Abstract
The aim of this study was the identification of novel biomarker candidates for the diagnosis of cholangiocellular carcinoma (CCC) and its immunohistochemical differentiation from benign liver and bile duct cells. CCC is a primary cancer that arises from the epithelial cells of bile ducts and is characterized by high mortality rates due to its late clinical presentation and limited treatment options. Tumorous tissue and adjacent non-tumorous liver tissue from eight CCC patients were analyzed by means of two-dimensional differential in-gel electrophoresis and mass-spectrometry-based label-free proteomics. After data analysis and statistical evaluation of the proteins found to be differentially regulated between the two experimental groups (fold change ≥ 1.5; p value ≤ 0.05), 14 candidate proteins were chosen for determination of the cell-type-specific expression profile via immunohistochemistry in a cohort of 14 patients. This confirmed the significant up-regulation of serpin H1, 14-3-3 protein sigma, and stress-induced phosphoprotein 1 in tumorous cholangiocytes relative to normal hepatocytes and non-tumorous cholangiocytes, whereas some proteins were detectable specifically in hepatocytes. Because stress-induced phosphoprotein 1 exhibited both sensitivity and specificity of 100%, an immunohistochemical verification examining tissue sections of 60 CCC patients was performed. This resulted in a specificity of 98% and a sensitivity of 64%. We therefore conclude that this protein should be considered as a potential diagnostic biomarker for CCC in an immunohistochemical application, possibly in combination with other candidates from this study in the form of a biomarker panel. This could improve the differential diagnosis of CCC and benign bile duct diseases, as well as metastatic malignancies in the liver.
Cholangiocellular carcinoma (CCC)1 is a malignant neoplasm that arises from the cholangiocytes, the epithelial cells lining the bile ducts. The tumors, consisting of a significant amount of fibrous stroma, are classified as intrahepatic, extrahepatic, or hilar according to their anatomic location. Most common are the Klatskin tumors, originating from the confluence of the right and left hepatic ducts (1). Compared with other types of cancer, CCC is a relatively rare disease, accounting for about 3% of all gastrointestinal malignancies (2). However, its incidence is increasing, and as a result of poor patient outcomes it has overtaken hepatocellular carcinoma as the main cause of death from a primary hepatobiliary tumor (3). Reasons for the high mortality rate (5-year survival rate of about 5%) (4) are the difficult diagnosis and limited treatment options. At present, extensive surgical resection of the extrahepatic bile ducts and parts of the liver or liver transplantation remain the only potentially curative treatment options, although most patients are considered inoperable at the time of diagnosis (5).
In general, the diagnosis of CCC is made based on histomorphological evaluation of core biopsies or cytological specimens. However, distinction between CCC and benign diseases such as reactive bile ductules or bile duct adenomas can be challenging when based on conventional histology alone. Additionally, it may be difficult to distinguish CCC from metastatic adenocarcinoma in the liver, especially when it originates from the pancreas like pancreatic ductal adenocarcinoma. Therefore, specific immunohistochemical tissue markers for CCC would be highly beneficial for further validation of the diagnosis. In routine immunohistochemical diagnosis of CCC, so far, the detection of p53 (a product of a tumor suppressor gene) has proven useful, although its application is limited because of low sensitivity (6). The cytokeratins Ck7, Ck8, Ck18, and Ck19 have been reported to have sensitivities of between 80% and 97% for CCC cells, but at low specificities and a similar expression as in non-tumorous cholangiocytes (7). In addition, the tumor marker carcinoembryonic antigen, which is a commonly applied serum marker, has been used for immunohistochemical staining of CCC tissue. Although this was reported to be positive in 100% of the tested CCC sections, it also was immunoreactive in 60% of hepatocellular carcinomas (8). Recently, it has been shown that the polycomb group protein EZH2 may be useful for differential diagnosis of cholangiolocellular carcinoma (a subtype of CCC), bile duct adenomas, and ductular reaction. This, however, applies only to this certain type of CCC (9). Establishing reliable immunohistochemical tumor markers specific for CCC therefore remains a challenge.
Several proteomic studies using different sample types and various techniques have been performed in order to identify CCC-specific proteins. The analysis of CCC cell lines, for example, has led to the identification of potential diagnostic and prognostic biomarker candidates (10–12). In addition, cell lines have been used to discover proteins predictive of the response to chemotherapy (13). Because results from cell culture experiments do not always reflect the actual conditions in the tumor, the use of patient samples can be advantageous. The most appropriate source of tumor-specific signals is tumor tissue, which in the past has been analyzed via two-dimensional electrophoresis (14) and mass-spectrometry-based proteomic approaches such as histology-directed MALDI-TOF-MS (15), Surface-enhanced laser desorption/ionization (SELDI) TOF-MS (16), or LC-MS/MS (17). So far, however, none of the potential biomarkers have been successfully implemented into clinical routine.
Recently, we demonstrated that the application of two complementary techniques, two-dimensional differential in-gel electrophoresis (2D-DIGE) and mass-spectrometry-based label-free LC-MS/MS, is an auspicious tactic for the discovery of novel biomarker candidates in hepatocellular carcinoma tissue (18). Here, we applied this well-established workflow as the initial step for the discovery of tissue markers that improve the differential diagnosis of intrahepatic CCC from benign bile duct diseases. In these experiments, CCC tumor tissue was compared with non-tumorous liver tissue (n = 8). Because this does not allow discrimination among different cell types such as hepatocytes, cholangiocytes, and tumor cells, an immunohistochemical determination of the cell-type-specific expression was subsequently performed for the most promising biomarker candidates. Stress-induced phosphoprotein 1, the protein showing the greatest specificity and sensitivity for CCC tumor cells, was verified as a suitable biomarker candidate for CCC in a larger patient cohort (n = 60).
EXPERIMENTAL PROCEDURES
Clinical Data
Non-tumorous liver tissue and cholangiocellular carcinoma tissue from 77 CCC patients (48 females and 29 males) was collected during surgery at the University Hospital of Essen, Department of General, Visceral and Transplantation Surgery, Germany. The age of the patients at the time of operation ranged from 28 to 81 years (mean 62) (supplemental material). Informed consent was obtained from each patient, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki. The local ethics committee approved the study (11-4839-BO).
Sample Set Composition
Three sample sets were created (supplemental material). Sample set 1, which was used for 2D-DIGE and label-free LC-MS experiments, contained non-tumorous liver tissue and CCC tissue samples from eight patients aged between 42 and 78. These samples were collected in the time period from 2002 to 2012. Sample set 2, used for immunohistochemical determination of cell-type-specific protein expression, contained samples from 14 patients, including 5 from sample set 1. The patients' ages ranged from 31 to 78 at the time of operation, with surgery performed between 2010 and 2012. For immunohistochemical verification, sample set 3, from an independent cohort of 60 patients, was analyzed. These patients were aged 28 to 81 and underwent surgery between 2001 and 2010.
Tissue Preparation
For pathological examination and immunohistochemical staining, non-tumorous liver tissue and CCC tumor tissue were fixed in buffered formalin and paraffin embedded. For the proteomics studies, the samples were placed on ice immediately after the resection, snap-frozen, and stored at −80 °C. Protein extraction was performed via sonication (six 10-s pulses on ice) in sample buffer (30 mm Tris-HCl, 2 m thiourea, 7 m urea, 4% CHAPS, pH 8.5) and subsequent centrifugation (15,000 × g, 5 min). The supernatant was collected, and the protein concentration was determined via protein assay (Bio-Rad, Hercules, CA).
2D-DIGE Analysis
Protein Labeling
For 2D-DIGE experiments, minimal labeling using 400 pmol cyanine dyes (GE Healthcare, Munich, Germany) per 50 μg of protein was performed according to the manufacturer's instructions. To avoid bias, tumorous and non-tumorous samples were labeled alternately with Cy3 and Cy5 dyes. A mixture of all samples was labeled with Cy2 for use as an internal standard.
Two-dimensional Gel Electrophoresis
For 2D-DIGE experiments, the appropriate Cy3- and Cy5-labeled sample pairs from each patient were mixed with the internal standard (ratio 1:1:1). Isoelectric focusing and second-dimension SDS-PAGE were performed as described previously (18).
Image Acquisition and Evaluation
2D-DIGE gels were scanned on a Typhoon 9400 (Amersham Biosciences) at a resolution of 100 μm. Images were preprocessed using ImageQuantTM (GE Healthcare) before intra-gel spot detection, inter-gel matching, and normalization of spot intensities to the internal standard in DeCyder 2DTM (GE Healthcare). A statistical analysis was performed with the Extended Data Analysis tool of DeCyder2DTM and resulted in a list of proteins meeting the following criteria: (i) protein spot present in at least 70% of all spot maps, (ii) p value of Student's t test (paired, two-sided) ≤ 0.05 (after adjustment for multiple testing, controlling the false discovery rate using the method of Benjamini and Hochberg), and (iii) absolute average ratio between experimental groups ≥ 1.5. These differentially expressed proteins were extracted from a preparative two-dimensional gel and identified via MALDI-TOF-MS.
Digestion and Protein Identification
Protein spots dissected from preparative gels were subjected to in-gel digestion with trypsin (Promega, Madison, WI), and the peptides were then extracted from the gel matrix. MALDI-TOF-MS analyses were performed on an UltraFlexTM II instrument (Bruker Daltonics, Bremen, Germany). Protein identifications were done with ProteinScape (v. 1.3 SR2) (Bruker Daltonics) using the UniProt database (release 2012 09, 538,010 entries) via Mascot (v. 2.3.0.2) (Matrix Sciences Ltd., London, UK). The database searches were run with propionamide (C) and oxidation (M) as variable modifications, a mass tolerance of 100 ppm, and a maximum of one missed cleavage. Proteins with a Mascot score > 64 were considered to be assigned correctly. Further details concerning the MALDI-TOF-MS analyses and protein identifications via ProteinScape have been published previously (19). PMF spectra and peak lists containing peptide annotations are listed in the supplemental material.
Label-free Analysis
Sample Preparation
Samples were loaded onto a 4–20% SDS-PAGE gel (TGXTM precast gels, Bio-Rad) and run for 1 min at 300 V. The proteins were stained with Coomassie Brilliant Blue and digested in-gel using trypsin (SERVA Electrophoresis, Heidelberg, Germany). The peptides were extracted via sonication on ice in 20 μl of 50% acetonitrile in 0.1% TFA. Acetonitrile was removed by means of vacuum centrifugation before the peptides were rehydrated in 0.1% TFA. The peptide concentration was determined via amino acid analysis on an ACQUITY-UPLC with an AccQ Tag Ultra-UPLC column (Waters, Eschborn, Germany) calibrated with Pierce Amino Acid Standard (Thermo Scientific, Bremen, Germany).
LC-MS/MS Analysis
Label-free MS-based analysis was performed on an Ultimate 3000 RSLCnano system (Dionex, Idstein, Germany) online coupled to an LTQ Orbitrap Elite (Thermo Scientific). For each analysis, 350 ng of tryptic peptides dissolved in 15 μl of 0.1% TFA were injected and pre-concentrated on a trap column (Acclaim® PepMap 100, 300 μm × 5 mm, C18, 5 μm, 100 Å) for 7 min with 0.1% TFA at a flow rate of 30 μl/min. The separation was performed on an analytical column (Acclaim® PepMap RSLC, 75 μm × 50 cm, nano Viper, C18, 2 μm, 100 Å) with a gradient from 5% to 40% solvent B over 98 min (solvent A, 0.1% formic acid; solvent B, 0.1% formic acid, 84% acetonitrile). The flow rate was set at 400 nl/min, and the column oven temperature was 60 °C. The mass spectrometer was operated in data-dependent mode. Full-scan MS spectra were acquired at a resolution of 60,000 in the Orbitrap analyzer, and tandem mass spectra of the 20 most abundant peaks were measured in the linear ion trap after peptide fragmentation by collision-induced dissociation.
Peptide Quantification and Filtering
The ion-intensity-based label-free quantification was done by evaluating the LC-MS data with Progenesis LC-MSTM (v. 4.0.4265.42984, Nonlinear Dynamics Ltd., Newcastle upon Tyne, UK). The generated .raw files were imported, and the most representative LC-MS run was selected as the reference to which the retention times of the precursor masses of all other runs were aligned. From the feature list containing m/z values of all peptides, only those charged positively 2-, 3-, or 4-fold were used for quantification. To correct experimental variation between the runs, the raw abundances of each feature were normalized. Details regarding the normalization have been published previously (18).
Protein Identification
Proteins from LC-MS runs were identified by Proteome Discoverer (v. 1.3) (Thermo Scientific) searching the UniProt database (release 2012 02, 534,695 entries) via Mascot (v. 2.3.0.2) (Matrix Sciences Ltd.). The following search parameters were applied: variable modifications, oxidation (M) and propionamide (C); tryptic digestion with up to one missed cleavage; precursor ion mass tolerance of 5 ppm; and fragment ion mass tolerance of 0.4 Da. The search results were filtered with a false discovery rate of less than 1% on the peptide level, which was calculated using the Percolator tool implemented in Proteome Discoverer before the data were imported into Progenesis LC-MS. In this way, each peptide was matched to a previously quantified feature. The mass spectrometry proteomics data of the label-free analysis have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (20) with the dataset identifier PXD000534 and DOI 10.6019/PXD000534.
Protein Quantification and Filtering
For the protein quantification, only peptides unique to one protein within the particular experiment were used. The normalized sum of all the unique peptide ion abundances identified as coming from a specific protein was used to calculate the p value of Student's t test (paired, two-sided) and the fold change for each protein. The protein grouping function of Progenesis LC-MS was disabled. Proteins showing a p value ≤ 0.05 after false discovery rate correction (21) and an absolute fold change greater than 1.5 were assumed to be differentially regulated. Proteins quantified with only one distinct peptide (unique by mass spectrometry) were removed from the experiment.
Annotation of Regulated Proteins
Previously generated lists of differential proteins were processed using Ingenuity Pathway Analysis software (v. 12402621, Ingenuity Systems, Redwood City, CA) in order to assign their cellular localizations. Computer-aided literature research was performed using SCAIView software (Fraunhofer Institute for Algorithms and Scientific Computing SCAI, Sankt Augustin, Germany) (22).
Immunohistochemistry
Preparation and Staining of Tissue Samples
Tissue microarrays with three cores per case (core diameter: 1 mm) from CCC and adjacent non-tumorous liver tissue were constructed. Paraffin-embedded 4-μm slides were dewaxed and pretreated in EDTA buffer (pH 9) at 95 °C for 20 min. All immunohistochemical stains were performed with an automated staining device (Dako Autostainer, Glostrup, Denmark). Both the source of the primary antibodies and the technical staining details of the automatically performed stainings are listed in the supplemental material. All stains were developed using a polymer kit (ZytoChemPlus (HRP), POLHRS-100, Zytomed Systems, Berlin, Germany). Primary antibodies were replaced by mouse or rabbit immunoglobulin for negative controls.
Evaluation of Immunohistochemical Stains
Using an immunoreactive score modeled after the work of Remmele and Stegner (23), stained tissue was graded into four categories regarding its staining intensity (0 = no, 1 = faint, 2 = moderate, and 3 = strong staining) and into five categories for the approximate proportion of positive cells (0 = no, 1 = up to 5%, 2 = 6% to 10%, 3 = 11% to 50%, and 4 = more than 50% positive cells). The examination was performed by two experienced independent pathologists whose results were averaged. The final immunoreactive score was calculated by multiplying the staining intensity by the number of positive cells (minimum 0, maximum 12).
To assess each marker's ability to separate tumor samples from hepatocytes and cholangiocytes, receiver operating characteristic analysis was performed using the R package pROC (24). AUC values were determined along with the corresponding 95% confidence intervals (with the variance of the AUC being computed as defined by DeLong et al. (25)). In order to compare the diagnostic characteristics of the candidates and choose the most promising one for validation in a larger sample set, sensitivity and specificity were assessed at the best cutoff for each curve. Sensitivity was defined as the percentage of samples with positive staining of the targeted cell type, and specificity was defined as the percentage of samples without positive staining of other cells. Candidate-specific optimization is important in order to account for different staining behaviors of antibodies and leads to higher optimal cutoff values for antibodies with more intense background staining and lower cutoffs with less background. For optimization, we used Youden's criterion, which is equivalent to the maximization of the sum of sensitivity and specificity. In the case of multiple cutoffs yielding an optimal Youden score, the one with the highest specificity was chosen. For the observed values of sensitivity and specificity at the optimized cutoffs, 95% confidence intervals were computed as well (using the Clopper–Pearson method). Although optimization leads to overoptimistic diagnostic values, these values are suitable for comparing different candidates. Optimized cutoff values were used for verification in sample set 3 to ensure unbiased results. For the putatively CCC-specific markers, separate receiver operating characteristic curves were derived for the comparisons with hepatocytes and cholangiocytes. For putatively hepatocyte-specific proteins, AUC values and diagnostic values were calculated with an expectation of lower values in CCC cells.
RESULTS
Quantitative Proteomic Analysis
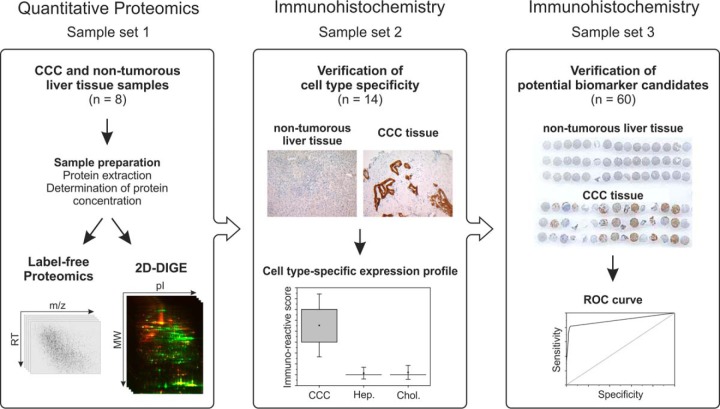
This study aiming at the identification of novel diagnostic biomarker candidates for cholangiocellular carcinoma combined two quantitative proteomics techniques, namely, 2D-DIGE and mass-spectrometry-based label-free proteomics, to analyze the protein expression profile of CCC tumor tissue (n = 8) in comparison to that of non-tumorous liver tissue (n = 8) (sample set 1). After an evaluation of the resulting data, biomarker candidates were verified by immunohistochemistry. The cell-type-specific expression was determined in a sample cohort from 14 patients (sample set 2) before the most promising candidate was verified in a large cohort of 60 patients (sample set 3). The overall workflow is shown in Fig. 1.
Fig. 1.
Schematic presentation of the workflow followed in the proteomic study.
Using the 2D-DIGE technique, we detected 1676 protein spots in at least 18 out of all 24 spot maps. Paired average ratios ranged from −30.54 to 30.19. In all, 678 spots were significantly differential between the two experimental groups (Student's t test, paired, two-sided, adjusted p values ≤ 0.05; paired average ratio ≥ 1.5). After extraction from a preparative gel, 183 protein spots, corresponding to 122 non-redundant proteins, were identified via MALDI-TOF-MS. Among these, 44 proteins were up-regulated and 78 were down-regulated in CCC tissue relative to controls. Two proteins, triosephosphate isomerase and α-enolase, showed differing regulation directions between multiple detected isoforms (supplemental material).
The same samples were also analyzed via label-free LC-MS/MS. Due to technical issues, the data of one control sample could not be evaluated. In order to still perform a paired sample comparison, we also removed the corresponding tumor sample from the study, leaving seven versus seven samples in this label-free experiment. Here, in total, 36,104 features with charge states of 2+, 3+, or 4+ were detected. After the database search, 14,206 features were assigned to peptide matches, leading to the identification of 2404 proteins (supplemental Fig. S1). After discarding proteins that were quantified with only one distinct peptide, we found 920 proteins to be significantly regulated (Student's t test, adjusted p values ≤ 0.05; fold change ≥ 1.5). Out of these, 516 were up- and 404 down-regulated in CCC tissue.
In the protein lists from both approaches, a total of 954 differential proteins were identified, with 34 found exclusively in the 2D-DIGE experiment and 832 identified only in the label-free study (supplemental Fig. S2). Thus, 88 proteins were found to be differential irrespective of the applied quantification method.
For most of the proteins from the overlap of both approaches, the same regulation directions were discovered, and the fold changes determined via 2D-DIGE and label-free proteomics were found to be highly correlated (Pearson's correlation coefficient r = 0.878) (supplemental Fig. S3). Nevertheless, two proteins (aminoacylase 1 and 3-hydroxyisobutyryl-CoA hydrolase) were reported with contrary regulation directions in the 2D-DIGE and label-free experiments.
The determination of protein localizations using Ingenuity Pathway Analysis software revealed a significantly greater amount of nucleic and plasma membrane proteins identified by label-free proteomics than by 2D-DIGE (supplemental Fig. S4). In the gel-based approach, therefore, a greater amount of cytoplasmic proteins was detected.
Selection of Potential Biomarkers for Further Verification
In order to select suitable candidates for the immunohistochemical experiments, we took different factors into account. The Euclidian distance, which for the label-free experiment was visualized by the volcano plot in supplemental Fig. S1, was calculated using the log2(fold change) and the log10(p value) of each protein (26). Further, the confidence of the identification (Mascot score and number of peptides) was observed. Manual and computer-aided literature research gave additional hints about which proteins might be appropriate candidates. This included evaluating which proteins have been described as being associated with CCC, other types of cancer, or other liver diseases. Finally, the availability of appropriate antibodies also was an important factor. After these considerations, 14 proteins, which are summarized in Table I, were chosen for determination of the cell type specificity via immunohistochemistry.
Table I. Potential biomarker candidates chosen for verification by immunohistochemistry.
The adjusted p values for paired Student's t test (two-sided) and fold changes are given. All p values shown are corrected for multiple testing, controlling the false discovery rate. In Progenesis, p values are obtained using the method of Benjamini and Hochberg (21); DeCyder uses the version Benjamini and Hochberg published in 2000 (52). In the case of two values for t test and fold change for one protein, this corresponds to different isoforms identified by 2D-DIGE.
| Number | Protein |
2D-DIGE |
Label-free proteomics |
||||
|---|---|---|---|---|---|---|---|
| Accession | Protein name | Gene name | Paired t test | Fold change | Paired t test | Fold change | |
| Up-regulated in CCC | |||||||
| 1 | P06727 | Apolipoprotein A-IV | APOA4 | 0.010 | 2.3 | 0.003 | 3.9 |
| 2 | O00299 | Chloride intracellular channel protein 1 | CLIC1 | 0.009 | 4.7 | 0.007 | 2.4 |
| 3 | P06396 | Gelsolin | GSN | 0.044 | 5.8 | 0.001 | 5.7 |
| 4 | P26038 | Moesin | MSN | 0.021 | 2.5 | – | – |
| 5 | P14618 | Pyruvate kinase isozymes M1/M2 | PKM2 | 0.025/0.047 | 8.8/2.6 | <0.001 | 7.3 |
| 6 | Q15181 | Inorganic pyrophosphatase | PPA1 | 0.001/0.011 | 2.1/1.8 | – | – |
| 7 | P50454 | Serpin H1 | SERPINH1 | – | – | 0.001 | 3.6 |
| 8 | P31947 | 14-3-3 protein σ | SFN | – | – | 0.01 | 8.2 |
| 9 | P31948 | Stress-induced phosphoprotein 1 | STIP1 | 0.006 | 1.7 | 0.007 | 1.7 |
| Down-regulated in CCC | |||||||
| 10 | P80404 | 4-aminobutyrate aminotransferase, mitochondrial | ABAT | 0.001 | 5.4 | <0.001 | 9.9 |
| 11 | P42765 | 3-ketoacyl-CoA thiolase, mitochondrial | ACAA2 | 0.001/0.001 | 11.7/8.0 | <0.001 | 8.7 |
| 12 | Q93088 | Betaine-homocysteine S-methyltransferase 1 | BHMT | 0.001/0.009 | 11.9/3.1 | <0.001 | 17.1 |
| 13 | P07148 | Fatty acid–binding protein, liver | FABP1 | 0.001 | 30.5 | <0.001 | 22.9 |
| 14 | P54868 | Hydroxymethylglutaryl-CoA synthase, mitochondrial | HMGCS2 | 0.016/0.032 | 2.7/1.8 | <0.001 | 14.0 |
Determination of the Cell Type Specificity
Non-tumorous liver tissue, which was used as control tissue in the previous experiments, is a mixture of different cell types including mainly hepatocytes but also, among others, hepatic stellate cells, vascular endothelial cells, smooth muscle cells, myofibroblasts, and cholangiocytes. The latter are the origin of CCC tumors. Therefore, this study required a determination of the cell-type-specific expression of each candidate between the identification and the actual verification. Here, the aim was to screen for those allowing the discrimination of tumorous cells from non-tumorous cholangiocytes and hepatocytes and consequently support the differentiation of CCC from benign bile duct diseases. Sample set 2, including tissues from 14 patients (including 5 from sample set 1), was stained immunohistochemically with antibodies against 14 candidate proteins. The evaluation of staining intensity and the amount of stained cells was performed regarding CCC tumor cells, hepatocytes, and non-tumorous cholangiocytes. The values for the AUC, optimized thresholds, and corresponding sensitivities and specificities are given in Table II, and a table including all confidence intervals and additional information can be found in the supplemental material. It is important to note that the given diagnostic values are overoptimistic because of the optimization process. However, they can be used to compare the different candidates. The cutoff that was derived from sample set 2 was fixed and used as a cutoff for the larger set. This ensured that the resulting diagnostic values for sample set 3 were unbiased.
Table II. Immunohistochemical verification of 14 potential biomarker candidates.
Tissue samples from set 2 (n = 14) were evaluated regarding positive antibody staining. Areas under the curve (AUC) from receiver operating characteristics were computed, and cutoff values for immunoreactive scores were optimized according to Youden's criterion. Infinitely high cutoff values (∞) indicate that there was no cutoff value at which groups could be separated meaningfully. In the case of proteins up-regulated in CCC tumor tissue (“putatively CCC-specific”), the sensitivity represents the proportion of samples positive for CCC cell staining. Specificities were determined for the ability to differentiate CCC tumor cells from normal hepatocytes (“vs. hepatocytes”), as well as for distinguishing between CCC cells and non-tumorous cholangiocytes (“vs. cholangiocytes”). AUCs and diagnostic values were derived with the expectation of greater values in tumor samples. For down-regulated candidates (“putatively hepatocyte-specific”), the sensitivity for detecting hepatocytes and the specificity for distinguishing these from CCC cells (“vs. CCC”) were determined. In this case, AUCs and diagnostic values were computed with the expectation of lesser values in tumor samples.
| Number | Gene name | Comparison | AUC | Cutoff | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Putatively CCC-specific | ||||||
| 1 | APOA4 | Vs. hepatocytes | 0.45 | ∞ | 0 | 100 |
| Vs. cholangiocytes | 0.87 | 3.5 | 79 | 92 | ||
| 2 | CLIC1 | Vs. hepatocytes | 0.99 | 7 | 100 | 93 |
| Vs. cholangiocytes | 0.59 | 10.5 | 79 | 42 | ||
| 3 | GSN | Vs. hepatocytes | 1 | 4.5 | 93 | 100 |
| Vs. cholangiocytes | 0.28 | ∞ | 0 | 100 | ||
| 4 | MSN | Vs. hepatocytes | 0.68 | 1 | 36 | 100 |
| Vs. cholangiocytes | 0.68 | 1 | 36 | 100 | ||
| 5 | PKM2 | Vs. hepatocytes | 0.97 | 3.5 | 100 | 86 |
| Vs. cholangiocytes | 0.58 | 10 | 43 | 100 | ||
| 6 | PPA1 | Vs. hepatocytes | 0.82 | 7 | 93 | 54 |
| Vs. cholangiocytes | 0.70 | 7 | 93 | 42 | ||
| 7 | SERPINH1 | Vs. hepatocytes | 0.99 | 10 | 86 | 100 |
| Vs. cholangiocytes | 0.98 | 10 | 86 | 100 | ||
| 8 | SFN | Vs. hepatocytes | 1 | 2.5 | 93 | 100 |
| Vs. cholangiocytes | 0.99 | 2.5 | 93 | 100 | ||
| 9 | STIP1 | Vs. hepatocytes | 1 | 3 | 100 | 100 |
| Vs. cholangiocytes | 1 | 3.5 | 100 | 100 | ||
| Putatively hepatocyte-specific | ||||||
| 10 | ABAT | Vs. CCC | 1 | 10 | 100 | 100 |
| 11 | ACAA2 | Vs. CCC | 0.79 | 10.5 | 100 | 57 |
| 12 | BHMT | Vs. CCC | 1 | 1.5 | 100 | 100 |
| 13 | FABP1 | Vs. CCC | 1 | 9 | 100 | 100 |
| 14 | HMGCS2 | Vs. CCC | 0.95 | 8.5 | 86 | 93 |
Out of the 14 tested proteins, 5 (chloride intracellular channel protein 1, gelsolin, moesin, pyruvate kinase isozymes M1/M2, and inorganic pyrophosphatase 1) were not considered for further experiments because of unsatisfactory sensitivity or specificity less than 50%, especially within the evaluation comparing CCC cells to cholangiocytes (Table II). In the case of APOA4, for the comparison of tumorous cells and hepatocytes, a sensitivity of 0% was observed at 100% specificity. Considering the AUC, which is close to 0.5, this means that hepatocytes and CCC cells were stained to a similar extent. In the comparison of tumorous to non-tumorous cholangiocytes, in contrast, promising diagnostic values were achieved.
The remaining proteins that were found to be up-regulated in CCC tissue—serpin H1, SFN, and STIP1—showed a specificity of 100% for the differentiation of CCC from both hepatocytes and cholangiocytes, along with high sensitivities between 86% and 100%.
Because candidates that were identified as down-regulated in CCC tissue in the proteomics study can be assumed to be hepatocyte-specific proteins, in these cases, the sensitivity refers to the staining quantity and intensity of hepatocytes. This was 100% for ABAT, mitochondrial 3-ketoacyl-CoA thiolase 2, BHMT, and FABP1 and 86% for mitochondrial hydroxymethylglutaryl-CoA synthase. The specificity in these cases indicates the proportion of samples in which tumorous cells were not positively stained (Table II: “versus CCC”). ABAT, BHMT, and FABP1 again reached 100%; mitochondrial hydroxymethylglutaryl-CoA synthase was slightly less specific with 93%, and mitochondrial 3-ketoacyl-CoA thiolase 2 reached only 57%.
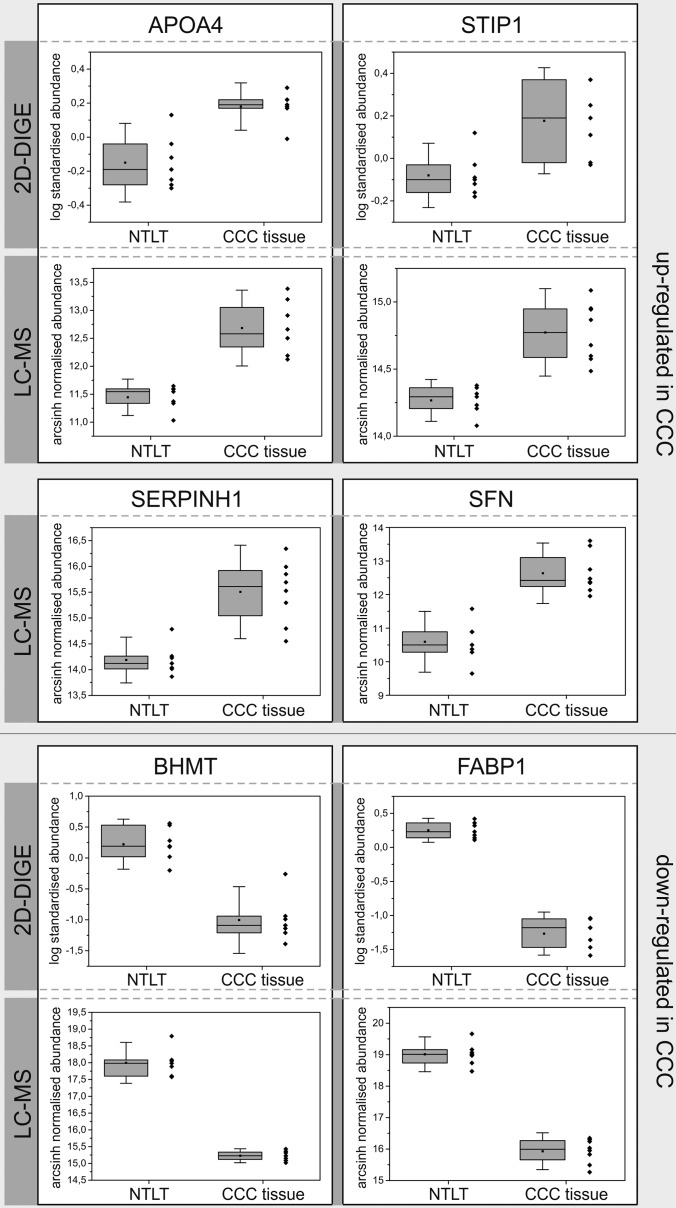
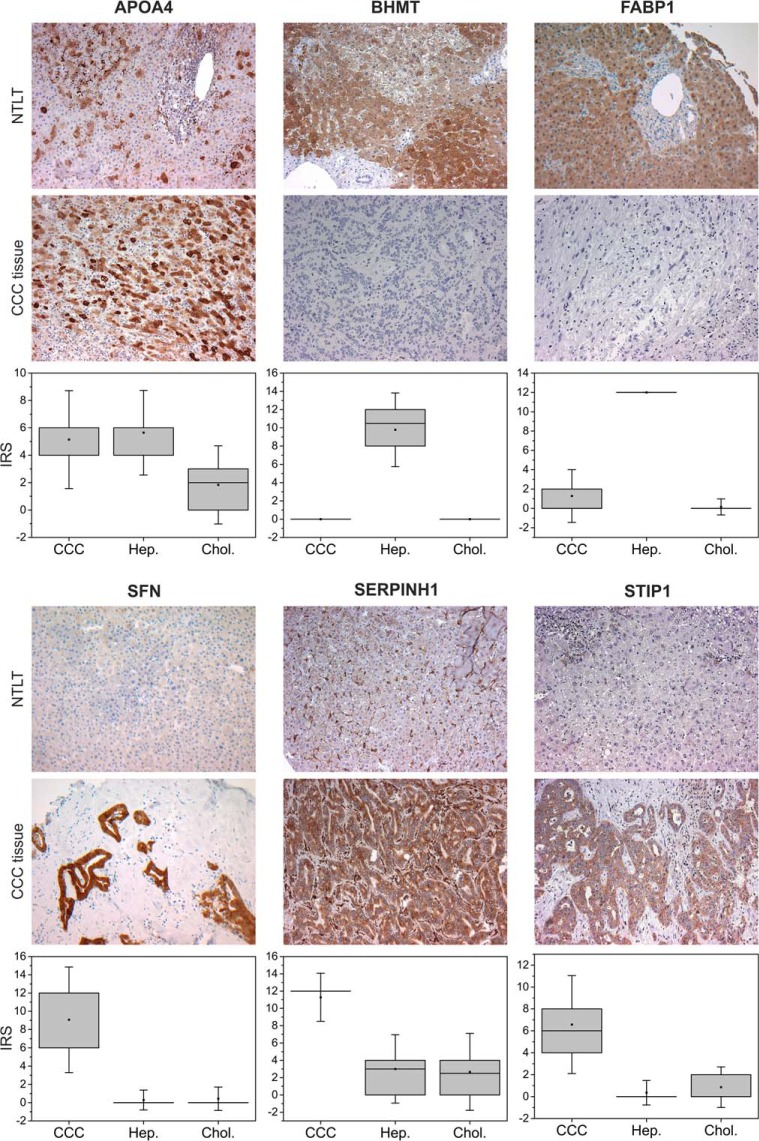
For six of the most promising candidates—APOA4, serpin H1, SFN, STIP1, BHMT, and FABP1—the regulation profiles obtained in the label-free and 2D-DIGE experiments and representative immunohistochemical staining patterns are presented in Figs. 2 and 3, respectively. In addition, Fig. 3 visualizes the expression levels of these proteins observed in the immunohistochemistry experiment across all 14 tested samples. For all other candidates, equivalent figures can be found in the supplemental material.
Fig. 2.
Regulation profiles of selected candidate proteins in the 2D-DIGE and the MS-based label-free experiments. Boxes represent 25th and 75th percentiles; whiskers indicate the standard deviation. The median is shown as a horizontal line, and the mean value as a square within the box. NTLT, non-tumorous liver tissue.
Fig. 3.
Verification of selected biomarker candidates by immunohistochemical staining of CCC tumor tissue and corresponding non-tumorous liver tissue (NTLT) from the same patient. When using an APOA4 antibody, an inhomogeneous regional staining of CCC tumor tissue and, in control tissue, of some hepatocytes and interstitial cells was observed, whereas non-tumorous cholangiocytes showed no signal. For BHMT and FABP1, hepatocytes displayed strong signal, whereas non-malignant portal fields including cholangiocytes and connective tissue and tumorous tissue remained unstained. SFN was not detectable in hepatocytes or non-neoplastic bile ducts, but it was observed in CCC cells. Serpin H1 was localized only to the cytoplasm of malignant cells and to sinusoidal cells of non-tumor liver tissue. In tumorous tissue, the antibody against STIP1 showed reactivity in CCC cells but not in tumorous connective tissue, while non-malignant tissue was not stained. Original magnification: ×200. Box plots represent the expression level of each candidate across all 14 tested patients (sample set 2) in CCC tumor cells, hepatocytes (Hep.), and cholangiocytes (Chol.) based on the immunoreactive scores.
Verification by Immunohistochemistry
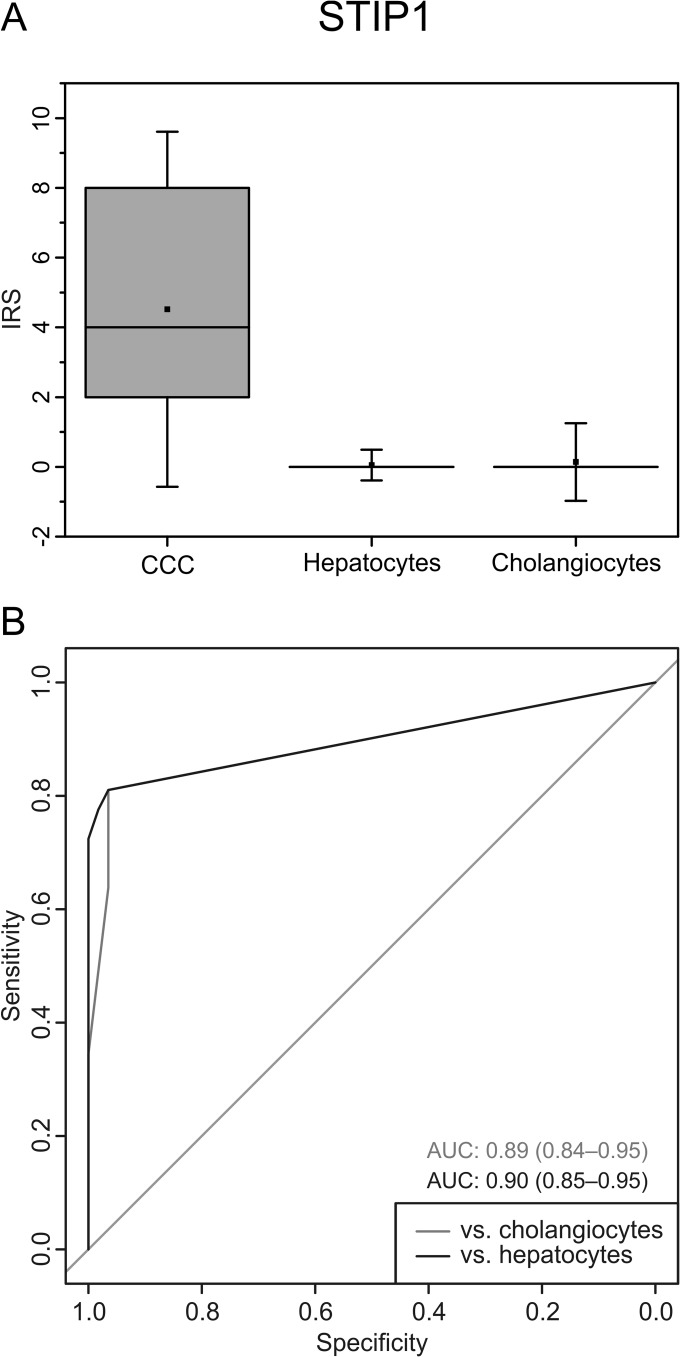
At this point, the most promising candidate for further analyses was STIP1, with 100% sensitivity and specificity for CCC cells in the 14 tested samples. Therefore, a larger cohort comprising 60 patient samples was assembled and tested for STIP1 expression by immunohistochemistry (Fig. 4). Here, cutoff values that were optimized using sample set 2 were applied to the evaluation of sample set 3 to ensure unbiased results. Of these 60 samples, 58 could be evaluated regarding tumor cell staining, and 57 regarding hepatocyte and cholangiocyte staining. Optimal cutoffs were determined separately for comparisons versus cholangiocytes (cutoff = 3.5) and hepatocytes (cutoff = 3), yielding two values each for sensitivity and specificity. In the comparison versus hepatocytes, specificity was 100% with a 95% confidence interval (CI) of 94% to 100% and a sensitivity of 72% (CI: 59% to 83%). In the case of cholangiocytes, specificity was again very high (96%; CI: 88% to 100%), and sensitivity reached 64% (CI: 50% to 76%).
Fig. 4.
Verification of STIP1 by immunohistochemistry in sample set 3 (n = 60). The box plot (A) visualizes the high expression of STIP1 in CCC tumor cells relative to hepatocytes and cholangiocytes. Boxes represent 25th and 75th percentiles; whiskers indicate the standard deviation. The median is shown as a horizontal line, and the mean value as a square within the box. The receiver operating characteristic (ROC) curves (B) illustrate the good sensitivity and specificity achieved with this biomarker candidate. AUC, area under the curve; ranges in brackets indicate the 95% confidence interval.
For clinical practice, a single cutoff would be more practical. In line with our above-mentioned optimization strategy, we chose the greater value of 3.5, resulting in a specificity of 98% (CI: 94% to 100%) and sensitivity of 64% (CI: see above) when comparing against both hepatocytes and cholangiocytes in combination.
DISCUSSION
To date, diagnosis of CCC remains problematic because patients develop symptoms only in an advanced stage of the disease. Furthermore, imaging modalities such as computed tomography and MRI that are being used in the evaluation of primary hepatic masses have low specificity. In some cases, the concentration of the serum marker CA 19–9 can give further confirmation, although its sensitivity and specificity for CCC are also low. For final validation, histological diagnosis is therefore often performed (27). Because this still is not always definitive, the identification of novel biomarkers for the immunohistochemical diagnosis of CCC is an important task. Such biomarkers could support the diagnosis of CCC and its differentiation from benign bile duct diseases such as reactive bile duct proliferations, as well as metastatic malignancies such as pancreatic ductal adenocarcinoma, which are so far difficult to distinguish from CCC. Therefore, in this study, tumorous and non-tumorous tissue samples were compared by means of the top-down proteomic method 2D-DIGE and a bottom-up label-free LC-MS approach.
The aim of combining these complementary methods for the discovery of novel biomarker candidates was to increase proteome coverage to improve the chance of identifying significant regulations, as well as to ensure greater reliability of candidates if they were found to be differentially expressed with both techniques. Regarding those proteins identified through both approaches, the correlation of these proteins' fold changes from the two experiments (Pearson's correlation coefficient r = 0.878) demonstrates the consistency of both techniques. Only 2 out of 88 proteins showed differing regulation directions when we compared both techniques. This could have been due to the detection of different isoforms in 2D-DIGE and LC-MS. With label-free proteomics, it is not possible to distinguish between different isoforms of one protein unless proteotypic peptides for a particular isoform are identified. If they are not, abundances of protein isoforms are averaged to calculate a shared fold change and p value.
In the 2D-DIGE and LC-MS experiments, bias due to tissue heterogeneity could not be avoided, and the ability to distinguish non-tumorous from tumorous cholangiocytes is a prerequisite for a useful CCC biomarker. Therefore, verification by immunohistochemistry was a critical step. The immunohistochemical examination was performed by two experienced pathologists who analyzed only the cell types of interest in the particular tissue (cancerous cells in the tumor tissue, hepatocytes and cholangiocytes in the non-tumorous tissue). Evaluation of immunohistochemical stains was performed using an immunoreactive scoring system. This is especially advisable for the analysis of inhomogeneous tumor tissue such as CCC. In contrast to the original recommendation for the immunoreactive score from Remmele and Steger, a more differentiated grading of the percentage of positively stained cells with five instead of four categories was chosen in order to reflect more subtle differences between staining patterns. As a result, from the immunohistochemical analysis, seven of the tested candidate proteins should be considered as potentially supportive for the diagnosis of CCC. These are STIP1, SFN, serpin H1, APOA4, ABAT, BHMT, and FABP1.
The most promising of these is STIP1, also known as Hsp70/Hsp90-organizing protein. This is a co-chaperone of Hsp70 and Hsp90 that participates in a large number of cellular processes such as RNA splicing, transcription, viral replication, protein folding and translocation, signal transduction, and cell cycle regulation (28). Recently, an immunohistochemical study of 330 tumor samples has shown that STIP1 acts as a prognostic biomarker in human ovarian cancer (29). It is thought to bind to the bone morphogenetic protein receptor ALK2 (activin A receptor, type II-like kinase 2) to activate the SMAD signaling pathway and transcription of inhibitor of DNA binding 3. This promotes cancer cell proliferation in ovarian malignancies (30). Considering the up-regulation of STIP1 in CCC cells revealed by our study, this mechanism might also be active in CCC tumor formation. Because STIP1 showed a very high specificity of 98% for CCC cells when comparing to both hepatocytes and non-tumorous cholangiocytes in the verification set including tissue sections from 60 patients and a moderate sensitivity of 64% (or 72%, depending on the cell type CCC was compared with), we conclude that STIP1 is a promising biomarker candidate for CCC. In comparison, for the tumor marker p53 protein, an immunohistochemical study in 2011 revealed positive staining in only 37% of intrahepatic and 46% of extrahepatic bile duct cancers. Although in this case no non-tumorous cholangiocytes were stained, 24% of dysplastic bile duct cells overexpressed p53 protein (6). The fact that STIP1 was not expressed in all CCC tumor samples again reflects the heterogeneity of this tumor. In practice, this might lead to a number of false negatives when using STIP1 as a stand-alone biomarker for CCC. Although, of course, its sensitivity and specificity need to be further substantiated in larger validation sets, an advisable implementation could be the integration of STIP1 in an immunohistochemical biomarker panel. Combining multiple biomarkers to create a single classification score can drastically improve the diagnostic accuracy relative to that of individual markers. Other proteins from this study could also contribute to such a biomarker panel.
SFN, for example, which is involved in a large spectrum of signaling pathways, showed high diagnostic values in the immunohistochemical evaluation of sample set 2 (n = 14). This protein is thought to be an important cell cycle protein in various cancer types (31–34). In 2007, an immunohistochemical study demonstrated its expression in 67.7% of 93 tested cases of intrahepatic CCC. Immunoreactivity was observed only in cancerous tissue, and not in normal bile duct cells (35). This is in line with our findings and confirms again the reliability of the methods and workflow applied during this study. Furthermore, Kuroda et al. (35) demonstrated that decreased SFN expression is a significant indicator of poor prognosis in intrahepatic CCC. In conclusion, this protein might be used as a prognostic biomarker for CCC, and given its connection to oncogenic processes in different malignancies, it might be a potential drug target worth further investigation.
In our current study, 100% sensitivity for CCC cells was furthermore revealed for the collagen-binding protein serpin H1, also known as HSP47 or colligin. Serpin H1 is thought to be involved in processing, glycosylation, and secretion of collagen and cross-linking of the three-dimensional assembly of type IV collagen molecules (36, 37). Its overexpression in fibrotic diseases with enhanced collagen biosynthesis such as glomerulosclerosis (38), pulmonary fibrosis (39), and liver cirrhosis (40, 41) has been demonstrated. In CCC, this reflects the usually dense desmoplastic stroma of the tumor entity. In conclusion, increased expression of serpin H1 is an indicator of strong collagen biosynthesis and, consequently, fibrotic changes in all kinds of tissue. Thus, it seems not to be specific for CCC, but it nevertheless might support the differential diagnosis of CCC and benign proliferations of liver or bile duct cells.
APOA4, a glycoprotein that is suggested to be involved in chylomicron assembly and pre–very low-density lipoprotein transport (42), was up-regulated in CCC tissue in our proteomics study. Although immunohistochemical verification in sample set 2 revealed no significant differences in staining of CCC cells and hepatocytes, based on its clear overexpression in CCC cells relative to normal cholangiocytes, APOA4 has good potential to prove useful in biomarker panels. As one main focus is the differentiation of CCC from benign alterations of cholangiocytes, discrimination of tumorous from non-tumorous bile duct cells is of greater importance than that of CCC cells from hepatocytes. Apart from that, the latter are in most cases distinguishable on the basis of morphological aspects. In combination with other biomarkers, however, even these uncertainties could be clarified, especially when combining CCC-specific with hepatocyte-specific proteins.
Three proteins that showed higher expression levels in normal liver tissue than in CCC tumors in our study are ABAT, BHMT, and FABP1, also named l-type or liver-type fatty acid-binding protein. In line with our findings, FABP1 has been described as expressed mainly in hepatocytes (43), but also in the small intestines (44) and the kidney (45). Also, ABAT, a major inhibitory neurotransmitter in the mammalian central nervous system, has previously been shown to be most abundant in the liver, at least on the transcript level (46). BHMT, which regenerates methionine from homocysteine via remethylation in the kidney and the liver (47), accounts for 0.6% to 1.6% of the total protein content in the liver (48). Decreased expression levels have been reported in hepatocellular carcinoma relative to normal liver tissue in several studies (18, 49–51). Nevertheless, the immunohistochemical staining of HCC tissue still shows a weak signal for BHMT (18), whereas CCC displayed none at all. In addition, ABAT, BHMT, and FABP1 might also be used to differentiate metastases deriving from hepatocytes from those of other origin such as the bile ducts.
In malignant cells, a wide range of metabolic pathways are dysregulated. The overexpressed proteins identified and verified in this study display some of the cell functions that are altered in tumorous bile duct tissue. With serpin H1, we have identified a marker for the fibrotic activity of the tumor cells that leads to the production of high amounts of extracellular matrix. Overexpression of APOA4 points to alterations in lipid metabolism, and the enhanced proliferation that generally characterizes tumor cells was here confirmed by the up-regulation of STIP1 and SFN. The applicability of these proteins as biomarkers for CCC will be tested in future experiments.
We strongly suggest the consideration of STIP1 as part of a biomarker panel to support the histopathological diagnosis of CCC in indistinct situations. The applicability of SFN, serpin H1, APOA4, ABAT, BHMT, and FABP1 is indicated by the results presented here, although it requires further verification. Especially if only small biopsies are available, the distinction of CCC from reactive bile ducts can be challenging when performed on morphological grounds only. In contrast to CCC, reactive bile ductules are often found in liver biopsies showing chronic cholestasis due to intrahepatic or extrahepatic bile duct diseases or found adjacent to malignant tumors due to bile flow alteration. A biomarker panel comprising all or some of the candidates identified here could help in this matter. Thus, the combination of proteins specific for CCC cells and those specific for hepatocytes can be especially advantageous.
Supplementary Material
Acknowledgments
We thank Kristin Rosowski, Birgit Korte, Stephanie Tautges, Dorothe Möllmann, Laura Malkus, and Don Marvin Voss for their excellent technical assistance, as well as Julian Uszkoreit for the kind support in converting and uploading the proteomics data to the PRIDE repository.
Footnotes
Author contributions: J.P., D.A.M., T.B., A.E.C., H.A.B., and B.S. designed research; J.P. and H.R. performed research; J.P., D.A.M., T.B., H.R., M.A., M.K., A.E.C., H.E.M., and H.A.B. analyzed data; J.P., D.A.M., T.B., M.A., H.A.B., and B.S. wrote the paper; H.R. and H.A.B. performed pathological diagnosis; M.A., M.K., and M.E. performed statistical analysis; J.F.S., F.W., and A.H. performed sample collection and study design; K.K. performed optimization of mass spectrometry tools.
* The PROFILE project is co-funded by the European Union (European Regional Development Fund - Investing in Your Future) and the German federal state North Rhine-Westphalia (NRW), Project No. z0911bt004e. A part of this study was funded by P.U.R.E. (Protein Unit for Research in Europe), a project of North Rhine-Westphalia, a federal state of Germany.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CCC
- cholangiocellular carcinoma
- 2D-DIGE
- two-dimensional differential in-gel electrophoresis
- AUC
- area under the curve
- ABAT
- 4-aminobutyrate aminotransferase, mitochondrial
- APOA4
- apolipoprotein A-IV
- BHMT
- betaine-homocysteine S-methyltransferase 1
- CI
- confidence interval
- FABP1
- fatty acid–binding protein 1
- SFN
- 14-3-3 protein σ
- STIP1
- stress-induced phosphoprotein 1.
REFERENCES
- 1. Khan S. A., Davidson B. R., Goldin R., Pereira S. P., Rosenberg W. M., Taylor-Robinson S. D., Thillainayagam A. V., Thomas H. C., Thursz M. R., Wasan H., and British Society (2002) Guidelines for the diagnosis and treatment of cholangiocarcinoma: consensus document. Gut 51 Suppl 6, VI1–VI9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patel T. (2006) Cholangiocarcinoma. Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 33–42 [DOI] [PubMed] [Google Scholar]
- 3. Khan S. A., Taylor-Robinson S. D., Toledano M. B., Beck A., Elliott P., Thomas H. C. (2002) Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J. Hepatol. 37, 806–813 [DOI] [PubMed] [Google Scholar]
- 4. Lazaridis K. N., Gores G. J. (2005) Cholangiocarcinoma. Gastroenterology 128, 1655–1667 [DOI] [PubMed] [Google Scholar]
- 5. Khan S. A., Thomas H. C., Davidson B. R., Taylor-Robinson S. D. (2005) Cholangiocarcinoma. Lancet 366, 1303–1314 [DOI] [PubMed] [Google Scholar]
- 6. Riener M. O. (2011) [Diagnosis of tumours of the liver and the biliary tract: new tissue and serum markers]. Pathologe 32 Suppl 2, 304–309 [DOI] [PubMed] [Google Scholar]
- 7. Langer F., von Wasielewski R., Kreipe H. H. (2006) [The importance of immunohistochemistry for the diagnosis of cholangiocarcinomas]. Pathologe 27, 244–250 [DOI] [PubMed] [Google Scholar]
- 8. Balaton A. J., Nehama-Sibony M., Gotheil C., Callard P., Baviera E. E. (1988) Distinction between hepatocellular carcinoma, cholangiocarcinoma, and metastatic carcinoma based on immunohistochemical staining for carcinoembryonic antigen and for cytokeratin 19 on paraffin sections. J. Pathol. 156, 305–310 [DOI] [PubMed] [Google Scholar]
- 9. Sasaki M., Matsubara T., Kakuda Y., Sato Y., Nakanuma Y. (2014) Immunostaining for polycomb group protein EZH2 and senescent marker p16INK4a may be useful to differentiate cholangiolocellular carcinoma from ductular reaction and bile duct adenoma. Am. J. Surg. Pathol. 38, 364–369 [DOI] [PubMed] [Google Scholar]
- 10. Yonglitthipagon P., Pairojkul C., Bhudhisawasdi V., Mulvenna J., Loukas A., Sripa B. (2012) Proteomics-based identification of alpha-enolase as a potential prognostic marker in cholangiocarcinoma. Clin. Biochem. 45, 827–834 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Yonglitthipagon P., Pairojkul C., Chamgramol Y., Loukas A., Mulvenna J., Bethony J., Bhudhisawasdi V., Sripa B. (2012) Prognostic significance of peroxiredoxin 1 and ezrin-radixin-moesin-binding phosphoprotein 50 in cholangiocarcinoma. Hum. Pathol. 43, 1719–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srisomsap C., Sawangareetrakul P., Subhasitanont P., Panichakul T., Keeratichamroen S., Lirdprapamongkol K., Chokchaichamnankit D., Sirisinha S., Svasti J. (2004) Proteomic analysis of cholangiocarcinoma cell line. Proteomics 4, 1135–1144 [DOI] [PubMed] [Google Scholar]
- 13. Morofuji N., Ojima H., Onaya H., Okusaka T., Shimada K., Sakamoto Y., Esaki M., Nara S., Kosuge T., Asahina D., Ushigome M., Hiraoka N., Nagino M., Kondo T. (2012) Macrophage-capping protein as a tissue biomarker for prediction of response to gemcitabine treatment and prognosis in cholangiocarcinoma. J. Proteomics 75, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 14. Darby I. A., Vuillier-Devillers K., Pinault E., Sarrazy V., Lepreux S., Balabaud C., Bioulac-Sage P., Desmouliere A. (2010) Proteomic analysis of differentially expressed proteins in peripheral cholangiocarcinoma. Cancer Microenviron. 4, 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeon Y. E., Lee S. C., Paik S. S., Lee K. G., Jin S. Y., Kim H. R., Yoo C. W., Park H. M., Han S. Y., Choi D. H., Kim H. K. (2011) Histology-directed matrix-assisted laser desorption/ionization analysis reveals tissue origin and p53 status of primary liver cancers. Pathol. Int. 61, 449–455 [DOI] [PubMed] [Google Scholar]
- 16. Scarlett C. J., Saxby A. J., Nielsen A., Bell C., Samra J. S., Hugh T., Baxter R. C., Smith R. C. (2006) Proteomic profiling of cholangiocarcinoma: diagnostic potential of SELDI-TOF MS in malignant bile duct stricture. Hepatology 44, 658–666 [DOI] [PubMed] [Google Scholar]
- 17. Kawase H., Fujii K., Miyamoto M., Kubota K. C., Hirano S., Kondo S., Inagaki F. (2009) Differential LC-MS-based proteomics of surgical human cholangiocarcinoma tissues. J. Proteome Res. 8, 4092–4103 [DOI] [PubMed] [Google Scholar]
- 18. Megger D. A., Bracht T., Kohl M., Ahrens M., Naboulsi W., Weber F., Hoffmann A. C., Stephan C., Kuhlmann K., Eisenacher M., Schlaak J. F., Baba H. A., Meyer H. E., Sitek B. (2013) Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol. Cell. Proteomics 12, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poschmann G., Sitek B., Sipos B., Ulrich A., Wiese S., Stephan C., Warscheid B., Kloppel G., Vander Borght A., Ramaekers F. C., Meyer H. E., Stuhler K. (2009) Identification of proteomic differences between squamous cell carcinoma of the lung and bronchial epithelium. Mol. Cell. Proteomics 8, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57 (1), 289–300 [Google Scholar]
- 22. Sahadevan S., Hofmann-Apitius M., Schellander K., Tesfaye D., Fluck J., Friedrich C. M. (2012) Text mining in livestock animal science: introducing the potential of text mining to animal sciences. J. Anim. Sci. 90, 3666–3676 [DOI] [PubMed] [Google Scholar]
- 23. Remmele W., Stegner H. E. (1987) [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue]. Pathologe 8, 138–140 [PubMed] [Google Scholar]
- 24. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J. C., Muller M. (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeLong E. R., DeLong D. M., Clarke-Pearson D. L. (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 [PubMed] [Google Scholar]
- 26. Kohl M., Megger D. A., Trippler M., Meckel H., Ahrens M., Bracht T., Weber F., Hoffmann A. C., Baba H. A., Sitek B., Schlaak J. F., Meyer H. E., Stephan C., Eisenacher M. (2013) A practical data processing workflow for multi-OMICS projects. Biochim. Biophys. Acta 1844, 52–62 [DOI] [PubMed] [Google Scholar]
- 27. Blechacz B., Komuta M., Roskams T., Gores G. J. (2011) Clinical diagnosis and staging of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 8, 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Odunuga O. O., Longshaw V. M., Blatch G. L. (2004) Hop: more than an Hsp70/Hsp90 adaptor protein. Bioessays 26, 1058–1068 [DOI] [PubMed] [Google Scholar]
- 29. Chao A., Lai C. H., Tsai C. L., Hsueh S., Hsueh C., Lin C. Y., Chou H. H., Lin Y. J., Chen H. W., Chang T. C., Wang T. H. (2013) Tumor stress-induced phosphoprotein1 (STIP1) as a prognostic biomarker in ovarian cancer. PLoS One 8, e57084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsai C. L., Tsai C. N., Lin C. Y., Chen H. W., Lee Y. S., Chao A., Wang T. H., Wang H. S., Lai C. H. (2012) Secreted stress-induced phosphoprotein 1 activates the ALK2-SMAD signaling pathways and promotes cell proliferation of ovarian cancer cells. Cell Rep. 2, 283–293 [DOI] [PubMed] [Google Scholar]
- 31. Ferguson A. T., Evron E., Umbricht C. B., Pandita T. K., Chan T. A., Hermeking H., Marks J. R., Lambers A. R., Futreal P. A., Stampfer M. R., Sukumar S. (2000) High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwata N., Yamamoto H., Sasaki S., Itoh F., Suzuki H., Kikuchi T., Kaneto H., Iku S., Ozeki I., Karino Y., Satoh T., Toyota J., Satoh M., Endo T., Imai K. (2000) Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene 19, 5298–5302 [DOI] [PubMed] [Google Scholar]
- 33. Suzuki H., Itoh F., Toyota M., Kikuchi T., Kakiuchi H., Imai K. (2000) Inactivation of the 14-3-3 sigma gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 60, 4353–4357 [PubMed] [Google Scholar]
- 34. Osada H., Tatematsu Y., Yatabe Y., Nakagawa T., Konishi H., Harano T., Tezel E., Takada M., Takahashi T. (2002) Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers. Oncogene 21, 2418–2424 [DOI] [PubMed] [Google Scholar]
- 35. Kuroda Y., Aishima S., Taketomi A., Nishihara Y., Iguchi T., Taguchi K., Maehara Y., Tsuneyoshi M. (2007) 14-3-3sigma negatively regulates the cell cycle, and its down-regulation is associated with poor outcome in intrahepatic cholangiocarcinoma. Hum. Pathol. 38, 1014–1022 [DOI] [PubMed] [Google Scholar]
- 36. Hogan B. L., Barlow D. P., Kurkinen M. (1984) Reichert's membrane as a model for studying the biosynthesis and assembly of basement membrane components. Ciba Foundation Symposium 108, 60–74 [DOI] [PubMed] [Google Scholar]
- 37. Kurkinen M., Taylor A., Garrels J. I., Hogan B. L. (1984) Cell surface-associated proteins which bind native type IV collagen or gelatin. J. Biol. Chem. 259, 5915–5922 [PubMed] [Google Scholar]
- 38. Razzaque M. S., Taguchi T. (1997) Collagen-binding heat shock protein (HSP) 47 expression in anti-thymocyte serum (ATS)-induced glomerulonephritis. J. Pathol. 183, 24–29 [DOI] [PubMed] [Google Scholar]
- 39. Razzaque M. S., Nazneen A., Taguchi T. (1998) Immunolocalization of collagen and collagen-binding heat shock protein 47 in fibrotic lung diseases. Mod. Pathol. 11, 1183–1188 [PubMed] [Google Scholar]
- 40. Masuda H., Fukumoto M., Hirayoshi K., Nagata K. (1994) Coexpression of the collagen-binding stress protein HSP47 gene and the alpha 1(I) and alpha 1(III) collagen genes in carbon tetrachloride-induced rat liver fibrosis. J. Clin. Invest. 94, 2481–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kawada N., Kuroki T., Kobayashi K., Inoue M., Nakatani K., Kaneda K., Nagata K. (1996) Expression of heat-shock protein 47 in mouse liver. Cell Tissue Res. 284, 341–346 [DOI] [PubMed] [Google Scholar]
- 42. Sundaram M., Yao Z. (2012) Intrahepatic role of exchangeable apolipoproteins in lipoprotein assembly and secretion. Arterioscler. Thromb. Vasc. Biol. 32, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 43. Pelsers M. M., Morovat A., Alexander G. J., Hermens W. T., Trull A. K., Glatz J. F. (2002) Liver fatty acid-binding protein as a sensitive serum marker of acute hepatocellular damage in liver transplant recipients. Clin. Chem. 48, 2055–2057 [PubMed] [Google Scholar]
- 44. Pelsers M. M., Namiot Z., Kisielewski W., Namiot A., Januszkiewicz M., Hermens W. T., Glatz J. F. (2003) Intestinal-type and liver-type fatty acid-binding protein in the intestine. Tissue distribution and clinical utility. Clin. Biochem. 36, 529–535 [DOI] [PubMed] [Google Scholar]
- 45. Maatman R. G., Van Kuppevelt T. H., Veerkamp J. H. (1991) Two types of fatty acid-binding protein in human kidney. Isolation, characterization and localization. Biochem. J. 273 (Pt 3), 759–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Biase D., Barra D., Simmaco M., John R. A., Bossa F. (1995) Primary structure and tissue distribution of human 4-aminobutyrate aminotransferase. Eur. J. Biochem. 227, 476–480 [DOI] [PubMed] [Google Scholar]
- 47. Delgado-Reyes C. V., Wallig M. A., Garrow T. A. (2001) Immunohistochemical detection of betaine-homocysteine S-methyltransferase in human, pig, and rat liver and kidney. Arch. Biochem. Biophys. 393, 184–186 [DOI] [PubMed] [Google Scholar]
- 48. Garrow T. A. (1996) Purification, kinetic properties, and cDNA cloning of mammalian betaine-homocysteine methyltransferase. J. Biol. Chem. 271, 22831–22838 [DOI] [PubMed] [Google Scholar]
- 49. Avila M. A., Berasain C., Torres L., Martin-Duce A., Corrales F. J., Yang H., Prieto J., Lu S. C., Caballeria J., Rodes J., Mato J. M. (2000) Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 33, 907–914 [DOI] [PubMed] [Google Scholar]
- 50. Liang C. R., Leow C. K., Neo J. C., Tan G. S., Lo S. L., Lim J. W., Seow T. K., Lai P. B., Chung M. C. (2005) Proteome analysis of human hepatocellular carcinoma tissues by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5, 2258–2271 [DOI] [PubMed] [Google Scholar]
- 51. Sun W., Xing B., Sun Y., Du X., Lu M., Hao C., Lu Z., Mi W., Wu S., Wei H., Gao X., Zhu Y., Jiang Y., Qian X., He F. (2007) Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol. Cell. Proteomics 6, 1798–1808 [DOI] [PubMed] [Google Scholar]
- 52. Benjamini Y., Hochberg Y. (2000) On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25, 60–83 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.