Abstract
We developed a new method for releasing viable cells from affinity-based microfluidic devices. The lumen of a microchannel with a U-shape and user-designed microstructures was coated with supported lipid bilayers functionalized by epithelial cell adhesion molecule antibodies to capture circulating epithelial cells of influx solution. After the capturing process, air foam was introduced into channels for releasing target cells and then carrying them to a small area of membrane. The results show that when the air foam is driven at linear velocity of 4.2 mm/s for more than 20 min or at linear velocity of 8.4 mm/s for more than 10 min, the cell releasing efficiency approaches 100%. This flow-induced shear stress is much less than the physiological level (15 dyn/cm2), which is necessary to maintain the intactness of released cells. Combining the design of microstructures of the microfluidic system, the cell recovery on the membrane exceeds 90%. Importantly, we demonstrate that the cells released by air foam are viable and could be cultured in vitro. This novel method for releasing cells could power the microfluidic platform for isolating and identifying circulating tumor cells.
I. INTRODUCTION
Affinity-based microfluidic devices are emerging as promising tools to efficiently isolate the rare cells from plurality such as circulating tumor cells (CTCs) and circulating fetal cells from peripheral blood, and stem cells from primary culture.1–7 Despite allowing for the rapid isolation of these clinically relevant cell types, microfluidic methods are facing the challenge of releasing and recovering the isolated target cells for further biological analysis.8 Due to the nature of low concentration of CTCs in whole cellular population after separation process, the acceptable efficiency of releasing cells should approach 100% for broader applications such as sensitively monitoring therapy response, diagnosis of early stage cancer, and analysis of genetic makeup. To date, the downstream analysis is limited on the imaging-based approaches after conducting the cell isolation. If the target cells captured by continuous flow affinity-based micro-channels can be released and recovered effectively and viably, the full genome wide studies, the culture in vitro, and pharmaceutical research of rare cell populations are made possible.
Potential approaches to release captured cells from affinity-based microfluidic devices include chemical methods such as gradient elution and mechanical methods such as the application of flow shear stress9–13 and air bubbles.14,15 For gradient elution methods, although a proteolytic enzyme combined with surfactant can release the captured cells, the surfactant would potentially disrupt the cell membrane and cause the degradation of surface markers, thus limiting the downstream biological analysis of cells.1,2,16 Moreover, exposed to chemical microenvironments different from physiological level, the phenotypic and functional information of the target cells may be altered.17 For flow-induced cell detachment, studies found in open literature show that the recovery efficiency of the captured cells ranges from 60% to 80%.10–12 Cheung et al.9 investigated the characteristics of the detachment of the isolated cells under flow acceleration in a bio-functionalized micro-channel; in this study, detachment efficiency of ∼90% of bounded cells required a high shear stress ∼200 dyn/cm2, which significantly decreased cell viability and caused changes in gene expression.3,18
Using bubble-induced detachment of affinity-adsorbed cells has been reported in recent years.14,15,19 Surface attached particles may be released by introducing an air-water interface, which creates a net surface tension upon particle. When the net force is larger than the surface adhesive force, the particle-surface linkage may be disrupted, thus releasing the particle. Wang et al.15 have shown the proof of concept using a sequence of air bubbles (0.1–0.5 ml in volume) to detach 99%+ blood cells captured with open-tubular capillary cell affinity chromatography. Barkley et al.14 also showed that air bubbles produced by a three-way valve combined with a syringe and then injected into the flow stream in a nylon tube could be used to effectively detach human erythrocytes affinity-adsorbed to the lumen of the nylon tube. While these works demonstrated the potential using air bubbles to release cells from the affinity surface in a straight capillary tube, the adaption to the chip-based microfluidic devices has not been demonstrated19 due to multiple technical challenges such as air bubble trapping, and irregular flow field of mixture of air bubbles and liquid resulting in low release efficiency. In addition, it was estimated the air-water interface could induce stress on the cell ranging from 20 to 300 dyn/cm2 according to the position of adhesive cell with respect to the air-water interface.14 The force exerted on the cells could severely damage the cells.
Supported lipid bilayer (SLB), an artificial mimic of the natural cell membrane, which is compatible with protein incorporation and possesses sustained lateral mobility,20–26 has been studied extensively as model system of biological membranes over past decades. Previously, many studies have demonstrated that this biomimetic multifunctional material has great bioinert property and lateral mobility, thus leading to effective resistance of non-specific proteins adsorption and cells adhesion.26–28 Although it is not entirely understood how lipid molecules interact with proteins and cells, these non-fouling behaviors have been demonstrated to be correlated with surface hydration, flexibility, and fluidity of the lipid molecules.23,25–29 Besides, the lateral mobility of SLB also has been demonstrated to allow lipid and proteins to re-organize their distribution on the surfaces.23,25,26,30,31 Our previous studies showed that when SLB modified with a particular antibody is used to bind cells by antibody-antigen reactions, its fluidity allows for the assembly of proteins and then increase the binding affinity and specificity of target cells.23,25,26 Owing to its biological inertness, resistance of non-specific binding of unwanted proteins and easy incorporation of biological functions, SLB is a great bio-sensing surface.26
This paper focuses on the development of new method for releasing rare cells like CTCs bound to the lumen of microfluidic devices that are immobilized with antibody functionalized SLB film. This novel method for releasing cells utilizes an air foam, a mass of air bubbles to sweep the micro-channel with user-designed micro-structures and to detach the cells with extremely high efficiency. Except for high efficiency, releasing cells through air foam renders viable cells to be collected and be cultured and is compatible with any design-complicated chip-based microfluidic devices. This novel ideal reaches a milestone for the field of rare cell isolation research.
II. MATERIALS AND METHODS
A. Microchannel fabrication
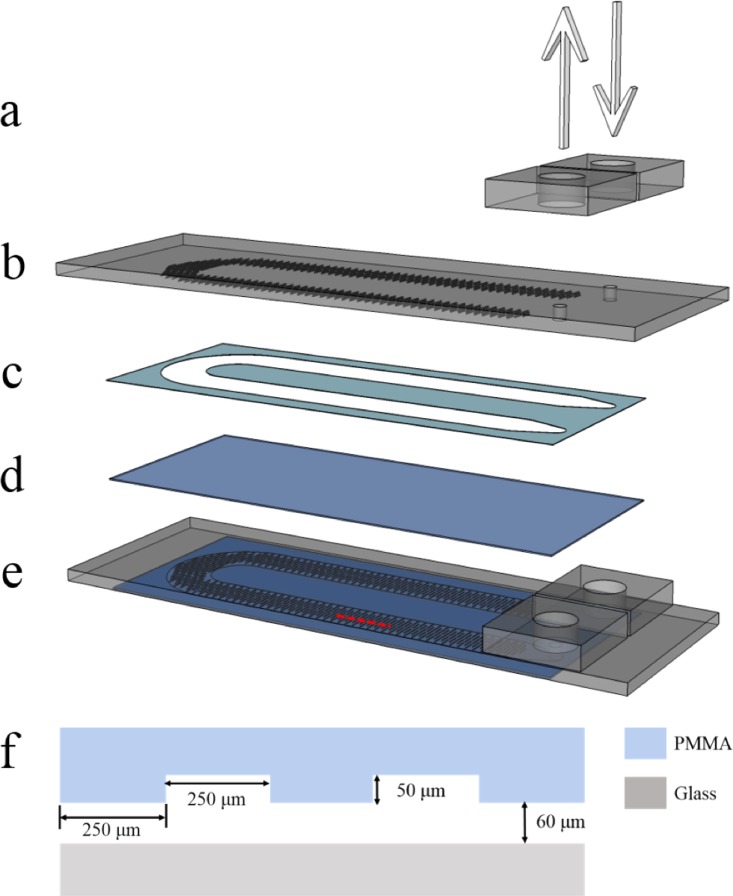
The microfluidic devices were fabricated with one inlet and outlet connected by a U-shape micro-channel using laser micro-machining technology. Shown in Figure 1 is the schematic diagram of the microfluidic device used in this study. The construction of the devices started with the manufacture of the PMMA top plate having two drilled holes, inlet and outlet, on it whose dimensions equal to a standard microscope slide. After carved into U channel shape, the spacer with double-coated acrylic adhesive on both sides was bounded to the PMMA top plate in one side and to the standard cover glass slide in the other side to form the microfluidic channel. The PMMA connectors bonded to the PMMA top plate by applying chloroform to the interface were used to join the outer tubes to the inner micro-channels. The micro-channel is then about 110 mm long, 5.5 mm wide, and 0.06 mm high. In order to disrupt the streamlines followed by cells of laminar flow that dominates the flow conditions in microfluidic-based devices and to increase the number of cell-surface interactions, the upper surface of the micro-channel was patterned with a line groove whose average height is about 0.05 mm, average width about 0.25 mm, and average length about 1 mm, as shown in Figure 1(f).
FIG. 1.
Exploded view of the microfluidic device: (a) PMMA connectors, (b) PMMA top plate, (c) spacer, (d) glass base slide, and (e) complete microfluidic device. (f) Dimension of micro-channel and microstructure.
B. Microchannel surface modification
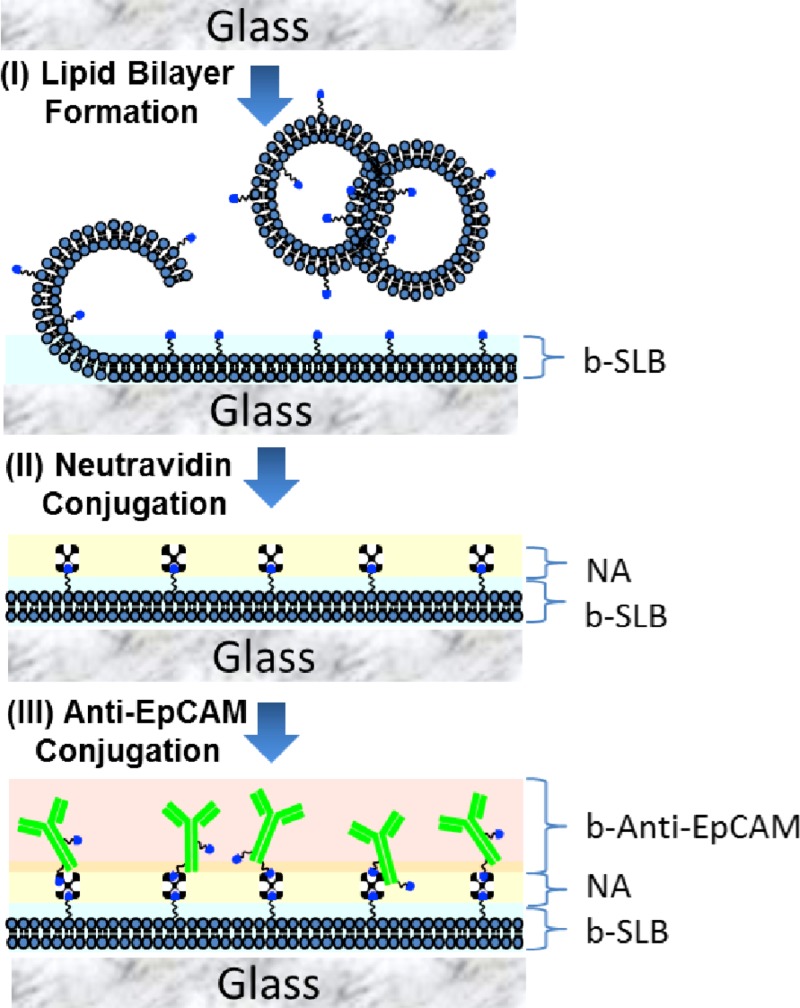
The immunoassay used to functionalize the lumen of microfluidic devices with particular antibodies is illustrated in Ref. 26. Briefly, to prepare an antibody-tethered SLB coating in the microfluidic channel, the sequential steps are followed (Figure 2): First, 0.2 mg/ml of lipid vesicles consisting of POPC/b-PE (95/5, 90/10, 85/15, and 80/20 mol/mol) was filled in the microfluidic channels and incubated for 1 h to form a complete lipid bilayer coating, followed by rinsing with 10 mM phosphate buffer saline (PBS) with 150 mM NaCl, pH = 7.2 to remove excess vesicles and unbound lipids. Second, 0.1 mg/ml solution of Neutravidin (NA) was flowed into SLB-bPE-coated micro-channel and incubated for 1 h, followed by rinsing with PBS buffer to remove excess NA. Then, 0.05 mg/ml solution of b-anti-EpCAM was introduced to conjugate with SA-SLB-bPE immobilized surface for 1 h, followed by rinsing with PBS buffer to remove excess b-anti-EpCAM.
FIG. 2.
A schematic illustration of surface immobilization of anti-EpCAM conjugated SLB in the microfluidic channel. (I) Lipid vesicles were absorbed and ruptured to form support lipid bilayer on the surface. (II) NA molecules were conjugated to the b-SLB film. (III) Biotinylated anti-EpCAM molecules were conjugated to create anti-EpCAM functionalized SLB film.
C. Colorectal cancer cell lines
Human colorectal cancer cell lines HCT 116 were maintained and grown in Dulbecco's modified Eagle medium (DMEM) (Gibco-RBL life Technologies, Paisley, UK) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic/antimycotic (100 U/ml penicillin sodium, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B; Gibco-RBL life Technologies, Paisley, UK), at 37 °C with 5% CO2 atmosphere in a humidified incubator. At approximately 90% confluence, cells were pre-stained with CellTracker Green CMFDA or CellTracker Red CMTPX (Invitrogen, CA, USA) for 1 h and followed by using 0.2% trypsin with 0.1% ethylenediaminetetraacetic acid (EDTA) (Sigma Aldrich, St. Louis, USA) to resuspend cells in the same medium for subsequent experiments.
D. Colorectal cancer cell capture, release, and recovery
Cell capture and release were characterized using HCT 116 cell lines. In order to demonstrate the ability of the SLB based microfluidic device to capture and then release target cells into a designed collection chip, around 300 to 2000 HCT 116 cells pre-stained with CellTracker green were spiked into 1 ml of the whole blood collected from healthy individuals and captured in the micro-channel, whose surfaces were immobilized with an anti-EpCAM antibody. After all the blood sample flowed through the functionalized micro-channel into the waste chamber (0.5 ml/h), following a PBS washing of about 300 μl (1 ml/h), Hoechst solution of 1 μg/ml was introduced into the micro-channel to stain the nuclear cells captured specifically and non-specifically for DNA content. Subsequently, the channel was fluorescently photographed and the total number of cells bound to the channel surface was counted. By identifying double positive cells (CellTracker+, DAPI+) as cancer cells and single positive cells (DAPI+) as leukocyte, the efficiency of the cell capture could be evaluated. The capture efficiency is defined as follows:
where Ec is the capture efficiency, Ns is the number of cells initially spiked into the whole blood, and Nc is the number of cells captured on the channel surface. In the phase of cell release, foam formed by trapping air bubbles in a liquid was flowed through the micro-channel so as to detach the specifically bound cancer cells and non-specifically bound white blood cells (WBCs). The released cells were carried to a collection device by the bubbly liquid, and then fluorescently imaged and counted on that device. Afterwards, the micro-channel was also inspected under the microscope to enumerate undetached cells. Based on the number of cells gathered on the collection device and remaining in the micro-channel, the efficiency of cell release and recovery of the system could be assessed. The release and recovery efficiencies are defined as
where Er is the release efficiency, Eo is the recovery efficiency, Nr is the number of cells remaining on the chip after releasing process, and Nm is the number of cells found on the collection membrane. The release efficiency focus on the number of cells carried by air foam while the recovery efficiency focus on the number of cells carried by air foam and arriving at the collection chip. Based on this definition, the recovery efficiency is smaller than or equal to the release efficiency. Through well-designed plumbing system of microfluidic devices, the recovery efficiency could approach the release efficiency.
In this study, a mass of air bubbles in the type of foam was used to detach cells bonded to the micro-channel surfaces. Those foams were produced by mechanically shaking the syringe in which the volume ratio of the culture medium to the air equal to 2. The resultant foam was then driven into the micro-channel for cell detachment by a syringe pump, as shown in Figure 3.
FIG. 3.
Photo of the cell-releasing process using medium foam.
Figure 4 shows the platform utilized to concentrate the released cells. By employing the spacer, the membrane with 2 μm pore size was concentrically attached to the drilled hole with 15 mm diameter on the PMMA plate, the dimensions of which correspond to the standard microscope slide. As the foam with detached cells passed through the membrane, the cells would be stopped in front of the membrane based on the size separation. Due to the small pore size of 2 μm diameter, all the cells, including cancer cells and WBCs, would be stayed on the membrane. In order to ensure that the foam could gently pass through the pores on the membrane and did not cause membrane clogging with un-expected debris, a slightly negative pressure was provided on the side of the membrane opposite to the side of cells. After completing cell-detaching experiments, the membrane was removed from the collection chip and mounted onto the standard microscope slide, and then could be imaged fluorescently under the microscope.
FIG. 4.
(a) Platform used to concentrate the released cells, (b) top view of collection chip, and (c) side view of collection chip.
E. Re-culture of colorectal cancer cell lines after isolation process
HCT 116 cells were pre-stained with CellTracker Red CMTPX (Life Technologies, CA, USA) and diluted in human blood to a final concentration of 500 cells ml−1. After flowing them through the chip using a syringe pump for aspiration, the captured cells were released by air-bubble and collected on 96 well tissue cultured plates at 37 °C under 5% CO2 in DMEM supplemented with 10% FBS and 1% antibiotic. At indicated time points, cells were photographed under phase contrast microscopy.
III. RESULTS AND DISCUSSION
A. Release of isolated cancer cells
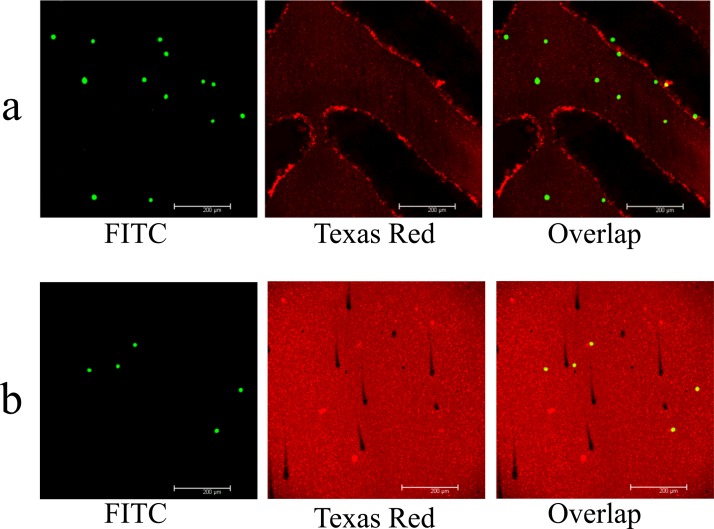
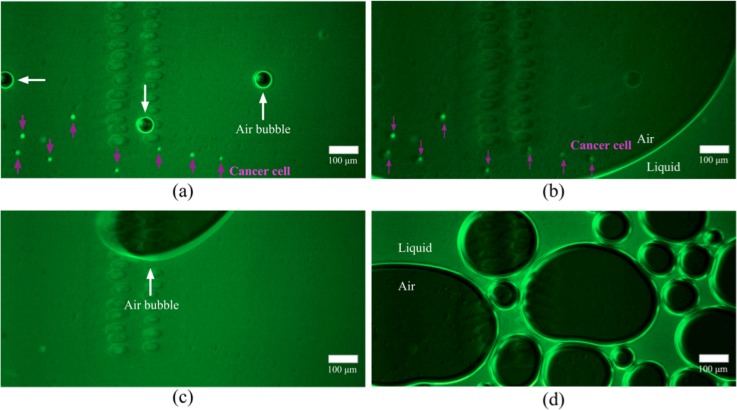
Through the process of surface modification described in Sec. II, the lipid layers might be coated both on the PMMA top plate and glass slide, as shown in Figure 5 by the fluorescent labeled liposome coating (in red). Figure 5 also shows that the functionalized PMMA surface could capture human colorectal cancer cell line Colo205 efficiently. In order to demonstrate the capacity for releasing cells captured with the lipid bilayer based microfluidic devices using the medium of the foam type, HCT 116 cells pre-stained with CellTracker green were first spiked into the PBS buffer and then flowed through the functionalized microfluidic channel. After conducting cell capture, the medium foam was introduced into the channel to release attached cells. Figure 6 shows the micrographs of the process in which the captured cells are released by having the foam pass through the micro-channel. When the foam enters into the channel and contacts the PBS buffer full of micro-channel after performing cell isolation, numerous small bubbles would merge into a large one due to the decrease in surface energy. As sweeping through the channel, these large bubbles would detach the cells bound to the surfaces near the channel center other than cells near the channel side walls where a liquid film is formed. In the latter stage of releasing cells, a mass of small bubbles gradually creeps all over the channel surfaces and the cells near the channel side walls would finally be detached. Figure 6(d) shows the snapshot of micro-channel filled with the foam after cell release. In the current experiment, there are 1297 cells captured in our microfluidic device (capture efficiency > 80%) and 3 cells remaining in the device after detaching cells by foam. The release efficiency is up to 99.7%.
FIG. 5.
Micrographs of micro-channels which were coated with SLB film conjugated with fluorescent dye Texas Red and functionalized by anti-EpCAM, and which were used to capture colorectal cancer cell line Colo 205 pre-stained with CellTrack Green (FITC): (a) PMMA top plate and (b) glass base slide.
FIG. 6.
Micrographs of the process of releasing cells using medium foam: after cell release for (a) 10.5 min, (b) 10.504 min, (c) 10.55 min, and (d) 12.08 min.
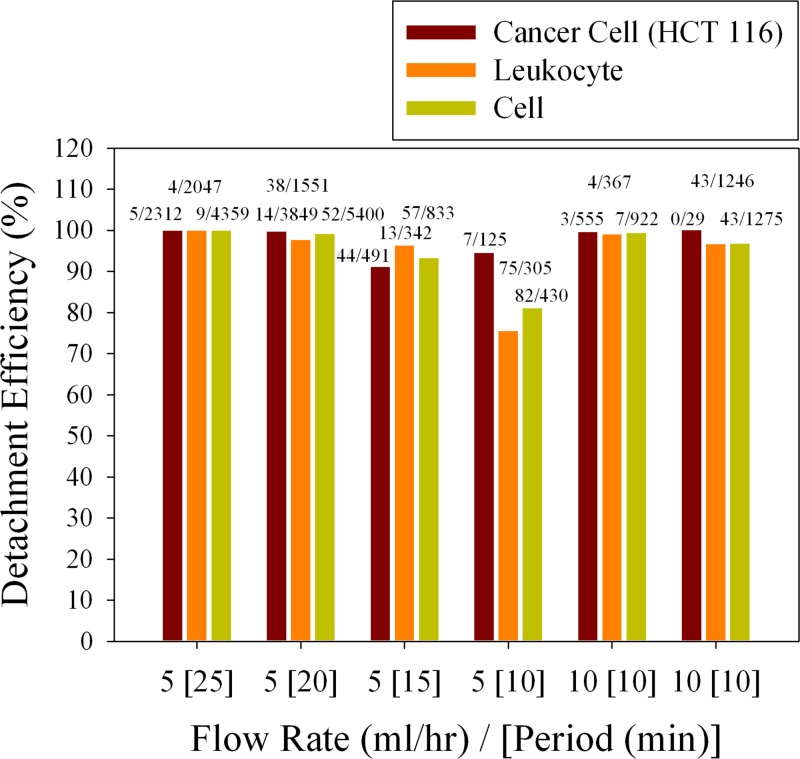
To further demonstrate the ability to release cells in the microfluidic devices, pre-stained HCT 116 cells were spiked into the whole blood collected from healthy donors and flowed through the micro-channel, followed by detaching cells including specifically binding cancer cells and non-specifically binding WBCs using the foam type medium. To reduce the influence of shear stress on the captured cells when performing cell detachment, the flow rate at which the foam sweeps through the micro-channel should be limited. Figure 7 shows the efficiency of cell release when varying the flow rate and the time during which the channel was washed by foam. It appears that the detachment efficiency of the cell line exceeds 90% in all test cases while the detachment efficiency of the leukocyte is about 75% if the medium foam is driven at the flow rate of 5 ml/h for 10 min. When considering the nuclear cells including cancer cells and leukocyte, the least efficiency of releasing cells is more than 80%. Moreover, the induced shear stress is about 4 dyn/cm2 at the flow rate of 5 ml/h and 8 dyn/cm2 at the flow rate 10 ml/h in the currently designed microfluidic channel. The shear stress is directly proportional to the flow rate given by9
where μ is the viscosity, Q is the flow rate, W is the channel width, and H is the channel height. One of noticeable features of SLB is the ability to resist the non-specific proteins adsorption and cells adhesion. Though using SLB with this non-fouling effect as base layer, there are unwanted cells non-specific bound to the channel surfaces including PMMA top plate and glass base slide. All the specific and non-specific bound cells would be detached by air foam, as shown in Figure 7. From the results above, the medium foam used to release cells in the lipid bilayer based microfluidic devices is considerately efficient and the foam-induced shear stress is much less than physiological level (15 dyn/cm2) observed in human blood vessels.32
FIG. 7.
Efficiency of cell release when varying the flow rate and the time during which the channel was washed by foam; the fifth and sixth experiments show repeated experimental results.
B. Recovery of released cancer cells
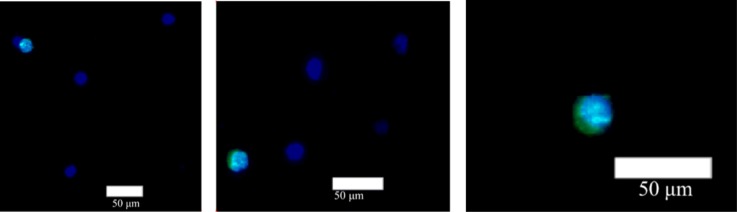
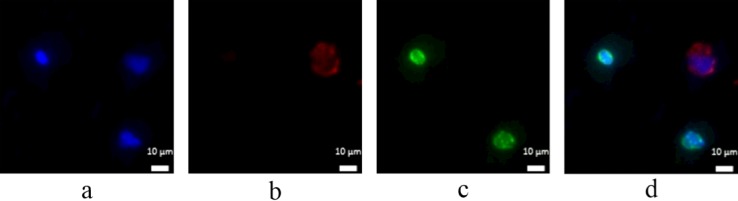
To evaluate the efficiency of the cell recovery, about 500 HCT 116 cells pre-stained with CellTracker green were spiked into the whole blood of 1 ml donated from healthy individuals and flowed through the lipid bilayer based microfluidic devices. After isolating cells, the medium foam was used to release cells to the collection devices, mainly consisting of the membrane with pore size of 2 μm diameter. Figure 8 shows the micrographs of concentrated cells including HCT 116 cells (CellTracker+, DAPI+) and WBCs (DAPI+) on the membrane. The morphology and the fluorescent signals relative to background noise of cells could be easily identified. One of the attractive features when cells released to the circular membrane with 15 mm diameter is that the time (20 min) needed to photograph the circular area (2 fluorescence channels) for counting cells is less than the time (2 h) to photograph the entire three-dimensional micro-channels otherwise. Table I shows the number of HCT 116 cells counted in the present experiments. It is likely that the cell-detaching efficiency using medium foam approaches 100% and the efficiency of cell recovery on the membrane exceeds 90%. The disparity between the detachment and recovery efficiency shows that some cells are lost when the medium foam carries the cells from the micro-channel to the membrane device. By detaching cells to the small area of the membrane, the performance of microfluidic devices could be easily and accurately evaluated, and out of focus images could be limited. In order to verify whether the cells are healthy enough to perform immunofluorescent staining after released from microfluidic devices using medium foam, a colorectal cancer cell line DLD1 was spiked into the whole blood drawn from healthy donors and then captured by the SLB based microfluidic device. After released into the collection chip, DLD1 underwent a series of steps of immunofluorescent staining. Figure 9 shows the micrographs of DLD1 after immune-staining phase, showing DLD1 could be labeled with antibodies directed against cytokeratins and the nuclear dye Hoechst (DAPI) while leukocyte could be labeled with antibodies against CD45 and the Hoechst (DAPI) for DNA content. All these results show that SLB based microfluidic devices combined with the method of foam-releasing cells could capture and detach viable cells for further analysis like immunostaining.
FIG. 8.
Micrographs of concentrated cells including HCT 116 cells: CellTracker+ (green)/DAPI+ (blue); and WBCs: CellTracker−/DAPI+ on the membrane.
TABLE I.
Number of counted HCT 116 cells and the efficiency evaluation of cell release and recovery.
| Number of cancer cells loaded (#) | Number of cells captured by the chip (#) | Number of cells remaining in the chip (#) | Number of cells acquired on the membrane (#) | Detachment efficiency (%) | Recovery efficiency (%) | |
|---|---|---|---|---|---|---|
| Experiment 1 | 587 | 117 | 1 | 116 | 99.15 | 99.15 |
| Experiment 2 | 480 | 72 | 0 | 67 | 100 | 93.1 |
| Experiment 3 | 447 | 111 | 0 | 110 | 100 | 99.1 |
FIG. 9.
Micrographs of DLD1 on collection device after the process of immunofluorescent staining: (a) image of DAPI signal, (b) image of cytokeratin signal, (c) image of CD45 signal, and (d) overlap image.
Although it is not fully understood what is the mechanism of using air foam to release cell bound to the lumen of microfluidic devices that are immobilized with antibody functionalized SLB film, the model of air-water interface detaching bacteria adhered to a surface helps us realize these multi-physical phenomena.33 Based on this model, the air-water interfacial force used to detach cells is closely related to interfacial tension and the position of a three-phase contact between a cell, air, and liquid, and it ranges from 0.02 dyn to 0.4 dyn. Exposed to the force, cell might be damaged in release phase. In this study, SLB was used as the base layer of functionalized surface, and the SLB assemblies are sensitive to air-water interfacial forces which could decrease the influence of the interfacial forces that would otherwise be applied to the cells directly. In order to demonstrate that cells are healthy when they are released from the SLB-based microfluidic devices using air foam, cell viability had been tested by live/dead kit. In the test case, 142 cells out of the 165 cells (HCT 116) collected on the membrane of the collection chip appear to be live. The viability rate is ∼86%.
C. In vitro culture of isolated colorectal cancer cells from blood
Foam composed of normal culture medium (DMEM contained with 10% FBS and 1% antibiotics) was used in this novel system to release the specific binding CTCs from the microfluidic device to the target area for further analysis and the substrate for culturing. To determine whether isolated colorectal cancer cell HCT116 can be cultured in vitro, the medium foam released cells were collected and seeded on 96 well tissue cultured plates at 37 °C under 5% CO2. After 1 day of seeding, cells adhered on the plate with round shape (Figure 10(a)). At day 3 of incubation, cells formed a small colony unit with the shape of flagstone (Figure 10(b)). To further observe the proliferation of air-bubble released cells on tissue cultured plates, the incubation time was extended to 7 days. Figure 10(c) shows that the size of colony unit increased with some accumulating cells overlay to the other cells (white arrow). These results illustrate that HCT116 could be released from chip and undamaged.
FIG. 10.
The captured HCT116 cells, pre-stained with CellTracker Red CMTPX, were released by air-bubble and cultured on 96 well tissue cultured plates for (a) 1 day, (b) 4 days, and (c) 7 days.
IV. CONCLUSION
To conclude, we have demonstrated a new method for releasing cells from affinity-based microfluidic devices whose lumen was coated with SLBs functionalized by anti-EpCAM antibody to capture colorectal cancer cells. After cells were adhered to the functional surface, air foam was used to release the whole cells in channels. The release efficiency is extremely high, approaching 100% at optimal running conditions. In addition, as we have demonstrated, the cells released through this way are healthy enough to be sustained in culture dish.
The microfluidic platform for CTC isolation has advantage of high capture efficiency while air-foam cell-releasing method introduced in this paper remedies its shortcoming: time-consuming CTC recognition on images and expands its application to downstream molecular analysis. With this novel method for releasing CTCs combined with microfluidic platform for capturing CTCs, the behavior of CTCs and mechanism of metastasis could be further studied consistently and routinely.
ACKNOWLEDGMENTS
The authors thank Dr. Han-Chung Wu (Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan) for providing anti-EpCAM. We thank National Science Council, Taiwan, for the funding support through Grant Nos. MOST 102-2321-B-001-060 and MOST 101-2113-M-001-021-MY2. We thank Academia Sinica, Taipei, Taiwan, for the funding support through Academia Sinica Summit Program. Jr-Ming Lai is supported by Academia Sinica Postdoctoral Fellowship 2013-2014.
References
- 1.Adams A. A., Okagbare P. I., Feng J., Hupert M. L., Patterson D., Göttert J., McCarley R. L., Nikitopoulos D., Murphy M. C., and Soper S. A., J. Am. Chem. Soc. 130(27), 8633–8641 (2008). 10.1021/ja8015022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dharmasiri U., Balamurugan S., Adams A. A., Okagbare P. I., Obubuafo A., and Soper S. A., Electrophoresis 30(18), 3289–3300 (2009). 10.1002/elps.200900141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang J. H., Krause S., Tobin H., Mammoto A., Kanapathipillai M., and Ingber D. E., Lab Chip 12(12), 2175–2181 (2012). 10.1039/c2lc40072c [DOI] [PubMed] [Google Scholar]
- 4.Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., and Toner M., Nature 450(7173), 1235–1239 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips J. A., Xu Y., Xia Z., Fan Z. H., and Tan W., Anal. Chem. 81(3), 1033–1039 (2009). 10.1021/ac802092j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stott S. L., Hsu C.-H., Tsukrov D. I., Yu M., Miyamoto D. T., Waltman B. A., Rothenberg S. M., Shah A. M., Smas M. E., Korir G. K., Floyd F. P., Gilman A. J., Lord J. B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L. V., Lee R. J., Isselbacher K. J., Maheswaran S., Haber D. A., and Toner M., Proc. Natl. Acad. Sci. U.S.A. 107, 18392–18397 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson M. N., Tsinberg P., Tang Z., Bischoff F. Z., Wilson T., and Leonard E. F., Biomicrofluidics 5(3), 034119 (2011). 10.1063/1.3623748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah A. M., Yu M., Nakamura Z., Ciciliano J., Ulman M., Kotz K., Stott S. L., Maheswaran S., Haber D. A., and Toner M., Anal. Chem. 84(8), 3682–3688 (2012). 10.1021/ac300190j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung L. S. L., Zheng X., Stopa A., Baygents J. C., Guzman R., Schroeder J. A., Heimark R. L., and Zohar Y., Lab Chip 9(12), 1721–1731 (2009). 10.1039/b822172c [DOI] [PubMed] [Google Scholar]
- 10.Dainiak M. B., Plieva F. M., Galaev I. Y., Hatti-Kaul R., and Mattiasson B., Biotechnol. Prog. 21(2), 644–649 (2005). 10.1021/bp049615g [DOI] [PubMed] [Google Scholar]
- 11.Kumar A., Plieva F. M., Galaev I. Y., and Mattiasson B., J. Immunol. Methods 283(1–2), 185–194 (2003). 10.1016/j.jim.2003.09.017 [DOI] [PubMed] [Google Scholar]
- 12.Ujam L. B., Clemmitt R. H., Clarke S. A., Brooks R. A., Rushton N., and Chase H. A., Biotechnol. Bioeng. 83(5), 554–566 (2003). 10.1002/bit.10703 [DOI] [PubMed] [Google Scholar]
- 13.Zheng X., Cheung L. S.-L., Schroeder J. A., Jiang L., and Zohar Y., Lab Chip 11(19), 3269–3276 (2011). 10.1039/c1lc20331b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkley S., Johnson H., Eisenthal R., and Hubble J., Biotechnol. Appl. Biochem. 40, 145–149 (2004). 10.1042/BA20030121 [DOI] [PubMed] [Google Scholar]
- 15.Wang K., Marshall M. K., Garza G., and Pappas D., Anal. Chem. 80(6), 2118–2124 (2008). 10.1021/ac702553w [DOI] [PubMed] [Google Scholar]
- 16.Panchision D. M., Chen H.-L., Pistollato F., Papini D., Ni H.-T., and Hawley T. S., Stem Cells 25(6), 1560–1570 (2007). 10.1634/stemcells.2006-0260 [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Riha G. M., Yan S. Y., Li M., Chai H., Yang H., Yao Q. Z., and Chen C. Y., Arterioscler., Thromb., Vasc. Biol. 25(9), 1817–1823 (2005). 10.1161/01.ATV.0000175840.90510.a8 [DOI] [PubMed] [Google Scholar]
- 18.Kang J. H., Choi S., Lee W., and Park J. K., J. Am. Chem. Soc. 130(2), 396 (2008). 10.1021/ja0770678 [DOI] [PubMed] [Google Scholar]
- 19.Li P., Gao Y., and Pappas D., Anal. Chem. 83(20), 7863–7869 (2011). 10.1021/ac201752s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson A. S., Glasmastar K., Sutherland D., Lidberg U., and Kasemo B., J. Biomed. Mater. Res., Part A 64A(4), 622–629 (2003). 10.1002/jbm.a.10442 [DOI] [PubMed] [Google Scholar]
- 21.Cremer P. S., Groves J. T., Kung L. A., and Boxer S. G., Langmuir 15(11), 3893–3896 (1999). 10.1021/la981240j [DOI] [Google Scholar]
- 22.Fahey P., Koppel D., Barak L., Wolf D., Elson E., and Webb W., Science 195(4275), 305–306 (1977). 10.1126/science.831279 [DOI] [PubMed] [Google Scholar]
- 23.Huang C.-J., Cho N.-J., Hsu C.-J., Tseng P.-Y., Frank C. W., and Chang Y.-C., Biomacromolecules 11(5), 1231–1240 (2010). 10.1021/bm901445r [DOI] [PubMed] [Google Scholar]
- 24.Kaizuka Y. and Groves J. T., Biophys. J. 86(2), 905–912 (2004). 10.1016/S0006-3495(04)74166-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tseng P.-Y. and Chang Y.-C., Biomacromolecules 13, 2254–2262 (2012). 10.1021/bm300426u [DOI] [PubMed] [Google Scholar]
- 26.Wu J.-C., Tseng P.-Y., Tsai W.-S., Liao M.-Y., Lu S.-H., Frank C. W., Chen J.-S., Wu H.-C., and Chang Y.-C., Biomaterials 34(21), 5191–5199 (2013). 10.1016/j.biomaterials.2013.03.096 [DOI] [PubMed] [Google Scholar]
- 27.Glasmastar K., Larsson C., Hook F., and Kasemo B., J. Colloid Interface Sci. 246(1), 40–47 (2002). 10.1006/jcis.2001.8060 [DOI] [PubMed] [Google Scholar]
- 28.Lewis A. L., Colloids Surf., B. 18(3–4),261–275 (2000). 10.1016/S0927-7765(99)00152-6 [DOI] [PubMed] [Google Scholar]
- 29.Lu J. R., Murphy E. F., Su T. J., Lewis A. L., Stratford P. W., and Satija S. K., Langmuir 17(11), 3382–3389 (2001). 10.1021/la0017429 [DOI] [Google Scholar]
- 30.Jonsson M. P., Jönsson P., Dahlin A. B., and Höök F., Nano Lett. 7(11), 3462–3468 (2007). 10.1021/nl072006t [DOI] [PubMed] [Google Scholar]
- 31.Triffo S. B., Huang H. H., Smith A. W., Chou E. T., and Groves J. T., J. Am. Chem. Soc. 134(26), 10833–10842 (2012). 10.1021/ja300374c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328(5986), 1662–1668 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Suarez C., Busscher H. J., and van der Mei H. C., Appl. Environ. Microbiol. 67(6), 2531–2537 (2001). 10.1128/AEM.67.6.2531-2537.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]