Abstract
The Mycobacterium tuberculosis Beijing genotype, consisting of the more ancient (atypical) and modern (typical) emerging sublineage, is one of the most prevalent and genetically conserved genotype families and has often been associated with multidrug resistance. In this study, we employed a 2D-LC-FTICR MS approach, combined with dimethylation of tryptic peptides, to systematically compare protein abundance levels of ancient and modern Beijing strains and identify differences that could be associated with successful spread of the modern sublineage. The data is available via ProteomeXchange using the identifier PXD000931. Despite the highly uniform protein abundance ratios in both sublineages, we identified four proteins as differentially regulated between both sublineages, which could explain the apparent increased adaptation of the modern Beijing strains. These proteins are; Rv0450c/MmpL4, Rv1269c, Rv3137, and Rv3283/sseA. Transcriptional and functional analysis of these proteins in a large cohort of 29 Beijing strains showed that the mRNA levels of Rv0450c/MmpL4 are significantly higher in modern Beijing strains, whereas we also provide evidence that Rv3283/sseA is less abundant in the modern Beijing sublineage. Our findings provide a possible explanation for the increased virulence and success of the modern Beijing sublineage. In addition, in the established dataset of 1817 proteins, we demonstrate the pre-existence of several, possibly unique, antibiotic efflux pumps in the proteome of the Beijing strains. This may reflect an increased ability of Beijing strains to escape exposure to antituberculosis drugs.
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB)1, is one of the most successful pathogens worldwide. Nowadays, still about 8–9 million new cases and 1.5 million deaths are recorded annually (1).
DNA fingerprinting of the M. tuberculosis complex has revolutionized studies on transmission of TB in the last two decades (2), but also disclosed the phylogeny of this important grouping of bacteria (3, 4). In essence, there are six major M. tuberculosis lineages. The first one was described in 1995 and designated the “Beijing” genotype family, which is highly prevalent in Asia, the Former Soviet Union states, and South Africa (5–8). Recently, the Beijing genotype family was identified as the major genotype family in the “East Asia clade” defined on single nucleotide polymorphism (SNP) typing (4). In the years after its disclosure, the Beijing genotype family drew attention because it seemed genetically highly conserved on basis of the available genetic markers, which could be related to active and recent spread, presumably because of selective advantages over other M. tuberculosis genotypes (5–8). Selective pressure on this group of bacteria could be induced by the introduction of mass BCG vaccination and treatment by anti-TB drugs (9). Consequently, in many geographic areas, Beijing strains were found significantly correlated with (multi)drug resistance, treatment failure, relapses after curative treatment, and transmission of resistant TB (6, 7, 10–12). In TB mouse models, Beijing strains revealed an up-regulated virulence and a higher ability to escape BCG vaccination (13–15). This is particularly alarming because there are indications that the Beijing genotype strains are emerging in multiple geographic areas, as they are associated with lower ages of patients and, hence, active transmission (6, 16, 17). Increased odds of M. tuberculosis Beijing are also observed for patients with a polymorphism in their natural resistance associated macrophage protein 1 (NRAMP1), encoded by the SLC11A1 gene (18). This supports the theory of a co-evolution of M. tuberculosis Beijing with their human host.
Initially, mechanisms underlying a higher adaptability were assumed to be related to alterations in DNA repair, but polymorphisms in related genes were also observed in strains of other genotype families (19). Although not yet explained in detail, a study published in 2012 revealed that a part of the Beijing strains revealed a much higher frequency of naturally occurring rifampicin resistant mutants than the traditionally measured 1 in 108 (20). This important observation was recently confirmed by Ford et al., who pointed out that the correlation between the Beijing genotype and resistance could indeed be explained by a higher mutation frequency (21). Not surprisingly, resistance against anti-TB drugs is especially a major problem in former Soviet Union States, China, and South Africa, where the Beijing genotype strains are highly prevalent (22–26).
In 2002, the Beijing genotype family was divided into the modern (typical) and ancient (atypical) lineage (27). Beijing strains without insertion of IS6110 in the NTF region were referred to as ancient Beijing strains and seem to resemble the ancestors of the Beijing family (27). In most countries, modern Beijing strains are much more prevalent than ancient ones, suggesting an active spread, except for Japan, where the ancient type of Beijing strains is more wide spread (28). The two Beijing lineages were also divided on basis of large genomic deletions, also referred to as Regions of Difference (RDs). RD105 is thought to be characteristic of all Beijing strains, whereas deletion of RD181 was found associated with modern Beijing strains (29). Although the modern and ancient Beijing strains seem to be closely related on the genetic level (30), differences in association with (multi)drug resistance and in the ability to cause and spread disease (31) have been described between both sublineages. Moreover, after several studies suggested enhanced ability of Beijing strains in general to circumvent BCG-vaccine induced immunity (13, 14), one study reported that modern Beijing strains were isolated more frequently from BCG-vaccinated patients than nonvaccinated individuals (9). The observation that the interaction of modern Beijing strains with the immune system is different than that of ancient strains was further confirmed by a recent study that described differences in pro-inflammatory cytokine induction for both strains (32). If modern Beijing strains indeed have selective advantages over other M. tuberculosis strains and spread more prosperously, this should be reflected in the degree of genetic conservation. This assumption was confirmed by whole genome sequencing (WGS) of Beijing strains from a wide spread geographic area. Three modern Beijing strains from China, Vietnam, and South Africa were found genetically highly conserved in comparison to three ancient strains from the same regions (30). This confirms the evolutionary advantage of the modern Beijing genotype over the closely related ancient Beijing and other M. tuberculosis strains. In fact, Schürch et al. traced only 31 nonsynonymous single nucleotide polymorphisms (nsSNPs) mutations characteristic of modern Beijing strains (30). However, the presence of these 31 nsSNPs is not sufficient to explain the consequences in terms of evolutionary development and adaptation. Therefore, we performed an in-depth comparison of the protein abundances of both sublineages. Comparative proteomic analysis of different M. tuberculosis strains has been reported before (33). So far, one study examined the proteomes of a single hyper- and hypovirulent M. tuberculosis Beijing strain by a label-free quantitative technique, and reported the differential regulation of virulence factors, such as Rv3875/ESAT-6 and other Esx-like proteins (34). However, biological variation limits the value of a traditional individual duplex (one-versus-one) experimental approach. In addition, individual characteristics that are not always representative of the entire grouping cannot be distinguished from structural differences between the two genotypes. To obtain a more reliable insight, multiple measurements of individual samples of each grouping are required. Furthermore, to limit the influence of biological and inter-strain variation, we generated a sample pool comprising five modern and five ancient Beijing strains, which were selected to optimally represent the full spectrum of both M. tuberculosis Beijing sublineages.
Sample pooling was previously successfully applied in proteomic experiments, and used in case there was insufficient individual sample material available (35, 36). Sample pooling has also been applied to create an internal reference sample in a so called “super-SILAC mix” (37, 38), that can be combined with clinical samples. Use of the super-SILAC approach resulted in a narrower distribution of protein abundance ratios. The narrower protein ratio distribution increased the significance of the altered protein ratios (35). Next to the pooled sample approach, we compared three ancient Beijing strains with three modern Beijing strains using traditional individual duplexes. We identified and quantified a cumulative number of 2392 proteins (±60% of the M. tuberculosis protein coding genome), of which 1817 were present in all of the analyses performed. We demonstrate the presence of multiple antibiotic extruding efflux pumps in the proteome of M. tuberculosis Beijing, notably without exposure of the pathogen to a drug. Three of these efflux pumps were previously not quantified in the proteome of M. tuberculosis H37Rv by a proteome-wide selected reaction monitoring approach (39).
Despite the highly uniform protein ratio distribution in both strains, we identified four proteins to be more abundant in the modern Beijing strains that can explain their emergence and perhaps higher virulence. We complemented our proteomic data with transcriptional analysis of selected genes in a larger cohort of 14 ancient Beijing and 15 modern Beijing strains. To verify the differential regulation of a particular virulence factor the experiments were supplemented with functional analysis.
MATERIALS AND METHODS
Molecular Typing Methods
M. tuberculosis Beijing strains were selected from a previously published selection of 259 Beijing strains (40). Fourteen ancient and 14 modern Beijing strains were included in the study to optimally represent the M. tuberculosis family. In addition, the successful modern Beijing strain B0/W148 was added to the selection (41–43). Using the M. tuberculosis Beijing characteristic marker RD105, we ensured that all the strains investigated indeed belong to the M. tuberculosis Beijing family. Additionally, mutT2/ogt were used to differentiate ancient from modern Beijing strains (29, 31). To exclude the possibility of contamination, or the selection of clonal isolates for proteomic analysis, we performed standard MIRU (mycobacterial interspersed repeat units) 24-loci VNTR (variable number of tandem repeats) with a few minor modifications (44). The in-house VNTR method based on the protocol of the MIRU-VNTR typing manual was used with minor modifications: the amount of DNA polymerase used was 0.75 units per multiplex PCR, and the initial concentration of labeled primers was increased to 8 μm for locus 2165 and locus 2163b. The amplicon sizes were determined by using the automated ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA).
Mycobacterial Culture Conditions
All M. tuberculosis isolates were derived from the reference database of clinical isolates at the National Institute for Public Health and the Environment (RIVM) in Bilthoven, the Netherlands. Strains were recultured from frozen stocks in 5 ml Tween-Albumin liquid culture broth (Tritium Microbiologie, the Netherlands) at 36 °C without shaking until an O.D. at 600 nm of 0.4 AU was reached. Of the mycobacterial pre culture, 1 ml was transferred to a 250 ml Erlenmeyer flask containing 100 ml Tween-Albumin broth and incubated under shaking conditions at 36 °C with constant aeration. Once the cultures reached an O.D. at 600 nm of 0.6 AU, representing the mid-log phase, the cells for proteome analysis were washed three times with ice cold PBS, dissolved in 5 ml Lysis-buffer (4% SDS, 100 mm Tris-HCl, pH 7.6) and heat-killed at 95 °C for 10 min. Lysates were stored at −20 °C until further usage. M. tuberculosis Beijng strains used for qPCR analysis were harvested at early log phase and stored in L6-buffer at −20 °C until use (45).
Minimal Inhibitory Concentration Determination
Susceptibility to the first line antimycobacterial drugs; rifampicin, isoniazid, ethambutol, and pyrazinamid, was determined according to Clinical and Laboratory Standards Institute guidelines (46), using the BACTEC MGIT-960 system (Becton, Dickinson and Co., Franklin Lakes, NJ).
Protein Sample Preparation and Dimethylation Isotope Labeling
Heat inactivated cells for proteomic experiments were mechanically lysed by bead-beating in a mini bead-beater 16 (BioSpec, Bartlesville, OK) for 5 min using glass beads. Thereafter, the cells were cooled down on ice for 5 min and the procedure was repeated twice. The cell lysates were cleaned from cell debris by centrifugation for 1 min at 14,000 × g and the supernatant was transferred to another tube. Proteins were digested using the filter aided sample preparation (FASP) method (47). In brief, 100 μg of DTT reduced proteins was loaded on a 30kDa filter. SDS was removed in three washes with 8 m urea. The proteins were reduced and carbamidomethylated, and the excess reagent was removed by three additional washes with 8 m urea. Proteins were then overnight digested using endoproteinase Lys-C (endoLysC) followed by a four hrs digestion using trypsin at RT. Tryptic peptides were desalted on C18 SepPak columns and derivatized on column by dimethyl labeling (48). Peptides derived from ancient Beijing strains were labeled with a light label (+28Da), whereas modern Beijing strains were labeled with heavy labels (+36Da). In addition, two sample pools consisting of five light labeled ancient Beijing strains and five heavy labeled modern Beijing strains were prepared by mixing the respective tryptic digests in a 1:1:1:1:1 manner before fractionation.
Strong Cation Exchange Chromatography
In total 100 μg of labeled peptides were fractionated by strong cation exchange (SCX) on a Agilent 1100 system equipped with an in-house packed SCX-column (320 μm ID, 15 cm, polysulfoethyl A 3 μm, Poly LC), run at 4 μl/min. The gradient started with a 10 min run at 100% solvent A 70/30/0.1 (water/acetonitrile/formic acid), after which a linear gradient reached 100% solvent B (250 mm KCl, 35% acetonitrile, and 0.1% formic acid) in 15 min, followed by 100% solvent C (500 mm KCl, 35% acetonitrile, and 0.1% formic acid) in the following 15 min. The gradient was held at 100% solvent C for 5 min to clean the column, then switched back to 100% solvent A. Thereafter, 15 fractions were collected in 1 min intervals, lyophilized and reconstituted in 30 μl 95:3:0.1 (water/acetonitrile/formic acid).
NanoLC-MS/MS
Dissolved fractions were analyzed by on-line nano-HPLC MS with a system consisting of a Agilent 1100 gradient HPLC system (Agilent, Waldbronn, Germany) as described previously (49), and a LTQ-FT Ultra mass spectrometer (Thermo, Bremen, Germany). Of each fraction, 5 μl was injected onto a home-made precolumn (100 μm×15 mm; Reprosil-Pur C18-AQ 3 μm, Dr Maisch, Ammerbuch, Germany) and eluted via a home-made analytical nano-HPLC column (15 cm × 50 μm; Reprosil-Pur C18-AQ 3 μm). The gradient was run from 0% to 30% solvent B (10:90:0.1 water/acetonitrile/formic acid) in 10–155 min. A tip of ∼5 μm was drawn at the tip of the nano-HPLC column to act as electrospray needle. Full scan mass spectra were acquired in the FT-ICR with a resolution of 25,000 at a target value of 5 × 106. The five most intense ions were selected and fragmented in the linear ion trap using collision-induced dissociation at a target value of 10,000.
Search Databases
MSMSpdbb was used to generate concatenated protein databases in FASTA format (50). Genomic sequences and annotational information of M. tuberculosis H37Rv (3996 entries) (51) were used, together with the genetic information of M. tuberculosis Beijing strain NITR203 (4071 entries) (52). This concatenated database contains all previously described mutations that are specific for modern Beijing strains, as well as the ancient Beijing “wild-type” sequence (4327 entries) (30). Protein products larger than 50 amino acids were considered for stop-to-stop translation. Peptides describing varying sequences or different translational start sites between both strains were only used when the sequence was longer than seven and shorter than 35 amino acids. Proteins that were clustered received the accession number and description of control strain M. tuberculosis H37Rv. Translated entries, which did not cluster with any of the annotated genes, were discarded. To use the database with MaxQuant software, the artificial J and O amino acid residues were replaced by a lysine residue. Alongside the concatenated data base we searched FASTA databases based on M. tuberculosis H37Rv (51) and M. tuberculosis Beijing NITR203 (52).
Data Interpretation
Peptide and protein identification and quantitation was accomplished using MaxQuant 1.4.0.3 (53). The false discovery rate (FDR) was set to 0.01 for both proteins and peptides. Minimal peptide length was set to six amino acids. The first search was performed using 20 ppm, whereas the main search was conducted with 10 ppm. Search of MS/MS spectra was performed with 20 ppm using the Andromeda search engine (54). In total 262 common contaminants were included in the searches by Andromeda. Enzyme specificity was set as C-terminal to arginine and lysine without proline restriction. A maximum of two missed cleavages was allowed. Variable modifications included N-terminal protein acetylation, methionine oxidation, and corresponding dimethyl labels.
Carbamidomethylation of cysteine was selected as a fixed modification. Proteins considered for quantification required a minimal peptide count of two, including unique and razor peptides. Proteins identified by site, matched against the reverse database or identified as a contamination, were excluded for further analysis. Statistical analysis of the outcomes was performed by Perseus using the significance B test with a Benjamini-Hochberg FDR <5%. Data files have been deposited in the publicly available ProteomeXchange Consortium (proteomecentral.proteomexchange.org) and can be accessed through the code: PXD000931. General properties of the proteome were examined using “Batch MW and pI Finder” tool and the gravy index was calculated using the “gravy-calculator.de” web tool, whereas PSORTb v3.0 (55) was used to determine protein localization and the TMHMM Server V.2.0 (56) for the determination of transmembrane helixes.
Transcriptional Analysis of Selected Genes by Quantitative Real Time PCR
RNA was isolated and purified from bacteria using the PureLink RNA Mini Kit (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. In addition, on column digestion was performed with RNase free DNase (Qiagen, Valencia, USA). The quality and quantity of RNA was examined by spectrophotometric measurements (260/280 nm). Thereafter, 0.5 μg of purified RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Life Technologies). Genomic DNA control samples were incubated without reverse transcriptase. Primers were designed using Primer3Plus (57) or derived from previous studies (34). The following forward and reverse primer sequences were used: 16s forward 5′-TCCCGGGCCTTGTACACA-3′; reverse 5′-CCACTGGCTTCGGGTGTTA-3′, Rv0450c/MmpL4 forward 5′-GTGTTCAAGGAAGGCGATTC-3′; reverse 5′-CAAGGGGTTGGTTACCCTCT-3′, Rv1269c forward 5′-CAAGGTGCTCACCAGTTTCA-3′; reverse 5′-CTGGTATGCCCTATCGTTGG-3′, Rv3137 forward 5′- GGTTGACCGATACCGTGTG-3′, reverse 5′-GCCACCAGGCAGTAAGACAG-3′, Rv3283/sseA forward 5′-ATACCTGGTTCGTGCTCACA-3′; and reverse 5′-AGCCGTCGTAGTTCCGTACA-3′. LightCycler 480 SYBR Green I Master (Roche Applied Science) was used as qPCR master mix. Samples were analyzed in a LightCycler 480 (Roche Applied Science, Penzberg, Germany). The following cycling conditions were used: The thermal cycler program was initiated by 10 min at 95 °C followed 35–40 cycles for 10 s at 95 °C, 10 s at 60 °C, and 10 s at 72 °C. High Resolution Melt analysis was performed after each program. mRNA quantities were determined in triplicate and normalized on 16s rRNA levels using the 2−ΔCt method. Mann-Whitney U test was used to determine p values.
Prediction of Protein Stability upon Mutation
Three independent algorithms were used to predict the effect of a previously described mutation (30) on the stability of Rv3283/sseA: PolyPhen-2 (58), I-Mutant3.0 (sequence mode) (59), and (Protein ANalysis THrough Evolutionary Relationships) PANTHER (60). The respective protein sequence was derived from tbdb.org (61), using the conversion of a glutamic acid residue to a lysine residue in position 276.
Quantitative Rhodanese Activity Assay
To perform quantitative rhodanese activity analysis, all chemicals were derived from Sigma-Aldrich Chemie B.V. (Zwijndrecht, Netherlands). Rhodanese activity was examined as described previously with minor modifications (62). In brief, cells were cultured to early-log phase and disrupted by bead-beating. Unbroken cells and cell debris was separated by centrifugation and discarded. The collected supernatant was filtered twice through a 0.2 μm filter and stored until use at −70 °C in PBS-containing 20% glycerol. The reaction mixture contained 50 μl of 125 mm sodium thiosulfate, 25 μl of 250 mm potassium cyanide, and 30 μl of 200 mm potassium phosphate buffer, pH 8.6. 20 μl of cell lysate was added to this reaction mixture. The reaction was carried out for 15 min at room temperature and was stopped by the addition of 25 μl of 38% formaldehyde. In the control set, formaldehyde was added prior to the addition of the cell lysate. The concentration of thiocyanate was determined by the addition of 125 μl 410 mm ferric nitrate in 14% (w/v) nitric acid. Rhodanese activity is reported as specific activity (unit/mg protein) of which the biological replicates were averaged for each of the isolates examined. One unit of enzyme activity was defined as micromoles of thiocyanate formed per min at pH 8.6.
RESULTS
Selection of M. tuberculosis Beijing Genotype Isolates
To quantify differences in the proteomes of modern and ancient M. tuberculosis Beijing sublineages we set out to generate two sample pools containing five strains of each genotype. Therefore, 14 ancient Beijing and 15 modern Beijing strains were selected from a published collection of 259 Beijing strains (40). The selected strains were representative of 13 countries on four continents i.e. the strains were isolated in that country or isolated from a patient born in the respective country. RD105 was determined to ensure all strains represent the M. tuberculosis Beijing genotype. MutT2/ogt were determined to differentiate between modern and ancient M. tuberculosis Beijing strains (31). MIRU-24 VNTR was determined to ensure a heterogeneous selection M. tuberculosis Beijing isolates; supplemental Fig. S1.
Qualitative Proteome Analysis of M. tuberculosis Beijing Strains
We set out to identify proteins that are differentially regulated in either modern Beijing or ancient Beijing strains. To this end, we utilized a pooled approach, in which we mixed the digests of five ancient Beijing strains and five modern Beijing strains. We also analyzed three individual duplexes, in which single ancient Beijing strains were compared with single modern Beijing strains. In each individual duplex, different strains were selected at random from our set of Beijing strains described above.
Three different search databases were used to analyze the peptides yielded; (1) a FASTA database based on the genome of M. tuberculosis H37Rv (51), a well annotated M. tuberculosis laboratory strain. (2) a database based on the genome of M. tuberculosis Beijing strain NITR203 (52), which contains all modern Beijing specific nsSNPs, and (3) a concatenated database comprising both H37Rv and NITR203; supplemental Fig. S2B. The combined analyses yielded a total of 30,748 unique peptides with a FDR <1% for all samples measured; see supplemental Table S1. None of the searched databases clearly outperformed the others in terms of peptide identifications. Therefore, we continued with the concatenated database, as it contains all major sequence variation caused by SNPs.
The cumulative number of unique protein identifications and quantifications was 2392; of which 1817 proteins were identified and quantified in all proteome experiments, constituting >75% of all the quantified proteins; see supplemental Fig. S2A and supplemental Table S2. These results represent the most comprehensive proteome description of a clinical M. tuberculosis genotype to date. When we compared the identified proteins of the individual duplexes with the pooled duplex, an overlap of >80% was achieved using our shotgun proteomics approach.
To confirm the quality of our proteomic dataset, we investigated if specific proteins were underrepresented on the basis of several physicochemical properties; see supplemental Fig. S3A–S3C. As can be concluded from the distribution of proteins based on mass, pI, or hydrophobicity (i.e. gravy index) which follows the theoretical database distribution. All categories are represented to the same extent. In addition, there is no bias on the basis of protein localization (55) and transmembrane proteins (56), even for proteins predicted to contain 13 predicted transmembrane helices; see supplemental Fig. S3D–S3E. In summary, from the above we conclude there is no skewing of the proteome we obtained.
Functional Classification of Proteins and Efflux Pumps
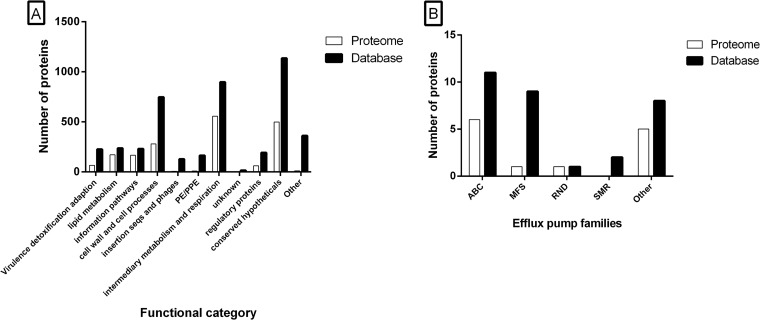
We categorized the proteins that were quantified in all four analyses, according to their functionality as given by Tuberculist (63). Proteins that were considered to be specific for M. tuberculosis Beijing NITR203 (52) by MSMSpdbb (50) were categorized as “other;” see Fig. 1A. The proteins of different functional categories are represented to the same extent, except for the PE/PPE family, the insertion sequences and phages and the proteins that we categorized as “other” (63).
Fig. 1.
Functional distribution of the proteome of M. tuberculosis Beijing. Proteins present in our MS-database (filled black bars) and our dataset (white bars) were categorized by: A, Functional class categories as given by Tuberculist. B, Bacterial efflux transporter families. ABC, ATP Binding Cassette; MFS, Major Facilitator Superfamily; RND, Resistance Nodulation cell Division; SMR, Small Multidrug Resistance.
Beijing strains are linked to (multi)drug resistance in many geographical areas (16, 22–26). Initially, the DNA repair mechanism was thought to be involved in the development of antibiotic resistance (19). Although not based on studying the DNA repair in detail, two recent studies confirmed this hypothesis by showing that several Beijing strains have a much higher frequency of rifampicin resistant mutants (20, 21). However, proteins such as efflux pumps can also play a crucial part in reduced susceptibility to antibiotics by lowering the intra bacterial concentration of drugs and hence creating a higher tolerance to particular compounds. To determine whether such pre-existing factors are present in the proteome of M. tuberculosis Beijing, we categorized putative efflux pump genes and transporters that play a role in drug resistance in M. tuberculosis, as previously listed (64). As presented in Fig. 1B, indeed several antibiotic transporting efflux pumps were disclosed in the proteome of in vitro cultured M. tuberculosis Beijing strains.
The pre-existence of these efflux pumps can potentially be caused by the analysis of multidrug resistant strains. We therefore determined the MICs for the first line antibiotics (rifampicin, isoniazid, ethambutol, and pyrazinamide) of the analyzed strains. Both the modern and ancient Beijing strains used in this study were susceptible to all first line drugs (data not shown).
The pre-existing presence of efflux pumps is not a unique characteristic of M. tuberculosis Beijing strains. Efflux pumps were also identified in the proteome of M. tuberculosis H37Rv by a proteome-wide selected reaction monitoring approach (39). We compared this comprehensive dataset of 2195 M. tuberculosis H37Rv proteins with the proteins we identified. Three of these proteins (Rv0341/iniB, Rv2688c, and Rv3728) were exclusively identified by our proteomic analysis of M. tuberculosis Beijing.
Quantitative Proteomic Profiling of Modern and Ancient M. tuberculosis Beijing Strains
WGS revealed that the modern and ancient Beijing strains are highly conserved on the genetic level (30). However, multiple studies have shown that modern Beijing strains are more “successful” in terms of transmission and development of antibiotic resistance (13, 14, 31, 65, 66). Moreover, in most geographic areas with a higher density of Beijing strains the modern Beijing strain are predominant, except for Japan (28). To determine whether bacterial factors on the protein level may be associated with the emergence of modern Beijing strains, we quantitatively compared the proteomes of five modern Beijing and five ancient Beijing strains. Therefore, we performed a pooled approach and three individual duplex analyses. The protein ratio distribution was only slightly narrower when the samples were pooled; see supplemental Fig. S4.
Forty-seven proteins were identified as differentially abundant using our pooled approach. The three individual duplex experiments identified respectively 74, 111, and 55 proteins of which the abundance was significantly different; see supplemental Table S3. We categorized all differentially abundant proteins according to their functionality, as given by Tuberculist (63). To identify proteins that are specifically more abundant in either the modern, or the ancient Beijing strains, we selected proteins that were over represented in the three individual approaches and the pooled approach; see Table I. Rv0450c/MmpL4 and Rv3137 were significantly more abundant in the modern Beijing strains, whereas Rv1269c, and Rv3283/sseA were identified to be more abundant in the ancient Beijing strains examined.
Table I. Differentially abundant proteins between modern Beijing and ancient Beijing strains.
| Rv identifier | Gene name | Description | PEP | Fold | Unique peptides |
|---|---|---|---|---|---|
| Rv0450c | MmpL4 | Transmembrane transport protein | 0 | 3,3–3,8 | 34 |
| Rv1269c | – | Conserved probable secreted protein | 3.96E-34 | <0,1–0,3 | 5 |
| Rv3137 | – | Probable monophosphatase | 8.69E-143 | 3,1–4,3 | 12 |
| Rv3283 | sseA | Probable thiosulfate sulfurtransferase | 0 | <0,1–0,2 | 22 |
Transcriptional Analysis of Selected Genes in a Large Cohort of M. tuberculosis Beijing Strains
To evaluate whether the mRNA levels of Rv0450c/MmpL4, Rv1269c, Rv3137, and Rv3283/sseA are differentially regulated throughout the entire modern Beijing or ancient Beijing genotypes, we complemented our MS-based proteomic observations with qPCR analysis in a larger cohort of M. tuberculosis Beijing strains by qPCR. Therefore, 14 ancient Beijing strains and 15 modern Beijing strains, including the strains used for the proteome analyses, as presented in supplemental Fig. S1, were analyzed.
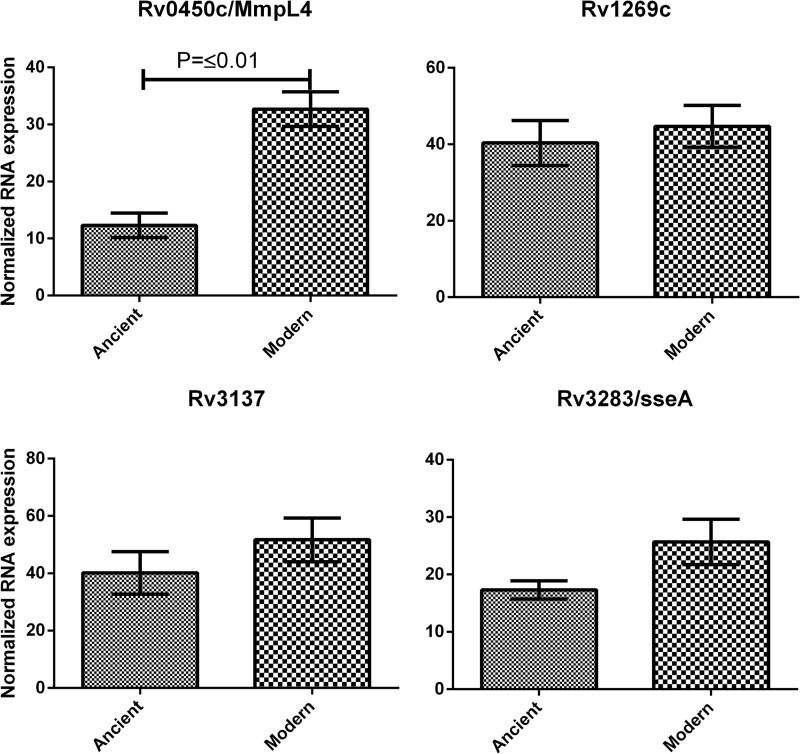
For Rv0450c/MmpL4 both the mRNA and the protein product were more abundant in the modern Beijing strains examined; see Fig. 2. In contrast, Rv1269c and Rv3137 did not show significant differences in the larger cohort of M. tuberculosis Beijing strains examined. Nevertheless, Rv3137 showed a trend in line with the obtained protein ratios, but the effect observed on the protein level was larger than the fold change observed on the mRNA level.
Fig. 2.
Observed mRNA levels for selected genes. mRNA levels of the selected protein candidates were analyzed in 14 ancient Beijing strains and 15 modern Beijing strains using qPCR. Each sample was analyzed in triplicate. Data are expressed as means ± S.E. of the mean.
An opposite trend between mRNA levels and protein levels was observed for Rv3283/sseA. Our proteome analysis showed a 5–10 fold higher abundancy of Rv3283/sseA in ancient Beijing strains. Contrary to these observations, higher transcriptional levels were observed for Rv3283/sseA in modern Beijing strains compared with ancient Beijing strains.
Functional Prediction of Amino Acid Residue Substitution in Rv3283/sseA
To investigate if the observed lower protein abundance of the protein Rv3283/sseA in modern Beijing strains could be caused by destabilization of the protein stucture by the altered amino acid at position 276, because of the nsSNP, reported to be specific for modern Beijing lineage strains (30), we applied the protein stability prediction programs, PolyPhen-2 (58) and I-Mutant3.0 (59) and the protein functionality prediction program PANTHER (60).
The 3D structure of 3HZU, a homolog of Rv3283/sseA was used by PolyPhen-2 among other information. Based on the calculation, PolyPhen-2 designates the mutation as being “benign,” “possibly damaging,” and “probably damaging.” The conversion of the glutamic acid residue toward a lysine residue in position 276 of Rv3283/sseA is considered to be “probably damaging” by PolyPhen-2.
As a second independent control, we also determined the impact of the mutation using the sequence option of I-Mutant3.0 (59), which evaluates changes in stability of proteins based upon a single site mutation. The amino acid substitution in the active domain of Rv3283/sseA was predicted to result in a free energy change of 1.02 Kcal/mol. This is calculated from the unfolding Gibbs free energy change of the mutated protein minus the unfolding free energy value of the native protein (Kcal/mol). A change of 1.02 Kcal/mol in predicted free energy is considerable, and therefore expected to largely decrease the stability of Rv3283/sseA.
Finally, we predicted the functionality of the mutant and wild-type version of Rv3283/sseA using PANTHER (60). This software considers site-specific variation between evolutionary related proteins to calculate substitution position-specific evolutionary conservation (subPSEC) score. When we calculated the functional consequence of a mutation in Rv3283/sseA the subPSEC score was −4.74 and the corresponding Pdeleterious (probability of functional impairment) was 0.85. Altogether, three independent prediction algorithms predict that the mutation in Rv3283/sseA will have a large impact on the proteins' function and stability.
To support the observation that Rv3283/sseA is less abundant in modern Beijing strains than in ancient Beijing strains, we determined its activity. Rv3283/sseA is one of the at least four rhodanese domain containing proteins in our proteomic dataset of M. tuberculosis Beijing. Two of the proteins with potential rhodanese activity, Rv0815c/cysA2 and Rv3117/cysA3, could not be distinguished from one another, because of their high sequence similarity, but could be distinguished from Rv3283/sseA in the proteome analysis. Rv0815c/cysA2, Rv3117/cysA3, and Rv2291/sseB were not identified to be differentially abundant in all of the ancient or modern Beijing strains examined. Rhodanese proteins can detoxify cyanide by converting thiosulfate to thiocyanate and sulfite (EC 2.8.1.1). This reaction can be monitored in a quantitative manner by the formation of a blood red iron(III) thiocyanate complex (62).
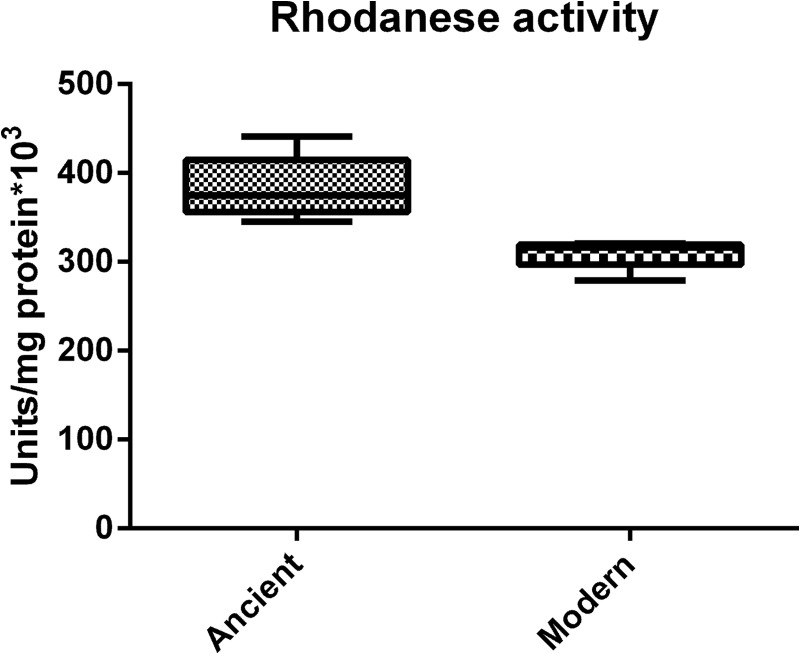
Rhodanese activity of cell lysates, containing all proteins with potential rhodanese activity, was determined in triplicate of the strains used for the proteomic analysis; see Fig. 3. The average rhodanese activity observed in the cell lysate of ancient Beijing strains was ∼23% higher than the rhodanese activity observed in the modern Beijing strains.
Fig. 3.
Rhodanese activity in M. tuberculosis Beijing sublineages. Rhodanese activity was determined in whole cell lysates of five ancient Beijing and five modern Beijing strains. Biological triplicates were taken and samples were analyzed in triplicate. The median is shown by the horizontal black line, with vertical whiskers representing the range.
DISCUSSION
The Beijing lineage of M. tuberculosis is one of the most successful M. tuberculosis genotypes worldwide, as indicated by its degree of genetic conservation and emergence (67, 68). Previous studies showed an increase of TB incidence caused by M. tuberculosis Beijing, combined with higher odds on drug resistance for these strains (6). Characterization of the Beijing strains revealed the presence of two sublineages, the modern and ancient Beijing strains (27). Although the two sublineages are highly conserved on the genetic level, epidemiological studies suggested a possible evolutionary advantage of the modern Beijing strains, as this lineage is most prevalent in many geographic areas, except for Japan. In addition, whole genome analysis demonstrated the presence of only 31 nsSNPs that are characteristic of modern Beijing lineage strains from China, Japan, and South Africa and this suggests a relative short time of divergence and successful spread (30). To understand the evolutionary success of these pathogens, it is essential to identify factors that contribute to their high transmissibility and propagation in the human population, possibly associated with enhanced antibiotic resistance and ability to circumvent BCG induced immunity (10, 13, 14, 31, 65, 66). We employed a pooled proteomics approach alongside three traditional individual duplex analyses to identify proteins that contribute to the successful phenotype of modern Beijing strains.
Selection of a Heterogeneous Set of M. tuberculosis Beijing Genotype Strains
To cover the full spectrum of the genotype family, we selected 14 ancient and 14 modern Beijing strains from a previous selection of 259 M. tuberculosis strains (40). Furthermore, we included the successful modern Beijing strain B0/W148, that comprises one-fourth of the M. tuberculosis Beijing isolates in different parts of Russia (41–43). The Beijing strains in our selection originated from 13 countries distributed over four continents. We determined RD105 to assure that all strains in our selection truly belonged to the M. tuberculosis Beijing genotype (29). A deletion of RD181 is considered to be characteristic of the modern sublineage (29). However, deletions of RD181 were also identified in “late” ancient Beijing strains (69, 70). Therefore, stringent differentiation between modern and ancient Beijing strains was accomplished using the M. tuberculosis modern Beijing genetic markers of putative DNA repair genes mutT2 and ogt (31). To exclude the possibility of mixed strains, contamination or the selection of clonal isolates we performed MIRU-24 VNTR-typing; see supplemental Fig. S1. Five modern and five ancient Beijing strains were randomly selected from this heterogeneous set of M. tuberculosis Beijing strains.
Description of the M. tuberculosis Proteome
The combined analysis of pooled and individual duplex approaches yielded a total of 2392 unique proteins, the highest reported number of quantified proteins in any clinical strain of M. tuberculosis to date. Comparison of the individual duplexes with the pooled duplex approach showed an overlap of 80–95% in identified proteins; see supplemental Fig. S2A. In total 1817 proteins were quantified in all four analyses using a concatenated database. A high overlap, achieved by a shotgun proteomics approach is typically achieved when one reaches high proteome coverage. Furthermore, this high overlap of protein identifications and quantifications in all four analyses suggests a low inter-strain and low biological variation in the strains examined.
In mycobacterial proteomics, the solid, thick cell wall of mycobacteria presents a major obstacle to full proteome recovery. In particular reproducible extraction of cell wall and membrane bound proteins can be difficult. Therefore, we examined the general physicochemical properties of the obtained proteome to assure a representative coverage of the proteome, and to prevent a bias toward easy-to-extract proteins. The concatenated search database was utilized to represent the theoretically maximal observable proteome; see supplemental Fig. S3. In the general properties that we assessed (Protein MW/pI/hydrophobicity), the observed proteome followed a distribution highly similar to that of the theoretically observable proteome, from which we conclude that our workflow is not biased regarding proteins with certain physiochemical properties. This was further confirmed by TMHMM (56) and protein localization analysis; see supplemental Fig. S3D, S3E (55).
Approximately 42% of the proteins present in our database were reproducibly quantified in the four analyses. Two third of the proteins that were predicted to be localized in the thick cell wall were included in this dataset. In addition, we were able to reproducibly quantify proteins with ≥13 transmembrane helixes. The high coverage of even the highly hydrophobic cell wall bound proteins suggests that our dataset provides a reliable representation of the M. tuberculosis Beijing proteome without discrimination of the more hydrophobic proteins.
Categorization of the Identified Proteins
Functional categorization, as given by Tuberculist (63), showed a homogeneous distribution of the 1817 proteins present in our dataset for most of the listed categories, except for the PE/PPE family and the insertion sequences and phages; see Fig. 1A. The categories containing the PE/PPE-family and insertion sequences and phages are notoriously difficult to identify. A recent study using “proteome-wide selected reaction monitoring” also suffered from an under representation of the PE/PPE-family (39). The PE/PPE family is named after their PE proline-glutamic acid and PPE proline-proline-glutamic acid N-terminal motif. Members of this family are characterized by a high number of glycine and alanine residues in combination with repetitive sequences. As a result, there are relatively few tryptic cleavage sites (39). This lack of tryptic digestion sites makes our workflow with endoLys-c and trypsin digestion less suitable for the detection of members from this protein family. A reported method for the optimal identification of PE/PPE proteins includes the extraction of proteins from the mycobacterial cell wall followed by a double tryptic digestion and a triple chymotrypsin digestion (71).
Identification of Pre-existing Efflux Pumps in the Beijing Genotype
Throughout many geographic areas, Beijing strains were reported to significantly correlate with (multi)drug resistance, treatment, relapses after curative treatment and transmission of resistant TB (6, 7, 10–12). Initially, altered DNA repair mechanisms in the M. tuberculosis Beijing genotype family were assumed to be responsible for the strong correlation with antibiotic resistance, because of mutations in three putative mutator genes in modern Beijing strains (19). More support for this hypothesis was gained after a recent study showed that Beijing strains in Vietnam have an increased frequency of rifampicin resistant mutants compared with strains of the East African Asian (EAI) genotype family (20). Unfortunately, the effects of variations in the DNA repair mechanism of M. tuberculosis Beijing has not been studied in full detail. Our systematic study did not reveal any quantitative difference for the genes involved in DNA repair, that is, mutT2, mutT4, and ogt. This suggests that mutations or post-translational mutations, but not protein abundance, are important for the aberrant DNA repair mechanism in M. tuberculosis Beijing.
The role of efflux pumps, which are able to transport antibiotics over the cell wall, has only been slightly touched upon in M. tuberculosis Beijing strains (72, 73). We examined whether efflux pumps are present in the proteome of M. tuberculosis Beijing strains in the absence of selective pressure of antibiotics. We therefore used a previously listed selection of M. tuberculosis efflux pumps to categorize the proteins included in our dataset (64). As presented in Fig. 1B, several types of efflux pumps are present in the proteome of in vitro cultured M. tuberculosis Beijing. To ensure that this observation is not caused by the presence of (multi)drug resistant strains potentially present in our selection, we determined the MICs of first line antibiotics for the strains analyzed. None of the M. tuberculosis strains examined were identified to be resistant for any of the tested drugs. This observation shows that the presence of the efflux pumps alone, present in the proteome of M. tuberculosis Beijing, is not sufficient to cause antibiotic resistance. However, expression of these antibiotic extruding proteins can potentially cause a higher tolerance to drugs or makes it more difficult to eliminate all pathogens by antibiotic therapy. Moreover, a reduced susceptibility to drugs such as rifampicin, as observed previously, may be responsible for more durable persistence and hence the selection of drug resistant bacteria (20). As recently reported, a much higher dosage of rifampicin was required to achieve a 100% killing of M. tuberculosis Beijing strains (20). Taken together, this suggests a potential role for efflux pumps in the development of (multi)drug resistance in M. tuberculosis Beijing strains. The pre-existing presence of efflux pumps is, however, not necessarily a unique characteristic for M. tuberculosis Beijing. Therefore, we compared the presence of efflux pumps in our dataset with those that were recently identified and quantified in the proteome of M. tuberculosis H37Rv (39). The used comprehensive M. tuberculosis H37Rv dataset was generated using proteome-wide selected reaction monitoring and contains 2195 proteins. Three efflux pumps, Rv0341/iniB, Rv2688c, and Rv3728, were exclusively identified in our dataset on Beijing strains. However, a proteogenomic approach that studied the proteome of M. tuberculosis H37Rv identified the respective proteins, Rv0341/iniB, Rv2688c, and Rv3728 (74). It should be noted that 123 LC-MS/MS analyses were performed, which resulted in the identification of Rv2688c by a single peptide. In contrast, we identified ≥4 unique peptides in each of the duplex analyses performed. This combined information is a strong indication that Rv2688c is more abundant in M. tuberculosis Beijing strains than in the proteome of M. tuberculosis H37Rv. Rv3728 was reported to be present in the proteome of M. tuberculosis H37Rv as well (74). Howbeit, peptides corresponding to Rv3728 were only identified by the Sequest algorithm, whereas Mascot and MassWiz failed to identify any peptide belonging to Rv3728. Therefore, as these proteins may preexist exclusively in the M. tuberculosis Beijing proteome, or be more abundant in the proteome of M. tuberculosis Beijing, they might contribute to the success of the Beijing family as a whole in the development of antibiotic resistance.
Identification of Differentially Abundant Proteins within the Beijing Genotype
With the pooled sample approach alongside three individual duplex approaches, we achieved reproducible high proteome coverage using shotgun proteomics. Furthermore, we demonstrated a highly uniform ratio distribution for a majority of the proteins in the M. tuberculosis Beijing strains analyzed. On average, the pooled approach and the individual comparisons yielded approximately the same number of differentially abundant proteins (pooled duplex 47, individual duplexes 74, 111, and 55, supplemental Table S3). Therefore, it is conceivable that not only the modern and ancient Beijing strains are highly similar on the genetic level (30), but that also protein regulation is conserved throughout the M. tuberculosis Beijing family as a whole; supplemental Fig. S4.
As expected, not all proteins identified as differentially abundant in the pooled approach were also identified as differentially abundant in the individual duplexes. A logical explanation is that very abundant proteins in single isolates cannot be averaged out. Furthermore, individual duplex approaches are prone to false positives caused by inter-strain and biological variation. It is important to keep in mind that both the pooled approach and the individual approaches can still yield false positives and are best used in parallel to identify sublineage specific differences.
From our dataset of 1817 proteins that were reproducibly quantified in each analysis, only four proteins showed to be differentially regulated in all strains examined; see Table I. This further confirms that most traits are highly conserved in the M. tuberculosis Beijing family. Therefore, our data suggests that the success of the modern Beijing strains compared with the ancient Beijing strains is caused by only a limited number of virulence factors.
Transcriptional Analysis of Differentially Abundant Proteins in a Large Cohort of M. tuberculosis Beijing Strains
Of the 1817 proteins present in our dataset only four proteins were differentially abundant in all four proteomic analyses (Rv0450c/MmpL4, Rv1269c, Rv3137, and Rv3283/sseA). To determine whether the mRNA levels of the corresponding proteins are differentially regulated throughout the entire modern Beijing or ancient Beijing genotypes, we complemented our MS-based proteomic observations with quantitative mRNA analysis in a larger cohort of M. tuberculosis Beijing strains by qPCR. Although mRNA does not always correlate with protein abundance levels present within the cell, it provides insight into the regulation of these specific genes (75–77). The strains listed in supplemental Fig. S1 were selected to represent the full repertoire of M. tuberculosis Beijing strains. This selection consisted out of 14 ancient Beijing and 15 modern Beijing strains and included the strains used for proteomic analysis.
The probable secreted protein Rv1269c was identified by MS to be >3-fold more abundant in ancient Beijing strains. However, we analyzed the cell lysate of M. tuberculosis Beijing, not the secretome. Quantitative mRNA analysis revealed that Rv1269c is equally transcribed by both modern and ancient Beijing strains; see Fig. 2. Therefore, we cannot rule out the possibility that Rv1269c is more efficiently secreted by modern Beijing strains as the proteomic data clearly pointed out that Rv1269c is less abundant in modern Beijing strains.
Subsequently, we analyzed the mRNA levels for Rv3137, a probable monophosphatase that is essential for the in vitro growth of M. tuberculosis H37Rv (78, 79). A trend similar to the ratios obtained by our proteomic experiments was detected. However, the average difference was much smaller on the mRNA level than observed on the protein level. Further separation of the modern Beijing strains from our selection with several genetic markers (31) will potentially allow us to identify a subspecies of modern Beijing strains that contains high levels of Rv3137. However, this indicates that Rv3137 is not a virulence factor that is up-regulated throughout the modern Beijing genotype as a whole.
The relative mRNA quantities of Rv0450c/MmpL4 were in agreement with the results obtained by our proteomic experiments. Previous studies identified Rv0450c/MmpL4 as a potential virulence factor of M. tuberculosis by signature-tagged transposon mutagenesis (80). In addition, this protein has been reported to be essential for growth in mouse models of TB (81). A recent study reported Rv0450c/MmpL4, together with Rv0451c/MmpS4 or Rv0677c/MmpS5 and Rv0676c/MmpL5, are needed for the secretion of iron-scavenging siderophores in M. tuberculosis (82). Our results demonstrate a clear up-regulation of both protein and mRNA throughout the modern Beijing genotype. Taken together with the previously reported modern Beijing specific mutation in Rv0676c/MmpL5 (30), this suggests that siderophores could be differentially expressed in the two Beijing sublineages. If the increased levels of Rv0450c/MmpL4 work as a compensating mechanism for the mutated Rv0676c/MmpL5 gene in modern Beijing strains remains to be determined. Nevertheless, the results point to differences in iron metabolism between both M. tuberculosis Beijing genotypes. This hypothesis is reinforced by a study that revealed that patients with a mutation in SLC11A1/NRAMP1, a divalent transition metal transporter involved in iron metabolism, have been associated with higher odds of TB caused by M. tuberculosis Beijing (18). The altered quantities of Rv0450c/MmpL4 in association with the modern Beijing specific mutation in Rv0676c/MmpL5 is possibly the missing link that explains the success of M. tuberculosis Beijing strains within individuals that possess a mutation in SLC11A1/NRAMP1. Therefore, our findings support the idea of M. tuberculosis coevolution with their human host.
Next to the secretion of siderophores, Rv0450c/MmpL4 and Rv0676c/MmpL5 has been identified as differentially regulated in rifampicin-resistant M. tuberculosis strains upon rifampicin exposure (83). Build around the already known function of these MmpL genes and the up-regulation of MmpL mRNA in a M. tuberculosis-rifampicin microarray model the authors suggested an involvement of Rv0450c/MmpL4 and Rv0676c/MmpL5 in the rifampicin efflux out of the cell (83).
For Rv3283/sseA, the mRNA levels showed a trend opposite to the results we obtained by proteomics. Where the protein appeared to be strongly up-regulated, and was quantitated by as much as 22 unique peptides on the protein level of ancient Beijing strains, we observed a slightly, but not significant, increase in the mRNA levels of modern Beijing strains. The discrepancy between the results obtained by proteomics and qPCR might be explained by the presence of a nsSNP in the genome of modern M. tuberculosis Beijing strains (30).
Enzymatic Activity of Rv3283/sseA
Several studies have linked the expression of Rv3283/sseA to a virulent M. tuberculosis phenotype. One study demonstrated the differential regulation of Rv3283/sseA by M. tuberculosis within the host cell (84). The association between the protein and an increase in virulence was further supported by a study that showed how a mutation in Rv3283/sseA leads to enhanced growth of M. tuberculosis in macrophages relative to in vitro growth of M. tuberculosis (85). Finally, the predicted rhodanese activity of Rv3283/sseA potentially helps to cope with oxidative stress, as the knock-out of a rhodanese domain containing protein in Azotobacter vinelandii altered their sensitivity toward oxidative damages (86).
In contrast to our proteomic data, the transcription levels of the Rv3283/sseA were higher, on average, in modern Beijing strains compared with ancient Beijing strains. We set out to determine whether a previously reported modern Beijing specific mutation could contribute to this discrepancy. This nsSNP confers a glutamic acid residue to a lysine residue, which has been shown to be specific for modern Beijing strains (30). Three independent bioinformatic algorithms were used to determine the effect of the mutation on the stability and function of the respective protein. PolyPhen-2, which uses the 3D crystal structure of Rv3283/sseA homolog 3HZU among other information, predicted that the mutation is “probably damaging” for the protein (58). Next to PolyPhen-2, we also used I-Mutant 3.0 in sequence mode (59). The calculated free energy change upon substitution of glutamic acid by a lysine residue was −1.02 kcal/mol, which indicates a significant loss of stability of Rv3283/sseA in modern Beijing strains. Analysis of the protein and the corresponding mutation by PANTHER pointed toward a functional impairment of the mutated protein (subPSEC = −4.74 and Pdeleterious = 0.85) (60). The predicted instability, and thereby a potential short half-life time of Rv3283/sseA in modern Beijing strains can explain the lower level of Rv3283/sseA observed in modern Beijing strains compared with ancient Beijing strains.
To further confirm our results we measured the rhodanese activity in whole cell lysates of the ten M. tuberculosis Beijing strains used for the comparative proteomic analysis. Besides Rv3283/sseA there are at least three additional proteins (Rv0815c/cysA2, Rv2291/sseB and Rv3117/cysA3) that contain a rhodanese domain. The presence of these proteins makes our rhodanese activity assay on whole cell lysate level not rhodanase enzyme specific. However, as the additional rhodanese domain containing proteins appeared to occur in equal quantities in most modern and ancient Beijing strains, this is not expected to yield any difference in the total rhodanese activity. We observed ∼23% more rhodanese activity in the ancient Beijing strains examined compared with modern Beijing strains. The relative predicted instability of the mutated Rv3283/sseA, together with the observed rhodanese activity, supports the results obtained by proteomics. In summary; our data suggests that modern Beijing strains possess, on average, less Rv3283/sseA than ancient Beijing strains.
Comparative Study
We compared the results of our systematic approach with a previous comparative proteomic study that described the differences between a hyper and a hypovirulent Beijing strain (34). In that study 23 proteins were reported to be more abundant in the hypo virulent strain. Of these 23 proteins, 17 were quantified in all four of our analyses, but none showed to be specifically more abundant in either the modern or the ancient Beijing strains; see supplemental Table S4. When we compared the 54 proteins that were reported to be more abundant in the previously reported hypervirulent strain, we identified 39 of those 54 proteins in all four of our analyses. Only one protein, Rv2680, showed to be more abundant in several modern Beijing strains. Two out of our three individual duplex approaches and the pooled approach demonstrated the up-regulation of Rv2680 in modern Beijing strains. Together these observations show that comparison of two individual isolates can be useful to identify virulence factors, but not to describe specific differences between the two Beijing sublineages. The proteins Rv0450c/MmpL4, Rv1269c, Rv3137, and Rv3283/sseA were identified as differentially abundant in both our pooled approach and our three individual duplex analyses. Of these proteins, Rv3137 was solely identified in the previously reported M. tuberculosis Beijing proteome dataset (34). However, Rv3137 was not reported to be up-regulated in the hypervirulent strain. This observation supports our hypothesis that Rv3137 is up-regulated in several, but not all modern Beijing strains.
Other Differentially Regulated Proteins
The W-Beijing strain, a distinct phylogenetic branch within the modern Beijing genotype mainly observed in the early 1990s in the USA (87), is known for the expression of a glycosylated variant of phthiocerol dimycocerosate (PDIM), the phenolic glycolipid (PGL-tb). W-Beijing strains are capable of synthesizing PGL-tb because of a mutation in their pks15/1 gene (88). Production of PGL-tb is thought to be one of the success factors of the W-Beijing strains (89). However, not all M. tuberculosis Beijing strains produce PGL-tb, even when they possess a functional pks15/1 gene (90). In our dataset, Rv2946c/pks1 is significantly less abundant in both the pooled approach and two out of three separate duplexes. This observation suggests that M. tuberculosis Beijing strains do not necessarily express equal amounts of PDIM and PGL-tb.
In this study, we identified Rv2688c in the proteome of M. tuberculosis Beijing, whereas this protein was not observed in the extensively studied proteome of M. tuberculosis H37Rv (39). Rv2688c is often described as a fluoroquinolones (FQ) export ATP-binding protein (91). The presence of Rv2688c was not only observed in the proteome of M. tuberculosis Beijing strains, but was also relatively high abundant in several of the modern Beijing strains studied. Interestingly, Beijing genotype isolates that are resistant to FQs occur significantly more than FQ-resistant isolates of other genotypes in Vietnam (92). A mutation in the gyrA gene was associated with this high-level FQ-resistance. However, an increased abundance of the FQ efflux pump, Rv2688c, possibly leads to a better tolerance of modern M. tuberculosis Beijing strains against FQs, which can eventually result in mutations that cause resistance.
Our study showed that Rv2946c/PKS1 and Rv2688c are not up-regulated in all modern Beijing strains. However, further characterization of the modern Beijing family might yield subfamilies in which this trait is conserved.
The DosR regulon, which is transcribed during the latent stage of infection, is reported to be constitutively up-regulated in W-Beijing strains (90). Our combined pooled and individual approach did not reveal differential abundance of proteins from the DosR regulon between modern and ancient Beijing strains. This suggests that up-regulation of DosR might be conserved throughout all M. tuberculosis Beijing strains in general compared with other M. tuberculosis lineages.
CONCLUSIONS
We identified an important virulence factor for modern Beijing strains in the form of Rv0450c/MmpL4. This observation is the first link between the proteome of modern M. tuberculosis Beijing strains and the higher odds of M. tuberculosis Beijing in individuals with a mutation in their SLC11A1/NRAMP1 gene. We further provide evidence for the lower abundance of Rv3283/sseA in modern Beijing strains, which seems to be a direct consequence of a previously described nsSNP. In addition, we listed several proteins previously reported as potential virulence factors, such as Rv2946c/PKS1 and FQ efflux pump Rv2688c, which could contribute to the success of modern Beijing strains. Taken together our observations contribute to a better understanding of the successful spread of modern Beijing strains, and may assist in combatting this serious threat in TB control. If the spread of Beijing strains is not controlled the current epidemic of TB may transform into an epidemic of resistant TB.
Supplementary Material
Footnotes
Author contributions: J.d.K., P.v.V., and D.v.S. designed research; J.d.K., P.d.H., and A.d.R. performed research; J.d.K., P.v.V., and D.v.S. analyzed data; J.d.K., P.v.V., and D.v.S. wrote the paper.
* This research was made possible by the National Institute for Public Health and the Environment (RIVM).
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S4.
1 The abbreviations used are:
- ABC
- ATP binding cassette
- endoLysC
- endoproteinase Lys-C
- FDR
- false discovery rate
- FQ
- fluoroquinolone
- MFS
- major facilitator superfamily
- MIC
- minimal inhibitory concentration
- NRAMP1
- natural resistance associated macrophage protein 1
- nsSNP
- nonsynonymous single nucleotide polymorphism
- PGL-tb
- phenolic glycolipid
- PDIM
- phthiocerol dimycocerosate
- PANTHER
- protein ANalysis THrough evolutionary relationships
- qPCR
- quantitative polymerase chain reaction
- RD
- regions of difference
- RND
- resistance nodulation cell division
- SNP
- single nucleotide polymorphism
- SMR
- small multidrug resistance
- SCX
- strong cation exchange
- subPSEC
- substitution position-specific evolutionary conservation
- TB
- tuberculosis
- MIRU-VNTR
- mycobacterial interspersed repeat units variable number of tandem repeats
- WGS
- whole genome sequencing.
REFERENCES
- 1. Organisation W. H. (2013) Global Tuberculosis Report 2013. 306 [Google Scholar]
- 2. Schurch A. C., van Soolingen D. (2012) DNA fingerprinting of Mycobacterium tuberculosis: from phage typing to whole-genome sequencing. Infect. Genet. Evol. 12, 602–609 [DOI] [PubMed] [Google Scholar]
- 3. Brosch R., Gordon S. V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Parsons L. M., Pym A. S., Samper S., van Soolingen D., Cole S. T. (2002) A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99, 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hershberg R., Lipatov M., Small P. M., Sheffer H., Niemann S., Homolka S., Roach J. C., Kremer K., Petrov D. A., Feldman M. W., Gagneux S. (2008) High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6, e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Soolingen D., Qian L., de Haas P. E., Douglas J. T., Traore H., Portaels F., Qing H. Z., Enkhsaikan D., Nymadawa P., van Embden J. D. (1995) Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33, 3234–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glynn J. R., Whiteley J., Bifani P. J., Kremer K., van Soolingen D. (2002) Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8, 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parwati I., Alisjahbana B., Apriani L., Soetikno R. D., Ottenhoff T. H., van der Zanden A. G., van der Meer J., van Soolingen D., van Crevel R. (2010) Mycobacterium tuberculosis Beijing genotype is an independent risk factor for tuberculosis treatment failure in Indonesia. J. Infect. Dis. 201, 553–557 [DOI] [PubMed] [Google Scholar]
- 8. Kremer K., Glynn J. R., Lillebaek T., Niemann S., Kurepina N. E., Kreiswirth B. N., Bifani P. J., van Soolingen D. (2004) Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42, 4040–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kremer K., van-der-Werf M. J., Au B. K., Anh D. D., Kam K. M., van-Doorn H. R., Borgdorff M. W., van-Soolingen D. (2009) Vaccine-induced immunity circumvented by typical Mycobacterium tuberculosis Beijing strains. Emerg. Infect. Dis. 15, 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devaux I., Kremer K., Heersma H., Van Soolingen D. (2009) Clusters of multidrug-resistant Mycobacterium tuberculosis cases, Europe. Emerg. Infect. Dis. 15, 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buu T. N., van Soolingen D., Huyen M. N., Lan N. T., Quy H. T., Tiemersma E. W., Kremer K., Borgdorff M. W., Cobelens F. G. (2012) Increased transmission of Mycobacterium tuberculosis Beijing genotype strains associated with resistance to streptomycin: a population-based study. PloS One 7, e42323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buu T. N., Huyen M. N., Lan N. T., Quy H. T., Hen N. V., Zignol M., Borgdorff M. W., Cobelens F. G., van Soolingen D. (2009) The Beijing genotype is associated with young age and multidrug-resistant tuberculosis in rural Vietnam. Int. J. Tuberc. Lung Dis. 13, 900–906 [PubMed] [Google Scholar]
- 13. Lopez B., Aguilar D., Orozco H., Burger M., Espitia C., Ritacco V., Barrera L., Kremer K., Hernandez-Pando R., Huygen K., van Soolingen D. (2003) A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abebe F., Bjune G. (2006) The emergence of Beijing family genotypes of Mycobacterium tuberculosis and low-level protection by bacille Calmette-Guerin (BCG) vaccines: is there a link? Clin. Exp. Immunol. 145, 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grode L., Seiler P., Baumann S., Hess J., Brinkmann V., Nasser Eddine A., Mann P., Goosmann C., Bandermann S., Smith D., Bancroft G. J., Reyrat J. M., van Soolingen D., Raupach B., Kaufmann S. H. (2005) Increased vaccine efficacy against tuberculosis of recombinant Mycobacterium bovis bacille Calmette-Guerin mutants that secrete listeriolysin. J. Clin. Invest. 115, 2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anh D. D., Borgdorff M. W., Van L. N., Lan N. T., van Gorkom T., Kremer K., van Soolingen D. (2000) Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6, 302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Spuy G. D., Kremer K., Ndabambi S. L., Beyers N., Dunbar R., Marais B. J., van Helden P. D., Warren R. M. (2009) Changing Mycobacterium tuberculosis population highlights clade-specific pathogenic characteristics. Tuberculosis 89, 120–125 [DOI] [PubMed] [Google Scholar]
- 18. van Crevel R., Parwati I., Sahiratmadja E., Marzuki S., Ottenhoff T. H., Netea M. G., van der Ven A., Nelwan R. H., van der Meer J. W., Alisjahbana B., van de Vosse E. (2009) Infection with Mycobacterium tuberculosis Beijing genotype strains is associated with polymorphisms in SLC11A1/NRAMP1 in Indonesian patients with tuberculosis. J. Infect. Dis. 200, 1671–1674 [DOI] [PubMed] [Google Scholar]
- 19. Ebrahimi-Rad M., Bifani P., Martin C., Kremer K., Samper S., Rauzier J., Kreiswirth B., Blazquez J., Jouan M., van Soolingen D., Gicquel B. (2003) Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9, 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Steenwinkel J. E., ten Kate M. T., de Knegt G. J., Kremer K., Aarnoutse R. E., Boeree M. J., Verbrugh H. A., van Soolingen D., Bakker-Woudenberg I. A. (2012) Drug susceptibility of Mycobacterium tuberculosis Beijing genotype and association with MDR TB. Emerg. Infect. Dis. 18, 660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ford C. B., Shah R. R., Maeda M. K., Gagneux S., Murray M. B., Cohen T., Johnston J. C., Gardy J., Lipsitch M., Fortune S. M. (2013) Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong H., Shi L., Zhao X., Sang B., Lv B., Liu Z., Wan K. (2012) Genetic diversity of Mycobacterium tuberculosis isolates from Tibetans in Tibet, China. PloS One 7, e33904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu Q., Su Y., Lu B., Ma Y., Zhao X., Yang X., Dong H., Liu Y., Lian L., Wan L., Wu Y., Wan K. (2013) Genetic diversity of Mycobacterium tuberculosis isolates from Inner Mongolia, China. PloS One 8, e57660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Casali N., Nikolayevskyy V., Balabanova Y., Ignatyeva O., Kontsevaya I., Harris S. R., Bentley S. D., Parkhill J., Nejentsev S., Hoffner S. E., Horstmann R. D., Brown T., Drobniewski F. (2012) Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 22, 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson R., Warren R. M., van der Spuy G. D., Gey van Pittius N. C., Theron D., Streicher E. M., Bosman M., Coetzee G. J., van Helden P. D., Victor T. C. (2010) Drug-resistant tuberculosis epidemic in the Western Cape driven by a virulent Beijing genotype strain. Int. J. Tuberc. Lung Dis. 14, 119–121 [PubMed] [Google Scholar]
- 26. European Concerted Action on New Generation Genetic, M., Techniques for the, E., Control of, T. (2006) Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg. Infect. Dis. 12, 736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mokrousov I., Narvskaya O., Otten T., Vyazovaya A., Limeschenko E., Steklova L., Vyshnevskyi B. (2002) Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res. Microbiol. 153, 629–637 [DOI] [PubMed] [Google Scholar]
- 28. Iwamoto T. (2009) Population structure analysis of Mycobacterium tuberculosis Beijing family in Japan. Kekkaku : Tuberculosis 84, 755–759 [PubMed] [Google Scholar]
- 29. Tsolaki A. G., Gagneux S., Pym A. S., Goguet de la Salmoniere Y. O., Kreiswirth B. N., Van Soolingen D., Small P. M. (2005) Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 43, 3185–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schurch A. C., Kremer K., Warren R. M., Hung N. V., Zhao Y., Wan K., Boeree M. J., Siezen R. J., Smith N. H., van Soolingen D. (2011) Mutations in the regulatory network underlie the recent clonal expansion of a dominant subclone of the Mycobacterium tuberculosis Beijing genotype. Infect. Genet. Evol.. 11, 587–597 [DOI] [PubMed] [Google Scholar]
- 31. Hanekom M., van der Spuy G. D., Streicher E., Ndabambi S. L., McEvoy C. R., Kidd M., Beyers N., Victor T. C., van Helden P. D., Warren R. M. (2007) A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 45, 1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Laarhoven A., Mandemakers J. J., Kleinnijenhuis J., Enaimi M., Lachmandas E., Joosten L. A., Ottenhoff T. H., Netea M. G., van Soolingen D., van Crevel R. (2013) Low induction of proinflammatory cytokines parallels evolutionary success of modern strains within the Mycobacterium tuberculosis Beijing genotype. Infect. Immun. 81, 3750–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jungblut P. R., Schaible U. E., Mollenkopf H. J., Zimny-Arndt U., Raupach B., Mattow J., Halada P., Lamer S., Hagens K., Kaufmann S. H. (1999) Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33, 1103–1117 [DOI] [PubMed] [Google Scholar]
- 34. de Souza G. A., Fortuin S., Aguilar D., Pando R. H., McEvoy C. R., van Helden P. D., Koehler C. J., Thiede B., Warren R. M., Wiker H. G. (2010) Using a label-free proteomics method to identify differentially abundant proteins in closely related hypo- and hypervirulent clinical Mycobacterium tuberculosis Beijing isolates. Mol. Cell. Proteomics 9, 2414–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neubauer H., Clare S. E., Kurek R., Fehm T., Wallwiener D., Sotlar K., Nordheim A., Wozny W., Schwall G. P., Poznanovic S., Sastri C., Hunzinger C., Stegmann W., Schrattenholz A., Cahill M. A. (2006) Breast cancer proteomics by laser capture microdissection, sample pooling, 54-cm IPG IEF, and differential iodine radioisotope detection. Electrophoresis 27, 1840–1852 [DOI] [PubMed] [Google Scholar]
- 36. Diz A. P., Truebano M., Skibinski D. O. (2009) The consequences of sample pooling in proteomics: an empirical study. Electrophoresis 30, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 37. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 38. Deeb S. J., D'Souza R. C., Cox J., Schmidt-Supprian M., Mann M. (2012) Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteomics 11, 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schubert O. T., Mouritsen J., Ludwig C., Rost H. L., Rosenberger G., Arthur P. K., Claassen M., Campbell D. S., Sun Z., Farrah T., Gengenbacher M., Maiolica A., Kaufmann S. H., Moritz R. L., Aebersold R. (2013) The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schurch A. C., Kremer K., Hendriks A. C., Freyee B., McEvoy C. R., van Crevel R., Boeree M. J., van Helden P., Warren R. M., Siezen R. J., van Soolingen D. (2011) SNP/RD typing of Mycobacterium tuberculosis Beijing strains reveals local and worldwide disseminated clonal complexes. PloS One 6, e28365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bifani P. J., Mathema B., Kurepina N. E., Kreiswirth B. N. (2002) Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10, 45–52 [DOI] [PubMed] [Google Scholar]
- 42. Narvskaya O. V., Mokrousov I. V., Otten T. F., Vishnevskii B. I. (1999) Genetic marking of polyresistant Mycobacterium tuberculosis strains isolated in the north-west of Russia. Probl. Tuberk. 39–41 [PubMed] [Google Scholar]
- 43. Narvskaya O. M. I. O., T, Vishnevsky B. (2005) Molecular markers: application for studies of Mycobacterium tuberculosis population in Russia. Trends in DNA fingerprinting research 15 [Google Scholar]
- 44. Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rusch-Gerdes S., Willery E., Savine E., de Haas P., van Deutekom H., Roring S., Bifani P., Kurepina N., Kreiswirth B., Sola C., Rastogi N., Vatin V., Gutierrez M. C., Fauville M., Niemann S., Skuce R., Kremer K., Locht C., van Soolingen D. (2006) Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boom R., Sol C. J., Salimans M. M., Jansen C. L., Wertheim-van Dillen P. M., van der Noordaa J. (1990) Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woods G. L., Brown-Elliott B. A., Desmond E. P., Hall G. S., Heifets L., Pfyffer G. E., Ridderhof J. C., Wallace R. J., Warren N. G., Witebsky F. G. (2011) Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, second edition. Clinical and Laboratory Standards Institute [PubMed] [Google Scholar]
- 47. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 48. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 49. Meiring H. D., van der Heeft E., ten Hove G. J., de Jong A. P. J. M. (2002) Nanoscale LC–MS(n): technical design and applications to peptide and protein analysis. J. Sep. Sci. 25, 12 [Google Scholar]
- 50. de Souza G. A., Arntzen M. O., Wiker H. G. (2010) MSMSpdbb: providing protein databases of closely related organisms to improve proteomic characterization of prokaryotic microbes. Bioinformatics 26, 698–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 52. Narayanan S., Deshpande U. (2013) Whole-Genome Sequences of Four Clinical Isolates of Mycobacterium tuberculosis from Tamil Nadu, South India. Genome Announc. 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 54. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 55. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 57. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., Sunyaev S. R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Capriotti E., Fariselli P., Rossi I., Casadio R. (2008) A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics 9, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brunham L. R., Singaraja R. R., Pape T. D., Kejariwal A., Thomas P. D., Hayden M. R. (2005) Accurate prediction of the functional significance of single nucleotide polymorphisms and mutations in the ABCA1 gene. PLoS Genet. 1, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reddy T. B., Riley R., Wymore F., Montgomery P., DeCaprio D., Engels R., Gellesch M., Hubble J., Jen D., Jin H., Koehrsen M., Larson L., Mao M., Nitzberg M., Sisk P., Stolte C., Weiner B., White J., Zachariah Z. K., Sherlock G., Galagan J. E., Ball C. A., Schoolnik G. K. (2009) TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 37, D499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vandenbergh P. A., Bawdon R. E., Berk R. S. (1979) Rapid test for determining the intracellullar rhodanese activity of various bacteria Int. J. Syst. Bacteriol. 29, 6 [Google Scholar]
- 63. Lew J. M., Kapopoulou A., Jones L. M., Cole S. T. (2011) TubercuList–10 years after. Tuberculosis 91, 1–7 [DOI] [PubMed] [Google Scholar]
- 64. Louw G. E., Warren R. M., Gey van Pittius N. C., McEvoy C. R., Van Helden P. D., Victor T. C. (2009) A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53, 3181–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mokrousov I., Jiao W. W., Sun G. Z., Liu J. W., Valcheva V., Li M., Narvskaya O., Shen A. D. (2006) Evolution of drug resistance in different sublineages of Mycobacterium tuberculosis Beijing genotype. Antimicrob. Agents Chemother. 50, 2820–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Iwamoto T., Yoshida S., Suzuki K., Wada T. (2008) Population structure analysis of the Mycobacterium tuberculosis Beijing family indicates an association between certain sublineages and multidrug resistance. Antimicrob. Agents Chemother. 52, 3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Parwati I., van Crevel R., van Soolingen D. (2010) Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10, 103–111 [DOI] [PubMed] [Google Scholar]
- 68. Borgdorff M. W., van Soolingen D. (2013) The re-emergence of tuberculosis: what have we learnt from molecular epidemiology? Clin. Microbiol. Infect. 19, 889–901 [DOI] [PubMed] [Google Scholar]
- 69. Faksri K., Drobniewski F., Nikolayevskyy V., Brown T., Prammananan T., Palittapongarnpim P., Prayoonwiwat N., Chaiprasert A. (2011) Genetic diversity of the Mycobacterium tuberculosis Beijing family based on IS6110, SNP, LSP and VNTR profiles from Thailand. Infect. Genet. Evol. 11, 1142–1149 [DOI] [PubMed] [Google Scholar]
- 70. Lu B., Zhao P., Liu B., Dong H., Yu Q., Zhao X., Wan K. (2012) Genetic diversity of Mycobacterium tuberculosis isolates from Beijing, China assessed by Spoligotyping, LSPs and VNTR profiles. BMC Infect. Dis. 12, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bell C., Smith G. T., Sweredoski M. J., Hess S. (2012) Characterization of the Mycobacterium tuberculosis proteome by liquid chromatography mass spectrometry-based proteomics techniques: a comprehensive resource for tuberculosis research. J. Proteome Res. 11, 119–130 [DOI] [PubMed] [Google Scholar]
- 72. Louw G. E., Warren R. M., Gey van Pittius N. C., Leon R., Jimenez A., Hernandez-Pando R., McEvoy C. R., Grobbelaar M., Murray M., van Helden P. D., Victor T. C. (2011) Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am. J. Respir. Crit. Care Med. 184, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Villellas C., Aristimuno L., Vitoria M. A., Prat C., Blanco S., Garcia de Viedma D., Dominguez J., Samper S., Ainsa J. A. (2013) Analysis of mutations in streptomycin-resistant strains reveals a simple and reliable genetic marker for identification of the Mycobacterium tuberculosis Beijing genotype. J. Clin, Microbiol. 51, 2124–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kelkar D. S., Kumar D., Kumar P., Balakrishnan L., Muthusamy B., Yadav A. K., Shrivastava P., Marimuthu A., Anand S., Sundaram H., Kingsbury R., Harsha H. C., Nair B., Prasad T. S., Chauhan D. S., Katoch K., Katoch V. M., Kumar P., Chaerkady R., Ramachandran S., Dash D., Pandey A. (2011) Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol. Cell. Proteomics 10, M111 011627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jungblut P. R., Holzhutter H. G., Apweiler R., Schluter H. (2008) The speciation of the proteome. Chemistry Central Journal 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schluter H., Apweiler R., Holzhutter H. G., Jungblut P. R. (2009) Finding one's way in proteomics: a protein species nomenclature. Chem. Cent. J. 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Low T. Y., van Heesch S., van den Toorn H., Giansanti P., Cristobal A., Toonen P., Schafer S., Hubner N., van Breukelen B., Mohammed S., Cuppen E., Heck A. J., Guryev V. (2013) Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 5, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 78. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 79. Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. (1999) Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34, 257–267 [DOI] [PubMed] [Google Scholar]
- 81. Domenech P., Reed M. B., Barry C. E., 3rd. (2005) Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wells R. M., Jones C. M., Xi Z., Speer A., Danilchanka O., Doornbos K. S., Sun P., Wu F., Tian C., Niederweis M. (2013) Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 9, e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. de Knegt G. J., Bruning O., ten Kate M. T., de Jong M., van Belkum A., Endtz H. P., Breit T. M., Bakker-Woudenberg I. A., de Steenwinkel J. E. (2013) Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberculosis 93, 96–101 [DOI] [PubMed] [Google Scholar]
- 84. Triccas J. A., Berthet F. X., Pelicic V., Gicquel B. (1999) Use of fluorescence induction and sucrose counterselection to identify Mycobacterium tuberculosis genes expressed within host cells. Microbiology 145, 2923–2930 [DOI] [PubMed] [Google Scholar]
- 85. Rengarajan J., Bloom B. R., Rubin E. J. (2005) Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102, 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cereda A., Carpen A., Picariello G., Tedeschi G., Pagani S. (2009) The lack of rhodanese RhdA affects the sensitivity of Azotobacter vinelandii to oxidative events. Biochem. J. 418, 135–143 [DOI] [PubMed] [Google Scholar]
- 87. Bifani P. J., Plikavtis B. B., Kapur V., Stockbauer K., Pan X., Lutfey M. L., Moghazeh S. L., Eisner W., Daniel T. M., Kaplan M. H., Crawford J. T., Musser B. N., Kreiswirth B. N. (1996) Origin and interstate spread of a New York multidrug-resistant Mycobacterium tuberculosis clone family. J. Am. Med. Assoc. 275, 6. [PubMed] [Google Scholar]
- 88. Constant P., Perez E., Malaga W., Laneelle M. A., Saurel O., Daffe M., Guilhot C. (2002) Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277, 38148–38158 [DOI] [PubMed] [Google Scholar]
- 89. Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N., Kaplan G., Barry C. E., 3rd. (2004) A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431, 84–87 [DOI] [PubMed] [Google Scholar]
- 90. Reed M. B., Gagneux S., Deriemer K., Small P. M., Barry C. E., 3rd. (2007) The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J. Bacteriol. 189, 2583–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pasca M. R., Guglierame P., Arcesi F., Bellinzoni M., De Rossi E., Riccardi G. (2004) Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 3175–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Duong D. A., Nguyen T. H., Nguyen T. N., Dai V. H., Dang T. M., Vo S. K., Do D. A., Nguyen V. V., Nguyen H. D., Dinh N. S., Farrar J., Caws M. (2009) Beijing genotype of Mycobacterium tuberculosis is significantly associated with high-level fluoroquinolone resistance in Vietnam. Antimicrob. Agents Chemother. 53, 4835–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. J.C., O. (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.