Abstract
A technique for microfluidic, pH modulated DNA capture and purification using chitosan functionalized glycidyl methacrylate monoliths is presented. Highly porous polymer monoliths are formed and subsequently functionalized off-chip in a batch process before insertion into thermoplastic microchannels prior to solvent bonding, simplifying the overall fabrication process by eliminating the need for on-chip surface modifications. The monolith anchoring method allows for the use of large cross-section monoliths enabling high flowrates and high DNA capture capacity with a minimum of added design complexity. Using monolith capture elements requiring less than 1 mm2 of chip surface area, loading levels above 100 ng are demonstrated, with DNA capture and elution efficiency of 54.2% ± 14.2% achieved.
INTRODUCTION
Nucleic acid capture and purification are often a necessary step prior to PCR amplification during genetic analysis to isolate the nucleic acids from other components of biological sample matrices such as cell lysate and blood plasma which could otherwise introduce components that inhibit PCR replication of target DNA sequences, degrading efficiency of the amplification process and resulting in poor assay reproducibility.1
Typically, modern laboratory scale DNA purification is achieved by silica-based solid phase extraction (SPE) where cell lysate is exposed to a silica surface in the presence of chaotropic agents.2 This strategy has been employed in a variety of microfluidic formats using packed beds of silica beads3 and polymer monoliths with embedded silica particles4–6 as the solid phase. The extraction efficiency of SPE methods is high (68%–80%); however, the chaotropic agents can be potent PCR inhibitors, thereby requiring copious washing to ensure that an inhibitor-free DNA solution is eluted as a final product.
An aqueous and PCR compatible alternate approach to chaotropic SPE is electrostatically driven, pH modulated nucleic acid capture on an amine-rich surface which can be controllably switched between cationic and neutral states. Such charge switching methods have been implemented in microfluidic systems, with various aminosilanes used to coat glass microchannels to yield a capture substrate with pH switchable surface charge.7
As an effective alternative to aminosilanes, the aminosaccharide biopolymer chitosan has also been employed as a pH modulated surface treatment for nucleic acid capture in microfluidic devices.8–10 While high loading levels and extraction efficiencies have been reported using chitosan as a charge-switching polymer for microfluidic DNA capture and release, reported methods typically require long channels distributed over large device areas to achieve this performance. This constraint is imposed by the need for sufficient surface area to achieve acceptable loading capacity. While high aspect ratio microstructures can be used to enhance surface area, this approach requires the application of complex fabrication methods that are undesirable for use in disposable sample preparation chips. Furthermore, long or wide channels are required so that the residence time during perfusion through the capture zone is significantly longer than the characteristic diffusion time for each sample component, ensuring sufficient interactions between DNA and the channel walls to promote efficient capture.
Here, we present a simple approach to microfluidic, pH-modulated nucleic acid capture in the form of a chitosan-functionalized porous polymer monolith. While monoliths have been used previously in various implementations of microfluidic silica-based SPE,4–6 the use of porous polymer monolith supports has not been previously explored for chitosan-enabled nucleic acid capture based on efficient charge switching. By employing monolith elements with high surface area and small pore size as chitosan supports, highly effective DNA capture with exceptionally high loading limits is achieved in a small on-chip footprint. The tortuous pore network inherent to the polymer monoliths also enables rapid release during the elution step. In addition to demonstrating the benefits of porous monoliths as high surface area substrates for efficient DNA capture, we further leverage a unique off-chip process that enables parallel batch scale preparation of chitosan-bearing monoliths, followed by integration of the pre-functionalized monolith elements into the disposable thermoplastic microfluidic devices. This process provides a scalable and low cost option for integrating nucleic acid capture, concentration, and release functionality to a range of microfluidic analytical platforms fabricated in thermoplastics. Using this approach, DNA loading levels above 100 ng are achieved using 1 mm long monolith capture elements, while repeatable recovery above 50% of the total loaded DNA sample is demonstrated.
MATERIALS AND METHODS
Glycidyl methacrylate (GMA), cyclohexanol, methanol, ethanol, 2,2-dimethoxy-2-phenylacetophenone (DMPA), sodium phosphate dibasic, N-[γ-maleimidobutyryloxy]succinimide ester (GMBS), and decahydronaphthalene (decalin) were purchased from Sigma–Aldrich (St. Louis, MO). Ethoxylated trimethylolpropane triacrylate (SR454) was received as a free sample from Sartomer (Warrington, PA). Cyclic olefin copolymer (COC; grade 1020R) was purchased from Zeon Chemicals (Louisville, KY). PUC 19 plasmid DNA was purchased from Fisher Scientific (Pittsburgh, PA).
Monolith preparation
GMA monolith elements were produced off chip and integrated into COC microchannels according to a previously reported method.11 Briefly, a solution consisting of 21.4% GMA, 14.2% SR-454, 53.4% Cyclohexanol, 10.7% methanol, and 0.4% DMPA was injected into a COC microchannel mold with a trapezoidal cross section and the access holes were sealed with tape. The COC mold was cut with a conical endmill to a depth of 640 μm. Photopolymerization was achieved with using a UV light source (Sun Ray 400 SM UV Flood Lamp) outputting 39 mW/cm2 for 600 s.
Direct chitosan functionalization of GMA monolith was achieved by soaking newly formed, washed monolith elements in a 1% solution of chitosan in water. Temperature was controlled over long reaction times by placing a vial of the monoliths and solution in an oven.
The bifunctional cross-linker GMBS was also used to attach chitosan to the GMA monoliths using an adaptation of a previously reported antibody anchoring procedure.12
Chip fabrication and bonding
Microchannels and access holes were directly milled in a 2 mm thick COC plaque using a Roland MDX650 CNC milling machine. Monolith receiving channels were patterned with the same end mill used for the mold, but milled to a depth of 580 μm to account for monolith shrinkage. Chip bonding was accomplished by first exposing both top and bottom portions of the chip to 25% (vol. %) decalin in ethanol for 10 min, then rinsing each thoroughly in ethanol and drying with nitrogen prior to monolith insertion. Functionalized monoliths were manipulated into position in the open channel with a wet fine-tipped paint brush before the two halve of the chip were mated together. The assembled chip was then pressed at 300 psi in a hot press at 60 °C for 10 min and released without cooling. Cryo-fracturing of chips for SEM imaging of the sealed monoliths was performed by making superficial cuts with a band-saw along the desired line of fracture, submersing the chip into liquid nitrogen, and cleaving the chip along the initial cut.
DNA capture and release experiments
Control solutions of DNA in loading buffer (50 mM MES pH 5) and elution buffer (10 mM Tris, 50 mM KCl, pH 10) were prepared by serial dilution. Prior to DNA loading, chitosan coated monolith chips were flushed with approximately 50 μl of the loading buffer. A solution of DNA in loading buffer was pumped through the monolith chip using a syringe pump, Hamilton gastight glass syringes, and capillary tubing from Polymicro Technologies with Upchurch PEEK fittings. Chip connections at inlet and outlet were made by pressing gauge 22 hollow stainless steel needles (Hamilton Syringe) into pre-drilled holes.13 Eluent was collected by bending or otherwise positioning the outlet needle to directly fill either a 96 well plate or individual Eppendorf tubes. A flowrate of 5 μl/min was throughout each test for all data shown in this paper with minimal observed back pressure.
DNA measurements
DNA concentrations were measured with a Quant-iT PicoGreen dsDNA Assay Kit from Invitrogen (Carlsbad, CA) using a SpectraMax M5 plate reader from Molecular Devices (Sunnyvale, California). Direct imaging of DNA on the capture monoliths was performed using Sybr Safe DNA gel stain (Invitrogen).
PCR was performed on a Roche Lightcycler 480 using LightScanner Master Mix from Idaho Technology, Inc. (Salt Lake City, UT) and the following primers:
Forward primer: 5′-TCCGACCCTGCCGCTTAC-3′.
Reverse primer: 5′-GACCTACACCGAACTGAGATACC-3′.
RESULTS AND DISCUSSION
Chitosan anchoring to GMA monoliths
Two different strategies were initially explored for functionalization of GMA monoliths with chitosan. In the first approach, a bifunctional cross-linker (GMBS) was used to couple amines from the chitosan polymer with epoxy groups on the GMA monolith. While we found this route to be effective in generating a dense layer of chitosan on the monolith surface, as determined by nucleic acid capture, it requires a complex multistep reaction process. To avoid this constraint, a second strategy based on direct reaction of chitosan with freshly formed monoliths was investigated. Mallik et al. reported a route for the direct attachment of proteins to GMA monoliths.14 In this approach, direct reaction between primary amines of the protein and the exposed reactive epoxy groups on the GMA monolith is achieved in the absence of a separate cross-linking agent, providing a simple and convenient route to monolith functionalization. However, due to the limited reaction efficiency, this technique required 6 days, room temperature perfusion of protein solution at pH 8 through the GMA monolith to achieve adequate conjugation. For conventional microfluidic applications, where monoliths are fabricated in situ within a microchannel by patterned photopolymerization, the extended reaction time renders the direct reaction route impractical, since each individual chip requires photolithographic fabrication of the monolith elements followed by extended reagent perfusion and incubation.
To overcome the limitation imposed by long incubation times required for direct chitosan attachment, we explored a preparation method in which large numbers of monolith elements are manufactured in parallel, functionalized off-chip in bulk solution, and individually integrated into the final microfluidic devices. Due to the parallel and bulk nature of this process, the long chitosan incubation time has little impact on the overall efficiency of the fabrication method. The reintegration method used relies on a triangular or trapezoidal channel cross-section in the monolith region.11 This characteristic shape, with sloping sidewalls allows for intimate sealing between the externally prepared monolith and the solvent-softened thermoplastic channel walls. In addition to facilitating large scale batch chitosan attachment, this technique allows for very short lengths of large cross-section monolith with excellent sealing at the interface between the monolith and microchannel walls, eliminating voids which would otherwise allow fluidic bypass and sample loss. Furthermore, compared with on-chip chitosan attachment using both the cross-linking and direct attachment routes, this off-chip functionalization strategy was found to be successful in avoiding clogging of the monolith which was commonly observed during on-chip attachment due to excessive chitosan build-up during active perfusion through the monolith element.
Reaction conditions for direct attachment of chitosan were optimized by varying both reaction time and temperature. Because chitosan has limited solubility at pH 8, as employed in previous work, this attachment method was performed in an unbuffered solution.14 A 1% chitosan solution in water was used, since higher concentrations were found to result in excessive solution viscosity. Best results were achieved using 3 days direct reaction with 1% chitosan solution at 40 °C.
DNA capture and elution performance of monoliths functionalized using on-chip crosslinking and off-chip direct reaction were found to be similar in initial tests. Based on this observation, direct functionalization was adopted as the standard method for further testing due to its simplicity and potential for large-scale batch production.
Nucleic acid loading, capture, and release
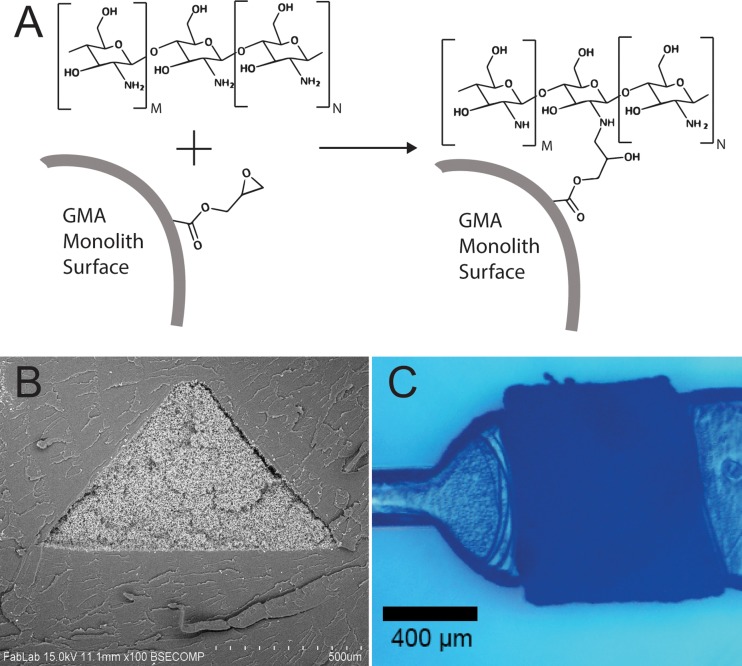
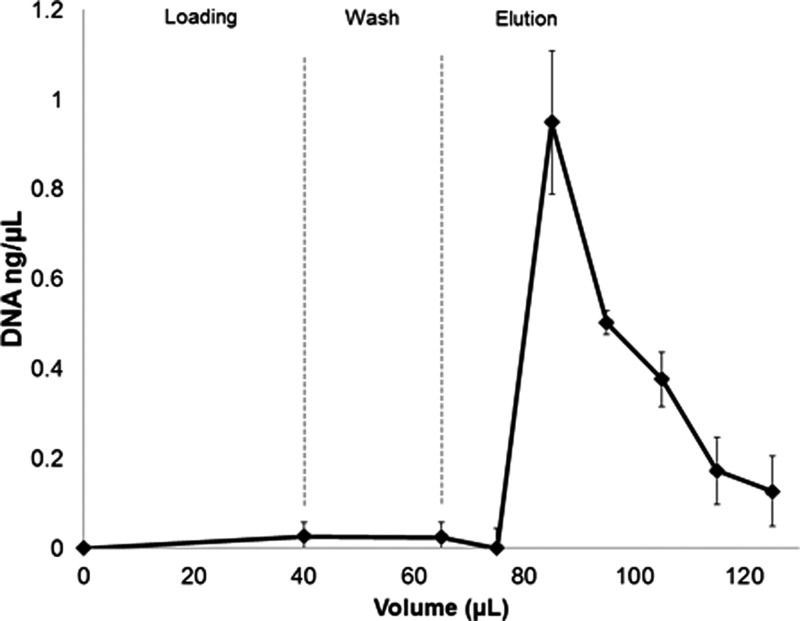
Chitosan functionalized monoliths were integrated into thermoplastic microchannels as shown in Figure 1 and characterized for DNA capture and recovery efficiency. The rough monolith surface and small void along the wall interface visible in Figure 1(b) are believed to result from the cryo-fracturing process used to open the chips for imaging. Data from a typical experiment are shown in Figure 2, which present average DNA concentrations at the chip outlet during loading (first point), washing (second point), and elution (following points). DNA concentrations at the outlet are negligible during loading and washing steps. DNA concentration reaches a peak after approximately 10 μl of elution buffer have been flushed through the system. Because of the strong pH dependence of Picogreen fluorescence, two separate serial dilutions are prepared to allow quantitation of DNA at pH 5 and at pH 10. The transitional first fraction after switching from pH 5 wash buffer to pH 10 elution buffer represents a potential undercounting of DNA. Using data collected from 3 separate batches of chips fabricated by three days direct-reaction attachment method, the chitosan-functionalized monoliths were effective at capturing and releasing DNA, with an average recovery of 54.2% ± 14.2%. While performance of the functionalized monoliths was reproducible, some fabricated devices in different batches were found to exhibit efficient DNA capture but low levels of release during elution. This behavior may be due to an excess of attached chitosan on the monolith surface in these experiments.
FIG. 1.
(a) Schematic of direct reaction between GMA monolith and chitosan. (b) SEM cross-section image of a GMA monolith in a COC microfluidic channel (cryo-fractured for imaging). (c) Bottom (flat side) view of a triangular monolith in a microfluidic channel.
FIG. 2.
Average (n = 3) DNA concentration in eluent from chitosan monolith chips from 3 batches fabricated with direct reaction method (3 days, 1% chitosan, 40 °C). 40 μl of 1 ng/μl DNA solution is loaded, followed by 30 μl of wash buffer and 60 μl of elution buffer, all at 5 μl/min. Average capture efficiency is (<99%) and recovery efficiency is 54.2% ± 14.2%.
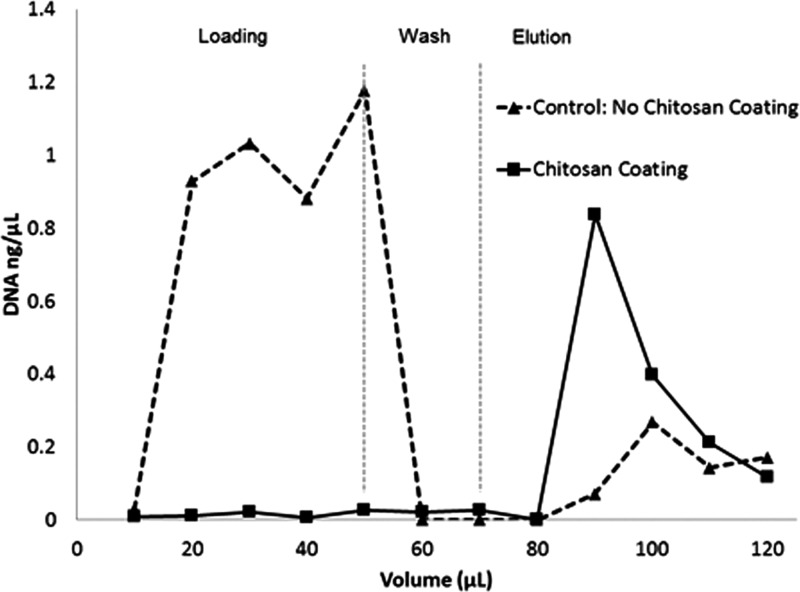
To ensure that DNA capture results from the chitosan modified monolith surface, rather than physical or chemical interactions with the GMA monolith itself, native monoliths without surface modification were used as controls. A particular concern was that DNA might become trapped between small submicron gaps formed at the interface between adjacent polymer nodules within the porous monolith matrix, resulting in physical retention of DNA within the matrix. Figure 3 shows a comparison between outlet DNA concentration for a control monolith and chitosan coated monolith during the capture, wash, and elution of 40 ng of DNA. Approximately 10% of the DNA was retained in the control monolith during loading and washing at pH 5. All of this, DNA is recovered in the elution buffer, confirming that the captured DNA is not physically trapped within the porous matrix. Loading DNA into a control monolith at high pH resulted in no measureable retention. Thus, while bare, unfunctionalized GMA monolith does exhibit weak pH dependent DNA capture capability, these tests demonstrate that the addition of a layer of chitosan is necessary for capturing a significant percentage of the DNA in solution.
FIG. 3.
DNA capture using an un-functionalized control monolith vs a chitosan coated monolith.
In addition to DNA entrapment within the monolith, a further potential concern is that proteins and other cell debris from biological samples may become entrapped or otherwise foul the monolith. In the case of proteins, due to the low hydrophobicity index of GMA no significant protein adsorption resulting from hydrophobic interactions is expected. We also note that monoliths based on butyl methacrylate and synthesized with similar morphology are commonly employed for proteins separations by liquid chromatography,15 where physical entanglement of proteins would degrade separation efficiency, and thus proteins entrapment within the monolith matrix is not a concern in this work.
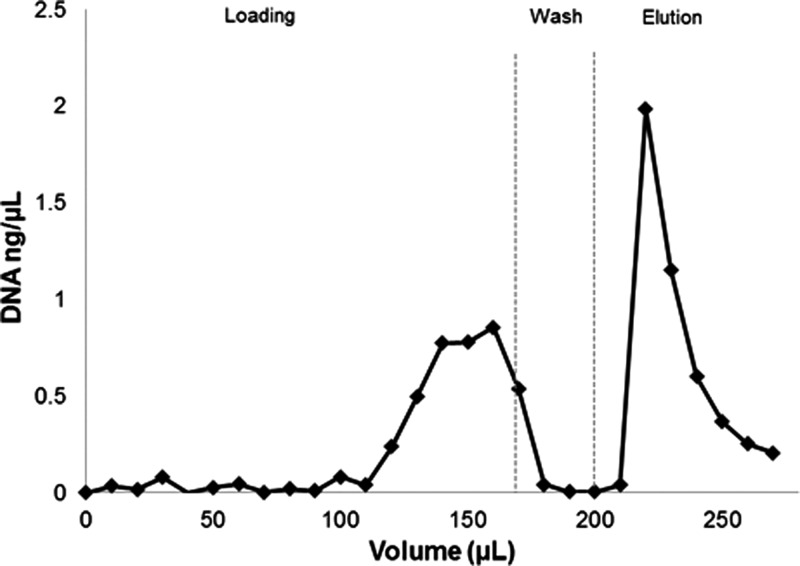
The maximum DNA loading capacity of a sample monolith was determined by pumping a solution of 1 ng/μl DNA at pH 5 through a 1 mm long monolith, occupying only 300 nl of channel volume, at 5 μl/min while collecting the eluent in 10 μl fractions. As shown in Figure 4, the concentration of DNA in the eluent remains negligible until the capture capacity of the monolith is reached, after which it rises to match the inlet concentration over the course of several fractions. A total of 110 ng DNA is captured before unbound analyte is observed to pass through the monolith during further loading. From the DNA bound to the monolith, approximately 40% is recovered after switching to the elution buffer; similar to the recovery fraction measured for other chips containing monoliths from the same processing batch. Eluted DNA was amplifiable by PCR in all cases and quantitative PCR measurements agreed well with fluorescence assays.
FIG. 4.
DNA capture capacity measurement. 160 μl of 1 ng/μl DNA solution is loaded, followed by 30 μl of washing buffer, and 70 μl elution buffer, all at a rate of 5 μl/min. The capture capacity of the 1 mm long monolith occupying 0.3 μl of channel volume is reached at approximately 110 ng and the concentration of DNA in the eluent rises to match the inlet concentration. After washing, approximately 40% of the captured DNA is eluted in a sharp peak with an elevated concentration.
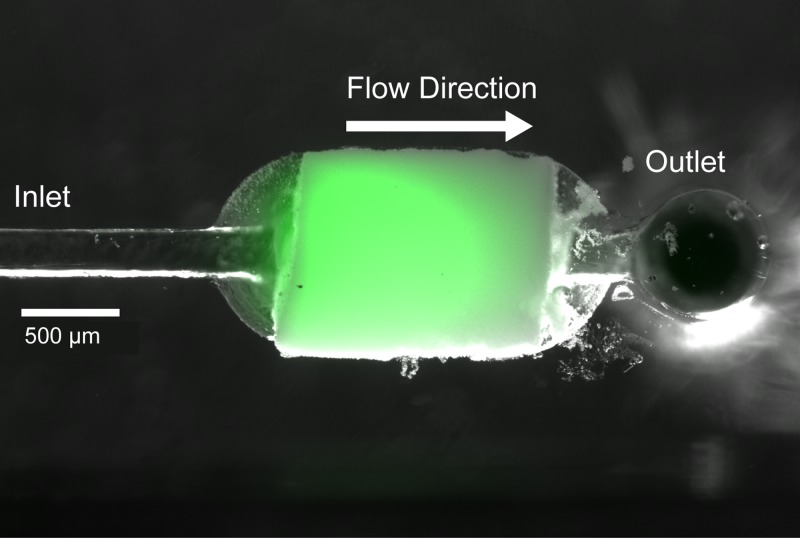
Any DNA not accounted for in the eluent is assumed to remain within the monolith, bound to the chitosan coated surface, where it may be visualized with a DNA intercalating dye as shown in Figure 5. Reversal of flow through a monolith after completion of the initial elution phase yielded negligible free DNA, implying that the remaining DNA is not merely physically trapped at the upstream end of the monolith, but rather tightly bound to the surface even after charge switching. Monoliths showed potential for repeat usage, with multiple DNA capture and release cycles successfully performed in sequential tests. However, for application to sample preparation for diagnostic assays, the potential for sample carryover between sequential samples within a single extraction element is a significant concern, making repeated usage of a single monolith impractical. However, noting that the high loading capacity demonstrated in this work enables efficient extraction within a small monolith footprint, large numbers of monolith capture elements may be fabricated to support multiple capture cycles on a single device without concern for carryover.
FIG. 5.
Monolith-bound DNA visualized with intercalating dye. Fluorescence image (green) overlayed on brightfield image (grayscale).
We note that Taylor dispersion caused by rapid flow of effluent through the open collection microchannel downstream of the porous monolith is likely responsible for the lack of sharp transitions each stage of the loading capacity test. The long tails of the elution peaks apparent in all of the figures are also presumed to be due to Taylor dispersion as a primary mechanism. While the effects of Taylor dispersion may be reduced by lowering the flow rates, allowing more concentrated recovery of DNA in a smaller volume, the benefits of this approach must be weighed against the increased elution time. A uniform flow rate of 5 μl/min was used for loading, washing, and elution steps in all the experiments, allowing the experiments detailed here to be performed in as little as 24 min. The average recovery of 54.2% ± 14.2% achieved in this work is similar to the recovery efficiency of previously reported chitosan-based nucleic acid capture employing functionalized planar surfaces.8–10 In contrast to prior work, however, the monolith based technique provides significantly higher loading capacity greater than 360 ng/μl of monolith volume, enabling exceptionally large amounts of DNA to be extracted within a small footprint. When combined with the inherent scalability of the monolith reintegration process, enabling low-cost manufacture of sample preparation chips for nucleic acid extraction, together with the compatibility of the monolith extraction elements with a simple flow-though capture and release process, the method reported here presents an attractive alternative for effective sample preparation in a disposable format.
CONCLUSION
Established methods for in-line nucleic acid cleanup in microfluidic systems require on-chip functionalization of the capture surfaces, increasing the complexity and cost of the overall fabrication process. Here, we have validated a simple approach to charge-switch based nucleic acid capture and purification using discrete reintegrated porous monolith elements that can be batch functionalized off-chip, followed by reintegrating individual elements into thermoplastic microchannels using standard device bonding protocols based on solvent bonding. The high surface area to volume ratio and large cross-sectional area to length ratio of the chitosan coated monolith elements allows for exceptionally high DNA capture capacity and efficient release in a small on-chip footprint and without introducing significant hydrodynamic resistance for effective flow-through operation. Because the total active surface area is defined by the volume of monolith used, this technique can be tailored to extract and release specific quantities of nucleic acid from a microfluidic fluid stream by defining only the length of monolith inserted. The flexibility and simplicity offered by the monolith-enabled method shows great promise for realizing effective nucleic acid capture, purification, and controlled release in thermoplastic micro total analysis systems using a highly manufacturable process.
ACKNOWLEDGMENTS
The authors would like to thank Niketa Jani and Keith Herold for assistance with PCR measurements and John Fisher for equipment access. This research was supported in part by funding from Canon U.S. Life Sciences, Inc., by NIH Grant No. R01AI096215, and by the DARPA N/MEMS S&T Fundamentals Program under Grant No. N66001-1-4003 issued by the Space and Naval Warfare Systems Center Pacific (SPAWAR) to the Micro/nano Fluidics Fundamentals Focus (MF3) Center. Electron microscopy was performed with facilities and support of the Maryland Nanocenter.
References
- 1.Breadmore M. C., Wolfe K. A., Arcibal I. G., Leung W. K., Dickson D., Giordano B. C., Power M. E., Ferrance J. P., Feldman S. H., Norris P. M., and Landers J. P., “ Microchip-based purification of DNA from biological samples,” Anal. Chem. 75(8), 1880–1886 (2003). 10.1021/ac0204855 [DOI] [PubMed] [Google Scholar]
- 2.Ivanova N. V., Dewaard J. R., and Hebert P. D. N., “ An inexpensive, automation-friendly protocol for recovering high-quality DNA,” Mol. Ecol. Notes 6(4), 998–1002 (2006). 10.1111/j.1471-8286.2006.01428.x [DOI] [Google Scholar]
- 3.Legendre L. A., Bienvenue J. M., Roper M. G., Ferrance J. P., and Landers J. P., “ A simple, valveless microfluidic sample preparation device for extraction and amplification of DNA from nanoliter-volume samples,” Anal. Chem. 78(5), 1444–1451 (2006). 10.1021/ac0516988 [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya A. and Klapperich C. M., “ Thermoplastic microfluidic device for on-chip purification of nucleic acids for disposable diagnostics,” Anal. Chem. 78(3), 788–792 (2006). 10.1021/ac051449j [DOI] [PubMed] [Google Scholar]
- 5.Wu Q., Bienvenue J. M., Hassan B. J., Kwok Y. C., Giordano B. C., Norris P. M., Landers J. P., and Ferrance J. P., “ Microchip-based macroporous silica sol-gel monolith for efficient isolation of DNA from clinical samples,” Anal. Chem. 78(16), 5704–5710 (2006). 10.1021/ac060390t [DOI] [PubMed] [Google Scholar]
- 6.Wen J., Guillo C., Ferrance J. P., and Landers J. P., “ Microfluidic-based DNA purification in a two-stage, dual-phase microchip containing a reversed-phase and a photopolymerized monolith,” Anal. Chem. 79(16), 6135–6142 (2007). 10.1021/ac0703698 [DOI] [PubMed] [Google Scholar]
- 7.Wen J., Guillo C., Ferrance J. P., and Landers J. P., “ DNA extraction using a tetramethyl orthosilicate-grafted photopolymerized monolithic solid phase,” Anal. Chem. 78(5), 1673–1681 (2006). 10.1021/ac051796t [DOI] [PubMed] [Google Scholar]
- 8.Cao W., Easley C. J., Ferrance J. P., and Landers J. P., “ Chitosan as a polymer for pH-induced DNA capture in a totally aqueous system,” Anal. Chem. 78(20), 7222–7228 (2006). 10.1021/ac060391l [DOI] [PubMed] [Google Scholar]
- 9.Reedy C. R., Price C. W., Sniegowski J., Ferrance J. P., Begley M., and Landers J. P., “ Solid phase extraction of DNA from biological samples in a post-based, high surface area poly(methyl methacrylate) (PMMA) microdevice,” Lab Chip 11(9), 1603–1611 (2011). 10.1039/c0lc00597e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagan K. A., Meier W. L., Ferrance J. P., and Landers J. P., “ Chitosan-coated silica as a solid phase for RNA purification in a microfluidic device,” Anal. Chem. 81(13), 5249–5256 (2009). 10.1021/ac900820z [DOI] [PubMed] [Google Scholar]
- 11.Kendall E. L., Wienhold E., Rahmanian O. D., and DeVoe D. L., “ Ex situ integration of multifunctional porous polymer monoliths into thermoplastic microfluidic chips,” Sens. Actuators, B Chem. 202, 866–872 (2014). 10.1016/j.snb.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Chen C.-F., Chang C.-W., and DeVoe D. L., “ Flow-through immunosensors using antibody-immobilized polymer monoliths,” Biosens. Bioelectron. 26(1), 182–188 (2010). 10.1016/j.bios.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C. F., Liu J., Hromada L. P., Tsao C. W., Chang C. C., and DeVoe D. L., “ High-pressure needle interface for thermoplastic microfluidics,” Lab Chip 9(1), 50–55 (2009). 10.1039/b812812j [DOI] [PubMed] [Google Scholar]
- 14.Mallik R., Jiang T., and Hage D. S., “ High-performance affinity monolith chromatography: Development and evaluation of human serum albumin columns,” Anal. Chem. 76(23), 7013–7022 (2004). 10.1021/ac049001q [DOI] [PubMed] [Google Scholar]
- 15.Liu J., Chen C.-F., Tsao C.-W., Chang C.-C., Chu C.-C., and DeVoe D. L., “ Polymer microchips integrating solid-phase extraction and high-performance liquid chromatography using reversed-phase polymethacrylate monoliths,” Anal. Chem. 81(7), 2545–2554 (2009). 10.1021/ac802359e [DOI] [PMC free article] [PubMed] [Google Scholar]