Abstract
With their advantages as molecular recognition elements, aptamers have been extensively studied and used for bioanalytical and biomedical applications. However, the process of enrichment and screening of aptamers remains a bottleneck for aptamer development. Recently, microfluidic methods have been increasingly used for rapid and efficient aptamer selection, showing their remarkable advantages over conventional methods. This review briefly introduces aptamers and their advantages. The conventional process of generating aptamers is discussed, followed by the analysis of the key obstacles to efficient aptamer selection. Microfluidic methods for highly efficient enrichment and screening of aptamers are reviewed in detail.
INTRODUCTION
Selective molecular recognition is always an important issue for chemical sensing, diagnosis and therapy. Antibodies have been generated for this purpose1 and have contributed significantly to a wide variety of applications.2–4 However, many limitations have been identified, such as difficult production, batch-to-batch variation, high cost, immunogenicity, temperature sensitivity, and irreversible denaturation.5,6 Since 1990, chemical antibodies called aptamers have emerged as potential alternatives for molecular recognition. Aptamers, first reported by Tuerk and Gold7 and Ellington and Szostak,8 are single-stranded oligonucleotides that can bind to different types of targets, such as small molecules, proteins, cells, and even tissues, with high affinity and selectivity.9–11
Aptamers have many advantages over antibodies.5 First, once the sequence is known, it can be easily and cheaply produced via standard solid phase synthesis techniques with no or little batch-to-batch variation. This solid-phase synthesis technique also enables the easy modification of aptamers with fluorophores, biotin, and other functional groups. Furthermore, aptamers are much smaller than antibodies, permitting rapid tissue penetration. Theoretically, aptamers can be in vitro selected against any molecule, including toxins, and the entire selection procedure can be controlled without physiological constraints. Finally, aptamers are quite stable even after exposure to extreme conditions and can be stored dry at room temperature.
There have been significant advances in the use of aptamers as recognition elements in many biosensor designs, and different approaches in this area have been reviewed extensively.11–16 The compatibility of aptamers with various detection schemes, such as electrochemical,13 fluorescence,17,18 colorimetric,19–22 chemiluminescence,23 field effect transistors,24 and surface plasmon resonance (SPR),25,26 has enhanced the progress of this field. Therapeutic application of aptamers became a reality in late 2004 when Macugen® was approved as a drug for age-related macular degeneration.27,28 Macugen® was developed using vascular endothelial growth factor hormone (VEGF) as the target molecule. Similarly, therapeutic aptamers are being developed for nucleolin,29,30 thrombin,31 platelet derived growth factor (PDGF),32 human neutrophil elastase (hNE),33 etc. In addition, many other aptamers have been used in different application platforms, such as in vivo tumor imaging, gene delivery, targeted drug delivery and as tools for cancer biomarker discovery.34–37 For example, the anti-prostate-specific membrane antigen (PSMA) aptamer has been extensively used for delivery of drugs38 and siRNA39 into PSMA-positive cells. The anti-epithelial cell adhesion molecule (EpCAM) aptamer has been applied for circulating tumor cell capture and enrichment.40 The aptamer sgc8 selected against leukemia cell line CCRF-CEM has been successfully applied for biomarker discovery to find tumor specific protein tyrosine kinase 7 (PTK7).41
Despite great promise and significant efforts in aptamer development over the past 20 years, it is troublesome that only a limited number of aptamer-target pairs have been intensively used, mainly for proof-of-principle of novel aptamer assays.15 For example, by far, the most frequently used aptamer is the anti-thrombin aptamer, which has been employed in >1000 publications, followed by aptamers targeting adenosine, cocaine, and platelet-derived growth factor, all of which account for over one-third of the total publications on aptamers. This lack of variation severely impedes the development and application of aptamers. Two reasons have been previously proposed to explain this problem. First, the intrinsic property of natural DNA with limited chemical diversity of interaction may lead to difficulty in selecting aptamers against their targets.42 This problem has been addressed to some extent by incorporating various artificial nucleobases with functional groups to increase the diversity of randomized libraries for improving the success rate of aptamer selection.42–44 Another reason arises from the intrinsic limitations of the conventional aptamer selection technique, known as SELEX (Systematic Evolution of Ligands by EXponential enrichment).7,8 In SELEX, two major steps are involved: the enrichment of candidates from a large initial library with 1014–1015 random sequences after many iterative rounds of selection, and the screening of tens to hundreds of aptamer candidates from the enriched library.45 The entire process is time-consuming, labor-intensive, inefficient, and expensive.
To overcome this limitation, great efforts have been put forth to develop innovative methods for rapid, efficient, and high-throughput generation of aptamers. Especially, due to the advantages of reduced reagent consumption, simple analytical methodology, high processing speed, high throughput, and automation potential, microfluidic technology has revolutionized the field of aptamer selection in terms of increased speed, reduced costs, improved resolving power, high throughput and automation.46–48 In this review, the conventional SELEX procedure will first be briefly introduced, bottlenecks in the conventional SELEX procedure will be discussed, and then, recent advances in aptamer selection using microfluidic platforms will be reviewed.
CONVENTIONAL APTAMER SELECTION PROCESS
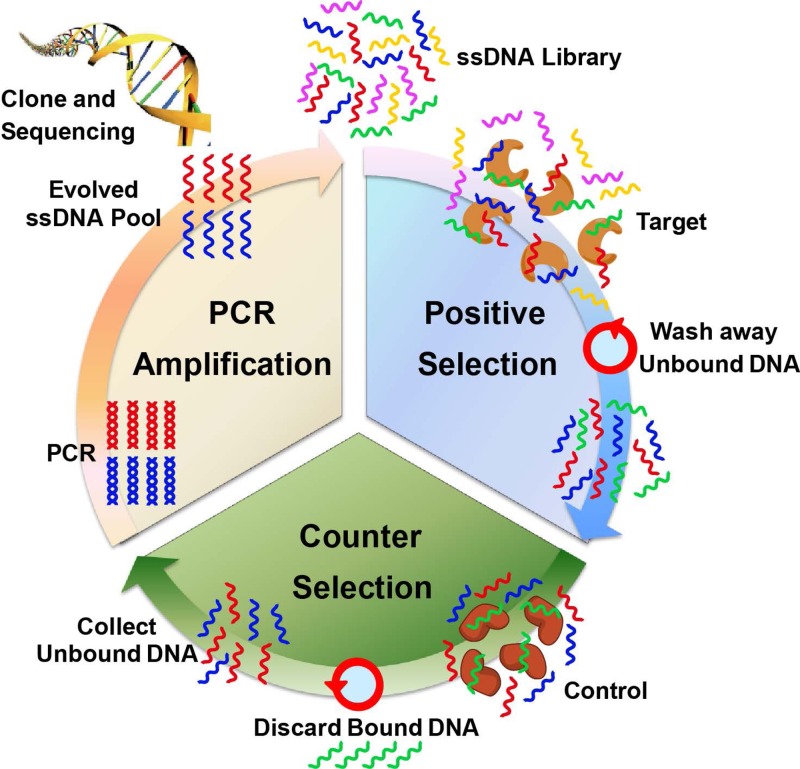
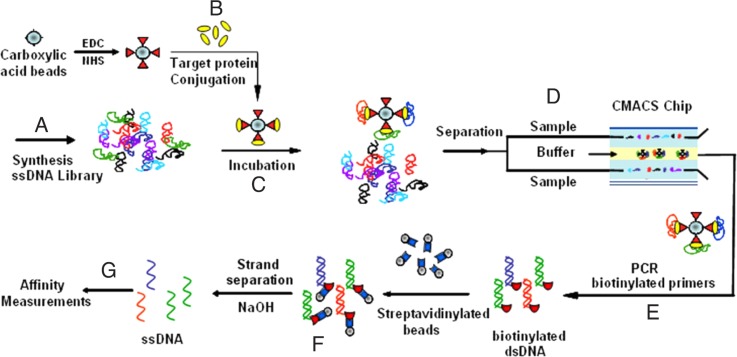
SELEX is an in vitro selection method designed to identify aptamers that are selectively bound to target molecules with high affinity. Typically, a combinatorial library of ssDNA or RNA sequences is generated containing a 30–50 nt randomized region flanked by ∼20 nt constant primer regions to facilitate polymerase chain reaction (PCR) amplification. This creates a library with a large number of random sequences (1014–1015), which is sufficiently diverse to contain a wide variety of potential binding ligands. In the enrichment process (Figure 1), the prepared initial library is incubated with the desired target molecule for binding. The unbound nucleic acids are removed from those bound specifically to the target molecule by various partitioning techniques, including filtration, affinity chromatography, or panning separations. These binding sequences are collected, PCR amplified, and purified, generating a new single-stranded nucleic acid pool suitable for further rounds of enrichment. Sometimes, counter selection is involved to remove the nonspecific binding sequences by incubating the library with negative controls and collecting the unbound sequences. The procedure is repeated iteratively several times (8–20 times in practice) with progressively stringent binding or washing conditions until the pool converges to one or a few sequence families. During the selection, the parameters can be easily manipulated to obtain aptamers optimal for a broad range of conditions, such as pH, temperature or buffer composition. After the library is enriched with sufficient affinity and selectivity, it is cloned into plasmids, which are then transfected into bacteria. Bacteria are grown and single colonies are picked and sequenced in large quantities to obtain aptamer candidates. After analysis of hundreds to thousands of sequences, the discovered consensus sequences with high repeats are chemically synthesized and their binding affinity and selectivity are tested individually.
FIG. 1.
Schematic illustration of the traditional SELEX process. A library of ssDNA is incubated with the target molecules for positive selection. The unbound sequences are removed by the chosen separation method. Then, the collected binding sequences are subjected to counter selection by incubating with control molecules to remove nonspecific sequences. The unbound sequences are collected and amplified by PCR to evolve an enriched ssDNA pool suitable for further rounds of selection. After the library is enriched, DNA cloning and sequencing are used to identify individual aptamer sequences for further tests.
While the conventional SELEX process is straightforward, it suffers from several intrinsic limitations. For example, in the enrichment step, the iterative procedure is tedious, time-consuming, and labor-intensive. Targets, such as proteins and small molecules, need to be immobilized on certain stationary phase for better separation. Care must be taken not to introduce bias during amplification or non-specific binding during selection. Typically, 8–20 rounds of selection are performed, which usually takes weeks to months to complete with no guarantee of success. In the screening step, always the sequences with high repeats are picked, and sequences with low percentage are often ignored, leading to loss of some high affinity sequences that have low abundance. Furthermore, chemical synthesis of long DNA sequences is an expensive process, especially with fluorophore labelling for affinity assessment. Therefore, considerable effort have been involved in streamlining the selection process, either by improving the separation efficiency to reduce the number of rounds of selection or by enabling direct evolution of enriched sequences to rapidly obtain aptamer sequences.
MICROFLUIDIC APPROACHES FOR IMPROVING SEPARATION EFFICIENCY
One of the most critical steps in SELEX is the separation of the bound species from molecules not bound to the target. Improvement of separation efficiency can dramatically reduce the number of rounds of selection and improve the selection throughput. Although affinity or filter separations are relatively straightforward, they do not offer particularly high resolving power in separation. Therefore, a wide variety of microfluidic techniques have been explored to enhance the separation efficiency, including capillary electrophoresis (CE),49–52 sol-gel isolation,53,54 and magnetic-channel-based selection.55,56
CE microfluidic SELEX
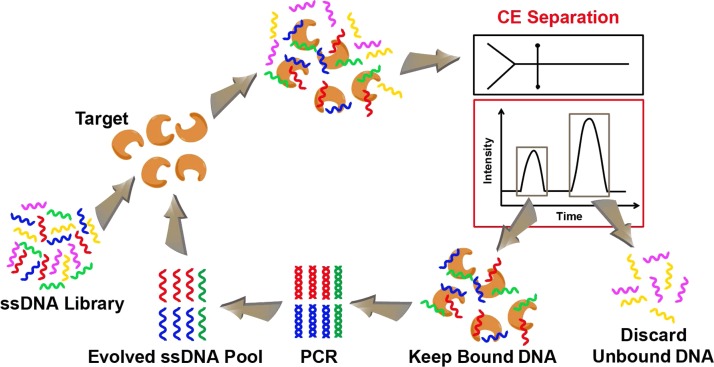
CE is a powerful analytical method that combines highly efficient separation and highly sensitive quantitative detection with the potential of automation and parallelism. Due to these advantages, Mendonsa and Bowser introduced CE into SELEX for isolating aptamers as CE-SELEX.49 The process of CE-SELEX is schematically shown in Figure 2. The random nucleic acid library is incubated with the target in free solution, which is then separated by CE. Unbound sequences migrate through the capillary with the same mobility, regardless of their sequence or length. Bound nucleic acids with altered size (due to the additional target moiety) migrate at a different mobility, allowing them to be collected as different fractions. Then, the collected sequences are PCR amplified, purified, and made single-stranded to generate a new pool suitable for the next round of selection. CE-SELEX has been widely used to successfully isolate aptamers for large targets, such as human immunoglobulin E (IgE),49,57 human immunodeficiency virus (HIV) reverse transcriptase,58 and protein kinase K.59 It has also been demonstrated to isolate aptamers against small targets, such as neuropeptide Y60 and N-methyl mesoporphyrin.61 The incubation and separation are performed in free solution, which greatly reduces the nonspecific binding and simplifies the process by eliminating target immobilization on a stationary phase. The high partitioning efficiency of the CE-SELEX allows it to significantly reduce the number of rounds to 1–4 and increase binding affinity and specificity of the selected aptamers.
FIG. 2.
Schematic illustration of the CE microfluidic SELEX process. A library of ssDNA is incubated with the target molecules. Capillary electrophoresis is used to separate bound sequences from unbound ones. Binding sequences are amplified by PCR and evolved as an enriched ssDNA pool suitable for further rounds of selection.
Krylov and his colleagues have developed a similar CE-SELEX named nonequilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) with exceptionally high selection efficiency.50 In the nonequilibrium condition, pure separation buffer containing neither library nor target is applied. Therefore, the equilibrium between DNA and DNA/target complex is no longer maintained and the DNA/target complex starts dissociating immediately after sample injection. The NECEEM can also accurately determine the binding parameters of the aptamer-target interaction during the selection, such as equilibrium dissociation constant (Kd), rate constants of complex formation (kon) and dissociation (koff). Furthermore, the Krylov group introduced equilibrium capillary electrophoresis of equilibrium mixtures (ECEEM) to CE-SELEX.51,62 In ECEEM, the electrophoresis running buffer contains the target at the concentration identical to that of incubated mixture. As a result, the dynamic equilibrium between DNA and DNA/target complex during capillary separation is maintained and aptamer candidates with different Kd values migrate with different mobilities. They applied ECEEM method for selection of smart aptamers with Kd values approaching theoretically predicted values in three rounds.
Later, a non-SELEX selection process for aptamers was also introduced by Krylov et al.52,63 By taking advantage of the low mass requirement of CE, the non-SELEX method eliminates PCR amplification from the aptamer selection process with only repeated partitioning steps. NECEEM has been used to partition the DNA-target complex from the free DNA. The advantage of non-SELEX is its speed and simplicity. Krylov et al. used h-Ras protein as the target, and completed the selection process within 1 h with affinity improved by more than 4 orders of magnitude. In contrast to the several days or several weeks required for traditional SELEX, the non-SELEX procedure can be automated using a single commercially available CE instrument.
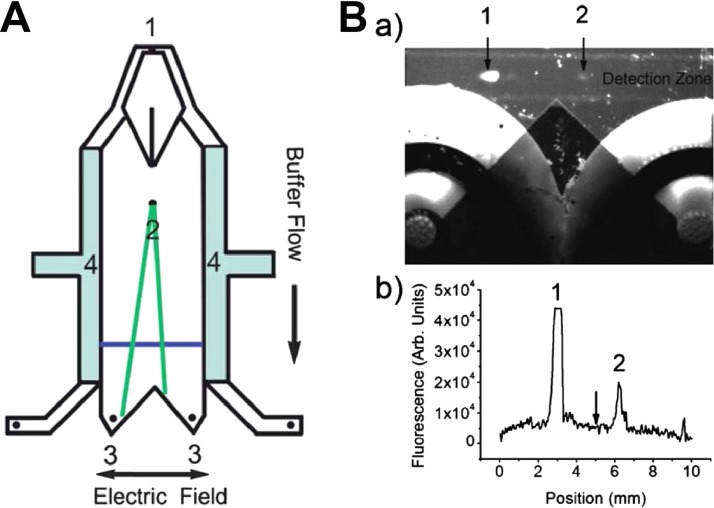
However, one of the significant drawbacks of CE-SELEX is the very small volume that can be injected, only several nanoliters. This limits the number of sequences that can be assessed and requires very high library concentration. In addition, the collection of complexes as they migrate from the end of the capillary is associated with complicated timing. To solve these problems, recently, Jing and Bowser developed a micro free flow electrophoresis (μFFE) device for aptamer selection against human IgE.64 As schematically shown in Figure 3(A), about 3 μl of the library-IgE mixture (∼1.8 × 1014 sequences) can be continuously streamed into a planar separation chamber within 30 min, allowing over 300-fold more volume than traditional CE-SELEX. Then, an electric field of 150 V/cm is applied perpendicular to the pressure driven flow, deflecting analytes laterally according to their mobilities. The unbound ssDNA sequences are deflected toward the anode due to the suppressed electroosmotic flow (EOF), while aptamer-IgE complexes are deflected only minimally and collected continuously, making the process much simpler than traditional CE-SELEX. Moreover, the transit time through the μFFE flow chamber is only 10–20 s, greatly decreasing the dissociation potential during the separation, compared with 5–15 min for traditional CE-SELEX. Although the separation efficiency of μFFE is expected to be lower than that of CE, the high rate of enrichment ensures the identification of high affinity aptamers after a single round of selection.
FIG. 3.
(A) Schematic of a μFFE device containing the buffer inlet (1), sample inlet (2), fraction collection outlets (3), and electrode channels (4). The blue line denotes the detection zone where the laser line is expanded across the separation channel. (B) (a) An image of a μFFE separation of free (1) and bound ssDNA (2) during the first round of selection. (b) A linescan across the detection zone imaged in (a). The arrow indicates the fraction cutoff point at the exit of the μFFE channel demonstrating clear separation. Reprinted with permission from M. Jing and M. T. Bowser, Lab Chip 11(21), 3703 (2011). Copyright 2011 Royal Society of Chemistry.
The CE-SELEX has the advantages of being performed in free solution, thereby reducing the opportunity for non-specific interactions and eliminating complicated immobilization strategies. However, it has been limited to targets that can induce sufficient changes of aptamers in the electrophoretic mobilities upon binding, while still maintaining size compatibility with the CE channel. Therefore, some small molecules that cannot cause a mobility shift when bound to aptamers, as well as cells or whole organisms that are larger than the CE channel, cannot be used to generate aptamers by CE-SELEX.
Sol-gel microfluidic SELEX
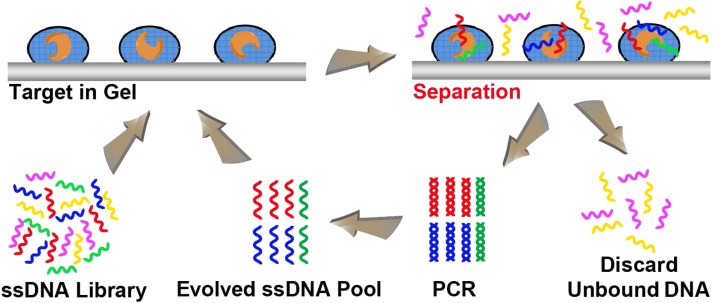
Many studies have described the encapsulation of a variety of biomolecules, including enzymes, antibodies, regulatory proteins, membrane receptors, and even whole cells, using a wide range of sol-gel derived nanocomposite materials.65–67 The proteins entrapped in sol-gels typically exhibit improved stability to thermal and chemical denaturation and increased storage capability over several months.53,68,69 The nanoporous structure of the sol-gels can allow the diffusion of some molecules, such as aptamers, while keeping others, such as proteins and chemicals, encapsulated in the pores. Based on this phenomenon, Kim et al. developed a sol-gel-based microfluidic SELEX for high throughput and efficient aptamer selection.54 Sol-gels are synthesized using silicate glasses with nanoporous structures. The encapsulation of proteins in sol-gels has the advantages of avoiding affinity capture tags or recombinant proteins for chemical immobilization on stationary phase in traditional SELEX, enabling the entrapped proteins to be maintained in their native states. Then, the sol-gels are spotted as droplets on an aluminum electrode-in-chip. The sol-gels can hold a large amount of active proteins in the nanoporous structure. The microfluidic chip allows sufficient binding between the nucleic acid library and the immobilized target proteins to trap the binding sequences with the target protein in the sol-gel droplets (Figure 4). After washing away non- or weak binding sequences, the electrode-in-chip acts as microheater to thermally elute the binding sequences with higher elution efficiency than that of other elution methods, such as ionic strength change. In addition, by aligning multiple sol-gel droplets containing different proteins on the chip, sol-gel SELEX has the ability to select aptamers to multiple proteins in a single cycle, significantly improving the throughput. The selected aptamers can be individually processed by localized heating of specific droplets. Meanwhile, these multiple proteins can serve as competitors for each other to improve the selection specificity.
FIG. 4.
Schematic illustration of the sol-gel microfluidic SELEX process. A library of ssDNA is incubated with sol-gel arrays of proteins in a microfluidic system for efficient selection of ssDNA aptamers against target molecules. The unbound sequences are discarded and bound sequences are amplified by PCR to evolve an enriched ssDNA pool suitable for further rounds of selection.
Sol-gel SELEX has been successfully applied to generate RNA aptamers with Kd in low nM range against recombinant yeast TATA binding protein and yeast transcription factor IIB protein with improved selection efficiency, which reduced the number of selection cycles to 5–8 rounds, compared with 8–13 rounds for conventional filter-based SELEX.54,70 Furthermore, Kim et al. optimized the formulation of the sol-gel composition for encapsulation of small molecules like bisphenol A and xanthine without chemical coupling.71,72 The sol-gels were microarrayed and tightly anchored to a porous silicon substrate for conveniently isolating high affinity aptamers against low molecular weight compounds. The sol-gel SELEX method allows for sufficient binding reactions between nucleic acids and targets and can lead to the efficient and multiplex isolation of aptamers specific to many targets, including proteins and small molecules. However, certain concerns still exist, such as the protein stability and integrity through the multiple selection cycles in sol-gel SELEX.
Magnetic bead-based microfluidic SELEX
Magnetic bead-based selection has been widely used for selecting aptamers against small molecules, proteins, and cells by immobilizing molecular targets on bead surfaces.73,74 However, the separation efficiency of magnetic selection has lagged far behind that of CE and other advanced separation methods, thus requiring multiple time-consuming selection rounds and delicate manual manipulation. To improve the separation efficiency, the Soh group has introduced microfluidics technology to integrate with magnetic bead-based SELEX (M-SELEX) for highly efficient isolation of aptamers.55 The process of M-SELEX is schematically shown in Figure 5. First, the target proteins are bound to the magnetic beads via bioconjugation. The number of proteins on each bead is precisely controlled, so as to control the ratio of targets and nucleic acids for highly stringent competition. Then, the prepared target-coated magnetic beads are incubated with oligonucleotide library in binding buffer. The mixture is separated in a continuous-laminar-flow, magnetically-activated, chip-based separation (CMACS) device to purify the DNA bound to target-coated beads. In the magnetic field, the magnetic beads travel along the chip at the center and elute through the middle product outlet, while the unbound nucleic acids elute through the side waste outlet. Using this M-SELEX method, an enriched aptamer pool was obtained that tightly bound to the light chain of recombinant Botulinum neurotoxin type A after a single round of selection with a Kd value of 33 nM. The M-SELEX system was demonstrated to be rapid, highly efficient, automatable, and applicable to a wide range of targets. However, it suffered from some practical disadvantages, such as the distortion of the flow streams by microbubbles and the low aptamer purity and recovery caused by bead aggregations in the microchannel.
FIG. 5.
Schematic illustration of the magnetic bead-based microfluidic SELEX process. The microfluidic selection process begins with the incubation of random ssDNA library (a) with target proteins conjugated to magnetic beads (b). After incubation (c), the separation of the target-bound sequences from the unbound ones is performed in the continuous-laminar-flow, magnetically-activated, chip-based separation (CMACS) device (d). Stringent washing conditions are then imposed in the microchannel to continuously elute weakly- and unbound sequences from the microfluidic chip. After the separation, the external magnets are removed, and the beads carrying the selected aptamers are released from the device. The bound sequences are amplified via PCR (e) and single-stranded products are generated (f). Finally, the binding affinities of the resulting aptamers are measured (g). Reprinted with permission from Lou et al., Proc. Natl. Acad. Sci. U.S.A. 106(9), 2989 (2009). Copyright 2009 National Academy of Sciences.
To overcome these problems, Soh et al. further improved the M-SELEX by fabricating the microchannel with ferromagnetic materials that can be magnetized with external magnets.75 During the separation, the magnetic beads are trapped in the channel by the magnetic field, and stringent washing condition can be imposed to continuously elute weakly- and unbound nucleic acids. After the separation, the external magnets are removed, and beads carrying aptamers are released. The usage of external magnets enables accurate control of the hydrodynamic and magnetophoretic trapping forces for high molecular partition efficiency. They performed three rounds of positive selection to select aptamers against streptavidin with strong binding affinity. In order to further increase the specificity of the selected aptamers, they performed another round of negative selection against bovine serum albumin (BSA). This is the first report to use both positive and negative selection in a microfluidic device for rapid isolation of aptamers with high affinity and specificity. Compared with conventional magnetic separation methods, M-SELEX is significantly more efficient, and the improved version has very high recovery of beads (∼99.5%) and partition efficiency (106). In addition, as relatively higher flow rates can be applied (>10 ml/h), the entire process of trapping, washing, and release can be achieved in less time (∼5 min). Aptamers against streptavidin75 and PDGF-BB56 were isolated in three rounds by this improved M-SELEX with Kd values of 25 nM and 3 nM, respectively.
Recently, Soh et al. combined the M-SELEX with next-generation sequencing and in situ-synthesized aptamer arrays to develop a quantitative parallel aptamer selection system (QPASS),76 which enables simultaneous characterization of the affinities and specificities of thousands of candidate aptamers in parallel (Figure 6). The target, human cancer biomarker angiopoietin-2 (Ang2), was immobilized on magnetic beads for four rounds of M-SELEX with moderate selection conditions. To quantitatively identify aptamer sequences that were enriched in each round of M-SELEX, they sequenced the all four rounds of pools with a next-generation sequencing technique and rank-ordered all sequences from each pool based on their copy number. The top 235 represented aptamer candidates from each pool were then synthesized on commercialized aptamer chips, with three copies of each sequence for triplicate measurement. The Kd values and binding specificities for thousands of aptamer candidates were quantitatively measured simultaneously. Such high-throughput aptamer characterization has relatively constant time and labor requirements, regardless of the number of molecules being analyzed. Using the QPASS method, they identified six high-affinity Ang2 aptamers with Kd < 30 nM and excellent specificity.
FIG. 6.
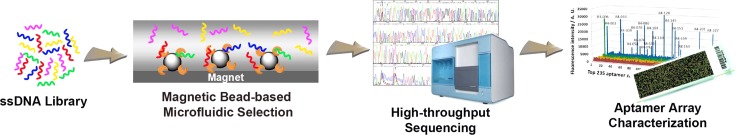
Schematic illustration of QPASS method for high-throughput aptamer characterization. Single DNA sequences of an enriched library are obtained by magnetic bead-based microfluidic selection. Then, the enriched pool from each round is sequenced by next-generation high-throughput sequencing technique. The top 235 from each pool (possible aptamer candidates) are in situ synthesized on aptamer chips to characterize their binding affinity and specificity in a high-throughput parallel manner.
M-SELEX is able to perform highly efficient positive and negative selection of aptamers with high affinity and specificity, significantly enhancing the sensitivity and specificity of the SELEX process. Still, several issues need to be addressed for M-SELEX methods. The chemical coupling between target molecules and magnetic beads may affect the structure and property of targets, and this possibility needs to be investigated case by case. Also, nonspecific interactions of nucleic acids to magnetic beads and the channel surface may occur during the selection. Several solutions have been proposed to resolve this problem, such as using negatively charged magnetic beads via electrostatic interactions55 or using ionic liquid modified channel surfaces77 to inhibit nonspecific interactions. Furthermore, so far only relatively simple and relatively low-throughput microfluidic chips have been used, and M-SELEX has not been carried out on a large scale.
INTEGRATED MICROFLUIDIC SYSTEM FOR APTAMER SELECTION
These above microfluidic SELEX systems have been applied only to the extraction step, not to the entire iterative SELEX process. The critical incubation step for ssDNA binding to targets and the subsequent PCR amplification still requires time-consuming and labor-intensive procedures in order to complete the entire SELEX process. Prolonged time and loss of the extracted aptamers may be caused by the later procedures. Microfluidics technology has the potential for automation of sample preparation, injection, manipulation, and filtration in a single chip. If the entire SELEX process can be moved to a chip-based microfluidic environment, SELEX could potentially be standardized with the advantages of increased speed and reduced cost.
Therefore, Hybarger et al. developed the first prototype of automated microfluidic SELEX system using LabView-controlled actuatable valves and a PCR machine.78 The prototype comprises reagent-loading microlines, pressurized reagent reservoir manifold, actuatable valves, and thermocycler. First, the target lysozyme is biotinylated and immobilized on streptavidin coated microline. The reagents are preloaded into respective microlines using the pressurized reagent delivery module. The delivery of reagents is controlled by actuatable valves via dye sensing sensors. Except the initial loading of DNA pool, the processes of in vitro transcription, incubation with targets, separation of bound sequences and reverse transcription (RT)/PCR amplification are all performed automatically on chip as programmed by LabView. Such an automatic microfluidic SELEX prototype has demonstrated an excellent example to generate integrated, self-contained and microfabricated platform for simple, rapid, and efficient aptamer selection.
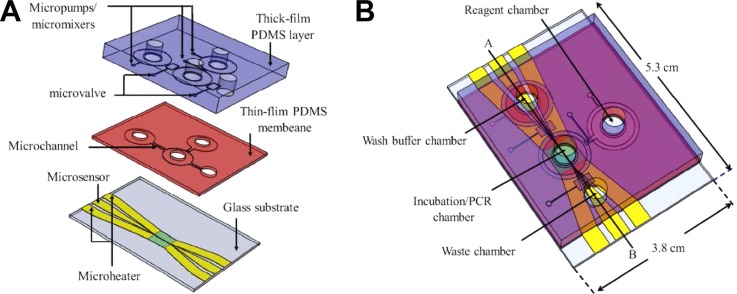
Later on, Huang et al. developed an automatic, magnetic bead-based microfluidic system integrated with a random ssDNA extraction device and an on-chip nucleic acid amplification device (micro-PCR) for rapid screening of C-reactive protein aptamers.79 The integrated microfluidic system was composed of three modules: a microfluidic control module for sample incubation and transportation processes, a magnetic bead-based ssDNA extraction module for aptamer screening, and a rapid nucleic acid amplification module (Figure 7). The entire process involving incubation, separation and amplification was performed on a single chip automatically within about 60 min for a single round, which is much faster than that of a traditional SELEX process. Later, Huang et al. improved the automatic microfluidic SELEX system by integrating it with a competitive assay chip,80 which can examine the affinity and specificity of selected nucleic acids before cloning and sequencing. With this approach, an aptamer specific to alpha-fetoprotein, a biomarker for liver cancer, was successfully selected by six rounds cycles with Kd value of 2.37 nM.
FIG. 7.
A schematic diagram of separated (A) and integrated (B) magnetic bead-based microfluidic system, comprised of three major modules: a microfluidic control module, a magnetic bead-based aptamer extraction module, and a rapid nucleic acid amplification module. Reprinted with permission from Huang et al., Biosens. Bioelectron. 25(7), 1761 (2010). Copyright 2010 Elsevier.
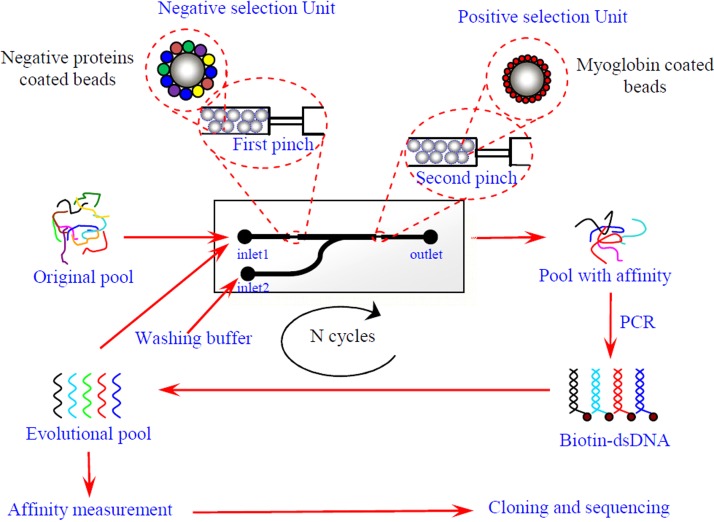
Very recently, Wang et al. developed an integrated microfluidic chip with both positive and negative selection.81 Taking cardiac markers, myoglobin (Myo), as an example, the working principle of microfluidic SELEX is illustrated in Figure 8. The target protein (Myo) and negative selection proteins, i.e., BSA, human serum albumin (HSA), and C reactive protein (CRP), were first coupled with polystyrene microbeads via passive adsorption, respectively. Next, the prepared beads were inhaled into the channel used the pump and blocked before two pinches, respectively. Then, the prepared DNA library was injected into channel at 0.5 μl/min from the inlet 1. The nonspecific sequences were retained in the first pinch, and the target bound sequences were kept in the second pinch. The washing buffer and eluting buffer were pumped into channel at 2 μl/min from inlet 2 in turn to wash away nonbinding sequences and elute the bound ones. The eluted ssDNA was collected from the outlet and amplified by PCR to generate the library for next round of selection. After 7 rounds of selection, the high affinity aptamers with Kd values at the nanomolar level were successfully obtained. This is the first example of microfluidic SELEX integrated with both positive and negative selection units, which may have the potential to be applicable to screening a wide variety of molecules against aptamers with high affinity and specificity.
FIG. 8.
Myoglobin-aptamer selection based on positive and negative selection units integrated microfluidic chip. Reprinted with permission from Wang et al., Anal. Chem. 86(13), 6572 (2014). Copyright 2014 American Chemical Society.
MICROFLUIDIC METHODS ENABLING DIRECT EVALUATION OF ENRICHED SEQUENCES
In contrast to the development of enrichment methods for improving the separation efficiency, rare attention has been paid to the screening process over the past two decades. In a normal screening process, the enriched DNA library has to be cloned into plasmids, and then the plasmids are transfected into bacteria for growing. Afterwards, the colonies are picked and sequenced in large quantities to obtain aptamer candidates. After bioinformatic analysis, possible candidates are then chemically synthesized, and their binding affinities are measured individually. Such a process is time-consuming, labor-intensive, inefficient, and expensive.
Agarose droplet emulsion PCR for efficient and cost-effective aptamer selection
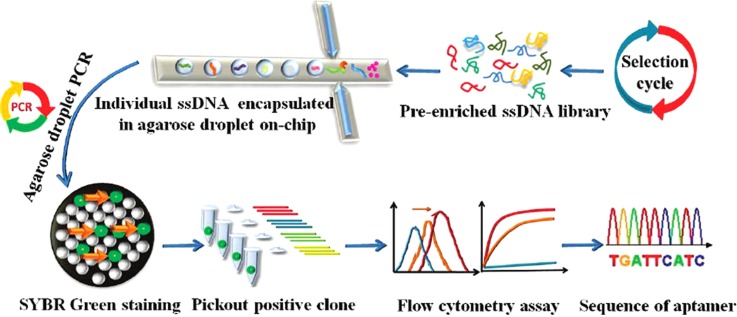
To address these problems, we have developed a novel method for efficiently screening aptamers from a complex ssDNA library by employing single-molecule emulsion PCR (ePCR) based on agarose droplet microfluidics.82 In ePCR, water-in-oil (W/O) emulsion is utilized to generate numerous droplets of reaction mixture in bulk oil phase, enabling the performance of millions of independent PCR amplification reactions in parallel. As the working-flow in Figure 9 shows, ssDNA of a pre-enriched library against cancer biomarker, SH2 domain-containing phosphatase (Shp2), was statistically diluted and mixed with agarose solution containing PCR reagents. The mixture was used as the water phase and injected into microfluidic chip with flow-focusing design. Agarose droplets were generated when the water phase are cut by the perpendicular oil phase. Then, agarose droplet ePCR was performed. The used ultralow gelling temperature agarose has a melting point about 56 °C and a gelling point around 16 °C with unique thermo-responsive sol-gel switching property. It remains in the liquid phase at all PCR temperatures, such that PCR can take place with high efficiency. After off-chip PCR amplification, the solution form of the agarose droplet can be switched to the solid gel phase by simply cooling the solution below the gelling point of agarose. Since the PCR forward primer is conjugated to agarose, amplicons can physically attach to the agarose matrix after PCR. As a result, DNA products amplified in the droplet can retain their monoclonality even after the oil phase is removed and afford flexible downstream processing and analysis.
FIG. 9.
Schematic illustration of direct evaluation of enriched sequences by agarose droplet microfluidics. Single DNA sequences of an enriched library obtained by traditional SELEX are encapsulated individually into agarose droplets for high-throughput single-copy DNA amplification. The resulting agarose droplets are cooled to become agarose beads and stained with SYBR Green to pick out highly fluorescent beads containing DNA colonies. The binding affinity of DNA in each fluorescent bead against the target molecule is screened. DNA sequences with low Kd values and good selectivity can be directly used as aptamers or can be sequenced and synthesized for further study. Reprinted with permission from Zhang et al., Anal. Chem. 84(1), 350 (2012). Copyright 2012 American Chemical Society.
After ePCR amplification and generation of agarose beads by cooling, the agarose beads were stained with SYBR Green. Only the bright clonal beads containing DNA were selected. The binding ability of amplified ssDNA from each clonal bead was then screened via high-throughput fluorescence flow cytometry. Only the amplified sequences with high binding affinity and high selectivity were chosen as aptamers and sequenced, or they could be directly used for downstream biomedical applications. Compared to the conventional cloning-sequencing-synthesis-screening work flow, this method takes advantage of the compartmentalization of microfluidic droplets and allows rapid molecular evolution of individual DNA sequences from an enriched library prior to knowing their exact sequence information, making the entire process more rapid, efficient, and cost-effective. This approach could also be further applied to other molecular evolution technologies including mRNA display and phage display.
Monoclonal surface display (MSD) SELEX for aptamer enrichment and identification
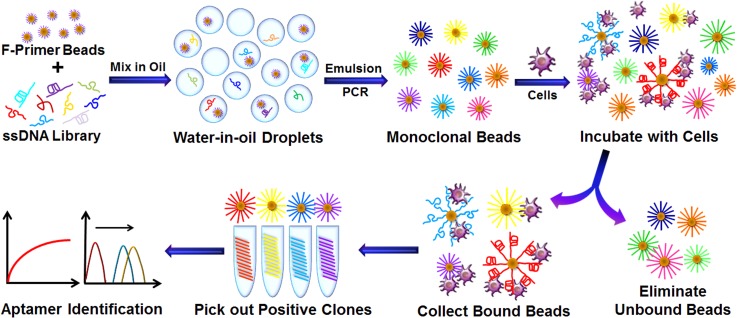
Although the agarose droplet microfluidic approach can evaluate the binding affinity of individual sequences before sequencing and DNA synthesis, the individual sequences from an enriched library still contains aptamer candidates having a broad distribution of binding affinity. A time-consuming screening process is still required to identify high-affinity ligands hidden in a large group of less desirable choices. Therefore, the SELEX methods allowing efficient enrichment and identification of high affinity aptamers without cloning, sequencing, chemical synthesis, and screening would greatly accelerate the selection process and speed up the development of aptamers. Towards this end, very recently, we have developed an advanced approach, named Monoclonal surface display SELEX (MSD-SELEX) for efficient enrichment and identification of aptamers from a library of monoclonal DNA-displaying beads produced via highly parallel single-molecule emulsion PCR.83
The working principle of MSD-SELEX is schematically shown in Figure 10. Random ssDNA library diluted at the single copy level is encapsulated into individual emulsion droplets containing a single forward-primer-coupled bead for emulsion PCR to generate monoclonal beads each displaying millions of unique DNA sequences. Therefore, the DNA library has been converted to monoclonal bead library. Large targets, such as cells, can be directly applied, while small targets, such as small molecules or proteins, need to be immobilized on microbeads. Then, the bead library is incubated with targets to form bead/target complexes, which can be directly visualized under the microscope. Because nonbinding or weak binding beads can be easily removed through washing process to enrich the library, only beads with high binding affinity sequences are retained and easily isolated to yield aptamer candidates, which can be further confirmed without the need of its sequence information because of the one-bead-one-sequence nature of the monoclonal nature of beads. The bead clones with low Kd values can be sequenced and synthesized for further application. The method has been successfully applied to rapidly isolate aptamers against cancer biomarker protein EpCAM and small toxin molecule aflatoxin B1 (AFB1) without the need for many rounds of selection, large-scale DNA sequencing, expensive and time-consuming DNA synthesis, and labor-intensive screening of a large population of aptamer candidates. The MSD-SELEX approach has been demonstrated to be a new way for simple, rapid, efficient and cost-effective molecular evolution of high affinity ligands.
FIG. 10.
The working principle of MSD-SELEX for aptamer enrichment and identification. Statistically diluted ssDNA library with forward primer-functionalized beads (F-Primer beads) was compartmentalized into water-in-oil droplets at the single-molecule level. Single copy emulsion droplet PCR was then performed to generate monoclonal beads each displaying millions of identical DNA sequences. A library of monoclonal beads is then incubated with target cells. By removing the unbound beads and collecting bound beads, the positive clones are isolated for subsequent sequence retrieval and aptamer identification. Reprinted with permission from Zhu et al., Anal. Chem. 86(12), 5881 (2014). Copyright 2014 American Chemical Society.
CONCLUSIONS
Owing to their advantages over antibodies, including quick and reproducible synthesis, easy and controllable modification, long-term stability, ability to sustain reversible denaturation, non-toxicity, lack of immunogenicity, and rapid tissue penetration, aptamers have been widely applied for target validation, drug delivery, biomolecule detection, therapeutics, diagnostics, and biosensing. Conventional methods for aptamer identification are time-consuming, expensive, labor intensive, and inefficient. With the advantages of reduced reagent consumption, simple analytical methodology, high processing speed, high throughput, and potential for automation, microfluidic technology is well suited to revolutionize the field of aptamer selection. As summarized in Table I, recently developed microfluidic methods have successfully demonstrated that microfluidic technology can effectively improve aptamer selection with remarkably increased speed, reduced cost, expanded resolving power, enhanced throughput and automated system design. With the increased demand for aptamer sequences in the fields of bioanalysis and biomedicine, more novel microfluidic methods for rapid and efficient aptamer screening are greatly anticipated to emerge in the near future.
TABLE I.
Summary of Microfluidic SELEX.
| Method | Target | Rounds | Kd | Reference | |
|---|---|---|---|---|---|
| CE-SELEX | CE-SELEX | Human IgE | 4 | 27 ± 8 nM | 49 |
| 4 | 23 ± 12 nM | 57 | |||
| Neuropeptide Y | 4 | 0.8 ± 0.3 μM | 60 | ||
| HIV reverse transcriptase | 4 | 180 ± 70 pM | 58 | ||
| rhVEGF165 | 4 | 22 ± 11 nM | 84 | ||
| N-methyl mesoporphyrin | 3 | 0.88 ± 0.12 μM | 61 | ||
| NECEEM-SELEX | Protein farnesyltransferase | 1 | 1 nM | 85 | |
| ECEEM-SELEX | MutS protein | 3 | 15 nM | 51 | |
| Kinetic CE-SELEX | MutS protein | 3 | 3.6 ± 0.5 nM | 62 | |
| non-SELEX | h-Ras protein | 3 | 0.2 μM | 52 | |
| μFFE | Human IgE | 1 | 22 ± 6 nM | 64 | |
| Sol-gel microfluidic SELEX | Yeast transcription factor IIB | 7 | 4 nM | 54 | |
| Xanthine | 7 | 4.2 μM | 72 | ||
| TATA-binding protein | 5–8 | 2.7 nM | 70 | ||
| Magetic bead-based microfluidic SELEX | Streptavidin | 3 | 25–65 nM | 75 | |
| BoNT/A-rLc | 1 | 34 nM | 55 | ||
| PDGF-BB protein | 3 | <3 nM | 56 | ||
| Angiopoietin-2 | 4 | <30 nM | 76 | ||
| Integrated microfluidic system | C-reactive protein | 5 | 3.51 nM | 79 | |
| Alpha-fetoprotein | 6 | 2.37 nM | 80 | ||
| A549shEcad | 15 | 15.2 nM | 86 | ||
| Myoglobin | 7 | 4.93 nM | 81 | ||
| Agarose droplet microfluidics | Shp2 | 10 | 24.9 ± 14.6 nM | 78 | |
| MSD-SELEX | EpCAM | 5 | 33 ± 3 nM | 83 | |
| AFB1 | 5 | 0.65 ± 0.11 μM | 83 | ||
ACKNOWLEDGMENTS
We thank the National Basic Research Program of China (Grant Nos. 2010CB732402 and 2013CB933703), the National Science Foundation of China (Grant Nos. 21205100, 21275122, and 21075104), National Instrumentation Program (Grant No. 2011YQ03012412), the Fundamental Research Funds for the Central Universities (No. 2012121025), and the National Science Foundation for Distinguished Young Scholars of China (Grant No. 21325522) for their financial support.
References
- 1.Kohler G. and Milstein C., Nature 256(5517), 495 (1975). 10.1038/256495a0 [DOI] [PubMed] [Google Scholar]
- 2.Kingsmore S. F., Nat. Rev. Drug Discovery 5(4), 310 (2006). 10.1038/nrd2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan D. J., O'Connor D. P., Rexhepaj E., Ponten F., and Gallagher W. M., Nat. Rev. Cancer 10(9), 605 (2010). 10.1038/nrc2902 [DOI] [PubMed] [Google Scholar]
- 4.Weiner L. M., Surana R., and Wang S., Nat. Rev. Immunol. 10(5), 317 (2010). 10.1038/nri2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayasena S. D., Clin. Chem. 45(9), 1628 (1999). [PubMed] [Google Scholar]
- 6.Hansel T. T., Kropshofer H., Singer T., Mitchell J. A., and George A. J., Nat. Rev. Drug Discovery 9(4), 325 (2010). 10.1038/nrd3003 [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C. and Gold L., Science 249(4968), 505 (1990). 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 8.Ellington A. D. and Szostak J. W., Nature 346(6287), 818 (1990). 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- 9.Shangguan D., Li Y., Tang Z., Cao Z. C., Chen H. W., Mallikaratchy P., Sefah K., Yang C. J., and Tan W., Proc. Natl. Acad. Sci. U. S. A. 103(32), 11838 (2006). 10.1073/pnas.0602615103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer G., Angew. Chem., Int. Ed. Engl. 48(15), 2672 (2009). 10.1002/anie.200804643 [DOI] [PubMed] [Google Scholar]
- 11.Mascini M., Palchetti I., and Tombelli S., Angew. Chem., Int. Ed. Engl. 51(6), 1316 (2012). 10.1002/anie.201006630 [DOI] [PubMed] [Google Scholar]
- 12.Sefah K., Phillips J. A., Xiong X., Meng L., Van Simaeys D., Chen H., Martin J., and Tan W., Analyst 134(9), 1765 (2009). 10.1039/b905609m [DOI] [PubMed] [Google Scholar]
- 13.Willner I. and Zayats M., Angew. Chem., Int. Ed. Engl. 46(34), 6408 (2007). 10.1002/anie.200604524 [DOI] [PubMed] [Google Scholar]
- 14.Cho E. J., Lee J. W., and Ellington A. D., Annu. Rev. Anal. Chem. 2, 241 (2009). 10.1146/annurev.anchem.1.031207.112851 [DOI] [PubMed] [Google Scholar]
- 15.Famulok M. and Mayer G., Acc. Chem. Res. 44(12), 1349 (2011). 10.1021/ar2000293 [DOI] [PubMed] [Google Scholar]
- 16.Iliuk A. B., Hu L., and Tao W. A., Anal. Chem. 83(12), 4440 (2011). 10.1021/ac201057w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C. J., Jockusch S., Vicens M., Turro N. J., and Tan W., Proc. Natl. Acad. Sci. U.S.A. 102(48), 17278 (2005). 10.1073/pnas.0508821102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rupcich N., Nutiu R., Li Y., and Brennan J. D., Angew. Chem., Int. Ed. Engl. 45(20), 3295 (2006). 10.1002/anie.200504576 [DOI] [PubMed] [Google Scholar]
- 19.Liu J. and Lu Y., Angew. Chem., Int. Ed. Engl. 45(1), 90 (2006). 10.1002/anie.200502589 [DOI] [PubMed] [Google Scholar]
- 20.Liu J. and Lu Y., Nat. Protoc. 1(1), 246 (2006). 10.1038/nprot.2006.38 [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z., Wu C., Liu H., Zou Y., Zhang X., Kang H., Yang C. J., and Tan W., Angew. Chem., Int. Ed. Engl. 49(6), 1052 (2010). 10.1002/anie.200905570 [DOI] [PubMed] [Google Scholar]
- 22.Yan L., Zhu Z., Zou Y., Huang Y., Liu D., Jia S., Xu D., Wu M., Zhou Y., Zhou S., and Yang C. J., J. Am. Chem. Soc. 135(10), 3748 (2013). 10.1021/ja3114714 [DOI] [PubMed] [Google Scholar]
- 23.Freeman R., Liu X., and Willner I., J. Am. Chem. Soc. 133(30), 11597 (2011). 10.1021/ja202639m [DOI] [PubMed] [Google Scholar]
- 24.Ohno Y., Maehashi K., and Matsumoto K., J. Am. Chem. Soc. 132(51), 18012 (2010). 10.1021/ja108127r [DOI] [PubMed] [Google Scholar]
- 25.Lee S. J., Youn B. S., Park J. W., Niazi J. H., Kim Y. S., and Gu M. B., Anal. Chem. 80(8), 2867 (2008). 10.1021/ac800050a [DOI] [PubMed] [Google Scholar]
- 26.Zhou W. J., Halpern A. R., Seefeld T. H., and Corn R. M., Anal. Chem. 84(1), 440 (2012). 10.1021/ac202863k [DOI] [PubMed] [Google Scholar]
- 27. E. T. Cunningham, Jr. , Adamis A. P., Altaweel M., Aiello L. P., Bressler N. M., D'Amico D. J., Goldbaum M., Guyer D. R., Katz B., Patel M., Schwartz S. D., and Macugen Diabetic Retinopathy Study Group, Ophthalmology 112(10), 1747 (2005). 10.1016/j.ophtha.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 28.Ng E. W., Shima D. T., Calias P., E. T. Cunningham, Jr. , Guyer D. R., and Adamis A. P., Nat. Rev. Drug Discovery 5(2), 123 (2006). 10.1038/nrd1955 [DOI] [PubMed] [Google Scholar]
- 29.Mongelard F. and Bouvet P., Curr. Opin. Mol. Ther. 12(1), 107 (2010). [PubMed] [Google Scholar]
- 30.Rosenberg J. E., Bambury R. M., Van Allen E. M., Drabkin H. A., P. N. Lara, Jr. , Harzstark A. L., Wagle N., Figlin R. A., Smith G. W., Garraway L. A., Choueiri T., Erlandsson F., and Laber D. A., Invest. New Drugs 32(1), 178 (2014). 10.1007/s10637-013-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Famulok M., Nat. Biotechnol. 22(11), 1373 (2004). 10.1038/nbt1104-1373 [DOI] [PubMed] [Google Scholar]
- 32.Sennino B., Falcon B. L., McCauley D., Le T., McCauley T., Kurz J. C., Haskell A., Epstein D. M., and McDonald D. M., Cancer Res. 67(15), 7358 (2007). 10.1158/0008-5472.CAN-07-0293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlton J., Sennello J., and Smith D., Chem. Biol. 4(11), 809 (1997). 10.1016/S1074-5521(97)90114-9 [DOI] [PubMed] [Google Scholar]
- 34.Fang X. and Tan W., Acc. Chem. Res. 43(1), 48 (2010). 10.1021/ar900101s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keefe A. D., Pai S., and Ellington A., Nat. Rev. Drug Discovery 9(7), 537 (2010). 10.1038/nrd3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan W., Wang H., Chen Y., Zhang X., Zhu H., Yang C., Yang R., and Liu C., Trends Biotechnol. 29(12), 634 (2011). 10.1016/j.tibtech.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan W., Donovan M. J., and Jiang J., Chem. Rev. 113(4), 2842 (2013). 10.1021/cr300468w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu T. C., Marks J. W. III, Lavery L. A., Faulkner S., Rosenblum M. G., Ellington A. D., and Levy M., Cancer Res. 66(12), 5989 (2006). 10.1158/0008-5472.CAN-05-4583 [DOI] [PubMed] [Google Scholar]
- 39.Dassie J. P., Liu X. Y., Thomas G. S., Whitaker R. M., Thiel K. W., Stockdale K. R., Meyerholz D. K., McCaffrey A. P., McNamara J. O. II, and Giangrande P. H., Nat. Biotechnol. 27(9), 839 (2009). 10.1038/nbt.1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Y., Zhu Z., An Y., Zhang W., Zhang H., Liu D., Yu C., Duan W., and Yang C. J., Anal. Chem. 85(8), 4141 (2013). 10.1021/ac400366b [DOI] [PubMed] [Google Scholar]
- 41.Shangguan D., Cao Z., Meng L., Mallikaratchy P., Sefah K., Wang H., Li Y., and Tan W., J. Proteome Res. 7(5), 2133 (2008). 10.1021/pr700894d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies D. R., Gelinas A. D., Zhang C., Rohloff J. C., Carter J. D., O'Connell D., Waugh S. M., Wolk S. K., Mayfield W. S., Burgin A. B., Edwards T. E., Stewart L. J., Gold L., Janjic N., and Jarvis T. C., Proc. Natl. Acad. Sci. U. S. A. 109(49), 19971 (2012). 10.1073/pnas.1213933109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaught J. D., Bock C., Carter J., Fitzwater T., Otis M., Schneider D., Rolando J., Waugh S., Wilcox S. K., and Eaton B. E., J. Am. Chem. Soc. 132(12), 4141 (2010). 10.1021/ja908035g [DOI] [PubMed] [Google Scholar]
- 44.Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E. N., Carter J., Dalby A. B., Eaton B. E., Fitzwater T., Flather D., Forbes A., Foreman T., Fowler C., Gawande B., Goss M., Gunn M., Gupta S., Halladay D., Heil J., Heilig J., Hicke B., Husar G., Janjic N., Jarvis T., Jennings S., Katilius E., Keeney T. R., Kim N., Koch T. H., Kraemer S., Kroiss L., Le N., Levine D., Lindsey W., Lollo B., Mayfield W., Mehan M., Mehler R., Nelson S. K., Nelson M., Nieuwlandt D., Nikrad M., Ochsner U., Ostroff R. M., Otis M., Parker T., Pietrasiewicz S., Resnicow D. I., Rohloff J., Sanders G., Sattin S., Schneider D., Singer B., Stanton M., Sterkel A., Stewart A., Stratford S., Vaught J. D., Vrkljan M., Walker J. J., Watrobka M., Waugh S., Weiss A., Wilcox S. K., Wolfson A., Wolk S. K., Zhang C., and Zichi D., PLoS One 5(12), e15004 (2010). 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sefah K., Shangguan D., Xiong X., O'Donoghue M. B., and Tan W., Nat. Protoc. 5(6), 1169 (2010). 10.1038/nprot.2010.66 [DOI] [PubMed] [Google Scholar]
- 46.Mosing R. K. and Bowser M. T., J. Sep. Sci. 30(10), 1420 (2007). 10.1002/jssc.200600483 [DOI] [PubMed] [Google Scholar]
- 47.Xu Y., Yang X., and Wang E., Anal. Chim. Acta 683(1), 12 (2010). 10.1016/j.aca.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 48.Weng C. H., Huang C. J., and Lee G. B., Sensors (Basel) 12(7), 9514 (2012). 10.3390/s120709514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendonsa S. D. and Bowser M. T., J. Am. Chem. Soc. 126(1), 20 (2004). 10.1021/ja037832s [DOI] [PubMed] [Google Scholar]
- 50.Berezovski M., Drabovich A., Krylova S. M., Musheev M., Okhonin V., Petrov A., and Krylov S. N., J. Am. Chem. Soc. 127(9), 3165 (2005). 10.1021/ja042394q [DOI] [PubMed] [Google Scholar]
- 51.Drabovich A., Berezovski M., and Krylov S. N., J. Am. Chem. Soc. 127(32), 11224 (2005). 10.1021/ja0530016 [DOI] [PubMed] [Google Scholar]
- 52.Berezovski M., Musheev M., Drabovich A., and Krylov S. N., J. Am. Chem. Soc. 128(5), 1410 (2006). 10.1021/ja056943j [DOI] [PubMed] [Google Scholar]
- 53.Kim S., Kim Y., Kim P., Ha J., Kim K., Sohn M., Yoo J. S., Lee J., Kwon J. A., and Lee K. N., Anal. Chem. 78(21), 7392 (2006). 10.1021/ac0520487 [DOI] [PubMed] [Google Scholar]
- 54.Park S. M., Ahn J. Y., Jo M., Lee D. K., Lis J. T., Craighead H. G., and Kim S., Lab Chip 9(9), 1206 (2009). 10.1039/b814993c [DOI] [PubMed] [Google Scholar]
- 55.Lou X., Qian J., Xiao Y., Viel L., Gerdon A. E., Lagally E. T., Atzberger P., Tarasow T. M., Heeger A. J., and Soh H. T., Proc. Natl. Acad. Sci. U. S. A. 106(9), 2989 (2009). 10.1073/pnas.0813135106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho M., Xiao Y., Nie J., Stewart R., Csordas A. T., Oh S. S., Thomson J. A., and Soh H. T., Proc. Natl. Acad. Sci. U.S.A. 107(35), 15373 (2010). 10.1073/pnas.1009331107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendonsa S. D. and Bowser M. T., Anal. Chem. 76(18), 5387 (2004). 10.1021/ac049857v [DOI] [PubMed] [Google Scholar]
- 58.Mosing R. K., Mendonsa S. D., and Bowser M. T., Anal. Chem. 77(19), 6107 (2005). 10.1021/ac050836q [DOI] [PubMed] [Google Scholar]
- 59.Tok J., Lai J., Leung T., and Li S. F., Electrophoresis 31(12), 2055 (2010). 10.1002/elps.200900543 [DOI] [PubMed] [Google Scholar]
- 60.Mendonsa S. D. and Bowser M. T., J. Am. Chem. Soc. 127(26), 9382 (2005). 10.1021/ja052406n [DOI] [PubMed] [Google Scholar]
- 61.Yang J. and Bowser M. T., Anal. Chem. 85(3), 1525 (2013). 10.1021/ac302721j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drabovich A. P., Berezovski M., Okhonin V., and Krylov S. N., Anal. Chem. 78(9), 3171 (2006). 10.1021/ac060144h [DOI] [PubMed] [Google Scholar]
- 63.Berezovski M. V., Musheev M. U., Drabovich A. P., Jitkova J. V., and Krylov S. N., Nat. Protoc. 1(3), 1359 (2006). 10.1038/nprot.2006.200 [DOI] [PubMed] [Google Scholar]
- 64.Jing M. and Bowser M. T., Lab Chip 11(21), 3703 (2011). 10.1039/c1lc20461k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avnir D., Coradin T., Lev O., and Livage J., J. Mater. Chem. 16(11), 1013 (2006). 10.1039/b512706h [DOI] [Google Scholar]
- 66.Brennan J. D., Acc. Chem. Res. 40(9), 827 (2007). 10.1021/ar6000268 [DOI] [PubMed] [Google Scholar]
- 67.Dickson D. J. and Ely R. L., Appl. Microbiol. Biotechnol. 97(5), 1809 (2013). 10.1007/s00253-012-4686-8 [DOI] [PubMed] [Google Scholar]
- 68.Frenkel-Mullerad H. and Avnir D., J. Am. Chem. Soc. 127(22), 8077 (2005). 10.1021/ja0507719 [DOI] [PubMed] [Google Scholar]
- 69.Pastor I., Ferrer M. L., Lillo M. P., Gomez J., and Mateo C. R., J. Phys. Chem. B 111(39), 11603 (2007). 10.1021/jp074790b [DOI] [PubMed] [Google Scholar]
- 70.Ahn J. Y., Jo M., Dua P., Lee D. K., and Kim S., Oligonucleotides 21(2), 93 (2011). 10.1089/oli.2010.0263 [DOI] [PubMed] [Google Scholar]
- 71.Ahn J. Y., Lee S., Jo M., Kang J., Kim E., Jeong O. C., Laurell T., and Kim S., Anal. Chem. 84(6), 2647 (2012). 10.1021/ac202559w [DOI] [PubMed] [Google Scholar]
- 72.Bae H., Ren S., Kang J., Kim M., Jiang Y., Jin M. M., Min I. M., and Kim S., Nucleic Acid Ther. 23(6), 443 (2013). 10.1089/nat.2013.0437 [DOI] [PubMed] [Google Scholar]
- 73.Wochner A., Menger M., Orgel D., Cech B., Rimmele M., Erdmann V. A., and Glokler J., Anal. Biochem. 373(1), 34 (2008). 10.1016/j.ab.2007.09.007 [DOI] [PubMed] [Google Scholar]
- 74.Murphy M. B., Fuller S. T., Richardson P. M., and Doyle S. A., Nucleic Acids Res. 31(18), e110 (2003). 10.1093/nar/gng110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qian J., Lou X., Zhang Y., Xiao Y., and Soh H. T., Anal. Chem. 81(13), 5490 (2009). 10.1021/ac900759k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cho M., Oh S. Soo, Nie J., Stewart R., Eisenstein M., Chambers J., Marth J. D., Walker F., Thomson J. A., and Soh H. T., Proc. Natl. Acad. Sci. U.S.A. 110(46), 18460 (2013). 10.1073/pnas.1315866110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Y. and Wang E., J. Chromatogr. A 1216(24), 4817 (2009). 10.1016/j.chroma.2009.04.024 [DOI] [PubMed] [Google Scholar]
- 78.Hybarger G., Bynum J., Williams R. F., Valdes J. J., and Chambers J. P., Anal. Bioanal. Chem. 384(1), 191 (2006). 10.1007/s00216-005-0089-3 [DOI] [PubMed] [Google Scholar]
- 79.Huang C. J., Lin H. I., Shiesh S. C., and Lee G. B., Biosens. Bioelectron. 25(7), 1761 (2010). 10.1016/j.bios.2009.12.029 [DOI] [PubMed] [Google Scholar]
- 80.Huang C. J., Lin H. I., Shiesh S. C., and Lee G. B., Biosens. Bioelectron. 35(1), 50 (2012). 10.1016/j.bios.2012.02.024 [DOI] [PubMed] [Google Scholar]
- 81.Wang Q., Liu W., Xing Y., Yang X., Wang K., Jiang R., Wang P., and Zhao Q., Anal. Chem. 86(13), 6572 (2014). 10.1021/ac501088q [DOI] [PubMed] [Google Scholar]
- 82.Zhang W. Y., Zhang W. H., Liu Z. Y., Li C., Zhu Z., and Yang C. J., Anal. Chem. 84(1), 350 (2012). 10.1021/ac2026942 [DOI] [PubMed] [Google Scholar]
- 83.Zhu Z., Song Y., Li C., Zou Y., Zhu L., An Y., and Yang C. J., Anal. Chem. 86(12), 5881 (2014). 10.1021/ac501423g [DOI] [PubMed] [Google Scholar]
- 84.Jing M. and Bowser M. T., Anal. Chem. 85(22), 10761 (2013). 10.1021/ac401875h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berezovski M. and Krylov S. N., J. Am. Chem. Soc. 124(46), 13674 (2002). 10.1021/ja028212e [DOI] [PubMed] [Google Scholar]
- 86.Weng C.-H., Hsieh I. S., Hung L.-Y., Lin H.-I., Shiesh S.-C., Chen Y.-L., and Lee G.-B., Microfluid. Nanofluid. 14(3–4), 753 (2013). 10.1007/s10404-012-1095-3 [DOI] [Google Scholar]