bonner J., Hu L., Boulianne G. L., Luo Z., Trimble W. S. (1997) Cingulate cortex synaptic terminal proteins and neural cell adhesion molecule in schizophrenia. Neuroscience 78, 99–110 [DOI] [PubMed] [Google Scholar]
- 24. Connell E., Darios F., Peak-Chew S., Soloviev M., Davletov B. (2009) N-terminal acetylation of the neuronal protein SNAP-25 is revealed by the SMI81 monoclonal antibody. Biochemistry 48, 9582–9589 [DOI] [PubMed] [Google Scholar]
- 25. Low P., Norlin T., Risinger C., Larhammar D., Pieribone V. A., Shupliakov O., Brodin L. (1999) Inhibition of neurotransmitter release in the lamprey reticulospinal synapse by antibody-mediated disruption of SNAP-25 function. Eur. J. Cell Biol. 78, 787–793 [DOI] [PubMed] [Google Scholar]
- 26. Brinkmalm G., Portelius E., Ohrfelt A., Mattsson N., Persson R., Gustavsson M. K., Vite C. H., Gobom J., Mansson J. E., Nilsson J., Halim A., Larson G., Ruetschi U., Zetterberg H., Blennow K., Brinkmalm A. (2012) An online nano-LC-ESI-FTICR-MS method for comprehensive characterization of endogenous fragments from amyloid beta and amyloid precursor protein in human and cat cerebrospinal fluid. J. Mass Spectrom. 47, 591–603 [DOI] [PubMed] [Google Scholar]
- 27. Bark I. C., Wilson M. C. (1994) Human cDNA clones encoding two different isoforms of the nerve terminal protein SNAP-25. Gene 139, 291–292 [DOI] [PubMed] [Google Scholar]
- 28. Prescott G. R., Chamberlain L. H. (2011) Regional and developmental brain expression patterns of SNAP25 splice variants. BMC Neurosci. 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aguado F., Majo G., Ruiz-Montasell B., Llorens J., Marsal J., Blasi J. (1999) Syntaxin 1A and 1B display distinct distribution patterns in the rat peripheral nervous system. Neuroscience 88, 437–446 [DOI] [PubMed] [Google Scholar]
- 30. Wiederhold K., Kloepper T. H., Walter A. M., Stein A., Kienle N., Sorensen J. B., Fasshauer D. (2010) A coiled coil trigger site is essential for rapid binding of synaptobrevin to the SNARE acceptor complex. J. Biol. Chem. 285, 21549–21559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mohrmann R., de Wit H., Connell E., Pinheiro P. S., Leese C., Bruns D., Davletov B., Verhage M., Sorensen J. B. (2013) Synaptotagmin interaction with SNAP-25 governs vesicle docking, priming, and fusion triggering. J. Neurosci. 33, 14417–14430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pertsinidis A., Mukherjee K., Sharma M., Pang Z. P. P., Park S. R., Zhang Y. X., Brunger A. T., Sudhof T. C., Chu S. (2013) Ultrahigh-resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ. Proc. Natl. Acad. Sci. U.S.A. 110, E2812–E2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang X. F., Cao P., Sudhof T. C. (2013) Deconstructing complexin function in activating and clamping Ca2+-triggered exocytosis by comparing knockout and knockdown phenotypes. Proc. Natl. Acad. Sci. U.S.A. 110, 20777–20782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Honer W. G. (2003) Pathology of presynaptic proteins in Alzheimer's disease: more than simple loss of terminals. Neurobiol. Aging 24, 1047–1062 [DOI] [PubMed] [Google Scholar]
- 35. Honer W. G., Barr A. M., Sawada K., Thornton A. E., Morris M. C., Leurgans S. E., Schneider J. A., Bennett D. A. (2012) Cognitive reserve, presynaptic proteins and dementia in the elderly. Transl. Psychiatry 2, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Minger S. L., Honer W. G., Esiri M. M., McDonald B., Keene J., Nicoll J. A., Carter J., Hope T., Francis P. T. (2001) Synaptic pathology in prefrontal cortex is present only with severe dementia in Alzheimer disease. J. Neuropathol. Exp. Neurol. 60, 929–936 [DOI] [PubMed] [Google Scholar]
- 37. Shimohama S., Kamiya S., Taniguchi T., Akagawa K., Kimura J. (1997) Differential involvement of synaptic vesicle and presynaptic plasma membrane proteins in Alzheimer's disease. Biochem. Biophys. Res. Commun. 236, 239–242 [DOI] [PubMed] [Google Scholar]
- 38. Bark C., Bellinger F. P., Kaushal A., Mathews J. R., Partridge L. D., Wilson M. C. (2004) Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J. Neurosci. 24, 8796–8805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bark I. C., Hahn K. M., Ryabinin A. E., Wilson M. C. (1995) Differential expression of SNAP-25 protein isoforms during divergent vesicle fusion events of neural development. Proc. Natl. Acad. Sci. U.S.A. 92, 1510–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bragina L., Giovedi S., Barbaresi P., Benfenati F., Conti F. (2010) Heterogeneity of glutamatergic and GABAergic release machinery in cerebral cortex: analysis of synaptogyrin, vesicle-associated membrane protein, and syntaxin. Neuroscience 165, 934–943 [DOI] [PubMed] [Google Scholar]
- 41. Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S., Aebersold R., Bennett K. L., Colinge J., Gstaiger M., Superti-Furga G. (2013) Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods 10, 307–314 [DOI] [PubMed] [Google Scholar]
- 42. Hong W. (2005) SNAREs and traffic. Biochim. Biophys. Acta 1744, 493–517 [PubMed] [Google Scholar]
- 43. Jahn R., Scheller R. H. (2006) SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.