Abstract
A new ultra-high-throughput screening assay for the detection of cellulase activity was developed based on microfluidic sorting. Cellulase activity is detected using a series of coupled enzymes leading to the formation of a fluorescent product that can be detected on a chip. Using this method, we have achieved up to 300-fold enrichments of the active population of cells and greater than 90% purity after just one sorting round. In addition, we proved that we can sort the cellulase-expressing cells from mixtures containing less than 1% active cells.
Cellulases are important enzymes with numerous applications across multiple industries, including biofuel, pulp, paper, textile and laundry, food, feed, brewing, and agriculture.1 Most cellulases have low activity and stability, so improving these properties would have substantial impact on numerous industrial processes.
Enzymatic properties can be improved by protein engineering2 but the limiting step is the screening process. Classical screening uses microtiter plates (MTPs), where each well contains cells expressing a single type of mutant enzyme. However, this type of screening is the bottleneck in directed evolution, because a maximum number of 105 clones can be screened over the course of weeks or even months3 and large quantities of reagents and consumables are needed. High-throughput screening methods based on either fluorescence activated cell sorting (FACS)4–7 or microfluidic devices8 increase the number of clones that can be screened and reduce the amount of consumables required. Here, we demonstrate the use of a high-throughput screening system for cellulases by combining lab-on-chip sorting devices with an emulsion-based fluorescent assay previously developed for use in flow cytometry.5
Water–in-oil emulsions are needed to maintain the connection between genotype and phenotype by compartmentalizing individual cells expressing a mutant enzyme together with the components of the fluorescence assay corresponding to the enzyme activity.7 For FACS, double emulsions (water-in-oil-in-water) are required because the instrument's mobile phase is an aqueous solution. Such double emulsions can be produced by stirring or agitation,9,10 but the resulting emulsions are polydisperse and multiple water droplets may be scattered within a single oil droplet. In addition, large droplets tend to produce more fluorescence because there are more substrate molecules available for conversion into the fluorescent product. The emulsions are produced in bulk, so each droplet will be detected at a different time point from the start of the reaction. This means that increased fluorescence may result because an enzyme has worked on the substrate for a longer amount of time, and the fluorescence of the droplet may plateau before sorting as the enzyme consumes all the available substrate. Cell loading is difficult to control because the average number of cells per droplet scales with droplet volume. Also, if several inner droplets, containing cells with different activities, are encapsulated within the same outer droplet, false positives may occur upon sorting. Consequently, it is impossible to differentiate fluorescence changes due to enzyme activity from those due to other effects using polydisperse double emulsions in FACS, but it is possible to achieve plus/minus screening,4 separating cells with activity from those without.
Droplet microfluidics overcomes many of the drawbacks of high-throughput enzyme sorting with FACS. Both the size and composition of the droplets can be tuned precisely. Furthermore, once the enzyme is mixed with the substrate, the incubation time can be controlled and all compartments will have the same conditions in terms of concentration and total number of substrate molecules. Although cell loading is still subject to Poisson statistics, the probability for cells to be loaded into a given droplet is the same and can be adjusted by tuning the input cell density. These characteristics make the microfluidic method more sensitive, flexible, and quantitative at detecting changes in enzyme activity than the FACS-based sorting of double emulsions.
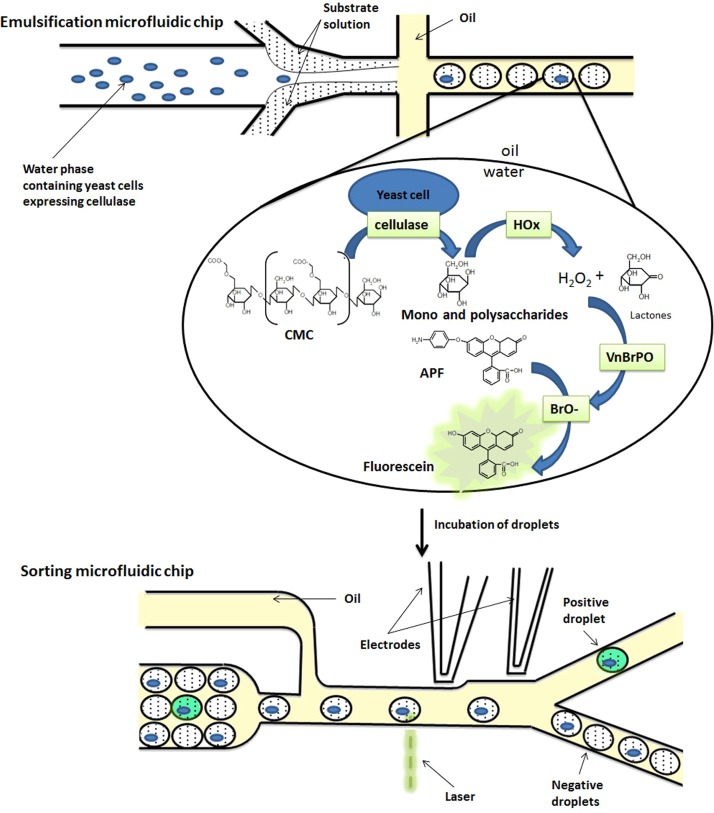
Here, we report a method in which droplet microfluidics is used to sort libraries containing different percentages of cells expressing cellulase activity and demonstrate enrichment of the cells expressing active cellulases. The entire process is summarized in Figure 1.
FIG. 1.
General overview of cellulase screening using droplet microfluidics. In the emulsification device, suspensions of yeast surface displayed libraries are co-flowed with the substrate solution at equal flow rates to a drop-forming junction where they mix. A stream of perfluorinated oil then breaks the aqueous mixture into monodisperse water-in-oil emulsions. Within each droplet, the cellulase reaction starts after compartmentalization and the fluorescent product is formed by a coupled enzymatic cascade in droplets containing cells that express the active enzyme. After a fixed incubation time, the emulsion droplets are re-injected into a microfluidic sorting device, where they are analyzed and sorted based on their fluorescence.
To detect cellulase activity, we designed an assay that uses a chain of coupled enzymatic reactions to yield fluorescence corresponding to cellulase activity without needing artificial substrates (which may lead to confounding effects, such as improved binding of the enzyme specifically to the artificial compound but not the natural substrate). In this method, cellulase hydrolyzes cellulose, its natural substrate, into monosaccharides and oligosaccharides that are further detected by the enzymatic cascade5 (Figure 1).
Based on previous FACS experiments, no difference in activity can be detected between the positive and the negative droplets before 2 h incubation time.5 Based on these observations, we expected the cells to require more than 2 h of incubation in droplets for the reaction to develop.
Emulsions were formed using a co-flow flow-focusing Polydimethylsiloxane device prepared by soft lithography as previously described8 and using fluorocarbon oil containing 1% (v/v) Krytox-PEG-Krytox detergent synthesized as reported in an earlier study.11,14 The solutions, one containing library cells (S. cerevisiae YPH500 cells, Agilent Technologies, Santa Clara, USA) and the other with the substrate,14 were mixed at the same flow rate, giving a one-to-one mixing ratio. The library cells were a defined mixture of cells transformed with cel5A pESC-Trp (positive cells) or empty pESC-Trp (negative cells). The two solutions therefore mixed just prior to encapsulation, minimizing the chance that fluorescent products would enter neighboring droplets. The substrate solution contained carboxymethyl cellulose (CMC), which has a high viscosity. To prevent fluctuations in the flow of substrate during the emulsification process, we optimized the flow rate and the concentration of CMC and found that a CMC concentration of 0.33% (w/v) produced monodisperse emulsions.
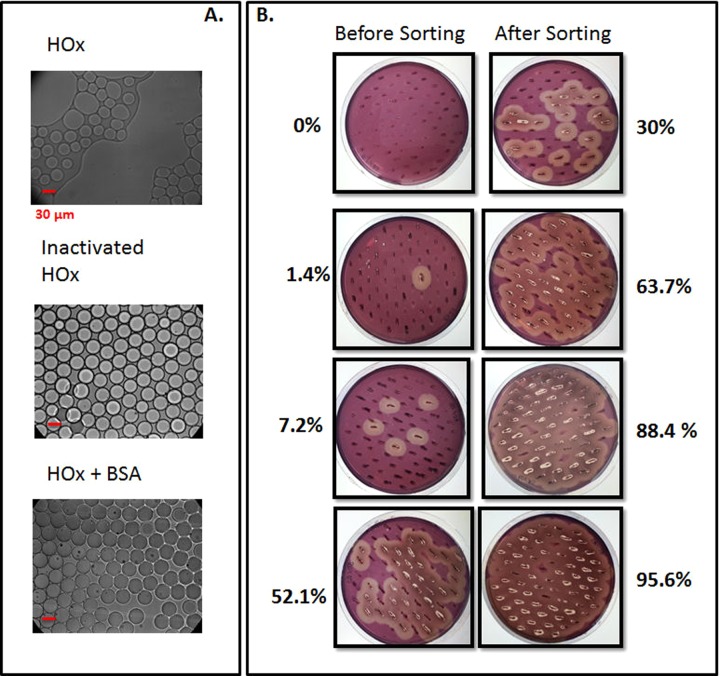
We discovered that the HOx required for the enzymatic cascade causes droplet coalescence. HOx alone was sufficient to cause the observed change in droplet stability because droplets containing only hexose oxidase in buffer exhibited the same amount of coalescence as those containing the full set of assay components. We hypothesized that the enzyme might be surface active, disturbing the emulsion interface, but emulsions of an inactivated form of the enzyme were stable (Figure 2(a)). One possible explanation is that active HOx may interact with the detergent through the active site. Adding bovine serum albumin (BSA), which is known to have a stabilizing effect,12 to the mixture improved droplet stability (Figure 2(a)). Emulsions of the assay mixture with BSA were stable for more than 1 day at room temperature.
FIG. 2.
(a) Transmission light micrographs of water-in-perfluorinated-oil emulsions produced using the microfluidic emulsification devices after 2 h incubation at room temperature. The emulsions contain 3 U/ml HOx either in its native form (left image), inactivated by heating at 99 °C for 20 min (middle image), or supplemented with 1 mg/ml BSA (right image). (b) Images of the results of the agar plate Congo Red cellulase assay before and after sorting, with the percentage of positive colonies indicated. The cells expressing cellulase activity show clear hallos.
The time required for the cellulase reaction to produce detectable quantities of fluorescent product was monitored using the droplet screening instrument. These devices proved to have a higher sensitivity than the FACS system because the optics are designed for the droplet size selected for the assay. We were able to detect cellulase activity just 20 min after the compartmentalization of cells. This shorter incubation time allowed us to couple the emulsification device directly to the droplet sorting device using a short piece of tubing. The rate of emulsion flow and the dimensions of the tube set the droplet incubation time.
Using the optimized conditions, we used droplet microfluidics to sort cellulase-expressing cells from a set of reference libraries. The reference libraries were created by mixing different concentrations of positive S. cerevisiae YPH500 cells expressing Cel5A cellulase and negative S. cerevisiae YPH500 cells transformed with the pESC-Trp empty vector. The mixed populations were emulsified together with the assay components in water-in-perfluorinated-oil emulsions and incubated at room temperature for 20 min. The gated population was sorted and the cells were spread on yeast nitrogen base casaminoacids (YNB CAA) Glu agar plates. An aliquot of the reference library was also plated on agar plates prior to sorting. Approximately, 100 cells before and after sorting were transferred to YNB CAA CMC Gal/Raf induction plates, and the Congo red assay13 was used to detect cells expressing cellulase. In this assay, colonies of positive cells developed transparent halos around them.14 The results before and after sorting are presented in Figure 2(b).
We enriched cellulase-expressing cells from a pool of negative cells, regardless of the starting concentration of positive cells. We were able to isolate the cellulase-expressing cells even when starting from a low percentage of active cells (0.1%). We obtained high enrichment factors of up to 300 when starting from low concentrations of positive cells, and we were able to sort to a purity of greater than 90%. These results exceed those obtained by comparable experiments using FACS.5
In conclusion, we developed a high-throughput screening system for cellulase activity based on droplet microfluidics. We optimized the emulsification conditions to produce highly stable and monodisperse droplets. The low dispersity of the emulsion enables the sensitive, tunable, and quantitative detection of cellulase activity. In addition, we substantially reduced the reaction time needed for the development of a fluorescent product from 2 h to 20 min. As a result, we sorted reference libraries of cellulases with various ratios of positive to negative cells, and regardless of the starting population of positive cells we were always able to enrich the active population to a higher purity than that obtained by FACS.
Acknowledgments
We are grateful to R. Sperling (School of Engineering and Applied Sciences (SEAS), Harvard University) for the kind gift of Krytox-PEG-Krytox surfactant. This work was partially supported by internal program MEF (125-600156) from the Fraunhofer Gesellschaft and by National Science Foundation (NSF) (DMR-1310266) and Harvard MRSEC (DMR-0820484). Dr. R. Ostafe would like to thank RWTH Aachen University for financial support during postdoctoral studies at Harvard University. Professor Dr. R. Prodanovic would further like to thank Fulbright Foundation for financial support. Professor Dr. Rainer Fischer would like to acknowledge the Cluster of Excellence “Tailor-made Fuels from Biomass,” which is funded through the Excellence Initiative by the German federal and state governments to promote science and research at German universities. This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF Award No. ECS-0335765. CNS is part of Harvard University. W.L.U. acknowledges the support from an NSERC PGS D.
REFERENCES
- 1.Kuhad R. C., Gupta R., and Singh A., Enzyme Res. 2011, 280696. 10.4061/2011/280696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuura T. and Yomo T., J. Biosci. Bioeng. 101, 449–456 (2006). 10.1263/jbb.101.449 [DOI] [PubMed] [Google Scholar]
- 3.Hann M. M. and Oprea T. I., Curr. Opin. Chem. Biol. 8, 255–263 (2004). 10.1016/j.cbpa.2004.04.003 [DOI] [PubMed] [Google Scholar]
- 4.Prodanovic R., Ostafe R., Blanusa M., and Schwaneberg U., Anal. Bioanal. Chem. 404, 1439–1474 (2012). 10.1007/s00216-012-6234-x [DOI] [PubMed] [Google Scholar]
- 5.Ostafe R., Prodanovic R., Commandeur U., and Fischer R., Anal. Biochem. 435, 93–98 (2013). 10.1016/j.ab.2012.10.043 [DOI] [PubMed] [Google Scholar]
- 6.Prodanovic R., Ostafe R., Scacioc A., and Schwaneberg U., Comb. Chem. High Throughput Screening 14, 55–60 (2011). 10.2174/1386207311107010055 [DOI] [PubMed] [Google Scholar]
- 7.Tawfik D. S. and Griffiths A. D., Nat. Biotechnol. 16, 652–656 (1998). 10.1038/nbt0798-652 [DOI] [PubMed] [Google Scholar]
- 8.Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J.-C., Marquez M., Klibanov A. M., Griffiths A. D., and Weitz D. A., Proc. Natl. Acad. Sci. U. S. A. 107, 4004–4009 (2010). 10.1073/pnas.0910781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aharoni A., Amitai G., Bernath K., Magdassi S., and Tawfik D. S., Chem. Biol. 12, 1281–1289 (2005). 10.1016/j.chembiol.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 10.Hai M., Bernath K., Tawfik D., and Magdassi S., Langmuir 20, 2081–2085 (2004). 10.1021/la035402+ [DOI] [PubMed] [Google Scholar]
- 11.Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angilè F. E., Schmitz C. H. J., Köster S., Duan H., Humphry K. J., Scanga R. A.et al. , Lab Chip 8, 1632–1639 (2008). 10.1039/b806706f [DOI] [PubMed] [Google Scholar]
- 12.Hai M. and Magdassi S., J. Controlled Release 96, 393–402 (2004). 10.1016/j.jconrel.2004.02.014 [DOI] [PubMed] [Google Scholar]
- 13.Nakazawa H., Okada K., Kobayashi R., Kubota T., Onodera T., Ochiai N., Omata N., Ogasawara W., Okada H., and Morikawa Y., Appl. Microbiol. Biotechnol. 81, 681–689 (2008). 10.1007/s00253-008-1667-z [DOI] [PubMed] [Google Scholar]
- 14. See supplementary material at http://dx.doi.org/10.1063/1.4886771E-BIOMGB-8-002404 for a detailed description of the experimental setup.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4886771E-BIOMGB-8-002404 for a detailed description of the experimental setup.