Abstract
One of the critical gaps in malaria transmission biology and surveillance is our lack of knowledge about Plasmodium falciparum gametocyte biology, especially sexual dimorphic development and how sex ratios that may influence transmission from the human to the mosquito. Dissecting this process has been hampered by the lack of sex-specific protein markers for the circulating, mature stage V gametocytes. The current evidence suggests a high degree of conservation in gametocyte gene complement across Plasmodium, and therefore presumably for sex-specific genes as well. To better our understanding of gametocyte development and subsequent infectiousness to mosquitoes, we undertook a Systematic Subtractive Bioinformatic analysis (filtering) approach to identify sex-specific P. falciparum NF54 protein markers based on a comparison with the Dd2 strain, which is defective in producing males, and with syntenic male and female proteins from the reanalyzed and updated P. berghei (related rodent malaria parasite) gametocyte proteomes. This produced a short list of 174 male- and 258 female-enriched P. falciparum stage V proteins, some of which appear to be under strong diversifying selection, suggesting ongoing adaptation to mosquito vector species. We generated antibodies against three putative female-specific gametocyte stage V proteins in P. falciparum and confirmed either conserved sex-specificity or the lack of cross-species sex-partitioning. Finally, our study provides not only an additional resource for mass spectrometry-derived evidence for gametocyte proteins but also lays down the foundation for rational screening and development of novel sex-partitioned protein biomarkers and transmission-blocking vaccine candidates.
Sexual stages represent only a small fraction of Plasmodium falciparum parasites that are present during human malaria infection, yet they alone are responsible for disease transmission (1). As such, the Malaria Eradication Research Agenda (malERA) has prioritized the need for studies that specifically address these transmission stages, with the hope of developing new transmission-blocking vaccines and drugs, as well as diagnostics that are specific for these sexual stages (2–4). In fact, one of the critical gaps in malaria transmission biology and surveillance centers on the lack of knowledge about the infectivity of symptomatic and asymptomatic gametocytemic individuals for mosquitoes. Many infected individuals harboring the Plasmodium falciparum sexual stage, or gametocyte, are asymptomatic carriers and they represent the primary reservoir for malaria transmission (5). Missing the opportunity to treat these carriers will increase the risk for epidemic malaria in regions that have approached the elimination phase. Thus, proper surveillance of gametocyte carriers is critical for evaluating ongoing malaria control and elimination programs. Surveillance is difficult, however, because gametocytes comprise only 0.1–2% of the total body parasite load during active infection (5), and are only observed in the bloodstream in their mature (Stage V) form, with the first four developing stages sequestered in tissues. Microscopy-based analysis for sex ratio determination and infectivity studies remains limited because of cost, training, and suitability for population-wide studies. Although light microscopy remains the gold standard for malaria diagnosis, the relatively low prevalence of circulating gametocytes makes it difficult to accurately detect much less quantify these stages. Moreover, because of variations in skill level of microscopists and inconsistency in method, exclusive use of light microscopy estimates of gametocyte carriage carries a high risk of error. Importantly, the presence of stage V gametocytes in the bloodstream alone, as determined by thick smear microscopy does not imply infectivity to mosquitoes. Ratios of male and female gametocytes in the blood circulation are skewed toward the female, but they can vary significantly based on co-infection, parasite and gametocyte density, and host environmental factors (6), and it is therefore hypothesized that this variation in sex ratios will influence mosquito infectivity. For example, mature gametocyte sex ratios can change during the course of infection in response to host cues or especially following antimalarial treatment resulting in an increase in the number of males (6, 7). However, it remains unknown whether the transmission potential to mosquitoes of the individuals in these studies fluctuated because of the changes in sex ratio.
There are currently no uncomplicated tools to distinguish male and female mature P. falciparum gametocytes (of which at least one of each is required for fertilization and ookinete development in the mosquito) at the molecular level. Although the proteome of Plasmodium gametocytes has been described (8–11), these previous analyses fell just short of providing the partitioned male and female proteomes for P. falciparum. Moreover, the availability of the genomes of human, primate, and rodent malaria parasites and the acquisition of sequence information for recent field isolates of P. falciparum have created the opportunity to understand gene diversity and conservation in sexual stage development across Plasmodia. Identifying markers that differ between male and female P. falciparum stage V gametocytes is critical in informing transgenic approaches aimed at separating the two. It has been argued that the inherent evolutionary differences between rodent and human malaria parasites, especially for the sexual stages, limit the utility of the P. berghei gametocyte proteome (11) in providing a priori knowledge of these markers. Several iterations and improvements to the P. berghei genome have been made available since 2005, whereas MS search engines have improved commensurately, further compounding the issue. However, we would also argue that the current evidence suggests a high degree of conservation in gametocyte gene complement across Plasmodium (12, 13), and therefore presumably in sex-specific genes - despite key differences such as gametocyte sequestration and morphology. Here, we report on our effort to address these scientific gaps to a certain extent and to test our gametocyte gene conservation hypothesis through the use of comparative protein bioinformatics analyses of the mature stage V gametocyte proteomes of two distinct P. falciparum strains with our update of the bioinformatic analysis of the P. berghei male and female gametocyte proteomes.
MATERIALS AND METHODS
Parasite Culture and Gametocyte Isolation
P. falciparum NF54 Gametocyte Culture
P. falciparum gametocytes were cultured in RPMI 1640 containing HEPES and glutamine and supplemented with 10% human serum and hypoxanthine as described earlier (14). P. falciparum NF54 strain was diluted to 0.5% mixed stage asexual parasites and 4% hematocrit in complete culture medium in six well plates. Plates were transferred to a 37 °C incubator and microaerophilic environment was created using desiccators candle jar. Media was exchanged every day without the addition of new blood from day one to day 17 (culture maturation). To remove asexual stage parasites 50 mm N-acetylglucosamine was added to the culture media from day eight (early stage gametocytes) until day 10. Blood smears were made every alternate day to monitor the progress of the culture and to determine gametocytemia on day 18. Stage V gametocytes were harvested from culture at day 17 postgametocytogenesis initiation and isolated by passage through a LS-25 Midi MACS column (CS Miltenyi). Synchronized trophozoite cultures that were used for Western blot analyses were generated from a recently thawed stabilate and harvested at 48 h post treatment of the mixed asexual culture with 5% sorbitol.
P. falciparum Dd2 Gametocyte Culture
The production of stage V gametocytes was performed using a modified version of a previously described protocol (15). Ten (10) ml cultures at 4% hematocrit and ∼5% ring parasitemia were sorbitol synchronized. After 24 h, trophozoite cultures were transferred to a T75 flask to which complete media and red blood cells were added to create a 30 ml culture with 2% hematocrit in each flask. After another 24 h, adding 50% old media and 50% new complete media stressed the newly reinvaded rings. Cultures were allowed to develop to late schizonts and then split into three T75 flasks evenly. Twenty (20) mLs of fresh media was then added to each flask. During sexual stage development, fresh media was added daily. At 48 h after invasion of a mixture of asexually and sexually committed merozoites, 1 ml of 1 m N-Acetyl-d-Glucosamine was added to all flasks in order to clear asexual parasites. Drug treatment was given during media changes for three consecutive days. On day nine of sexual development, the cultures were MACS column separated to purify late stage gametocytes. Purified cultures were washed in PBS and snap frozen. Preparation of the synchronized trophozoite protein lysate for Western blot analyses is as described above for NF54.
Protein Extraction
The GiRBC1 eluate from the MACS column was washed with cold PBS three times prior to protein extraction. The freeze-thaw method was applied to extract the soluble proteins by adding 120 μl 5 mm phosphate buffer containing 0.5 mm PMSF, 1 mm EDTA, and 1 mm protease inhibitors mixture (Sigma, St. Louis, MO) to 1 × 106 GiRBC pellets. A total of four freeze-thaw cycles were used. The supernatant was collected as the soluble protein fraction after centrifugation at 20,000 × g for 30 min at 4 °C. To get the membrane proteins, the pellet was washed with cold PBS for three times prior to being dissolved in 95 μl SDT-lysis buffer composed of 4% (w/v) SDS, 100 mm Tris/HCl, and 0.1 m DTT, pH 7.6, and then boiled at 95 °C for 5 min. The supernatant was collected as the membrane protein fraction after centrifugation at 20,000 × g for 5 min at 4 °C.
Multi-Lane Combined In-gel Digestion (MLCID)
We used a Multi-Lane Combined In-gel Digestion (MLCID) strategy to reduce the impact of nonspecific absorption during the process of in-gel tryptic digestion and to avoid losing SDS-PAGE separation power. For NF54 parasites, we used three lanes for the soluble protein fraction and four lanes for the membrane fraction, respectively, and each lane was loaded with 20 μl of sample under reducing conditions. After resolving on a 4–20% precast gradient gel (BioRad, Hercules, CA), the proteins were stained with Coomassie. GiRBC soluble and membrane fractions were cut into 14 slices by combining three lanes (soluble) and 16 slices by combining four lanes (membrane). Both the soluble and membrane fractions from Dd2 were cut into 14 slices by combing three lanes. Gel slices were cut into 1 × 1 mm pieces prior to de-staining, reduction and alkylation, tryptic digestion and peptide extraction. The extracted peptides were lyophilized and then resuspended in 2% acetonitrile, 97.9% water, and 0.1% formic acid buffer for LC-MS/MS analysis.
LC-MS/MS
Biological in-gel digestion replicates were analyzed independently as follows. One third of the MLCID sample of all the fractions, were injected onto an Agilent LC-MS system comprised of a 1200 LC system coupled to a 6520 Q-TOF via an HPLC Chip Cube interface. The only exception to this process was made for the first low molecular weight fraction, which consisted primarily of hemoglobin, and only 1/50th of this fraction was injected. The sample was trapped and analyzed using an Agilent Polaris-HR-Chip-3C18 chip (360 nL, 180 Å C18 trap with a 75 μm i.d., 150 mm length, 180 Å C18 analytical column). Peptides were loaded onto the enrichment column automatically by autosampler using 97% solvent A (0.1% formic acid in water) and 3% solvent B (0.1% formic acid in 90% acetonitrile) at a flow rate of 2 μl/min. Elution of peptides from the analytical column was performed using a gradient starting at 97% A at 300 nL/min. The mobile phase was 3–10% B for 4 min, 10–35% B for 56 min, 35–99% for 2 min, and maintained at 99% B for 6 min, followed by re-equilibration of the column with 3% B for 10 min. Data dependent (autoMS2) mode was used for MS acquisition by Agilent 6520 Q-TOF at 2 GHz. Precursor MS spectra were acquired from m/z 315 to 1700 and the top 4 peaks were selected for MS/MS analysis. Product scans were acquired from m/z 50 to 1700 at a scan rate of 1.5/second. A medium isolation width (∼4 amu) was used, and a collision energy of slope 3.9 V/100 Da with a 2.9 V offset was applied for fragmentation. A dynamic exclusion list was applied, with precursors excluded of 0.50 min after two MS/MS spectrum was acquired.
Mass Spectrometry Data Search and Analysis
Each sample was further fractionated into 14 membrane and 14 soluble fractions. Raw data from Dd2 sample runs (two biological replicates, 217,165 MS/MS total spectra) and NF54 GiRBC sample runs (three biological replicates, 497,006 MS/MS total spectra) was converted to mzXML format using Trapper (Institute for Systems Biology, Seattle, Washington). A merged search was performed on the mzXML data for each fraction using the PepArML metasearch engine (16), which automatically conducts target and decoy searches using the following: Mascot (17), OMSSA (18), and Tandem (19) with native, K-score (20) and S-score pluggable scoring modules (21), and Inspect (22) with MS-GF spectral probability scoring (23). The results were then combined using an unsupervised machine-learning strategy, and the peptide identification false discovery rates (FDR) were estimated using identifications from the reversed decoy searches (24).
The data was searched by a combined database of SwissProt Human and Plasmodium falciparum sequences from GeneDB (2013.02), which consists of 28,960 entries with the following parameters; fixed modification: carbamidomethyl cysteine and variable modification: oxidized methionine; mass tolerance: 30 ppm and 20 ppm respectively for precursor and fragment ions; one missed cleavage. The results from the metasearch were combined and the results were parsed into the MASPECTRAS 2 data analysis system (25) with data filters of 1% spectra FDR and 5% peptide FDR, and protein identifications were then clustered to remove redundancy. Proteins were clustered together if there was a peptide identification shared between them, because this indicates substantial sequence similarity, and the protein with the greatest number of peptides identified was considered the unique protein identification from that group. Throughout this paper we report only the unique identifications. Proteins identified by single peptides were manually validated. The data analysis pipeline meets all MIAPE standards (26) and the proteomics data have been deposited in the ProteomeExchange via the PRotein IDEntifications database (PRIDE) partner repository with the data set identifier PXD000813 (27). The protein lists have also been uploaded to PlasmoDB (plasmodb.org).
For the reanalysis of the Khan et al. data set (11), the individual MS raw files from Male (113,213 total MS/MS spectra) and Female data set (243,468 total MS/MS spectra) were searched against a combined database of SwissProt Human, Mouse, and P. berghei. Using these results we determined the male/female partitioned proteomes for P. falciparum gametocytes through a subtractive bioinformatics proteomics approach. Briefly, in our approach, we take protein identification lists and use set comparisons to generate protein lists that are enriched for biological states, with those protein lists clustered to remove redundancy. Therefore, we took the NF54 and Dd2 gametocyte-infected red blood cell lysate proteome and subtracted out all host proteins, generating the NF54 and Dd2 gametocyte proteomes. Putative male-specific, female-specific, and sex-unspecific proteomes were generated by taking protein identifications unique to NF54 and Dd2, respectively. These putative proteomes were then BLAST searched against the two previous data sets of Khan et al. and Silvestrini et al. (8, 11).
Identified proteins were annotated by GeneDB (02, 2013); specifically, the Gene Ontology database was searched by BLAST homology for annotations. The surface expressed (S.E.) proteins were predicted by searching for canonical signal peptides with the SignalP 4.1 Server (28). Transmembrane domain information was obtained on all identified proteins by the transmembrane protein prediction tool TMHMM Server v. 2.0 (29).
Analysis of Diversity and Divergence for Male- and Female-enriched P. falciparum Gametocyte Proteins
The SNP πN diversity statistic, representing mean pairwise nonsynonymous SNP diversity per site, was calculated for previously generated data within each genic region for a given population using the VCFtools-site-pi utility (30). For each pair of populations, the Fst divergence statistic was calculated for each gene with the VCFtools implementation of Fst and weighted Fst estimators as described in Weir and Cockerham (31).
Expression of Recombinant Proteins and Generation of Polyclonal Antibodies for Proteomic Validation
The selection of predicted immunogenic domains for each protein were based on physiochemical properties of each gene (PF3D7_0906100; PF3D7_1218800; PF3D7_0309100; and PF3D7_0422000) using the Bcepred server (32) and Immune epitope Database (IEDP) (33). Each codon-optimized gene or gene fragment (GenScript) was used as a template for PCR along with the following primer sets (all 5′ to 3′): PF3D7_0906100, F-CACCATGGGTAACAAAATTAGC, R-TTTCAGGTTTTTGATACGTTCC; PF3D7_1218800, F-CACCAAAATCGTGCTGTCCA, R-ACCGAAGTAAATAAAACTCGGTTC; PF3D7_0309100, F-CACCGACCTGAGCGGCCT, R-CAGTTCTTCGTTTTTGATGAACACG. Each 20 μl PCR reaction consisted of 0.25 μl of DNA (200 ng/μl), 4.0 μl of 5x iProof DNA polymerase buffer, 0.4 μl of dNTPs (10 mm each), 0.4 μl each of forward and reverse primers (10 μm), 0.1 μl of iProof DNA polymerase (2 U/μl), and 14.85 μl of sterile deionized water. Reaction conditions were 98 °C for 2 min, followed by 40 cycles of 98 °C for 15 s, 62 °C for 25 s, and 72 °C for 25 s. All forward primers were appended with the nucleotides CACC on the 5′ end to facilitate directional insertion of each amplicon into the E. coli expression vector pBAD202/d-TOPO. Ligation and transformation steps were carried out according to the manufacturer's protocols, and clones were grown on selective LB agar plates with kanamycin (50 μg/ml) overnight at 37 °C. For each gene, colonies were picked and screened by PCR and then sequenced to confirm proper orientation and reading frame. Prior to induction, positive clones were grown in LB media + kanamycin (50 μg/ml) overnight at 37 °C in a shaking incubator (200 RPM). Each overnight culture was then used to seed 50 ml of fresh LB + kanamycin (50 μg/ml), grown to an O.D.600 ≈ 0.4, and then induced with arabinose (0.01%) for 6 - 8 h at 37 °C. Cells were harvested by centrifugation at 10,000 × g for 10 min, and the presence of recombinant protein from each expression was confirmed by Western blot using mouse anti-His monoclonal Ab (Sigma). The cell pellets were then processed for recombinant protein using BugBuster Reagent (EMD Millipore, Billerica, MA) following the manufacturer's protocol for both soluble and insoluble fractions. Recombinant protein was then purified by immobilized metal affinity chromatography (ProBond, Invitrogen, Carlsbad, CA) following the manufacturer's “hybrid” protocol for inclusion bodies and the “native” protocol for soluble protein. Following elution from the column, eluates positive for protein by Western blot were pooled and dialyzed overnight (3500–10,000 Da MWCO) against imadozole-free elution buffer and then concentrated using diafiltration (3000 Da MWCO).
To generate polyclonal antibodies, Swiss Webster mice were immunized with purified recombinant protein emulsified with incomplete Freund's adjuvant (Sigma) following a prime and three boosts at two week intervals. The mice were exsanguinated at the end of the immunization regimen to collect serum.
Immunofluorescence Microscopy Assays
P. falciparum NF54 gametocyte and trophozoite samples were fixed with 4% paraformaldehyde/0.0075% glutaraldehyde and prepared for fluorescence microscopy by washing three times with PBS. The cells were permeabilized with 0.2% Triton-X 100/PBS for 10 min and then washed as before. After washing, samples were blocked with 3% BSA in PBS overnight at 4 °C. The samples were then incubated with mouse antigametocyte protein serum (1:50) for 1 h at RT. Cells were washed with PBS as before and detected with Alexa Fluor(R) 488 Goat Anti-mouse IgG (H+L), highly cross-adsorbed (Molecular Probes®, 1:1000) in 0.02% Evans blue for 1 h at RT. Following incubation, the cells were washed three times with PBS, resuspended in PBS, spotted on slides and allowed to air dry. Samples were mounted using Slow Fade Gold antifade reagent with DAPI (Molecular Probes, Eugene, OR) or Aqua Poly/mount (Polysciences Inc., Warrington, PA). Samples were imaged using a Nikon Upright E800 microscope equipped with SPOT camera and software and a Nikon 90i light microscope (Nikon Corp., Tokyo, Japan) connected to a Hamamatsu ORCA high sensitivity monochrome CCD camera.
RESULTS
The Plasmodium falciparum Stage V Gametocyte Proteome: NF54 versus Dd2
We selected two P. falciparum strains for proteomic analysis of stage V gametocytes: a transmission-competent reference isolate, NF54 (West Africa) (34) and a Southeast Asian clone Dd2 (35). Importantly, Dd2 has a defect in male gametocyte development resulting in arrested male stages of rectangular and teardrop shape that are usually not found in culture. The genetic basis of this defect is at least in part a mutation in the male development 1 (MDV-1) gene (36). Assuming that the stage V proteome of Dd2 will be enriched for female proteins, we can then characterize the male- and female-enriched P. falciparum proteomes by comparative analysis of Dd2 and NF54.
We produced three biological replicates of P. falciparum (NF54 isolate) stage V gametocytes in the presence of GlcNAc to block asexual replication at the onset of gametocytogenesis and therefore ensure pure Stage V gametocyte populations at the day of collection, that is, 17 days post initiation of gametocytogenesis. Microscopic analysis of thin-blood smears from the day 17 culture suggested a predominantly stage V culture (∼6–7% gametocytemia), with few stage IV gametocytes and a sex ratio of ∼1:4 for male versus female (Fig. 1A, Table I). In parallel, we cultured the P. falciparum Dd2 mutant clone and induced gametocytogenesis in the presence of GlcNAc to produce two biological replicates of preferentially female-enriched stage V (day 17) gametocyte samples. We determined by microscopy the number of prestage V gametocytes in two replicate cultures with low and high gametocytemias to be 14.5% and 27.3%, respectively (Table II). The proportion of male gametocytes that are arrested developmentally and with altered morphology (35), were 0.6% and 1.2%, respectively, for the teardrop forms and 0.4% and 1.6%, respectively, for the rectangular forms. Although far fewer in number these arrested male stages 1–4 were considered a potential source of contaminating male proteins.
Fig. 1.
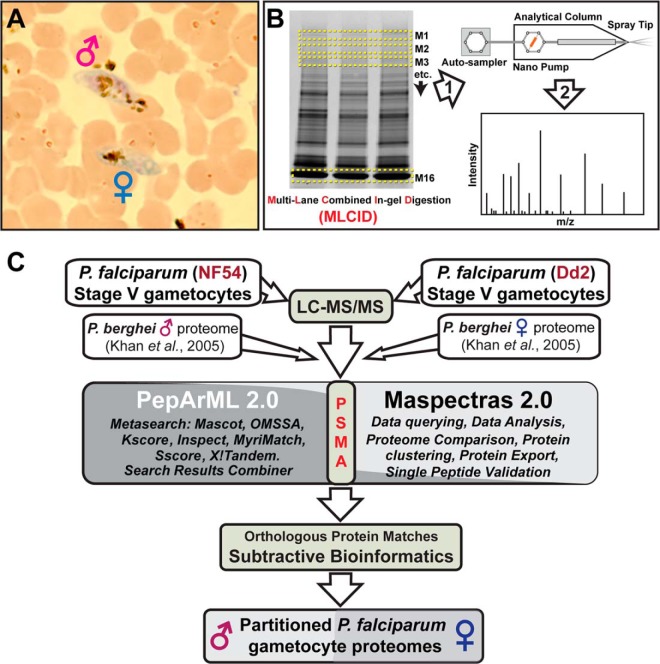
Proteomic and bioinformatic strategy to sex-partition Plasmodium falciparum stage V gametocyte proteins. A, Gametocyte sex ratios were determined using differential staining and microscopy. Improved R66 Giemsa is diluted 1:40 in Sorensen buffer (pH 7.2) and used to stain a methanol-fixed, thin smear of stage V (mature) gametocytes from a gametocyte culture. Gametocytes can be differentiated according to five classical parameters (35, 55) as well as by differential Giemsa staining patterns. Males (M) appear pink with no distinct nucleus and females (F) appear blue with a distinct nucleus. B, Proteomics workflow. Soluble and membrane proteins were extracted for Multi-Lane Combined In-gel Digestion (MLCID, see methods) prior to Chip-based nano-HPLC MS/MS analysis. C, Bioinformatics workflow. LC MS/MS data from NF54 and Dd2 is searched on PepArML meta search engine with six search engines. P. berghei MS/MS male proteome and MS/MS female proteome is also uploaded to PepArML and Maspectras2. The result is combined in PepArML by unsupervised machine learning and stored in Maspectras2 and queried for analysis. Proteome results are then compared with the P. berghei MS/MS male proteome and MS/MS female proteome as shown (refer to supplemental Fig. S1 for additional detail). This comparison is done by orthologous protein matches and a subtractive bioinformatics approach (see Methods), producing partitioned proteomes for male and female gametocytes.
Table I. Microscopic verification of the Stage V gametocyte male-to-female ratio for the Plasmodium falciparum NF54 used in this study. Several thin-smear slides taken from each N-acetylglucosamine (GlcNAc) treated culture of NF54 used for MS analysis was examined by oil-immersion microscopy (1000X) at day 17 post-initiation of gametocytogenesis by 1/40 dilution of Giemsa in Sorensen's buffer. Reads 1–3 correspond to biological replicates.
| Read 1 | Read 2 | Read 3 | Pooled | |
|---|---|---|---|---|
| Total # RBCs counted | 1204 | 1212 | 1130 | 3546 |
| Number of males (pink) | 16 | 15 | 17 | 48 |
| Number of females (blue) | 88 | 58 | 40 | 186 |
| Total # gametocytes (Stage V) | 104 | 73 | 57 | 234 |
| Gametocytemia (Stage V) | 0.09 | 0.06 | 0.05 | 0.07 |
| Male/Female Ratio | 0.18 | 0.26 | 0.43 | 0.26 |
Table II. Microscopic verification of purified Pre- and Mature Stage V gametocytes for the Plasmodium falciparum Dd2 line used in this study. The Dd2 clone has a defect in male gametocyte development (35). However, the presence of Pre-Stage V gametocytes would complicate the partitioning of Dd2 proteins because male-specific markers in P. falciparum, for example, alpha-tubulin II are expressed in pre-stage V females. In addition to determining the number of prestage V's we also ascertained the number of “tear drop” and “rectangular” forms, which have been hypothesized to be malformed male gametocytes (35). The contamination level with prestage V as well as these malformed parasites helped explain our Dd2 gametocyte proteome.
| Gametocytemia |
||
|---|---|---|
| HIGH | LOW | |
| Total # RBCs counted | 536 | 758 |
| Number of stage V (female) | 354 | 425 |
| Number of prestage V | 138 | 73 |
| Number of arrested male: Rectangular | 8 | 2 |
| Number of arrested male: Teardrop | 6 | 3 |
| Total # Gametocytes | 506 | 503 |
| Gametocytemia (%) | 94.4 | 66.3 |
We demonstrated highly consistent acquisition of the mixed-sex stage V gametocyte proteome (Fig. 2A, supplemental Table S1), with greater than 95% reproducibility in protein identifications across biological replicates (Fig. 2A–2B). We identified 1703 mature NF54 stage V proteins, including all the known and well-studied gametocyte surface markers such as P230, P48/45, P47, and the LCCL domain-containing (CCp) protein family of molecules [Table I, supplemental Table S1A; reviewed in reference (37)]. Of these, 449 (26%) proteins were consistently and exclusively identified in the membrane fraction of each sample. In parallel, a total of 1337 proteins were identified reproducibly from the two Dd2 biological replicates (Table II, supplemental Table S1B). Of these, 726 (54%) proteins were identified consistently and exclusively in the membrane fraction of each sample. For both NF54 and Dd2 it remains unclear if these proteins localize to the parasite plasmalemma or RBC membrane. We observed that NF54 and Dd2 have 1105 proteins in common (supplemental Table S1C), 598 NF54-enriched proteins (supplemental Table S1D), and 232 Dd2-enriched proteins (supplemental Table S1E), which may include proteins emanating from the malformed, arrested male gametocytes (Table II). We compared our NF54 gametocyte stage V proteome (1703 proteins), which will be referred to in the text as DTgV, with the published P. falciparum 3D7/NF54 gametocyte stage V proteomes, gametocyte stage I-II and trophozoite protein lists (8), the data sets of which we refer to in our study here on as FSgV (2031 proteins, supplemental Table S1F), FSgI-II (1427 proteins, supplemental Table S1G), and FS Asx (1345 proteins, supplemental Table S1H), respectively. For these three data sets, we converted the previous and now obsolete accession numbers for each protein to the new protein identifiers as described in the P. falciparum 3D7 databases in GeneDB (Version 3) and PlasmoDB (Version 9.3. 11 Mar 11, 2013). The NF54 stage V proteome was comparable to the 3D7/NF54 gametocyte stage V proteome (8), with 1274 proteins found to be in common between the two NF54 data sets. We also noted the addition of 429 NF54 proteins to the gametocyte proteome (Fig. 2D). Therefore, we conclude that these two data sets represent the most complete P. falciparum NF54 stage V-enriched gametocyte proteome to date. The Dd2 stage V proteome was also comparable to the FSgV proteome, with 1106 proteins in common (Fig. 2E). Similar to the results with our NF54 stage V comparison, we noted that ∼230 Dd2-enriched proteins had partitioned from FSgV. The acquisition of the NF54 and Dd2 stage V gametocyte proteomes (summary MS statistics are provided in Table III) represents the first step in our Systematic Subtractive Protein Bioinformatics analysis (SSB) approach, which is outlined in Fig. 1C.
Fig. 2.
Summary of protein identifications from the reanalysis of Plasmodium berghei ANKA 2. Summary of protein identifications from the reanalysis of Plasmodium berghei ANKA 2.34 Male and Female Gametocyte Proteomes and the current Stage V Plasmodium falciparum gametocyte proteomes for the NF54 isolate and Dd2 strain. Reproducibility of the analyses of the P. falciparum NF54 samples is shown. Three biological replicates for P. falciparum NF54 preparations were analyzed by MS. The 1090 proteins identified from analysis of all replicates were considered for statistical analysis. A, Peptides per identified protein for each biological replicate B, and spectra per identified protein for each biological replicate. Normalized peptide count and spectral count were analyzed by one-way ANOVA with Geisser-Greenhouse correction method. Average values are plotted with error bars of 95% CI. C, P. falciparum NF54 and Dd2 protein identification. There were 1105 proteins in common between 1703 Pf NF54 and 1337 Pf Dd2 total protein identification at 1% spectra FDR (minimum one unique peptide for a protein, see methods for single peptide validation details) and 5% peptide FDR for a protein. We observed that 598 proteins partitioned to the NF54 samples and 232 proteins to the Dd2samples. D–E, Proteins found in common between the NF54 orDd2 protein lists and the FSgV data set (8). We observed that 74% NF54 and 82% of the Dd2 proteins were proteins were present in the Silvestrini gametocyte stage V data set (8). F, Male and Female P. berghei protein identification following the PSMA re-analysis of the published data set (11). Out of 1546 total unique protein identification 762 proteins were in common between P. berghei PSMA Male and P. berghei PSMA Female at 1% spectra FDR (minimum one unique peptide for a protein, see methods for single peptide validation details) and 5% peptide FDR for a protein. We found that 366 proteins were specific for male P. berghei and 418 proteins were P. berghei female-specific. G, A comparison of PSMA Male with the original ANKA-Male protein list found only 136 proteins in common between the two data sets. H, A comparison of PSMA Female with the original ANKA-Female protein list found only 70 proteins in common between the two data sets.
Table III. Mass spectrometry data summary statistics for P. falciparum NF54 and Dd2 stage V gametocytes. *783 proteins are shared between membrane and soluble NF54 proteins and *621 proteins are shared between Dd2 membrane and soluble proteins. **1105 proteins are shared between Dd2 and NF54 proteins.
| Strain | Total # spectra | # Unique spectra | Unique proteins identified |
|---|---|---|---|
| P. falciparum NF54 | |||
| Membrane fraction | 97,128 | 96,167 | 1455 |
| Soluble fraction | 87,308 | 86,390 | 1031 |
| Total NF54 | 184,436 | 182,557 | 1703* |
| P. falciparum Dd2 | |||
| Membrane fraction | 80,127 | 36,064 | 1065 |
| Soluble fraction | 77,364 | 49,184 | 893 |
| Total Dd2 | 157,491 | 85,248 | 1337* |
| Combined total | 341,927 | 267,805 | 1935** |
Re-analysis of the Rodent Malaria Sex-specific Proteomes
To define and characterize the subset of conserved male and female Plasmodium gametocyte markers, we sought to compare the Dd2 and NF54 data with the available male and female proteomes from the rodent malaria parasite, P. berghei ANKA 2.34 (11) (see Fig. 1C for the strategy). However, given the number of iterations of the P. berghei genome since 2005, as well as further refinement of MS search engines capabilities, we first performed a PepArML-Search MASPECTRAS2 Analysis (PSMA) of the original peptide spectral data from that study, to allow for appropriate and updated comparisons of protein identities (25, 38).
Our re-analysis resulted in updated P. berghei male (1128 clustered proteins) and female (1180 clustered proteins) gametocyte proteomes (Table IV, supplemental Table S2A, 2B). From these two data sets we identified 762 clustered proteins (Fig. 2F) that were common between male and female gametocytes, as compared with the 51 shared proteins reported between berghei males and females (11). The re-analyzed (PSMA) sex-specific protein lists increased from 278 to 366 male proteins (Fig. 2G, supplemental Table S2D) and from 171 to 418 female proteins (Fig. 2H, supplemental Table S2E). We found that only 136 proteins were conserved between the original and current male-specific protein lists (Table 2F) and 70 proteins between the original and current Female-specific protein lists (Table 2G). Taken altogether, the combined data sets represent the most complete ANKA sex-specific gametocyte proteomes to date.
Table IV. Comparative mass spectrometry data summary statistics. Summary of the re-analyses of the Plasmodium berghei ANKA 2.34 Male and Female Gametocyte Proteomes using an updated P. berghei genome database (version 2013–01) and the PepArML Search-MASPECTRAS2 Analysis (PSMA) platform (25, 38).
| Total # spectra | # Unique peptide | Unique proteins identified | |
|---|---|---|---|
| P. berghei MALE | |||
| Previousa | – | – | 650 |
| Currentb | 30,786 | 9389 | 1128 |
| P. berghei FEMALE | |||
| Previous | – | – | 541 |
| Current | 41,719 | 9465 | 1180 |
| MALE enriched | |||
| Previous | – | 698 | 278 |
| Current | 5076 | 2158 | 366 |
| FEMALE enriched | |||
| Previous | – | 216 | 171 |
| Current | 3405 | 1359 | 418 |
| Shared Male/Female | |||
| Previous | – | 758 | 69 |
| Current | 64,024 | 10,407 | 762 |
a Search engine used in previous analyses: Mascot (11).
b Search engine used in current analyses: Mascot, OMSSA, X!Tandem, Kscore, Sscore, Inspect and MyriMatch.
Apart from the increased resolution of the data set through the use of the PSMA approach, we also noted that seven proteins that were described previously to be male-specific (11), actually partitioned to the PSMA female-specific protein list and that two proteins that were reported initially to be female-specific were now found to be in the PSMA male-specific list (Table V, supplemental Table S2H). We also observed that several proteins were putatively shared between males and females in the current PSMA analyses with differential enrichments in one sex over the other (Table VI, supplemental Table S2I).
Table V. A PSMA reanalysis of the existing Plasmodium berghei male and female gametocyte proteome using the current iteration of the genome revealed significant changes in the assignment of male and female proteins.
| Acc. number | Protein description | Previousα |
Currentβ |
||
|---|---|---|---|---|---|
| M | F | M | F | ||
| PBANKA_050450 | Cytoplasmic dynein intermediate chain, putative | • | • | ||
| PBANKA_080230 | Ubiquitin transferase, putative | • | • | ||
| PBANKA_090620 | Conserved Plasmodium protein, unknown function | • | • | ||
| PBANKA_093990 | U6 snRNA-associated Sm-like protein LSm4, putative (LSM4) | • | • | ||
| PBANKA_103870 | Conserved Plasmodium protein, unknown function | • | • | ||
| PBANKA_113640 | Conserved Plasmodium protein, unknown function | • | • | ||
| PBANKA_120490 | Conserved Plasmodium protein, unknown function | • | • | ||
| PBANKA_123400 | Vacuolar ATP synthetase, putative | • | • | ||
| PBANKA_141440 | Exportin-T, putative | • | • | ||
Table VI. A PSMA reanalysis of the existing Plasmodium berghei male and female gametocyte proteome using the current iteration of the genome revealed that many sex-enriched proteins were shared between males and females. We examined the relative enrichment of unique peptides in the respective male and female batches from the original study to determine the degree of expression of these shared proteins.
| Batch data |
Accession number | Protein description | Seq.Cov. (%) | Score | Nr. proteins | Nr. peptides | Nr. spectra |
Pb Male (M) |
Pb Female (F) |
M/F ratio | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M2 | M3 | F1 | F3 | Nr. peptides | Nr. spectra | Nr. peptides | Nr. spectra | ||||||||
| • | • | • | • | PBANKA_021400 | Dynein heavy chain, putative | 13.39 | 996 | 1 | 68 | 130 | 1 | 4 | 67 | 126 | 0.015 |
| • | • | • | • | PBANKA_040150 | Exportin 1, putative | 9.56 | 137 | 1 | 10 | 20 | 4 | 7 | 7 | 13 | 0.575 |
| • | • | • | PBANKA_050430 | Flagellar outer arm dynein-associated protein, putative | 48.52 | 80 | 1 | 3 | 12 | 3 | 11 | 1 | 1 | 3 | |
| • | • | • | • | PBANKA_050730 | Dynein heavy chain, putative | 26.85 | 2556 | 1 | 150 | 373 | 149 | 371 | 2 | 2 | 74.5 |
| • | • | • | • | PBANKA_061400 | Conserved Plasmodium protein, unknown function | 4.61 | 51 | 1 | 2 | 12 | 1 | 7 | 2 | 5 | 0.5 |
| • | • | • | • | PBANKA_091740 | Armadillo repeat protein PF16 | 29.47 | 298 | 1 | 15 | 60 | 15 | 51 | 6 | 9 | 2.5 |
| • | • | • | • | PBANKA_113320 | cdc2-related kinase 2 | 19.8 | 65 | 1 | 5 | 8 | 4 | 5 | 2 | 3 | 2 |
| • | • | • | PBANKA_130070 | CCp1/LCCL domain-containing protein | 43.66 | 1185 | 1 | 63 | 292 | 1 | 1 | 63 | 291 | 0.016 | |
| • | • | • | PBANKA_135960 | 6-cysteine protein P48/45 | 29.9 | 250 | 1 | 15 | 38 | 13 | 31 | 4 | 7 | 3.25 | |
| • | • | PBANKA_142440 | CS domain protein, putative | 16.3 | 95 | 1 | 5 | 6 | 2 | 2 | 7 | 13 | 0.286 | ||
| • | • | • | • | PBANKA_143220 | Male development gene 1 | 50.47 | 320 | 1 | 16 | 129 | 12 | 43 | 15 | 86 | 0.8 |
| • | • | • | PBANKA_145880 | Kinesin, putative | 26.88 | 519 | 1 | 37 | 71 | 36 | 70 | 1 | 1 | 36 | |
| • | • | • | • | PBANKA_146300 | Osmiophilic body protein | 59.41 | 4011 | 1 | 186 | 1804 | 7 | 9 | 186 | 1795 | 0.038 |
Importantly, two proteins that were highlighted [cf. original Table I in (11)] as male-specific based on separation of gametocytes expressing GFP under the control of a sex-specific promoter, the Dynein heavy chain (PBANKA_092540) and Dynein heavy chain (PBANKA_041610) retained the same partitioning (supplemental Table S2A-E). The 6-cysteine protein, P230p (PBANKA_030600), was described previously to be male-specific and we find this to hold true following PSMA re-analysis as well. There were two proteins, the transmission-blocking vaccine candidate, P48/45 (PBANKA_135960) and the male development gene 1 (PBANKA_143220), which were not reported previously in the original proteomic study of P. berghei gametocytes, but following PSMA, we noted that these two previously described male-specific proteins (39, 40) were indeed present in the original raw data set and in our reanalysis did not exhibit sex-specific partitioning. Further interrogation of the data suggests that P48/45 appear to be enriched (3.25-fold based on spectral counts) in P. berghei males as opposed to females (supplemental Table S6). Interestingly, despite its name, male development gene 1 (MDV1) appeared to be less enriched in males as opposed to females based on our PSMA reanalysis, which is consistent with the report by Lal et al., (2009), wherein it was shown that MDV1 is in both males and females but is particularly important for female gametocyte development in the rodent malaria parasite (41). Although it was shown recently that the egress of male gametes is partly dependent on MDV1 (42). We also noted that a dynein heavy chain protein (PBANKA_050730) that did not partition to either sex appears to have a 74.5-fold enrichment in males. Conversely, we also determined that another dynein heavy chain protein (PBANKA_021400) appears to be enriched 67-fold in females. A kinesin (PBANKA_145880), which has not been fully described to date, was enriched 36-fold in the male fraction. The CCp1/LCCL1 domain containing protein (PBANKA_130070), which was hypothesized to be female-specific (37, 43), was also found in both male and female fractions, but was 63-fold enriched in the male fraction. The osmiophilic body protein (PBANKA_146300), a putative female-specific protein was found in both male and female fractions and is 27-fold enriched in the female fraction. These sex-specific enrichments are significant and support the notion that while MS sensitivity enables their detection in the fraction, these fractions are likely unable to identify completely male- or female-specific proteins. However, evidence from the literature is derived from both P. berghei and P. falciparum studies, and it remains to be seen whether falciparum-specific expression and sex-partitioning mirrors that of P. berghei.
We anticipated differential partitioning following PSMA, because we had also observed that a direct comparison of the Pb PSMA lists with the published Pb sex-specific lists resulted in only 136 shared male-proteins (Fig. 2E and supplemental Table S2H) and only 70 shared female-proteins (Fig. 2H and supplemental Table S2I). Moreover, we observed that there are 100 proteins from the original male-specific (ANKA-M) data set (278 proteins) that are also found in the PSMA conserved male and female proteins (762 proteins). We could therefore account for only 178 male proteins of the original 278 male-specific proteins. In addition to the 70 shared female-specific proteins, we found that 85 proteins from the original female-specific (ANKA-F) data set were also in the PSMA conserved male and female protein list (762 proteins). We could therefore account for only 86 female-enriched proteins of the original 171 female-specific proteins. Altogether, re-analysis of the available P. berghei mature gametocyte proteome data by PSMA has significantly deepened the analysis of this data set. We used this new annotation for comparison with the P. falciparum NF54/Dd2 stage V proteome data.
Systematic Subtractive Protein Bioinformatics Analysis (SSB) to Partition Male and Female Stage V Gametocyte Proteomes
To define the male and female proteomes of P. falciparum stage V gametocytes we employed a SSB work-flow using the re-analyzed P. berghei mature gametocyte proteome data as a reference (Fig. 1C, supplemental Fig. S1). This approach is based on the fundamental assumption that sexual development is conserved evolutionarily across the genus, although not all proteins will sex-partition similarly between the different lineages. Thus, the re-analyzed P. berghei male/female gametocyte PSMA data sets along with the proteome of a mutant P. falciparum clone, Dd2, which is defective in mature male gametocyte development, permit the sex-partitioning of our mixed-sex NF54 stage V gametocyte proteome. At this juncture, it is important to note that because we are using a bioinformatic approach that is contingent on successful protein identification employing our specific methodology and instrumentation (see Methods), we refer to all male and female protein classifications in our analysis as “male- or female-enrichments.” However, we refer to the sex-specific proteomes generated by Khan et al., (11) and including our PSMA proteomes as “male/female-specific” and the stage-specific proteomes assembled by Silvestrini et al., (8) as “stage X-specific” protein lists, because these were produced independently and the categorizations are well-supported. Furthermore, integral in this process is use of known or well-supported P. falciparum male/female gametocyte proteins as positive control markers (e.g. P47). This will allow us to track their partitioning throughout the SSB workflow (Fig. 3B–3G, supplemental Fig. S1A–S1C), as described below.
Fig. 3.

Results from the Systematic Subtractive Protein Bioinformatic Approach (SSB). A, Male and Female P. berghei protein identification following the re-analysis of the published data set (11). (Refer to Fig. 2) B–G, Detail overlap results from supplemental Fig. S1A-C. H, Partitioned P. falciparum male- (174) and female- (258) enriched proteins.
Step 1
We curated the Dd2 proteome by first identifying Dd2 proteins that are conserved in the male P. berghei PSMA Male data set and generating a new list of male-subtracted proteins, that is, Dd2 female-enriched proteins (Dd2-FE; 1258 proteins). The male proteins that are shared with Dd2 are set aside (Dd2-M; 79 proteins) (Fig. 3B, supplemental Table S3A–S3B).
Step 2
This set was then combined with the mixed-sex protein list from NF54 to identify proteins that are conserved between Dd2-FE and NF54 and are likely to be enriched in female- proteins (Dd2-FE/NF54; 1045 proteins; Fig. 3C, supplemental Table S3C1). The proteins that are excluded from the overlap are then used to generate a list of likely NF54 male-enriched proteins (NF54-ME; 658 proteins) and Dd2-FE-specific proteins (Dd2-FE-SP; 213 proteins) (Fig. 3C, supplemental Table S3C2).
Step 3
The NF54-ME protein list is then used to search the P. berghei male-specific gametocyte protein list (PSMA Male; 366 proteins) to identify orthologs by BLAST (Fig. 3D). P. falciparum NF54 proteins with syntenic matches in the male P. berghei list are likely to be genus conserved and NF54 male specific candidates (NF54-M; 155 proteins). The NF54-ME proteins that do not have matches are likely to be NF54-specific proteins (NF54-SP; 503 proteins, supplemental Table S3F1).
Step 4
The Dd2-FE/NF54 (1045) protein list was used to search the P. berghei female-specific gametocyte protein list (PSMA Female; 418 proteins) to identify orthologs by BLAST. This analysis identified 181 proteins conserved between the two data sets, and 864 proteins that are Dd2-FE/NF54-specific (supplemental Table S3F2) and 237 proteins that are specific to P. berghei females (Fig. 3E).
Step 5
The NF54-ME list (male-enriched, 658 proteins) was used to determine if there are proteins that are conserved with the PSMA Female (418 proteins) list. We were surprised to find 60 proteins that are conserved between PSMA Female and the putative male-enriched NF54-ME protein subset (supplemental Table S3G). Following the comparison, we noted that 598 proteins are likely to be enriched with candidate male-partitioning P. falciparum proteins (supplemental Table S3H).
Although we did not anticipate missing proteins that do not partition to each of the subsequent protein lists, we nonetheless performed a set of additional data filtering steps as follows:
• Determine if There are Any Overlapping Proteins between NF54 F (181 Proteins) and PSMA Male (366 Proteins)
This filtering step was performed to ensure that we had not assigned erroneously a female protein that was actually found in the PSMA Male list. As expected no overlapping proteins were observed (data not shown).
• Determine the Overlap between Dd2-M or “M1” Protein List (79 Proteins) and the NF54-M or “M2” Protein List (155 Proteins) to Assess Dd2 versus NF54 Specific Protein Subsets
We observed 60 conserved proteins between these two lists (Supplemental Table S3I), and these conserved proteins represent the putative P. falciparum-specific male gametocyte proteins. Proteins that are conserved between the two P. falciparum lines that have diverged both geographically and phylogenetically (44) were categorized as conserved P. falciparum male proteins (conserved Pf-M; 60 proteins). Interestingly, 19 proteins were found to be specific to only Dd2 (Dd2-M-SP) and 95 proteins were NF54-specific (NF54-M-SP).
• Determine if There are Proteins that are Conserved between NF54-M or “M2” List (Male-specific Candidates, 155 Proteins) and the PSMA Female (Female, 418 Proteins) List
The NF54-M list was predicted to be highly male-partitioned and we observed, as we had expected in this quality check of the data, no overlap with P. berghei female proteins.
• Determine if There are Proteins That are Conserved between NF54-SP (503 Proteins) with the PSMA Female (418 Proteins) List
We identified 60 proteins, and these were set aside and categorized as NF54-SP-female proteins (NF54-SP-F) for subsequent inclusion in the berghei-driven female-enriched protein list. The remaining 443 proteins, which were not shared with Dd2 or present in the P. berghei data set, were considered NF54-specific proteins, nonsex partitioning (or potentially P. falciparum male-specific) (Fig. 3F, supplemental Table S3J).
• Determine if There are Proteins That are Conserved between Dd2-FE-SP (213 Proteins) and the PSMA Female (418 Proteins) List
We identified 17 female proteins that appear to be completely Dd2-specific (Dd2-SP) in our analysis (Fig. 3G, supplemental Table S3K). Because we expected that all of the Dd2-FE-SP were gametocyte female-enriched proteins, we hypothesized that the remaining 196 proteins from this list are Dd2-specific, non-(sex) partitioning.
• Determine if There are Proteins That are Conserved between Dd2-FE/NF54 (1045 Proteins) and PSMA Male (366 Proteins) List?
As expected, we did not identify any contaminating males in this protein list because, in theory, the Dd2-FE is female enriched and thus any matching proteins from the mixed NF54 gametocyte stage V proteome, should be female-enriched (data not shown).
The systematic steps described above allowed us to assemble a list of putative cross-strain (NF54-Dd2) P. falciparum male (Pf-M) and female (Pf-F) stage V gametocyte protein lists. Our initial Pf-M list, which was a grouping of NF54-M and Dd2-M proteins, identified 174 proteins; whereas our Pf-F list, which was a grouping of NF54-F, Dd2-SPF, and NF54-SPF proteins, identified 258 proteins. Of note there were marked differences in the composition of the Pf-M and Pf-F protein lists from either NF54 or Dd2, with the latter contributing only 11% of the proteins for Pf-F and 33% of the proteins for Pf-M.
Partitioning of Male- and Female-enriched Gametocyte Proteins in P. falciparum
Pf-MALES
Of those described to be male specific in P. berghei (11), we noted that the Mitogen-activated protein kinase 2 (MAP2; PF3D7_1113900), the Dynein light chain type 2 (PF3D7_1114000) were also only found in the male proteome in P. falciparum (supplemental Table S4A). The ortholog of the second P. berghei dynein heavy chain (PF3D7_0905300), which was described to be male-specific as well, was found to be nonsex partitioning. The 6-cysteine protein, P230p (PF3D7_0208900), which was described previously to be male-specific in P. berghei and confirmed by our PSMA analysis was also found to be male-enriched in P. falciparum. The same was noted for the NIMA-related kinase 1 (NEK1; PF3D7_1228300), which was also male-enriched in P. falciparum. The ortholog of the P. cynomolgi and P. vivax sperm-specific protein Don Juan (PF3D7_1413200) was found to be a male-specific candidate. This protein was not found in the FS Asx and FSgI-II databases, although Florens et al., (2002) detected successfully the protein in highly synchronized trophozoites (9). One of the top male-partitioning proteins in P. berghei (PBANKA_050730) (11) was detected in our total NF54 gametocyte stage V proteome but the ortholog (PF3D7_1023100) did not partition to either sex. PBANKA_050730 was already found to be shared in berghei males and females so it would naturally not partition in P. falciparum, because the male- and female-specific berghei lists were used to guide the assembly of the male and female lists in P. falciparum.
Pf-FEMALES
Our data set supports the argument that both the NIMA related kinase-4 (NEK4, PF3D7_0719200) and LCCL/CCp3 (PF3D7_1407000) proteins are conserved between P. berghei and P. falciparum female gametocytes (supplemental Table S4B). We also noted that NIMA related kinase 2 (NEK2, PF3D7_0525900) partitioned to females. A functional NEK2 has been shown to be essential for human/murine parasite development in the mosquito and its expression appears to be enriched in female gametocytes (45). The ortholog of the P. berghei female-specific (11) Dynein heavy chain (PF3D7_0729900) did not partition to either sex. Interestingly, P28, which is expressed as a transcript in gametocytes and only translated during gametogenesis, was identified in the female proteome. Although there are clear differences in the conditions of gametocyte cultures as opposed to in vivo development, which may lead to the misexpression of P28 protein, it is also possible that female gametocyte activation may have occurred during cell harvest. Our data however, supports the current hypothesis that P28 is female-specific. Approximately 46% of the 299 proteins (138) are conserved proteins with unknown function and represent a rich set of potential female markers for subsequent study.
We then took the Pf-M (174 proteins) and Pf-F (258 proteins) proteomes that were generated above to identify proteins that are conserved in the asexual trophozoite and the earlier gametocyte stages I-II, as described above, to further refine our P. falciparum male and female Stage V proteomes (Pf-M' and Pf-F', respectively). We also sought to identify any potential overlapping proteins that may have partitioned “artificially” according to the P. berghei PSMA lists but in fact are nonsex partitioning in P. falciparum and found none (data not shown).
Comparison of Predicted Functions of Gametocyte Proteins that Sex-partition in P. berghei and P. falciparum
To identify potential functional differences in male versus female gametocytes based on the proteomics data, we measured enrichment of molecular function based on Gene Ontology (GO) terms in the male and female protein data sets from both P. berghei (Fig. 3A) and P. falciparum (Fig. 3H). Indeed we identified enrichment of GO terms relating to cell motility and movement as well as microtubule cytoskeleton in the male gametocyte proteins, reflecting the exflagellation process during male gamete maturation (Fig. 4A). Conversely, the analysis identified GO term enrichment in female proteins for RNA binding and processing as well as for protein translation, relating to the translational repression mechanism that allows for just-in-time protein expression in the female gamete (46). The relevance of other enriched functions such as particular enzymatic activities in male (kinase, phosphatase) and female gametocyte proteins (transferase, isomerase, and oxidoreductase) remains to be determined.
Fig. 4.
Analyses of GO molecular function and signatures of selection in male and female P. falciparum proteins. A, Enrichment analysis of GO molecular function terms in Plasmodium berghei and Plasmodium falciparum. Z-scores are shown as representation of enrichment, color coded as the key in top left corner. Histogram shows general distribution of enrichments across all samples. Histograms along columns show exact enrichment amounts. Column normalization for total proteins detected was performed by calculating total number of GO terms divided by total proteins detected. For simplicity, GO terms were reduced to 22 categories and enrichment calculated over these. B, Genetic diversity within a parasite population is represented by PiN across 25 parasite isolates from Senegal and divergence between parasite populations is represented by Fst when comparing the Senegalese isolates with a set of parasite strains from Papua New Guinea (PNG). Several as yet uncharacterized sex-specific genes (i.e. PF3D7_1430800 and PF3D7_0131600) show high levels of divergence suggesting that they are under strong diversifying selection.
Signatures of Natural Selection in Male and Female Gametocyte Proteins
All the major transmission blocking vaccine candidates are not exposed to the human immune system during in viable gametocytes: Pfs25 is not expressed as a protein until gamete formation and others such as P47 and P230 are present on the gametocyte surface before gamete formation. Most gametocytes are cleared though and not taken up by a mosquito and thereby able to induce transmission-reducing immunity in human populations. P47 has an important role in immune evasion that contributes to the differential susceptibility of Anopheles gambiae M and S strains to P. falciparum infection (47). The immunomodulatory role of P47 is reflected in unusual population structure with fixed genetic differences between African and non-African parasite populations (48, 49). Likewise, we anticipated that Stage V gametocyte proteins that are either exposed on the infected RBC surface or on the gamete surface would show signatures of selection. To systematically test for signatures of natural selection in the genes encoding the sets of male and female P. falciparum gametocyte proteins identified in our study, we determined the rate of single nucleotide polymorphisms (SNPs) within a parasite population in Senegal (West Africa), and between this West African parasite population and a population from Papua New Guinea (48) (supplemental Table S5, Fig. 4B). As a measure of balancing selection within the Senegalese population, we calculated SNP π (N) for each gene based on a sequence comparison of a total of 25 sequenced parasite isolates (44). SNP π (N) quantifies the number of pairwise nonsynonymous differences among the set of strains analyzed. As a measure for positive selection between populations, we also calculated Fst (F-statistic or fixation index) for each gene by comparing the genetic diversity within the African parasite population with the diversity in the PNG parasite population. As predicted, P47 showed one of the highest Fst levels among all the male and female proteins (Fig. 4B). Interestingly, the female gametocyte marker and major vaccine candidate Pfs25 has an even higher value, reflecting strong positive selection potentially caused by selection of gamete recognition and compatibility. In addition, several hypothetical proteins are encoded by genes with high Fst such as the male-enriched gene PF3D7_1430800 and the female-enriched gene PF3D7_1358400. Except for one gene encoding a conserved hypothetical male protein PF3D7_0313600, the sex-partitioned gene set shows significantly lower diversity within the African population than known markers of balancing selection such as MSP1 or SERA5. This would suggest that none of the male and female-enriched proteins described here are subject to selection by their interaction with the human immune system, either because these proteins are not present on the infected RBC surface or because of their low abundance during human infection.
Identification of P. falciparum-enriched Gametocyte Sex-partitioned Proteomes
Once we had assembled the male- and female-enriched protein lists based on conservation with P. berghei, we sought to determine if we can derive, through the SSB approach, the P. falciparum-specific sex partitioned protein lists. We first performed a principal components analysis (PCA) using male and female-enriched P. falciparum stage V proteomes (Pf-M and Pf-F, respectively), replicate Dd2 and NF54 stage V gametocyte proteomes, as well as FSgV and FSgI-II protein lists (Fig. 5A). As expected the PCA demonstrated a clear enrichment for stage V proteins in NF54. To derive the putative male-enriched stage V protein list, we compared the NF54-enriched male list (443 proteins, supplemental Table S3J) with the FS Asx protein list and identified 354 proteins that likely represent the nonconserved falciparum-enriched male proteome (supplemental Table S6A, Fig. 5B). To derive the putative female-specific stage V protein list, we first removed 573 proteins that are shared between Dd2FE/NF54-SP (864 proteins) and the ANKA Male-Female Common (762 proteins, supplemental Table S2C) protein lists. This step was effective in filtering out proteins that would still be present in the Dd2-FE/NF54-SP list and that are conserved between falciparum and berghei. From this P. falciparum female-enriched protein list (291 proteins, supplemental Table S6B, Fig. 5C), we filtered out proteins that are also found in the FS Asx (1345 proteins) set to identify 177 gametocyte-enriched female proteins (supplemental Table S6C). We were cognizant that the FS Asx data set may not have captured all the late trophozoite or schizont proteins that may be present in our samples, because GlcNAc treatment does not remove completely all persisting asexual stages from a gametocyte culture. These “missed” asexual proteins are highlighted in the respective male and female lists (supplemental Tables S6A and S6C). Annotation of proteins to be asexual-specific in current P. falciparum genome remain simply predictions based on earlier proteomics studies. It is possible that the annotated asexual proteins are in fact shared with gametocyte stages. These proteins may be expressed in mature stages or represent relic proteins that are expressed in early gametocyte stages but persist in stage V gametocytes. However, confirming this “shared” attribute falls outside the scope of the current study. Considering the presence of such putative merozoite/schizont proteins, we assembled a conservative list of 339 male- and 174 female stage V gametocyte-enriched proteins.
Fig. 5.

Principal components analysis (PCA) and SSB analyses for the identification of P. falciparum male-enriched proteins that are lost as a result of the Dd2 male development defect. A, PCA was performed on male- and female-enriched P. falciparum stage V proteomes (Pf-M and Pf-F, respectively), two biological replicates (BR1 and BR2) from Dd2, three biological replicates (BR1–3) from NF54, FSgV (3D7/NF54 stage V), and FSg I-II (3D7/NF54 stages I-II). Proteins with at least 10 spectral counts averaged across all proteomes were used for qualitative protein analysis. B–C, Stage-specific analyses identified 354 putative P. falciparum- male- B, and 177 female- C, enriched gametocyte proteins. (D–G) Stage-specific analysis using published data sets (8) identifies 843 gametocyte-specific proteins in the NF54 proteome D, of which 216 are also present in FSg I-II and 627 are only expressed in more mature gametocyte stages E. The same analysis for Dd2 identifies 533 gametocyte-specific proteins F, of which 189 are present in FSg I-II, whereas 344 are only expressed in Stage III and later G. H–I, Stage-specificity of male and female-enriched Dd2 proteins reveals limited overlap between FSg I-II and conserved, male-enriched P. falciparum proteins H. However, we noted significant overlap between Dd2 Stage III-V and P. falciparum-enriched female proteins I.
We further determined the degree of NF54 stage V specificity of proteins to stage V gametocytes by comparing DTgV with the FS Asx protein lists and noted that 860 proteins were conserved (Fig. 5D). These 860 proteins comprise 50% of the total DTgV proteome, and may represent proteins that are simply conserved between trophozoites and stage V gametocytes. Although possible, it is unlikely that these are contaminating asexual proteins that are simply present in the culture, especially following treatment with GlcNAc. Of the remaining 843 proteins from DTgV (supplemental Table S6D), 216 proteins were found to be conserved in the FSgI-II data set (supplemental Table S6E), indicating that these proteins are expressed early in gametocytogenesis and likely remain present in stage V gametocytes, given the apparent absence of stage I-II in the culture, as determined by microscopy (see above). In our collective experience, the lack of gametocyte stages I-II is a direct indication of the complete absence of asexual stages. The remaining 627 proteins that did not match with either the FS Asx or FSgI-II data sets, represents a potential pool of stage III-IV gametocyte markers in addition to those that are present in stage V (supplemental Table S6F). Of these 627 proteins, 422 proteins (67%) were identified in the membrane fraction and supported by GO predictions for cellular component and biological process. Of the 627, 188 proteins (30%) have predicted transmembrane domains and 52% (325/627 proteins) of which are conserved hypothetical proteins. Several proteins that have been described previously appear in this putative list of Stage III-V markers, including four LCCL domain-containing proteins CCp2 (PF3D7_1455800), CCp3 (PF3D7_1407000), CCp5 (PF3D7_0109100), and FNPA (PF3D7_1451600), which all have hypothesized adhesive functions (40). CCp3 and CCp5 were found to be female-enriched proteins in our analyses. The NIMA related kinases, NEK2 (PF3D7_0525900) and NEK4 (PF3D7_0719200) are also found in this list and categorized as female-specific proteins as well. There are 95 male-enriched proteins present in this list, including the two dynein heavy chain proteins (PF3D7_1122900 and PF3D7_0905300). This short list of proteins represents a pool of targets that may be used to select specifically stage III gametocytes from culture.
Identification of the Stage-enriched Male/Female Protein Markers as Indicators for Stage Transition and to Investigate the Stage-enriched Dd2 Defect
Since Dd2 is defective in the production of mature, stage V male gametocytes (35), we hypothesized that the Dd2 proteome is more enriched proportionately in stage I-II markers, representing the small subset of rectangular and tear-drop forms that we had described from the Dd2 culture (Table II). Following PCA analysis, we observed that Dd2 clustered with gametocyte stages I-II proteins, which likely reflects the arrested male gametocytes in this strain. Moreover, the male- and female-enriched stage V proteomes appear to be distinct from all other clusters. The PCA also shows biological reproducibility of our proteome, as NF54 and Dd2 replicates cluster clearly.
To identify the putative arrest of male gametocytes in Dd2 candidate male protein markers, we also sought to further narrow the Dd2 stage V gametocyte data set into stage-specific proteins, and thus compared the Dd2 gametocyte stage V proteome (1337 proteins), referred to as Dd2gV, with the FS Asx and FSgI-II data sets. We noted that 804 proteins were conserved between FS Asx and Dd2gV data sets (Fig. 5F, supplemental Table S7A). Of these, 603 Dd2 trophozoite-conserved proteins are also found in the NF54 trophozoite-conserved subset (supplemental Table S7B). These 603 proteins comprise 56% of the total Dd2gV proteome, and may similarly represent proteins that are conserved between trophozoite and stage V gametocytes, or contaminating proteins that remain present in the GlcNAc-treated culture. Of the remaining 533 proteins from Dd2gV, 189 proteins were found in the FSgI-II data set as well (Fig. 5C, supplemental Table S7C), indicating that these proteins are expressed early in gametocytogenesis and remain present on female stage V gametocytes. For Dd2, we found 344 proteins that are stage V-enriched (supplemental Table S7D). Of these stage V proteins, 227 proteins (66%, 227/344) were identified in the membrane fraction, 62% (142/227 proteins) are conserved hypothetical proteins with diverse predicted GO functions, and 89% (127/142) of these proteins have predicted transmembrane domains.
Because Dd2 is defective in producing morphologically distinct stage V males, we examined whether the 189 Dd2 proteins shared with FSgI-II are enriched in male gametocytes (Fig. 5H). We found that only 12 proteins were common between the Pf-M protein list (174 proteins) and the Dd2gI-II set (supplemental Table S7E). These 12 proteins are also shared between the Pf-M and NF54-SP protein list (supplemental Table S3J), which is potentially male-enriched, and the Dd2gI-II set (data not shown). Of these 12 proteins, two proteins PF3D7_0508200 and PF3D7_1215100 had corresponding transcript markers for immature gametocytes (50), and were also thus absent from the stage V-enriched Pf-M' protein list (109 proteins, supplemental Table S4C). As expected, 46 proteins were found in Dd2gI-II list that are orthologs of proteins that were shared between males and females in P. berghei (supplemental Table S7F). Twelve of these proteins have ∼2-fold (or greater) enrichment in males in P. berghei. Although we cannot be certain that the same fold-enrichment exists for falciparum, based on these analyses, only 24/189 (13%) of the Dd2gI-II proteins are male; suggesting no enrichment of male partitioned proteins. This is not unexpected given that the 162 remaining Pf-M proteins may be expressed only at stage III or IV during gametocytogenesis.
We also examined whether the 344 Dd2 proteins that are candidates for Stage III-V are enriched in female proteins (Fig. 5I). To test this hypothesis we looked for proteins in the Dd2gV stage-V -enriched protein list that are also found in the Pf-F (291 P. falciparum female-enriched proteins) list that we had generated previously, which we then refined to only stage V-enriched female proteins, Pf-F' (81 proteins). We observed that 28 proteins (supplemental Table S7G) were not found in the Dd2gV stage-V-enriched protein list, and 53 proteins were conserved (supplemental Table S7H), suggesting that only 15% (53/344) of the Dd2gIII-V-enriched protein list were in the combined Dd2/NF54 Female protein list (supplemental Table S6B). It is possible that the number of mature stage V females in Dd2 with the full protein complement is low; falling below the identification criteria threshold used in this study. There are 113 proteins that are shared between Dd2/NF54-SP females and Dd2 (supplemental Table S7I), which gives a total of 164 female-enriched proteins in the Dd2gIII-V list. The lack of enrichment in female proteins in this list may suggest that a significant proportion of the 178 Dd2gIII-V proteins (supplemental Table S7J) are male-female shared proteins or stage III males. In fact, only 26/178 (15%) of these proteins have orthologs in the PSMA (P. berghei) male-female shared protein list. Moreover, expanding the Pf-Female protein list to include those that are not stage V-enriched (to capture putative stage III-IV proteins) does not result in additional “female hits” in the Dd2gIII-V list (supplemental Fig. S2A). Moreover, of the 291 proteins that remain partitioned to Dd2gIII-V, only 10% (30/291) have orthologs in the male-female shared protein list. Assuming that the developmental defect in Dd2 extends beyond the presence of mature males and also results in slower maturation of females, the 178 Dd2/NF54-SP female proteins may represent the remaining complement of mature stage V proteins (supplemental Table S7K).
A comparison of the Dd2gV stage-V-enriched protein list with the Pf-M (P. falciparum male-enriched) protein list identified 40 conserved proteins (supplemental Fig. S2B, supplemental Table S7L), which may include stage III male candidate markers. We found that two conserved proteins of unknown function, PF3D7_1413200 and PF3D7_1235800, had corresponding stage-specific transcript markers for immature gametocytes, that is, stages II-IV (31) and another conserved protein, PF3D7_1404200 had a corresponding mature (stage V) gametocyte transcript marker. The remaining 69 proteins (supplemental Table S7M) may represent a pool of candidate stage IV-V male gametocyte markers. Indeed, we found that of the 10 proteins with corresponding stage-specific-enriched transcript markers, five were mature stage V gametocyte proteins, and three corresponded to young (stage I) or immature (II-IV) gametocytes. Two of the 10 proteins were predicted to be ring-stage specific, which are likely contaminants in the sample and the last protein did not have a predicted profile that was captured by the defined stages used in the Joice et al. (2013) study (50).
Sex Partitioning for a Subset of Putative Female Protein Markers is not Conserved between P. berghei and P. falciparum
We successfully generated mouse antibodies to three proteins (PF3D7_1218800; PF3D7_0906100; PF3D7_0309100). PF3D7_1218800, and PF3D7_0309100 were selected based on their predicted female-specificity using the ANKA male and female proteomes (supplemental Table S3). PF3D7_0906100 was also selected because it appeared to be female-partitioning in P. berghei. However, it was also found in the P. falciparum 3D7/NF54 Trophozoite (8) and Stage I-II gametocytes (8) and “Dd2-alone” stage V protein list. We tested the stage specificity of these antibodies by staining NF54 male and female gametocytes taken (Fig. 6) and noted that antibodies for all three proteins stained fully “falciform” stage V gametocytes from day 17 cultures. Interestingly, PF3D7_0309100 appears to differentiate falciform from stage IV-V transitioning gametocytes, i.e. elongated gametocytes, from those gametocytes with more pronounced curvature but not completely falciform (indicated as V and V' stages in Fig. 6) (51). The antibodies were further validated by Western blot analysis to assess stage specificity (Fig. 7A). We found that the anti-PF3D7_0906100 antibodies recognized a ∼28 kDa protein band (predicted Mr = 21.9 kDa) in both day 17 gametocyte and synchronized trophozoite lysates from NF54 and Dd2. However, the staining intensity appeared to be more pronounced for NF54 gametocytes and less so for Dd2 trophozoites. Anti-PF3D7_1218800 antibodies recognized a ∼37 kDa protein band (predicted Mr = 39.6 kDa) and a lower Mr protein band around 26 kDa in NF54 and Dd2 gametocytes only. However, the antibodies also recognized a similar lower Mr band, ∼26 kDa in the Dd2 trophozoite sample. The exact nature and identity of this protein band is not clear at present, but given the developmental defect in Dd2 during gametocytogenesis, we cannot rule out the possibility of dysregulated protein expression in the asexual stages of Dd2. Moreover, we cannot completely rule out the presence of stage I/II gametocytes in the sample, although none were observed by microscopy and PF3D7_1218800 was not found in the FsgI-II protein list. Anti-PF3D7_0309100 antibodies appear to be highly specific to gametocytes in both NF54 and Dd2.
Fig. 6.
Immunofluorescence analysis of antibodies elicited against putative male- and female-enriched gametocyte stage V proteins. Antibodies were raised in mouse against PF3D7_1218800, PF3D7_0390100 and PF3D7_0906100. The lower panel is a cartoon depicting the various stages of NF54 gametocyte development, especially stages IV and V, which we have observed as the primary stage present in a day 18 gametocyte culture. Gametocytes that are labeled V' are considered mature stage V parasites, and those labeled V, are considered complete “falciform” (crescent) shaped stage V gametocytes. EB, Evans blue counter stain for protein, and appear red. GV (gametocyte stage V marker antibodies) are detected with Alexa488-conjugated secondary antibodies and appear green. DAPI stains DNA and appears blue. Scale bar = 10 μm.
Fig. 7.
Western blots and immunohistochemistry using NF54 and Dd2 strains validate female stage V gametocyte specificity for two of three candidate protein markers. A, Western blots of total day 17 stage V gametocyte and synchronized trophozoite protein lysates using the respective antibodies along with anti-P. falciparum aldolase antibodies used as a loading control. Western blots showed that PF3D7_1218800 and PF3D7_0309100 were present in female gametocytes (Dd2 is defective in mature male gametocyte development), yielding bands in gametocyte but not trophozoite samples for both NF54 and Dd2. Antibodies to PF3D7_0906100 stain gametocytes and trophozoites from NF54 but primarily stains Dd2 gametocytes. B, Representative results for fluorescence immunohistochemistry of strain NF54. PF3D7_1218800 and PF3D7_0906100 were found to be female-specific (green). PF3D7_0309100, which is an ortholog of a putative female-specific protein in P. berghei is not sex-specific in P. falciparum. C, Representative results for fluorescence immunohistochemistry of strain Dd2. PF3D7_1218800, PF3D7_0906100, and PF3D7_0309100 were all found to stain female parasites (green), as Dd2 fail to produce mature stage V male gametocytes. Parasite sex was microscopically determined using published morphological characteristics of male and female gametocytes (35, 55) or spatially distinct DAPI staining (blue) of DNA. Female gametocytes have condensed DAPI staining whereas male gametocytes have diffused DAPI staining (35, 55). Isotype control antibody staining of mature stage V female gametocytes for both Dd2 and NF54 are shown in the lower right set of panels.
We further determined sex-specific staining by the antibodies using the morphological characteristics of male and female gametocytes (35) along with strong DAPI costaining of DNA (Fig. 7B–7C). Of the three antisera tested, only two (PF3D7_1218800 and PF3D7_0906100) were found to be female-specific by fluorescence microscopy for both NF54 (Fig. 7B) and Dd2 (Fig. 7C), sex-partitioning that is conserved between P. berghei and P. falciparum. We also consistently noted that Dd2 female stage V gametocytes appeared more bulbous as compared with NF54. PF3D7_0906100 also referred to as a putative Developmental Protein partitioned initially with the female-enriched protein list. However, because we had identified it in both our Dd2 stage V gametocyte (Dd2gV) protein list and the Trophozoite (FS Asx) protein list (8), we did not include this protein in the stage V-enriched male and female protein lists. Although the ortholog in P. berghei (PBANKA_041520) was included in the female-enriched protein list for P. berghei, a close examination of the spectral data revealed that it was identified by a single peptide only in a single replicate batch of purified females (supplemental Table S2E). It is likely that this protein exists in both males and females in P. berghei. Antibodies to PF3D7_0309100, which is found in the putative female-enriched “NF54-Dd2 Common Proteins” list (supplemental Table S1C), surprisingly stained both males and females (Fig. 7B–7C). We also observed several examples of antibody labeling of Dd2 (day 17) gametocyte that is in essence rectangular or cuboidal (Fig. 7C). These rectangular/cuboidal and teardrop (data not shown) forms exhibit a DNA stain distribution indicative of a male form. PF3D7_0309100 was detected in FSgI-II and FSgV protein lists (8), suggesting that Dd2 male gametocyte maturation reaches stage II but arrests prior to progression to stage III.
DISCUSSION
One of the major caveats of the SSB approach is that the analysis is limited by the proteome size and accuracy of categorization. We recognized, a priori, that we are likely to miss out on a few potential male or female proteins, as we had noted in the comparison of our data set with the previously published, FSgV mixed-sex proteome. However, it is also clear that given the differential efficiencies in proteome acquisition across MS approaches, the community must consider using the combined databases in their proteome mining studies for proteins of interest. For example, we were able to corroborate the identification of the non-(sex) partitioning, PIESP2 (PF3D7_0501200) in stage I-II and stage V gametocytes. It is a putative exported parasite-infected erythrocyte surface protein, with a predicted PEXEL motif, in stage V gametocytes. PIESP2 was notable in that transcript for this protein is virtually absent in stages I-II and stage V gametocytes (PlasmoDB) and it was also found to be potentially on the RBC membrane surface in asexuals (52) and during liver stage development (53).
We had also observed the consistent partitioning of Dd2-enriched proteins following our comparisons with the NF54 (DTgV) and the 3D7/NF54 (FSgV) stage V proteomes. Proteomic analyses of Dd2 has not been performed to date, but this observed partitioning is really unexpected as we would expect that the differences in gene content between the two P. falciparum strains should probably be minimal. Although we have begun a partial analysis of the Dd2 developmental defect by identifying the enrichment and expression of stage and male-partitioned proteins by SSB and fluorescence microscopy, the morphological differences observed even for the mature female Dd2 gametocytes suggest that the defect may also result in dysregulated expression of proteins that would not normally be present during “normal” gametocytogenesis.
From a translational perspective, we have provided sex-partitioned information for several stagespecific gametocyte transcript markers (51) that can be used to analyze blood samples taken during surveillance studies in malaria endemic countries. In the same vein, the antibodies that we have generated in this study may ultimately represent novel diagnostic markers for gametocyte carriage in the blood or other body fluid (e.g. urine or saliva) of infected but asymptomatic individuals. PF3D7_0906100 (conserved hypothetical protein), PF3D7_0309100 (a putive development gene), and PF3D7_1218800 (putative secreted ookinete protein, PSOP17) have not been well characterized to date. Previous attempts to knock out PSOP17 in P. berghei were unsuccessful and highlight the importance of this protein in parasite development in the asexual blood stages as well (54). The complete characterization of each of these proteins is the focus of current efforts in our laboratories, as they have potential as new transmission-blocking vaccine targets and/or sex-enriched biomarkers of gametocyte carriage.
Supplementary Material
Acknowledgments
We thank Shahid M. Khan (Khan, S. M. et al. (2005) Proteome analysis of separated male and female gametocytes reveals novel sex-specific plasmodium biology. Cell 121, 675–687) for providing the raw data from the original Plasmodium berghei gametocyte study, Daniel E. Neafsey for valuable suggestions and facilitating analyses, Heidi Sinsel for artwork and the PRotein IDEntifications (PRIDE) database team for their assistance in uploading the data.
Footnotes
Author contributions: D.T., M.M., and R.R.D. designed research; D.T., C.U., D.K.M., J.G.K., R.P., A.T., I.G., and R.R.D. performed research; D.T., C.U., D.K.M., J.G.K., R.P., A.T., D.G., E.M., M.M., and R.R.D. contributed new reagents or analytic tools; D.T., C.U., D.K.M., J.G.K., R.P., D.G., E.M., M.M., and R.R.D. analyzed data; D.T., C.U., D.K.M., J.G.K., R.P., I.G., D.G., E.M., M.M., and R.R.D. wrote the paper.
* Financial support: This work was funded by the Bill & Melinda Gates Foundation grant OPP1054413 (R.R.D.), the Bloomberg Family Foundation and the Johns Hopkins Malaria Research Institute (JHMRI) (to R.R.D., A.T., and R.P.M.), and the Calvin and Helen Lang Postdoctoral Fellowship (D.K.M.). This research was also supported in part by the Human Frontiers Science Program (R.R.D.), the National Institutes of Health (NIH) Grants R01AI08258 (R.R.D.), R01A1077558 (M.M.), NIH contract TAS:75 0872::TAS (D.R.G.) and UL1TR001079 from the NIH Institute for Clinical and Translational Research.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S7.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S7.
Conflict of Interest: The authors declare that no conflicts of interest exist that can be perceived to influence the conduct of the study.
1 The abbreviations used are:
- GiRBC
- gametocyte infected red blood cell
- GcV
- gametocyte stage V
- Asx
- asexual blood stages
- SCX
- strong cation exchange
- ppm
- parts per million
- FA
- formic acid
- PSMA
- PepArML Search MASPECTRAS 2 Analysis
- Pb
- Plasmodium berghei
- Pf
- Plasmodium falciparum
- SSB
- Systematic Subtractive Protein Bioinformatic analysis.
REFERENCES
- 1. Babiker H. A., Schneider P., Reece S. E. (2008) Gametocytes: insights gained during a decade of molecular monitoring. Trends Parasitol. 24, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alonso P. L., Brown G., Arevalo-Herrera M., Binka F., Chitnis C., Collins F., Doumbo O. K., Greenwood B., Hall B. F., Levine M. M., Mendis K., Newman R. D., Plowe C. V., Rodriguez M. H., Sinden R., Slutsker L., Tanner M. (2011) A research agenda to underpin malaria eradication. PLoS Med. 8, e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. malERA Consultative Group on Drugs. (2011) A research agenda for malaria eradication: drugs. PLoS Med. 8, e1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. malERA Consultative Group on Vaccines. (2011) A research agenda for malaria eradication: vaccines. PLoS Med. 8, e1000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bousema T., Drakeley C. (2011) Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 24, 377–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paul R. E., Brockman A., Price R. N., Luxemburger C., White N. J., Looareesuwan S., Nosten F., Day K. P. (1999) Genetic analysis of Plasmodium falciparum infections on the north-western border of Thailand. Trans. R. Soc. Trop. Med. Hyg. 93, 587–593 [DOI] [PubMed] [Google Scholar]
- 7. Robert V., Read A. F., Essong J., Tchuinkam T., Mulder B., Verhave J. P., Carnevale P. (1996) Effect of gametocyte sex ratio on infectivity of Plasmodium falciparum to Anopheles gambiae. Trans. R. Soc. Trop. Med. Hyg. 90, 621–624 [DOI] [PubMed] [Google Scholar]
- 8. Silvestrini F., Lasonder E., Olivieri A., Camarda G., van Schaijk B., Sanchez M., Younis Younis S., Sauerwein R., Alano P. (2010) Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics 9, 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 10. Hall N., Karras M., Raine J. D., Carlton J. M., Kooij T. W., Berriman M., Florens L., Janssen C. S., Pain A., Christophides G. K., James K., Rutherford K., Harris B., Harris D., Churcher C., Quail M. A., Ormond D., Doggett J., Trueman H. E., Mendoza J., Bidwell S. L., Rajandream M. A., Carucci D. J., Yates J. R., 3rd., Kafatos F. C., Janse C. J., Barrell B., Turner C. M., Waters A. P., Sinden R. E. (2005) A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 [DOI] [PubMed] [Google Scholar]
- 11. Khan S. M., Franke-Fayard B., Mair G. R., Lasonder E., Janse C. J., Mann M., Waters A. P. (2005) Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687 [DOI] [PubMed] [Google Scholar]
- 12. Sinden R. E. (2009) Malaria, sexual development, and transmission: retrospect and prospect. Parasitology 136, 1427–1434 [DOI] [PubMed] [Google Scholar]
- 13. Sinden R. E., Carter R., Drakeley C., Leroy D. (2012) The biology of sexual development of Plasmodium: the design and implementation of transmission-blocking strategies. Malar. J. 11, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ponnudurai T., Meuwissen J. H., Leeuwenberg A. D., Verhave J. P., Lensen A. H. (1982) The production of mature gametocytes of Plasmodium falciparum in continuous cultures of different isolates infective to mosquitoes. Trans. R. Soc. Trop. Med. Hyg. 76, 242–250 [DOI] [PubMed] [Google Scholar]
- 15. Fivelman Q. L., McRobert L., Sharp S., Taylor C. J., Saeed M., Swales C. A., Sutherland C. J., Baker D. A. (2007) Improved synchronous production of Plasmodium falciparum gametocytes in vitro. Mol. Biochem. Parasitol. 154, 119–123 [DOI] [PubMed] [Google Scholar]
- 16. Edwards N., Wu X., Tseng C. (2009) An unsupervised, model-free, machine-learning combiner for peptide identifications from tandem mass spectra. Clin. Proteomics 5, 23–36 [Google Scholar]
- 17. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 18. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 19. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 20. MacLean B., Eng J. K., Beavis R. C., McIntosh M. (2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 21. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: Identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 23. Kim S., Gupta N., Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 25. Ubaida Mohien C., Hartler J., Breitwieser F., Rix U., Remsing Rix L., Winter G. E., Thallinger G. G., Bennett K. L., Superti-Furga G., Trajanoski Z., Colinge J. (2010) MASPECTRAS 2: an integration and analysis platform for proteomic data. Proteomics 10, 2719–2722 [DOI] [PubMed] [Google Scholar]
- 26. Taylor C. F., Paton N. W., Lilley K. S., Binz P. A., Julian R. K., Jr., Jones A. R., Zhu W., Apweiler R., Aebersold R., Deutsch E. W., Dunn M. J., Heck A. J., Leitner A., Macht M., Mann M., Martens L., Neubert T. A., Patterson S. D., Ping P., Seymour S. L., Souda P., Tsugita A., Vandekerckhove J., Vondriska T. M., Whitelegge J. P., Wilkins M. R., Xenarios I., Yates J. R., 3rd, Hermjakob H. (2007) The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 25, 887–893 [DOI] [PubMed] [Google Scholar]
- 27. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 29. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 30. Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R., and 1000 Genomes Project Analysis Group. (2011) The variant call format and VCFtools. Bioinformatics 27, 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weir B. S., Cockerham C. C. (1984) Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 [DOI] [PubMed] [Google Scholar]
- 32. Saha S., Raghava G. P. (2007) Prediction methods for B-cell epitopes. Methods Mol. Biol. 409, 387–394 [DOI] [PubMed] [Google Scholar]
- 33. Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P. E., Nielsen M., Peters B. (2012) Immune epitope database analysis resource. Nucleic Acids Res. 40, W525–W530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delemarre B. J., van der Kaay H. J. (1979) Tropical malaria contracted the natural way in the netherlands. Ned. Tijdschr. Geneeskd. 123, 1981–1982 [PubMed] [Google Scholar]
- 35. Guinet F., Dvorak J. A., Fujioka H., Keister D. B., Muratova O., Kaslow D. C., Aikawa M., Vaidya A. B., Wellems T. E. (1996) A developmental defect in Plasmodium falciparum male gametogenesis. J. Cell Biol. 135, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Furuya T., Mu J., Hayton K., Liu A., Duan J., Nkrumah L., Joy D. A., Fidock D. A., Fujioka H., Vaidya A. B., Wellems T. E., Su X. Z. (2005) Disruption of a Plasmodium falciparum gene linked to male sexual development causes early arrest in gametocytogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16813–16818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pradel G. (2007) Proteins of the malaria parasite sexual stages: expression, function and potential for transmission blocking strategies. Parasitology, 1–19 [DOI] [PubMed] [Google Scholar]
- 38. Ubaida Mohien C., Colquhoun D. R., Mathias D. K., Gibbons J. G., Armistead J. S., Rodriguez M. C., Rodriguez M. H., Edwards N. J., Hartler J., Thallinger G. G., Graham D. R., Martinez-Barnetche J., Rokas A., Dinglasan R. R. (2013) A bioinformatics approach for integrated transcriptomic and proteomic comparative analyses of model and nonsequenced anopheline vectors of human malaria parasites. Mol. Cell. Proteomics 12, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Schaijk B. C., van Dijk M. R., van d. V., van Gemert G. J., van Dooren M. W., Eksi S., Roeffen W. F., Janse C. J., Waters A. P., Sauerwein R. W. (2006) Pfs47, paralog of the male fertility factor Pfs48/45, is a female specific surface protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 149, 216–222 [DOI] [PubMed] [Google Scholar]
- 40. van Dijk M. R., van Schaijk B. C., Khan S. M., van Dooren M. W., Ramesar J., Kaczanowski S., van Gemert G. J., Kroeze H., Stunnenberg H. G., Eling W. M., Sauerwein R. W., Waters A. P., Janse C. J. (2010) Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 6, e1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lal K., Delves M. J., Bromley E., Wastling J. M., Tomley F. M., Sinden R. E. (2009) Plasmodium male development gene-1 (mdv-1) is important for female sexual development and identifies a polarised plasma membrane during zygote development. Int. J. Parasitol. 39, 755–761 [DOI] [PubMed] [Google Scholar]
- 42. Ponzi M., Siden-Kiamos I., Bertuccini L., Curra C., Kroeze H., Camarda G., Pace T., Franke-Fayard B., Laurentino E. C., Louis C., Waters A. P., Janse C. J., Alano P. (2009) Egress of Plasmodium berghei gametes from their host erythrocyte is mediated by the MDV-1/PEG3 protein. Cell. Microbiol. 11, 1272–1288 [DOI] [PubMed] [Google Scholar]
- 43. Simon N., Scholz S. M., Moreira C. K., Templeton T. J., Kuehn A., Dude M. A., Pradel G. (2009) Sexual stage adhesion proteins form multi-protein complexes in the malaria parasite Plasmodium falciparum. J. Biol. Chem. 284, 14537–14546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van Tyne D., Park D. J., Schaffner S. F., Neafsey D. E., Angelino E., Cortese J. F., Barnes K. G., Rosen D. M., Lukens A. K., Daniels R. F., Milner D. A., Jr., Johnson C. A., Shlyakhter I., Grossman S. R., Becker J. S., Yamins D., Karlsson E. K., Ndiaye D., Sarr O., Mboup S., Happi C., Furlotte N. A., Eskin E., Kang H. M., Hartl D. L., Birren B. W., Wiegand R. C., Lander E. S., Wirth D. F., Volkman S. K., Sabeti P. C. (2011) Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet. 7, e1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reininger L., Tewari R., Fennell C., Holland Z., Goldring D., Ranford-Cartwright L., Billker O., Doerig C. (2009) An essential role for the Plasmodium nek-2 nima-related protein kinase in the sexual development of malaria parasites. J. Biol. Chem. 284, 20858–20868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mair G. R., Braks J. A., Garver L. S., Wiegant J. C., Hall N., Dirks R. W., Khan S. M., Dimopoulos G., Janse C. J., Waters A. P. (2006) Regulation of sexual development of Plasmodium by translational repression. Science 313, 667–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molina-Cruz A., Dejong R. J., Ortega C., Haile A., Abban E., Rodrigues J., Jaramillo-Gutierrez G., Barillas-Mury C. (2012) Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of anopheles gambiae mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 109, E1957–E1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manske M., Miotto O., Campino S., Auburn S., Almagro-Garcia J., Maslen G., O'Brien J., Djimde A., Doumbo O., Zongo I., Ouedraogo J. B., Michon P., Mueller I., Siba P., Nzila A., Borrmann S., Kiara S. M., Marsh K., Jiang H., Su X. Z., Amaratunga C., Fairhurst R., Socheat D., Nosten F., Imwong M., White N. J., Sanders M., Anastasi E., Alcock D., Drury E., Oyola S., Quail M. A., Turner D. J., Ruano-Rubio V., Jyothi D., Amenga-Etego L., Hubbart C., Jeffreys A., Rowlands K., Sutherland C., Roper C., Mangano V., Modiano D., Tan J. C., Ferdig M. T., Amambua-Ngwa A., Conway D. J., Takala-Harrison S., Plowe C. V., Rayner J. C., Rockett K. A., Clark T. G., Newbold C. I., Berriman M., MacInnis B., Kwiatkowski D. P. (2012) Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anthony T. G., Polley S. D., Vogler A. P., Conway D. J. (2007) Evidence of nonneutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol. Biochem. Parasitol. 156, 117–123 [DOI] [PubMed] [Google Scholar]
- 50. Joice R., Narasimhan V., Montgomery J., Sidhu A. B., Oh K., Meyer E., Pierre-Louis W., Seydel K., Milner D., Williamson K., Wiegand R., Ndiaye D., Daily J., Wirth D., Taylor T., Huttenhower C., Marti M. (2013) Inferring developmental stage composition from gene expression in human malaria. PLoS Comput. Biol. 9, e1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aingaran M., Zhang R., Law S. K., Peng Z., Undisz A., Meyer E., Diez-Silva M., Burke T. A., Spielmann T., Lim C. T., Suresh S., Dao M., Marti M. (2012) Host cell deformability is linked to transmission in the human malaria parasite Plasmodium falciparum. Cell. Microbiol. 14, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Florens L., Liu X., Wang Y., Yang S., Schwartz O., Peglar M., Carucci D. J., Yates J. R., 3rd, Wub Y. (2004) Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol. Biochem. Parasitol. 135, 1–11 [DOI] [PubMed] [Google Scholar]
- 53. Trieu A., Kayala M. A., Burk C., Molina D. M., Freilich D. A., Richie T. L., Baldi P., Felgner P. L., Doolan D. L. (2011) Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol. Cell. Proteomics 10, M111.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ecker A., Bushell E. S., Tewari R., Sinden R. E. (2008) Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol. Microbiol. 70, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carter R., Graves P. M. (1988) Gametocytes. In Malaria: Principles and Practice of Malariology, 253–305 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.