The ability to utilize physical cues such as nanotopographical features,[1] substrate stiffness,[2] geometry and the dimension of extracellular matrix (ECM) protein patterns[3] to control stem cell fate has great potential in regenerative medicine. In particular, biomaterials that are used to fabricate scaffolds and implantable substrates for stem cell-based regenerative medicine are now being investigated intensively in order to elicit specific behaviors from stem cells, including differentiation, migration and proliferation. For instance, in spinal cord and peripheral nerve injuries, the specific response of neuronal cells to nanotopographical cues is reported as one of the critical factors that must be achieved, as it is the specific guidance of axons that would lead to enhanced therapeutic effects within the injured spinal cord.[4] In particular, if the nerve gap resulting from an injury is too large, the distal and proximal sides of the damaged nerves will not be able to communicate efficiently, thus impeding the natural regeneration process.[5] As a result, a significant amount of effort has been invested in developing biomaterials that can result in axonal guidance and the growth of transplanted neurons within the injured spinal cord.[5,6] For this purpose, neural stem cells (NSCs), which can differentiate into neurons and glial cells, have been investigated for transplantation within injured spinal cords as they hold great promise for hastening functional recovery.[6,7] We and other groups have previously shown that the growth, differentiation and polarization of NSCs are strongly influenced by cell-cell and cell-extracellular matrix (ECM) interactions.[3a,3c] For example, ECM protein patterns and patterned nanotopographical features can be employed to control the polarity, directional growth and influence the neuronal differentiation of NSCs.[3c,8] The challenge, however, is to provide an engineered microenvironment to the NSCs, through the development and application of novel nano materials, that can specifically control the axonal alignment and growth of NSC-derived neurons for the development of more effective treatments for spinal cord injuries. Here we report the fabrication of arrays of graphene-nanoparticle hybrid structures for the differentiation and growth of adult hNSCs. More importantly, these graphene-nanoparticle hybrid structures resulted in the formation of highly aligned axons from the differentiating hNSCs.
Graphene, which consists of a monolayer of carbon atoms arranged in a 2D honeycomb lattice,[9] has been shown to be a very useful nanomaterial in biomedical applications[10] due to its excellent flexibility, thermal properties, electrical conductivity, high strength, stiffness and biocompatibility.[11] Recently, the physicochemical properties of graphene and its biocompatibility have inspired scientists to utilize this material for stem cell-based tissue engineering.[12] For instance, graphene has been shown to support the proliferation and differentiation of adult and pluripotent stem cells.[10c,13] In the case of neural tissue, it has been demonstrated that the physicochemical properties of graphene can facilitate excellent integration.[10c,14] On the other hand, it has also been demonstrated that nanotopographical features that are generated using arrays of silica microbeads can lead to the acceleration of axonal growth of hippocampal neurons in vitro.[15] We thus hypothesized that substrates, which consist of nanoparticle-based nanotopographical features modified with graphene, could be an excellent platform to further enhance the differentiation of hNSCs into neurons and could be used to control axonal growth of the differentiating hNSCs.
To this end, we generated arrays of graphene-nanoparticle hybrid structures using positively charged silica nanoparticles and graphene oxide (GO), which is a chemically versatile nanomaterial containing oxygen functional groups attached to the graphene basal plane. This is particularly advantageous as the oxygen functional groups allow the GO nanosheets to attach readily to molecules or surfaces – in our case the GO nanosheets were used to coat the surface of 300 nm silica nanoparticles (SiNPs) to form graphene-silica nanoparticle hybrids (SiNP-GO). The control and test substrates used to grow and differentiate hNSCs in this study are shown in Figure 1a [See Experimental Methods in Supporting Information for additional details about substrate fabrication (Section 1), graphene synthesis (Section 2) and characterization (Figure S1)]. All of the substrates were treated with the ECM protein laminin (10 μg/mL for 4 h), which is essential for the adhesion, growth, and differentiation of hNSCs. Human NSCs were then seeded onto these substrates and proliferated in culture media containing basic fi broblast growth factor (bFGF, 20 ng/mL) and epidermal growth factor (EGF, 20 ng/mL). After 24 h, differentiation was initiated by withdrawing the culture medium and replacing it with basal medium lacking growth factors. Immunocytochemistry and quantitative polymerase chain reaction (qPCR) were performed on the differentiated hNSCs after 14 days to investigate the influence of SiNP, GO and SiNP-GO on neuronal differentiation.
Figure 1.
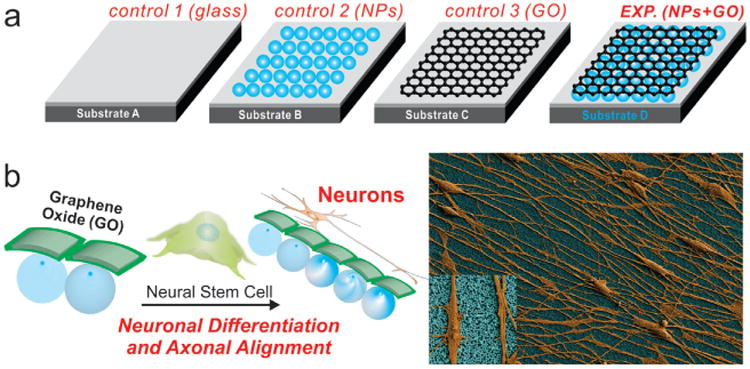
Schematic diagram depicting the influence of nanoparticle (NP) monolayers coated with graphene oxide (GO) on the alignment of the axons extending from hNSCs, and the differentiation of hNSCs into neurons. a) Different control and experimental conditions for differentiating hNSCs into neurons where Substrate A is a glass substrate having a positively charged surface, Substrate B is a glass substrate having a monolayer of positively charged NPs, Substrate C is a glass substrate having a positively charged surface and coated with GO, Substrate D is a glass substrate having a monolayer of positively charged NPs coated with GO. b) hNSCs cultured and differentiated on Substrate D having a monolayer of NPs coated with GO show enhanced neuronal differentiation and axonal alignment. The differentiated hNSCs (orange) and the NPs-coated with GO (blue) in the SEM image have been pseudocolored to enhance the contrast. (Inset) Zoom-in image showing the axons aligned on a monolayer of NPs coated with GO.
On Day 2 after the removal of growth factors, the hNSCs on all substrates were observed to have attached well and were growing. Typically, the axons of hNSCs grow in random directions when cultured on most substrates, unless the substrate contains patterned proteins.[3a,3c] In all of our conditions, the hNSCs also grew and extended in random directions until Day 5 (Figure S2). However, after Day 5, we observed that the extending axons began aligning only on the GO and SiNP-GO substrates and not on glass and SiNP substrates (Figure S2). Finally, on Day 14, the differentiated hNSCs on the GO and SiNP-GO substrates exhibited very well aligned and well-extended axons (Figure S2 and Figure 2a). On the other hand, the control hNSCs, which were differentiated on the SiNP and glass substrates, also had extended axons, but showed no alignment. To quantify this, we calculated the variation in the angle of orientation of the axons extending from differentiated hNSCs on substrates containing GO and compared it with the orientation of the axons from hNSCs differentiated on the control SiNP and glass substrates. Analysis of our data confirmed that the variation in the angle of orientation of the axons from differentiated hNSCs on the GO and SiNP-GO substrates was ±17.8° and ±9.16° respectively (Figure 2b), while the axons from the differentiated hNSCs on glass and SiNP substrates extended randomly, having a much wider variation of ± 42° and ±46.11°, respectively, in the angle of their orientation (Figure 2b). The images in Figure 2a clearly show that the axons extending from differentiated hNSCs aligned exclusively on substrates having GO as a component of the ECM. We also investigated the influence of nanotopographical features on the length of the axons extending from hNSCs. Recently, self-assembled silica microbeads were shown to significantly accelerate the extension of axons from hippocampal neurons in vitro.[15] We thus analyzed the lengths of the extending axons from the hNSCs differentiated on the different substrates on Day 14. The average length of the axons extending from differentiated hNSCs cultured on SiNPs was 20.76% more than the average length of those cultured on glass, and 11.3% more than those cultured on GO (Figure 2b). We therefore confirmed that the alignment of axons is exclusively due to the presence of GO within the ECM while the presence of the underlying SiNP monolayer can lead to an increase in the average length of the axons from hNSCs differentiated on SiNP-GO. This hNSC behavior was also confirmed using SEM (Figure 3a).
Figure 2.
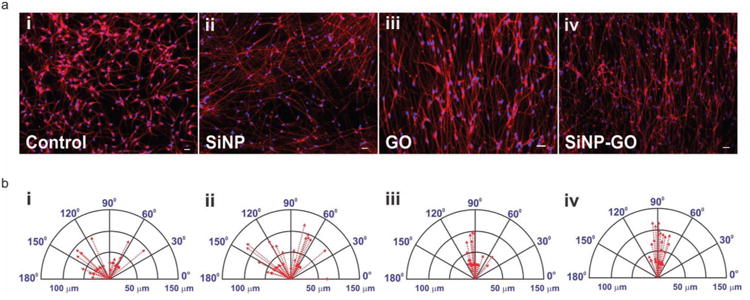
Aligned growth and extension of axons from differentiated hNSCs and compass plots showing the variation in the angle of orientation and the lengths of the axons. a) Differentiated hNSCs are immunostained with TuJ1 (red). The axons show no alignment on glass and SiNPs, whereas the axons are significantly aligned on GO and SiNP-GO. Scale bar: 10 μ m b) The compass plots show a large variation in the angle of orientation of axons on glass (±42°) and SiNPs (±46.11°) and minimal variation on GO (±17.8°) and SiNP-GO (±9.16°). The compass plot also shows that axons extending on SiNP and SiNP-GO are longer than those extending on glass and GO.
Figure 3.
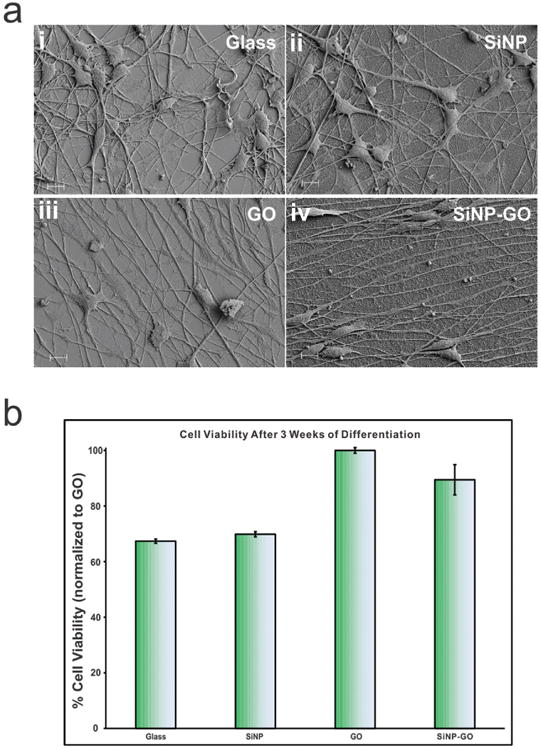
Scanning electron microscopy (SEM) showing the behavior of hNSCs and MTS assay for biocompatibility and long term survival of differentiated hNSCs on GO and SiNP-GO. a) SEM images confirm that the axons do not align on control and SiNP substrates and they align on GO and SiNP-GO substrates. Scale bar is 10 μ m. b) MTS assay results show that GO and SiNP-GO is biocompatible and aids in the long term survival of hNSCs as compared to glass and SiNPs. The results have been normalized to hNSC viability on GO.
We then went on to investigate whether the alignment of axons from the differentiating hNSCs on GO and SiNP-GO could be due to crowding of hNSCs and thus be dependent on the seeding density as it has previously been demonstrated that differences in cell density can yield a noticeable difference in cell alignment.[16] To this end, we reduced the cell density by 50% and observed the behavior of the hNSCs over a period of two weeks. We found that the cells behaved in the same manner as described above, even at the lower cell density (Figure S3). This result confirmed that the axonal alignment of differentiating hNSCs on the SiNP-GO substrates is not dependent on the cellular density of the hNSCs but only on the presence of GO. This is a remarkable finding as it suggests that the only factor determining the alignment of axons from differentiating hNSCs, is the presence of GO. This result could be very useful, especially for the development of scaffolds to restore neuronal function within damaged regions of the central nervous system.
In order for our platform to be used to control cell behavior or develop scaffolds, they should be biocompatible. In the case of regenerative medicine, the materials not only have to be bio-compatible but must also support stem cell differentiation and survival over long periods of time.[17] To this end, as a potentially advantageous material for tissue engineering, graphene has already been shown to support the long-term survival and induce neuronal differentiation of hNSCs.[10c] As such, we used a standard cell viability assay (MTS assay) which confirmed that the GO and SiNP-GO substrates significantly enhanced cells survival after 3 weeks of differentiation as compared to the control SiNP and glass substrates (Figure 3b). This is particularly advantageous for stem cell biology and regenerative medicine as the differentiating stem cells are required to grow, differentiate and survive to have beneficial and lasting effects.
Having established that the SiNP-GO substrates could promote cell survival and differentiation for extended periods, we sought to explore and quantify the effects of SiNP-GO on neuronal differentiation of hNSCs. To this end, we investigated the expression of immature and mature neuronal markers in the differentiated hNSCs after two weeks. Our immunostaining data demonstrated that most of the aligned axons from differentiated hNSCs were characterized by the expression of the neuronal marker TuJ1, and also the presence of mature neuronal markers such as MAP2 and synapsin (Figure 4a). We also confirmed the expression of axonal marker, GAP43. Next, to quantify the expression levels of these neuronal markers, we performed qPCR analyses on mRNA collected from the hNSCs differentiated on GO and SiNP-GO substrates and compared them to hNSCs differentiated on SiNP and glass substrates. While the expression levels of neuronal and axonal markers were up-regulated on all substrates as compared to the control glass substrates, we found that the hNSCs differentiated on SiNP-GO substrates showed the highest expression levels for all neuronal markers such as TuJ1, MAP2 and synapsin (Figure 4b). Thus, we can conclude that the combined effect of having SiNP and GO on a single platform shows increased neuronal differentiation and remarkable alignment of differentiated hNSCs.
Figure 4.
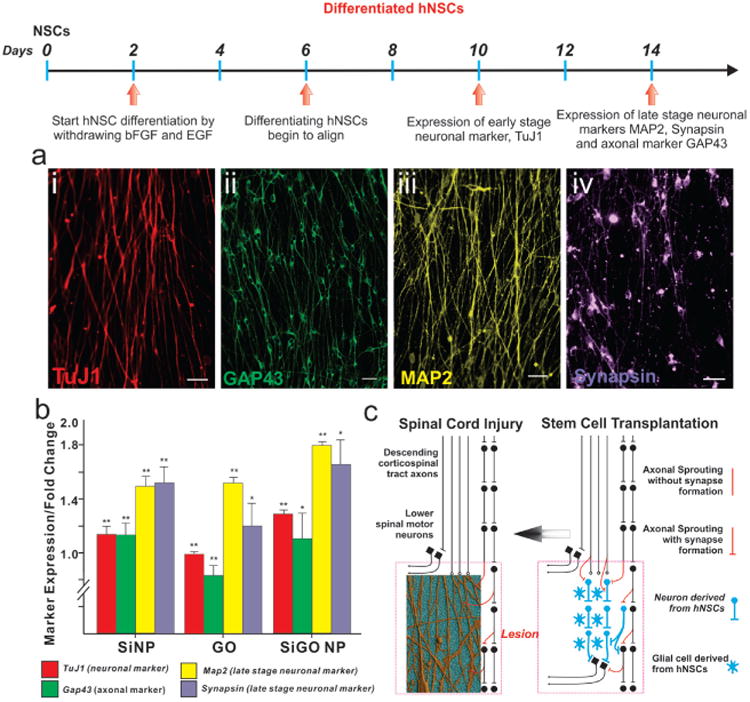
Enhanced neuronal differentiation of hNSCs on a SiNP-GO substrate. a) hNSCs spontaneously differentiated on SiNP-GO show the presence of early stage neuronal marker TuJ1 (red) and late stage neuronal markers MAP2 (pseudocolored yellow) and synapsin (pseudocolored purple). They also highly express the axonal marker GAP43 (green). Scale bar: 10 μm. b) Quantitative RT-PCR (qPCR) results for early and late stage neuronal markers expressed by the hNSCs differentiated on the different substrates. The results are normalized to the expression levels of the neuronal markers in hNSCs differentiated on glass. *P < 0.05, **P < 0.01 compared to hNSCs differentiated on glass using student t -test analysis (n = 4). The results clearly show that hNSCs differentiated on the SiNP-GO substrate show significantly enhanced expression of early and late stage neuronal markers. The expression of axonal marker GAP43 increases due to the presence of SiNP monolayers. c) Scheme depicting the significance of alignment and growth of axons from differentiating hNSCs. The hNSCs which can be transplanted into the injured region (lesion) of a spinal cord differentiate into neurons and glial cells (image on right). The axons from the neurons (derived from hNSCs) if aligned can hasten the recovery process. Our SiNP-GO hybrid structures can provide the ideal microenvironment to align axons which could potentially improve communication leading to rapid recovery of the injured spinal cord (image on left). The schematic diagram was adapted from a commentary by T Ben-Hur.[20]
To determine if the axons from differentiating hNSCs align exclusively on GO, we also used pristine graphene deposited on glass, using chemical vapor deposition (CVD). Although this behavior has not been reported previously, we did observe axonal alignment on pristine graphene, similar to the axonal alignment observed on GO (Figure S4). However, the water solubility of GO and the presence of functional groups allows positively charged SiNP monolayers to be readily coated with GO, by simply dipping the substrate into a solution of GO. Another factor that has to be considered is the interaction of the ECM protein, laminin, with pristine graphene. Proteins have been shown to have higher and more rapid immobilization on GO, as compared to pristine graphene due to the abundant surface oxygen-containing groups such as epoxide, hydroxyl and carboxyl groups present on GO.[18] The presence of these polar functional groups on GO makes the GO-coated substrates very hydrophilic as compared to the pristine graphene-coated substrates, which significantly affects the adsorption of proteins.[13] We thus believe that laminin, which is dissolved in water, more readily assembles on GO as it is water soluble in contrast to pristine graphene, which is hydrophobic. Considering these factors, as compared to graphene, GO is more advantageous for coating SiNPs, assembling ECM proteins, and aligning the axons from differentiating hNSCs. Next, we further investigated whether the axonal alignment was due to the unique chemical structure of graphene, which is composed of carbon atoms in a hexagonal lattice. For this purpose, we chose another nanomaterial, molybdenum disulfide (MoS2), which is from the family of two dimensional layered transition metal dichalcogenides and has a physical structure similar to that of graphene. Nanoflakes of MoS2 were deposited on glass substrates, onto which laminin was assembled. We then grew and differentiated hNSCs on MoS2. Importantly, we observed that while the hNSCs grew well and differentiated on MoS2, they did not showed any axonal alignment, thus confirming that the unique chemical structure of graphene causes the axons to align (Figure S5).
Finally, for potential future therapeutic applications in regenerative medicine, it would be crucial to demonstrate the alignment and enhanced neuronal differentiation using flexible and biocompatible polymeric substrates, which can be transplanted in vivo. We thus reproduced our results using flexible and biocompatible polymeric substrates made from polydimethylsiloxane (PDMS), a polymer which has been widely used for implantable neural devices such as flexible microelectrodes and three-dimensional scaffolds for tissue engineering.[19] We prepared monolayers of SiNPs on thin, flexible PDMS substrates by stamping the PDMS substrates on monolayers of SiNPs generated on glass cover slips. The stamping led to clean and complete transfer of the SiNP monolayers onto the PDMS surface (Figure 5a, b). We then dipped the PDMS substrates having SiNPs into a solution of GO and dried the substrates using a stream of pure nitrogen gas. In this way, we achieved high-quality SiNP monolayers coated with GO using PDMS. Control substrates were similarly prepared using PDMS polymer instead of glass. We then coated these substrates with laminin and differentiated the hNSCs as before. We observed that the differentiating hNSCs showed excellent alignment of axons on the PDMS substrates containing GO and SiNP-GO. SEM image analysis confirmed that the presence of SiNP-GO on PDMS led to the alignment of axons as previously observed (Figure 5c). Immunostaining confirmed the presence of neuronal marker TuJ1 and axonal marker GAP43 (Figure 5d). We thus believe that our results using flexible implantable polymeric substrates further demonstrates the potential of using SiNP-GO as a new hybrid material for enhancing neuronal differentiation and aligning axons, thus hastening the functional recovery of injured spinal cords.
Figure 5.
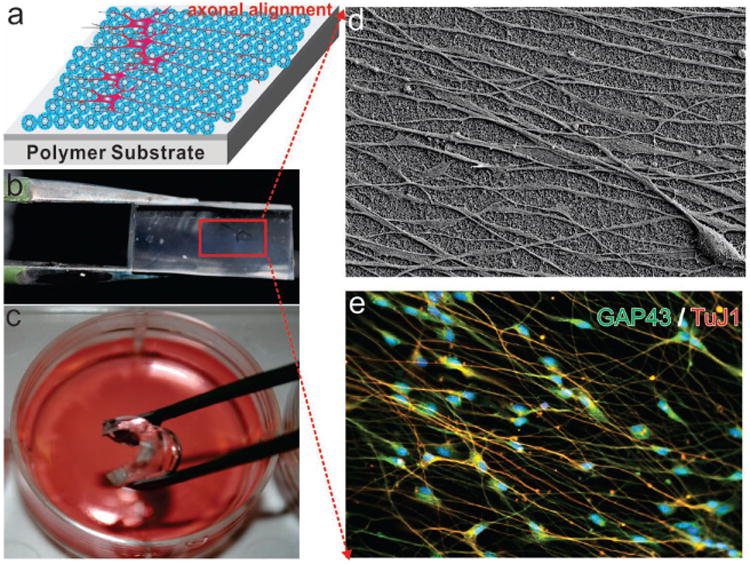
Axonal Alignment of differentiated hNSCs on SiNP-GO on flexible and biocompatible substrates made from polydimethylsiloane (PDMS). (a) schematic diagram of axonal alignment of differentiated hNSCs on SiNP-GO on polymer substrates. b) SiNP-GO monolayer on PDMS. c) Flexible PDMS substrate with SiNP-GO in media for culturing hNSCs. d) SEM image of SiNP-GO on PDMS substrate showing highly aligned axons from hNSCs on Day 14. e) Immunocytochemistry results showing the expression of neuronal marker TuJ1 and axonal marker GAP43 in hNSCs.
We have demonstrated that the engineered microenvironment consisting of nanotopographical features modified with GO provides instructive physical cues that lead to enhanced neuronal differentiation of hNSCs along with significant axonal alignment. We have also demonstrated the alignment of differentiating hNSCs on implantable, flexible polymeric substrates, which has tremendous potential in regenerative medicine. We currently do not understand the mechanism governing axonal alignment. However, we are in the process of investigating the underlying principles that govern the alignment of hNSCs due to graphene-nanoparticle hybrid structures. Nevertheless, we envision that the alignment of axons from the differentiating hNSCs using SiNP-GO can potentially be applied to developing GO-based materials for transplanting hNSCs into injured sites of the central nervous system in order to efficiently repair impaired communication. Overall, we believe our hybrid nanostructures comprised of a nanoparticle monolayers coated with GO have tremendous implications for the potential use of GO as an ECM component especially in the field of neurobiology.
Supplementary Material
Acknowledgments
The authors wish to thank Mr. Valentine Starovoytov for his help in the preparation of SEM samples. This work was supported by the NIH Director's Innovator Award [(1DP20D006462-01)] and the N.J. Commission on Spinal Cord grant [(09-3085-SCR-E-0)].
Footnotes
Supporting Information: Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Aniruddh Solanki, Department of Chemistry and Chemical Biology Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA, Fax: (+1) 732-445-5312.
Sy-Tsong Dean Chueng, Department of Chemistry and Chemical Biology Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA, Fax: (+1) 732-445-5312.
Perry T. Yin, Department of Biomedical Engineering Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
Rajesh Kappera, Department of Materials Science and Engineering Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Manish Chhowalla, Department of Materials Science and Engineering Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
Ki-Bum Lee, Email: kblee@rutgers.edu, Department of Chemistry and Chemical Biology Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA, Fax: (+1) 732-445-5312, http://chem.rutgers.edu/–kbleeweb; Department of Biomedical Engineering Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
References
- 1.a) Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, Jin S. Proc Natl Acad Sci USA. 2009;106:2130. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Solanki A, Shah S, Yin PT, Lee KB. Sci Rep. 2013;3:1553. doi: 10.1038/srep01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophys J. 2008;95:4426. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Park SY, Choi DS, Jin HJ, Park J, Byun KE, Lee KB, Hong S. ACS Nano. 2011;5:4704. doi: 10.1021/nn2006128. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ruiz SA, Chen CS. Stem Cells. 2008;26:2921. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Solanki A, Shah S, Memoli KA, Park SY, Hong S, Lee KB. Small. 2010;6:2509. doi: 10.1002/smll.201001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geller HM, Fawcett JW. Exp Neurol. 2002;174:125. doi: 10.1006/exnr.2002.7865. [DOI] [PubMed] [Google Scholar]
- 5.Miller C, Jeftinija S, Mallapragada S. Tissue Eng. 2002;8:367. doi: 10.1089/107632702760184646. [DOI] [PubMed] [Google Scholar]
- 6.Teng YD, Lavik EB, Qu X, Park KI, Ourednik J, Zurakowski D, Langer R, Snyder EY. Proc Natl Acad Sci USA. 2002;99:3024. doi: 10.1073/pnas.052678899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Sandner B, Prang P, Rivera FJ, Aigner L, Blesch A, Weidner N. Cell Tissue Res. 2012;349:349–362. doi: 10.1007/s00441-012-1363-2. [DOI] [PubMed] [Google Scholar]; b) Abematsu M, Tsujimura K, Yamano M, Saito M, Kohno K, Kohyama J, Namihira M, Komiya S, Nakashima K. J Clin Invest. 2010;120:3255. doi: 10.1172/JCI42957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Kim DH, Provenzano PP, Smith CL, Levchenko A. J Cell Biol. 2012;197:351. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kumar G, Tison CK, Chatterjee K, Pine PS, McDaniel JH, Salit ML, Young MF, Simon CG., Jr Biomaterials. 2011;32:9188. doi: 10.1016/j.biomaterials.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geim AK, Novoselov KS. Nat Mater. 2007;6:183. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 10.a) Li JL, Bao HC, Hou XL, Sun L, Wang XG, Gu M. Angew Chem Int Ed. 2012;51:1830. doi: 10.1002/anie.201106102. [DOI] [PubMed] [Google Scholar]; b) Yang K, Zhang S, Zhang G, Sun X, Lee ST, Liu Z. Nano Lett. 2010;10:3318. doi: 10.1021/nl100996u. [DOI] [PubMed] [Google Scholar]; c) Park SY, Park J, Sim SH, Sung MG, Kim KS, Hong BH, Hong S. Adv Mater. 2011;23:H263. doi: 10.1002/adma.201101503. [DOI] [PubMed] [Google Scholar]; d) Myung S, Solanki A, Kim C, Park J, Kim KS, Lee KB. Adv Mater. 2011;23:2221. doi: 10.1002/adma.201100014. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Park JK, Jung J, Subramaniam P, Shah BP, Kim C, Lee JK, Cho JH, Lee C, Lee KB. Small. 2011;7:1647. doi: 10.1002/smll.201100012. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Myung S, Yin PT, Kim C, Park J, Solanki A, Reyes PI, Lu Y, Kim KS, Lee KB. Adv Mater. 2012;24:6081. doi: 10.1002/adma.201202961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Murali S, Cai W, Li X, Suk JW, Potts JR, Ruoff RS. Adv Mater. 2010;22:5226. doi: 10.1002/adma.201001068. [DOI] [PubMed] [Google Scholar]
- 12.Some S, Ho SM, Dua P, Hwang E, Shin YH, Yoo H, Kang JS, Lee DK, Lee H. ACS Nano. 2012;6:7151. doi: 10.1021/nn302215y. [DOI] [PubMed] [Google Scholar]
- 13.Chen GY, Pang DWP, Hwang SM, Tuan HY, Hu YC. Bio-materials. 2012;33:418. doi: 10.1016/j.biomaterials.2011.09.071. [DOI] [PubMed] [Google Scholar]
- 14.Kotov NA, Winter JO, Clements IP, Jan E, Timko BP, Campidelli S, Pathak S, Mazzatenta A, Lieber CM, Prato M, Bellamkonda RV, Silva GA, Kam NWS, Patolsky F, Ballerini L. Adv Mater. 2009;21:3970. [Google Scholar]
- 15.Kang K, Choi SE, Jang HS, Cho WK, Nam Y, Choi IS, Lee JS. Angew Chem Int Ed. 2012;51:2855. doi: 10.1002/anie.201106271. [DOI] [PubMed] [Google Scholar]
- 16.Lam MT, Sim S, Zhu X, Takayama S. Biomaterials. 2006;27:4340. doi: 10.1016/j.biomaterials.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 17.a) Solanki A, Kim JD, Lee KB. Nanomedicine. 2008;3:567. doi: 10.2217/17435889.3.4.567. [DOI] [PubMed] [Google Scholar]; b) Shah B, Yin PT, Ghoshal S, Lee KB. Angew Chem Int Ed. 2013 doi: 10.1002/anie.201302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Zhang F, Yang H, Huang X, Liu H, Guo S. Langmuir. 2010;26:6083. doi: 10.1021/la904014z. [DOI] [PubMed] [Google Scholar]
- 19.a) Cooke DF, Goldring AB, Yamayoshi I, Tsourkas P, Recanzone GH, Tiriac A, Pan T, Simon SI, Krubitzer L. J Neurophysiol. 2012;107:3543. doi: 10.1152/jn.01101.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hallfors N, Khan A, Dickey MD, Taylor AM. Lab Chip. 2013;13:522. doi: 10.1039/c2lc40954b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Hur T. J Clin Invest. 2010;120:3096. doi: 10.1172/JCI43575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


