Abstract
Metastasis is the end product of an evolutionary process in which diverse interactions between cancer cells and their microenvironment yield alterations that allow these cells to transcend their programmed behavior. Tumor cells thus populate and flourish in new tissue habitats and, ultimately, cause organ dysfunction and death. Understanding the many molecular players and processes involved in metastasis could lead to effective, targeted approaches to prevent and treat cancer metastasis.
The tumor–node–metastasis (TNM) staging system used for most solid tumors considers the tumor size and degree of local invasion (T), the number, size, and location of lymph nodes (N), and the presence or absence of distant metastases (M).1 Metastases of tumors originating in different sites, such as the breast or lung, are treated differently because they are thought to behave like the tissue of origin, with characteristic patterns and kinetics of spread, and distinct profiles of chemosensitivity. Lymph nodes are of paramount importance in current staging practices, but it is hard to interpret the clinical significance of the distance of metastases from the primary site (e.g., a supraclavicular N3 vs. a mediastinal N2 lymph node in lung cancer). Indeed, the distance from the primary tumor to the organ of metastasis does not affect staging. For this reason, the real value of staging is to serve as an indicator of the primary cancer’s composite capability to metastasize, rather than to ensure that the tumor lies within the prescribed limits of a local intervention. Recent advances bring hope for characterizing the metastatic behavior of cancer cells beyond the simplistic TNM stage. In the future, staging could include identification of subpopulations of tumor cells that have different metastatic behavior. A deeper understanding of the molecular and genetic concepts and processes involved in metastasis may pave the way toward new prognostic models and ways of planning treatment.
BASIC CONCEPTS OF METASTASIS
ORIGINS OF CELLULAR HETEROGENEITY
Primary tumors consist of heterogeneous populations of cells with genetic alterations that allow them to surmount physical boundaries, disseminate, and colonize a distant organ. Metastasis is a succession of these individual processes2–4 (Fig. 1), and fully metastatic cells are rare clones in the primary tumor. In animal models, 0.01% or fewer of the cancer cells entering the circulation develop into metastases.5,6 The intrinsic genomic instability of cancer cells increases the frequency of alterations necessary to acquire metastatic capacity. The genomic instability and heterogeneity of tumor cells are apparent in the chromosomal gains, losses, and rearrangements associated with cancer. DNA integrity can be compromised by aberrant cell-cycle progression, telomeric crisis (i.e., telomere dysfunction characterized by cytogenetic abnormalities and chromosomal instability), inactivation of DNA repair genes (see the Glossary), and altered epigenetic control mechanisms. For example, 50% of cancers have lost the tumor-suppressor protein p53, which responds to DNA damage by inducing apoptosis or arresting cell growth.7 Loss of p53 allows the accumulation of cells with DNA damage.8
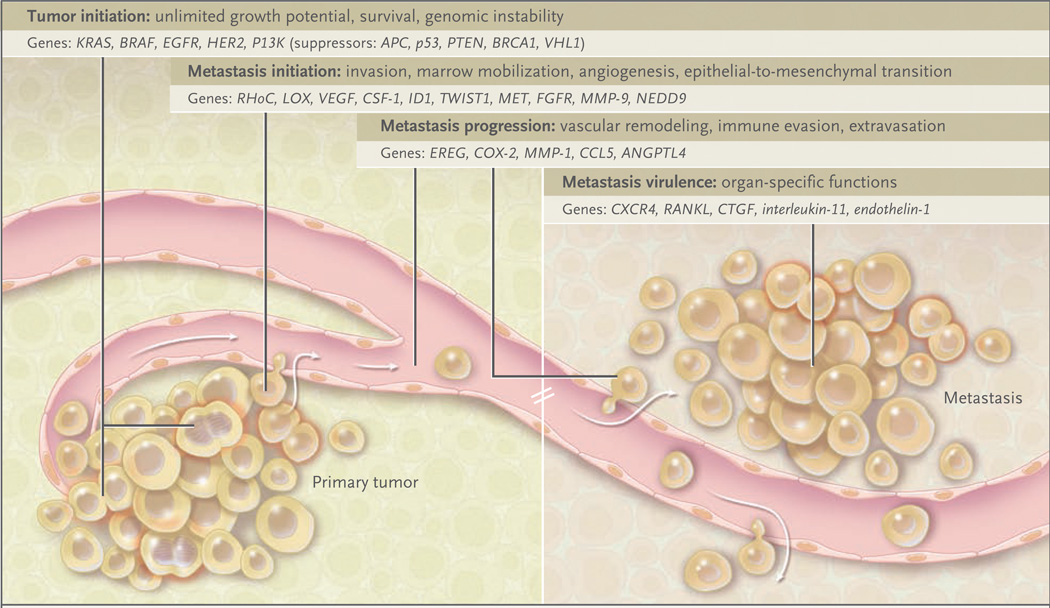
Figure 1. Tumor Initiation and Metastasis.
The initiation and progression of tumors depend on the acquisition of specific functions by cancer cells at both the primary and metastatic sites. Functions associated with tumor initiation are provided by oncogenic mutations and inactivation of tumor-suppressor genes. Functions associated with the initiation of metastasis include functions to which tumor cells resort for local invasion and for circumventing hypoxia and other limitations facing a growing tumor. Most functions for the initiation of both the tumor and metastasis remain essential for cancer cells to continue their metastatic development. Functions for metastasis progression provide a local advantage in a primary tumor and a distinct and sometimes organ-specific function during metastasis. Cancer cells that are endowed with these three sets of functions still depend on functions associated with metastasis virulence; these functions confer a selective advantage solely during the adaptation and takeover of a specific organ microenvironment. Genes associated with each of these functions have been identified in recent years.
SELECTIVE PRESSURES OF THE TUMOR MICROENVIRONMENT
Each tissue has a physical structure and an established functional anatomy complete with compartmental boundaries, a vascular supply, and a characteristic extracellular milieu of nutrients and stroma. Cancer cells that circumvent this organization become exposed to environmental stresses, including a lack of oxygen or nutrients, a low pH, reactive oxygen species, and mediators of the inflammatory response. Such pressures can select tumor cells with the capability of growth despite these challenges and in the process can cause them to acquire an aggressive phenotype. For example, hypoxia stabilizes hypoxia-inducible factor (HIF), which cues a program of gene expression that leads to changes in anaerobic metabolism, angiogenesis, invasion, and survival.9,10 HIF boosts the expression of lysyl oxidase; lysyl oxidase regulates the activity of focal adhesion kinase in a way that enhances cell-matrix adhesion and invasion.11 High levels of lysyl oxidase correlate with shorter metastasis-free survival and a poor prognosis in head and neck cancer, as well as in estrogen-receptor–negative breast cancer.12 Another product of HIF-induced gene activation, the chemokine (C-X-C motif) receptor CXCR4, together with its ligand, the chemokine stromal-cell–derived factor 1 (SDF-1, also called CXC chemokine ligand 12 [CXCL12]), facilitates the survival of cancer cells at sites of metastasis in breast cancer and renal-cell cancer.13
CANCER STEM CELLS AND METASTASIS
The question of the extent to which self-renewing cancer stem cells initiate and sustain cancers of different types is a subject of intense investigation, and there are probably different answers according to different tumor types. Such cells are envisioned as a subpopulation of cancer cells that — by one mechanism or another — have the capacity to act as tumor-propagating cells.14 These cells might resist apoptosis and DNA damage caused by drugs; they might also require a niche or specific microenvironment in order to grow.15 Such attributes would support the establishment of both primary and metastatic tumors. The SDF-1–CXCR4 axis is thought to function in support of cancer cells and stem cells or precursor cells.16 A “premetastatic” niche has been described in animal models in which bone marrow–derived progenitor cells home to specific distant sites before the formation of a metastasis.17,18 The ability of stem cells to evade destruction and survive in distant sites, including the bone marrow, may explain why micrometastases can remain dormant after removal of the primary tumor, only to recur years later.
THE ENVIRONMENT OF THE PRIMARY TUMOR
INVASION AND EPITHELIAL-TO-MESENCHYMAL TRANSITION
In many primary tumors with invasive properties, intercellular adhesion is reduced, often because of a loss of E-cadherin, a direct mediator of cell–cell adhesive interactions. The cytoplasmic tail of E-cadherin is tethered, via α-catenin and β-catenin, to the actin cytoskeleton; one of actin’s properties is to maintain cell junctions. The importance of maintaining intercellular adhesion was shown in a mouse model of pancreatic cancer in which disruption of the expression of E-cadherin led to early invasion and metastasis.19 Various mechanisms can cause a loss of E-cadherin: mutations resulting in an inactive protein, gene silencing by promoter methylation, or down-regulation stimulated by growth factor receptors (e.g., epidermal growth factor receptor [EGFR], fibroblast growth factor receptor [FGFR], insulin-like growth factor I [IGF-I] receptor, and MET) or SRC family kinases.19,20 Expression of the E-cadherin gene (CDH1) is also inhibited by several transcriptional repressors.21,22 Loss of E-cadherin function is necessary, though not sufficient for epithelial-to-mesenchymal transition, a process whereby epithelial cells switch to a mesenchymal progenitor-cell phenotype, enabling detachment and reorganization of epithelial-cell sheets during embryonic development, as well as tumor invasion and metastasis.23
MOTILITY AND EXTRACELLULAR-MATRIX REMODELING
The extracellular matrix serves as a scaffold along which cells attach and move by means of contacts between cell-surface receptors called integrins and extracellular-matrix components such as fibronectin, collagen, and laminin. Integrins also interact in a cytoplasmic complex consisting of focal adhesion kinases and SRC family kinases to mediate attachment to the actin cytoskeleton. Through calcium-dependent guanosine triphosphatases (GTPases), extracellular-matrix signals cause cytoskeletal changes that form individual cytoplasmic extensions called filopodia, which coalesce into larger lamellipodia, structures that are important in migratory movement. Expression profiling of melanoma cell lines obtained by means of in vivo selection has shown that the calcium-dependent GTPase RhoC is important in lung metastasis.24 Homozygous RhoC-deficient mice have normal formation of primary tumors but impaired cancer-cell mobility and almost no lung metastases.25 NEDD9, a scaffolding protein involved in cell adhesion, colocalizes in focal contacts with focal adhesion kinase and can promote cell motility and invasion.26 Various members of the matrix metalloproteinase (MMP) family (e.g., MMP-2 and MMP-9) are also implicated in cancer-cell invasion.27–29 Independent screens for genes that mediate bone or lung metastasis in breast cancer have identified MMP-1 as being necessary for spread to the bone and lungs.30,31 The metastasis-suppressor microRNA miR335, which inhibits metastasis to the lungs and bones in human breast-cancer xenografts, suppresses the expression of two mediators of cancer-cell invasion, the transcription factor SOX-4 and tenascin-C, a matrix glycoprotein.32 A low level of miR335 in breast-cancer cells is associated with relapse.
STROMAL INTERACTIONS
Not only are cancer cells able to traverse the structural boundaries of the primary tumor, but they can also co-opt local and bone marrow–derived stromal-cell responses to their advantage. At points of basement-membrane invasion in mouse tumors, tumor-associated macrophages proliferate in response to tumor-derived colony-stimulating factor 1 and produce growth factors (e.g., fibroblast growth factor, EGFR ligands, and platelet-derived growth factor [PDGF]) and proteases (e.g., MMPs and cathepsins).33,34 In addition, tumor-associated macrophages activate a particular type of carcinoma-associated mesenchymal cell, the myofibroblast, which secretes the cytokine SDF-1; this cytokine enables the myofibroblast to recruit endothelial progenitor cells.35 Impaired metastases of breast-cancer cells to the lungs occur in mice with genetic defects in macrophages.36 The stroma-derived cytokine, transforming growth factor β (TGF-β), induces the expression of genes such as ANGPTL4 in breast-cancer cells; TGF-β enhances metastatic activity and is associated with increased metastases to the lungs in estrogen-receptor–negative breast cancer.37 In short, several types of stromal cells and their secreted factors provide selective prometastatic advantages.
ORGAN-SPECIFIC METASTASIS
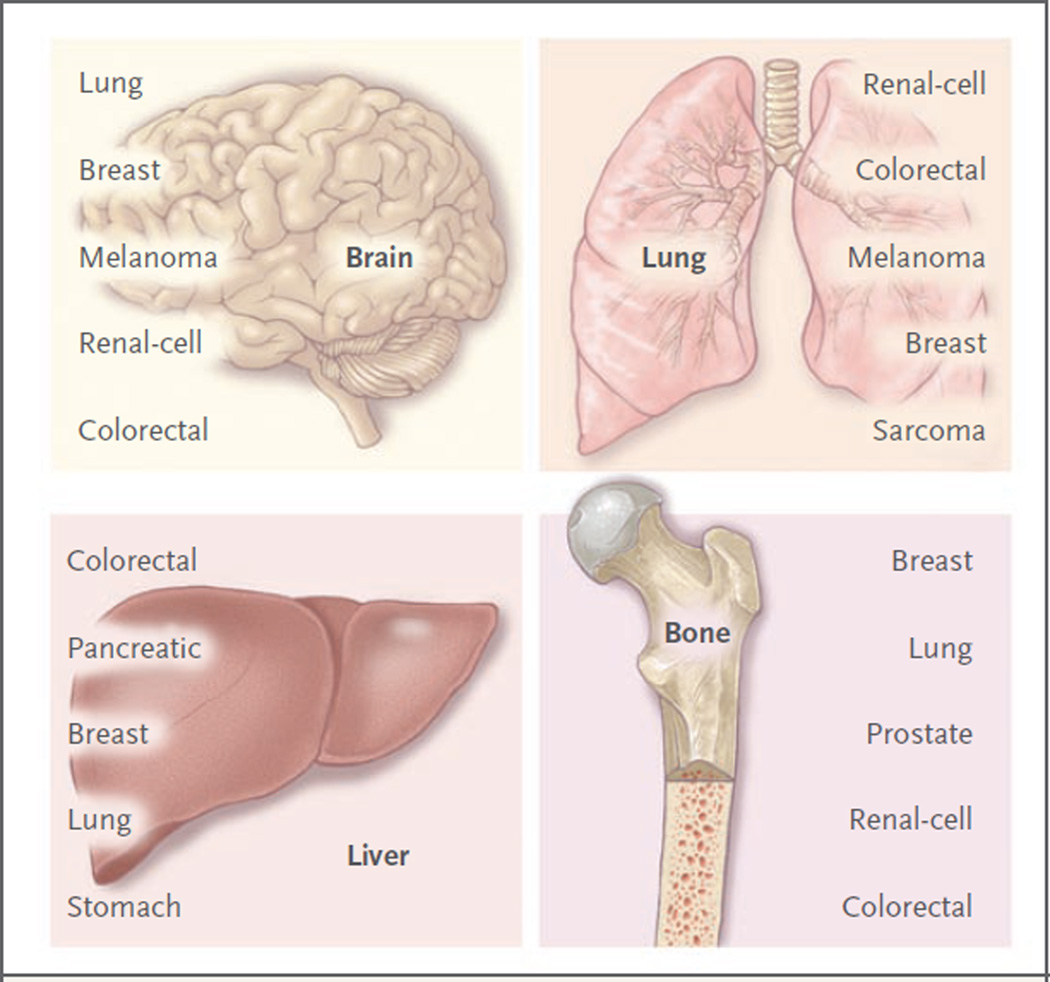
Some types of cancers have a characteristic proclivity to metastasize to certain organs, but not to others (Fig. 2).38–42 Breast cancer spreads to the bones, lungs, brain, and liver; distant metastases of prostate cancer occur most prominently in bone. Breast-cancer and prostate-cancer cells can both spread to and colonize the bone, but they form osteolytic or osteoblastic metastases, respectively. According to Paget’s “seed” vs. “soil” hypothesis, perceived compatibilities between disseminated cancer cells (the seed) and certain distant sites (the soil) have long influenced our view of the metastatic process.43 The formation of bone metastases alters bone homeostasis — the balance of action of osteoclasts in degrading bone against the constant rebuilding of bone by osteoblasts. Breast-cancer cells preferentially cause osteolytic lesions by inducing osteoclasts to secrete PTHrP (parathyroid hormone–related peptide), tumor necrosis factor α (TNF-α), and cytokines such as interleukin-1, interleukin-6, interleukin-8, and interleukin-11. These factors cue osteoblasts to release RANKL (the ligand for the receptor activator of nuclear factor-κB [RANK]), which stimulates osteoclast differentiation (Fig. 3). Osteoclasts demineralize bone, thereby causing the release of growth factors such as bone morphogenetic proteins, IGF-I, and TGF-β from the exposed bone matrix; all these growth factors support cancer-cell proliferation and induce further release of PTHrP. In a breast-cancer xenograft model, breast-cancer cells that preferentially colonized bone had up-regulated expression of genes encoding CXCR4, osteopontin, CTGF, MMP-1, and interleukin-11.30 By contrast, prostate-cancer cells secrete osteoblaststimulating factors such as Wnt family ligands, bone morphogenetic proteins, PDGF, and endothelin-1; these factors stimulate formation of the hallmark osteoblastic metastases of prostate cancer. Tumor-derived signals suppress the ability of osteoblasts to secrete osteoprotegerin, a RANKL antagonist that blocks RANKL–RANK interaction and resulting osteoclast activation. Thus, factors secreted by cancer cells, or “seeds,” can influence the type of metastases formed.
Figure 2. Patterns of Metastatic Spread of Solid Tumors.
Brain metastases may occur as a result of hematogenous spread late in the course of a widely metastatic tumor, or as a result of secondary metastasis from a primary or a metastatic tumor that can access the arterial circulation through the pulmonary venous circulation to seed the brain.38 Tumors with the highest incidence of brain metastases include lung carcinoma, breast carcinoma, melanoma, and to a lesser extent, renal-cell and colorectal carcinomas. Leptomeningeal disease may develop through the spread of cancer cells through perineural lymphatic vessels, and it is a sign of advanced disease.38 Some tumors have a strong proclivity for dissemination to the lungs; for example, in one study, the rate of dissemination associated with sarcoma was 23%.39 Other tumors that frequently spread to the lungs include renal-cell, colorectal, melanoma, and breast carcinomas.40,41 Gastrointestinal tumors easily access the liver circulation through the portal-vein system. The incidence of liver metastases is highest among patients with colorectal or pancreatic cancer, followed by breast and lung cancers.40 Estrogen-receptor–negative breast-cancer tumors more often metastasize to visceral organs, including the liver, whereas estrogen-receptor–positive breast cancer more often metastasizes to the bone.42 Bone metastasis occurs in patients with primary tumors associated with breast, lung, prostate, renal-cell, and colon cancer, in this order of frequency.40 Bone metastases may be primarily osteolytic or osteoblastic, depending on the tumor of origin.
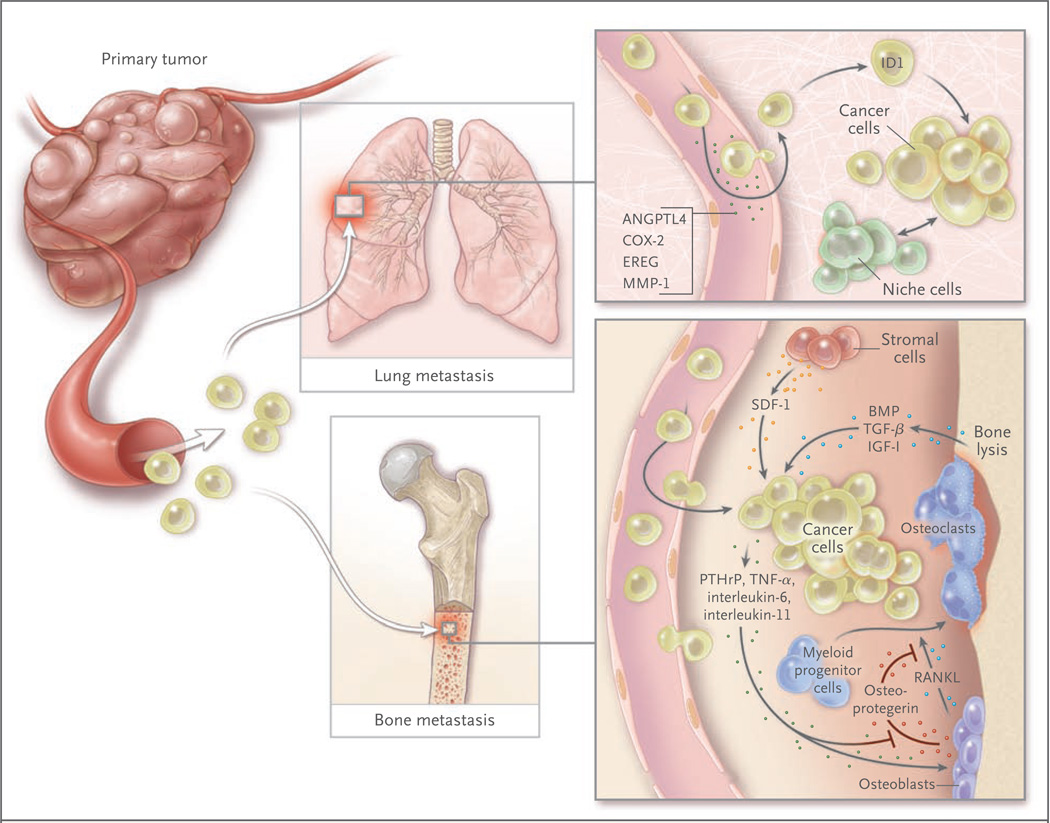
Figure 3. Genes, Functions, and Cellular Players in Organ-Specific Metastasis.
Organ-specific metastasis of breast-cancer cells involves different molecular players during colonization of the lungs and the bones. In the lung, cancer cells producing EREG, COX-2, MMP-1, and ANGPTL4 are better equipped to exit the pulmonary vasculature, since these factors alter the integrity of lung microcapillary endothelia; this function is less important for infiltration into the bone marrow because of the naturally fenestrated structure of the bone marrow sinusoid vasculature. In the lung parenchyma, the activity of the antidifferentiation gene ID1 and interactions with still unknown “niche” factors promote tumor reinitiation. In the bone marrow, stromal-cell–derived factor 1 (SDF-1), acting through its chemokine (C-X-C motif) receptor (CXCR4) on cancer cells, is thought to provide cell-survival functions. The secretion of parathyroid hormone–related peptide (PTHrP), interleukin-6, tumor necrosis factor α (TNF-α), interleukin-11, and other factors by cancer cells stimulates osteoblasts to release the ligand for the receptor activator of nuclear factor-κB (RANKL), which in turn stimulates osteoclast differentiation from myeloid progenitor cells. Other cancer cell–derived factors suppress the production of the RANKL antagonist osteoprotegerin, augmenting the efficacy of RANKL. The lytic action of osteoclasts releases bone matrix–associated growth factors, including transforming growth factor β (TGF-β), insulin-like growth factor I (IGF-I), and bone morphogenetic proteins (BMPs). IGF-I is a survival factor, and TGF-β incites cancer cells to further release PTHrP, interleukin-11, and other prometastatic factors, establishing a vicious cycle.
Cancer cells may regulate the expression of other molecules to target colonization in other organs.44 Such molecules include the gene encoding ezrin (an intracellular protein needed for early survival of metastatic osteosarcoma cells in the lung), serine–threonine kinase 11 (STK11, also known as LKB1) (a metastasis-suppressor gene regulating NEDD9 in lung cancer45), and genes in an 18-gene breast-to-lung metastatic gene-expression signature including the EGFR ligand EREG, COX-2, MMP-1, ANGPTL4, and other mediators of infiltration and colonization by cancer cells in the lung.46
The soil, or distant metastatic site, is a largely nonpermissive environment, as evidenced by the rarity of metastatic clones arising after injecting millions of cells into circulation in experiments in animals. In humans, also, thousands of circulating tumor cells have been found in the absence of metastases. Certain seed–soil interactions can support the cancer cell’s ability to survive in the metastatic microenvironment, including the RANKL–RANK interaction. Another example involves the SDF-1 chemokine in the bone marrow, which recruits breast-cancer and prostate-cancer cells and enhances their survival.47 Whereas the mechanisms of metastasis to bone and lung have been extensively studied and are partially understood, there is a dearth of information about the molecular basis for metastasis to other organs, such as the liver and brain.
AN INTEGRATED MODEL OF METASTASIS
In the past decade, our view of metastasis has changed from snapshots detailing specific biologic processes to a moving picture of how various cancer cells acquire functions and co-opt stromal signals for spread and encampment in a distant site. Random genetic and epigenetic alterations in cancer cells in combination with a plastic and responsive microenvironment support the metastatic evolution of tumors. Moreover, genes needed at individual steps along the metastatic process have been identified.
These genes have been classified into three categories: initiation, progression, and virulence48 (Fig. 1). Genes that are associated with metastatic progression give the cancer cell particular advantages at multiple points during its sojourn to a distant site. These advantages can influence the cell’s metastatic destination. Genes associated with the initiation of metastasis and virulence operate in the earliest and latest stages of invasion and growth in the primary tumor and different metastatic habitats, respectively. The use of such a framework to organize specific genes and their functions allows a multidimensional picture (including locale and time) of metastasis and may aid in the development of rational antimetastatic strategies.
MODELS OF METASTASIS AND TUMOR PROGRESSION
Early theories of metastasis pitted models of genetic predetermination against those of orderly anatomic progression. The advent of molecular genetics has refashioned the model of tumor progression in which somatic mutations were thought to accumulate sequentially, resulting in rare cells capable of metastasis.49 Other models emphasize dynamic heterogeneity and clonal selection, principles that suggest that an unstable metastatic variant can expand and prevail in the population of cells.50,51 The presence of metastasis genes in gene-expression signatures of primary tumors would seem to challenge the traditional tumor-progression model of somatic evolution in which metastatic cells would be too rare to influence a population-averaged gene-expression profile of the primary tumor. This finding, however, probably reflects an abundance of partially competent cancer cells that have accumulated a sufficient number of malignant functions to promote expansion of the primary tumor, and which may be necessary but not sufficient for forming metastases. By contrast, genes associated with metastatic virulence provide an aggressive edge in survival and proliferation solely during colonization of the metastatic site (Fig. 1). Many of these genes do not give the primary tumor a selective advantage, and thus they would not be represented in gene signatures of the primary tumor.
METASTASIS-PROGRESSION GENES
Genes that are necessary for certain functions such as vascular remodeling can participate in both the primary tumor and the metastatic environment; these genes are metastasis-progression genes, and they could be enriched in primary tumors (Fig. 1). An 18-gene lung-metastatic signature derived from selected in vivo breast-cancer cells that efficiently spread to the lungs includes EREG, COX-2, and MMP-1. These genes cooperate in remodeling the vasculature in sites of mammary tumors and lung metastasis. In the breast, they allow neoangiogenesis and intravasation of cancer cells; in the lung, they mediate extravasation of circulating cancer cells from capillaries into the parenchyma.52 Breast cancers with the lung-metastatic signature have a high risk of lung metastases, but not of metastases to the bones or liver. A likely explanation is that extravasation is not essential for passage through the fenestrated vasculature of the bone marrow and liver sinusoids. Thus, metastasis-progression genes may couple the tissue-specific features of the microenvironment in a particular organ to a matching role in the progression of a primary tumor. Expression of the lung-metastatic signature gene ANGPTL4 is a bystander event in mammary tumors, but when cancer cells expressing ANGPTL4 reach lung capillaries, its role in mediating extravasation by disrupting endothelial cell–cell contacts becomes manifest.37
Both the cells of primary tumors and metastatic cells require the ability to initiate self-renewal and bypass senescence. ID1 (inhibitor of differentiation-1) is the sole transcriptional regulator in the lung-metastatic signature, and it can be found in clusters of cancer cells within breast tumors of the basal or triple-negative (i.e., estrogen-receptor–negative, progesterone-receptor–negative, and human epidermal growth factor receptor type 2 [HER2]–negative) subtype. Suppression of ID1 expression inhibits the initiation of mammary tumors and metastases in the lungs.53 Thus, ID1 may promote micrometastatic outgrowth from dormancy at the metastatic site. Related to this function, in mouse models of metastatic breast cancer, ID1 cooperates with activated RAS oncogenes to avert cell senescence.54
METASTATIC DISSEMINATION
Cancer cells can disseminate from a tumor very early in the life of a tumor. They have been detected in the bone marrow of patients with breast cancer with early-stage disease. Such cancer cells were genetically distinct from the matched primary tumors, but bone marrow–derived cancer cells in patients with overt metastatic disease were less genetically disparate.55,56 This finding may reflect differences between the departure time from the primary neoplasm and the duration of exposure to selective pressures. Dormant cancer cells isolated from the bone marrow of transgenic mice with preinvasive breast cancer and patients with ductal carcinoma in situ became activated when transplanted into the bone marrow and caused the growth of lethal tumors.57 Many mechanisms for metastatic dormancy have been postulated.58 In patients with advanced metastatic disease, breast cancer cells that are competent in vascular entry can efficiently exit at a distant organ and perhaps reenter to repeat the process. Tumor infiltration by means of its own circulating progeny of metastatic cancer cells has been raised as a possible mechanism for the later rapid expansion of tumor growth.59 According to this hypothesis, large primary tumors may also be the end product of aggressive reseeding. This would be a new perspective on the long-standing observation that metastatic relapse correlates with tumor size.60
CLINICAL IMPLICATIONS
MOLECULAR SIGNATURES OF METASTASIS
Gene-expression signatures of primary breast cancers that predict clinical outcome61–65 generally do not overlap and range from a 70-gene “poor-prognosis” signature (detected with the use of the MammaPrint test) to a hand-picked set of 21 “recurrence” genes (detected with the use of the Oncotype Dx test) that includes estrogen-receptor, HER2, and proliferation markers. Other signatures consist of genes with expressions that are associated with a process or pathway, such as the response to serum mitogens,66 hypoxia,67 activation of specific oncogenes (e.g., RAS, MYC, and SRC),68,69 stimulation with a growth factor (e.g., TGF-β),37 or treatment with specific chemotherapeutic drugs to establish a drug-sensitivity profile.70 To specifically identify genes that mediate metastasis, animal models have been used to select in vivo for highly metastatic and organspecific derivatives of human cancer cell lines.3 Such signatures can correlate with bone-specific and lung-specific spread.30,46 The lung-metastasis signature further correlates with clinical outcome, including the recurrence of disease in the lungs, in primary breast-cancer tumors.60 Functional validation approaches (e.g., overexpression or knockdown experiments in culture or xenograft experiments) have confirmed that these genes, particularly in combination, are critical for metastatic functions.52
TARGETS OF THERAPY
In principle, each metastasis-specific gene is a potential target for a treatment. Ongoing clinical trials target the metastatic initiation gene c-MET (e.g., the small-molecule inhibitor ARQ 197, in phase 1–2 trials) and two metastatic virulence genes, RANK ligand (e.g., denosumab, in phase 3 trials) and TGF-β (e.g., monoclonal antibody GC1008, in phase 1 trials). Combination therapy may be needed to overcome the intrinsic biologic redundancy in metastasis and to target sequential steps in metastasis. In one series of preclinical experiments, only combination (not single-agent) therapy with the drugs celecoxib and cetuximab, meant to target two metastatic progression genes, was effective in blocking lung metastases by highly lung-metastatic breast-cancer cells.52 If cancer cells are constantly on the move between sites of metastasis in the lung, treatment with these drugs could prevent further reseeding and growth of metastatic sites. Cancer treatment may need to combine multiple antimetastatic drugs with cytotoxic chemotherapy. For example, bevacizumab, an antibody targeting vascular endothelial growth factor, is being studied in combination with chemotherapy in the adjuvant setting for colorectal, ovarian, and non–small-cell lung cancers. Therapies that target the mechanisms that keep dormant micrometastases alive are also needed.
CLINICAL TRANSLATION
Clinical trials involving antimetastatic agents face a number of obstacles. Any adjuvant trial to assess the recurrence of metastatic disease requires many patients because of the infrequency and lag time to progression of metastatic disease in many types of cancer. Measuring response rates beyond stable disease will further increase the number of patients in a trial. Moreover, correlative studies of tissue obtained from metastatic sites are essential to understand the results of such trials.
These barriers are sobering, and they perhaps underscore the conceptual shifts that will be needed for the development of new cancer therapies. What changes can we envision? The profile of a tumor could include not only histopathological or genetic determinants, or both, but also a molecular snapshot that would indicate a “metastasis quotient.” The metastasis quotient could be a measure of how adept the cells are with respect to metastatic functions, and it could serve as a prognostic framework. By focusing on metastatic progression and virulence functions, cancer therapy might be dictated by the metastatic site and not only by the specific tissue of origin. A current example of a treatment targeting a metastatic organ is the use of bisphosphonates or denosumab (an anti-RANK antibody), or both, to treat bone metastases originating from the breast, lung, and even multiple myeloma. Drug regimens for patients with cancer might include multiple drugs targeting different metastatic sites and seeding among sites. There is now hope for achieving the ultimate goal — curing metastatic disease.
Acknowledgments
Dr. Massagué reports receiving consulting fees from Acceleron Pharmaceuticals, lecture fees from Eli Lilly, Genzyme, and Eisai.
We thank D. Nguyen, G. Riely, K. Politi, and D. Spriggs for insightful discussions.
Glossary of Selected Genes
- ANGPTL4
Angiopoietin-like 4
- APC
Adenomatous polyposis coli
- BRAF
(Also known as V-raf murine sarcoma viral oncogene homologue B1)
- BRCA1
Breast-cancer gene 1
- COX-2
Cyclooxygenase-2
- CSF-1
Colony-stimulating factor 1
- CTGF
Connective-tissue growth factor
- CXCR4
CXC chemokine receptor 4
- EGFR
Epidermal growth factor receptor
- EREG
Epiregulin
- FGFR
Fibroblast growth factor receptor
- HER2
Human epidermal growth factor receptor type 2
- ID1
Inhibitor of differentiation-1
- MMP-1
Matrix metalloproteinase 1
- MMP-9
Matrix metalloproteinase 9
- NEDD9
Neural precursor cell expressed, developmentally down-regulated 9
- P13K
Phosphoinositide 3-kinase
- PTEN
Phosphatase and tensin homologue
- RANKL
Ligand for the receptor activator of nuclear factor-κB
- RHoC
Ras homologue gene family, member C
- STK11
Serine–threonine kinase 11 (also known as LKB1)
- TWIST1
Twist homologue 1
- VEGF
Vascular endothelial growth factor
- VHL1
von Hippel–Lindau 1
Footnotes
No other potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.DeVita VT Jr, Lawrence TS, Rosenberg SA, editors. DeVita, Hellman, and Rosenberg’s Cancer: principles & practice of oncology. 8th ed. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 2.Weinberg RA. The biology of cancer. New York: Garland Science; 2007. [Google Scholar]
- 3.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 6.Luzzi KJ, MacDonald IC, Schmidt EE, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 9.Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 10.Bristow RG, Hill RP. Hypoxia and metabolism: hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 11.Higgins DF, Kimura K, Bernhardt WM, et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J Clin Invest. 2007;117:3810–3820. doi: 10.1172/JCI30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 13.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 14.Clarke MF, Fuller M. Stem cells and cancer: two faces of Eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kucia M, Reca R, Miekus K, et al. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23:879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan RN, Riba RD, Zacharoulis S, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev. 2008;22:559–574. doi: 10.1101/gad.1636908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 20.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 21.Cano A, Pérez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Yang MH, Wu MZ, Chiou SH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 24.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [Erratum, Nature 2001;411:974.] [DOI] [PubMed] [Google Scholar]
- 25.Hakem A, Sanchez-Sweatman O, You-Ten A, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 28.López-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 29.Martin MD, Matrisian LM. The other side of MMPs: protective roles in tumor progression. Cancer. Metastasis Rev. 2007;26:717–724. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
- 30.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 31.Minn AJ, Kang Y, Serganova I, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavazoie SF, Alarcón C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 34.Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 2005;7:513–520. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 36.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Padua D, Zhang XH, Wang Q, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavrilovic IT, Posner JB. Brain metastases: epidemiology and pathophysiology. J Neurooncol. 2005;75:5–14. doi: 10.1007/s11060-004-8093-6. [DOI] [PubMed] [Google Scholar]
- 39.Billingsley KG, Burt ME, Jara E, et al. Pulmonary metastases from soft tissue sarcoma: analysis of patterns of diseases and postmetastasis survival. Ann Surg. 1999;229:602–610. doi: 10.1097/00000658-199905000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess KR, Varadhachary GR, Taylor SH, et al. Metastatic patterns in adenocarcinoma. Cancer. 2006;106:1624–1633. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- 41.Leiter U, Meier F, Schittek B, Garbe C. The natural course of cutaneous melanoma. J Surg Oncol. 2004;86:172–178. doi: 10.1002/jso.20079. [DOI] [PubMed] [Google Scholar]
- 42.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13:1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 43.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–573. [PubMed] [Google Scholar]
- 44.Khanna C, Wan X, Bose S, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 45.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 46.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Loberg R, Taichman RS. The pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis. Cancer Metastasis Rev. 2006;25:573–587. doi: 10.1007/s10555-006-9019-x. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 49.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 50.Ling V, Chambers AF, Harris JF, Hill RP. Quantitative genetic analysis of tumor progression. Cancer Metastasis Rev. 1985;4:173–192. doi: 10.1007/BF00050694. [DOI] [PubMed] [Google Scholar]
- 51.Waghorne C, Thomas M, Lagarde A, Kerbel RS, Breitman ML. Genetic evidence for progressive selection and overgrowth of primary tumors by metastatic cell subpopulations. Cancer Res. 1988;48:6109–6114. [PubMed] [Google Scholar]
- 52.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 53.Gupta GP, Perk J, Acharyya S, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swarbrick A, Roy E, Allen T, Bishop JM. Id1 cooperates with oncogenic Ras to induce metastatic mammary carcinoma by subversion of the cellular senescence response. Proc Natl Acad Sci U S A. 2008;105:5402–5407. doi: 10.1073/pnas.0801505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein CA, Blankenstein TJ, Schmidt-Kittler O, et al. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet. 2002;360:683–689. doi: 10.1016/S0140-6736(02)09838-0. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt-Kittler O, Ragg T, Daskalakis A, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hüsemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Aguirre-Ghiso JA. The problem of cancer dormancy: understanding the basic mechanisms and identifying therapeutic opportunities. Cell Cycle. 2006;5:1740–1743. doi: 10.4161/cc.5.16.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norton L, Massagué J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 60.Minn AJ, Gupta GP, Padua D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bertucci F, Cervera N, Birnbaum D. A gene signature in breast cancer. N Engl J Med. 2007;356:1887–1888. doi: 10.1056/NEJMc070393. [DOI] [PubMed] [Google Scholar]
- 62.Buyse M, Loi S, van’t Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 63.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression–based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 64.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 65.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 66.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2(2):e7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chi JT, Wang Z, Nuyten DS, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3(3):e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–741. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 69.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 70.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]