Abstract
Genotoxic events have been known as crucial step in the initiation of cancer. To assess the risk of cancer, genotoxicity assays, including comet, micronucleus (MN), chromosomal aberration, bacterial reverse, and sister chromatid exchange assay, can be performed. Compared with in vitro genotoxicity assay, in vivo genotoxicity assay has been used to verify in vitro assay result and definitely provide biological significance for certain organs or cell types. The comet assay can detect DNA strand breaks as markers of genotoxicity. Methods of the in vivo comet assay have been established by Japanese Center for the Validation of Alternative Methods (JaCVAM) validation studies depending on tissue and sample types. The MN can be initiated by segregation error and lagging acentric chromosome fragment. Methods of the in vivo MN assay have been established by Organization for Economic Co-operation and Development (OECD) test guidelines and many studies. Combining the in vivo comet and MN assay has been regarded as useful methodology for evaluating genetic damage, and it has been used in the assessment of potential carcinogenicity by complementarily presenting two distinct endpoints of the in vivo genotoxicity individual test. Few studies have investigated the quantitative relation between in vivo genotoxicity results and carcinogenicity. Extensive studies emphasizes that positive correlation is detectable. This review summarizes the results of the in vivo comet and MN assays that have investigated the genotoxicity of carcinogens as classified by the International Agency for Research on Cancer (IARC) carcinogenicity database. As a result, these genotoxicity data may provide meaningful information for the assessment of potential carcinogenicity and for implementation in the prevention of cancer.
Keywords: In vivo genotoxicity, Carcinogenicity, Comet assay, Micronucleus assay
INTRODUCTION
Cancer is the leading cause of human mortality all over the world.1 Most cancer tissues show a number of complex chromosomal aberrations.2,3 The induction and accumulation of genetic damage can cause genomic instability, and it is known as a crucial step in the generation of cancer.4 Oncogenicity studies of carcinogenic potential using genotoxicity assays are on the rise. Altered gene expression, abnormal cell growth, and disruption of normal cell function may be related with the genotoxic effects of industrial carcinogens or other potential genotoxic agents. These phenomenon can result in the genomic instability and possibly carcinogenesis.1 For evaluating risk of cancer, genetic damage can be determined by genotoxicity assays, including comet assay, micronucleus assay, chromosome aberration assay, gamma-H2AX, and bacterial reverse testing. In this review, the micronucleus assay and the comet assay are focused.5 Since comet assay takes advantages of speediness, high sensitivity and flexibility for measuring capacity of DNA-strand breakage at the level of individual cells, and micronucleus assay exhibits highly reliable, rapid, and broad-spectrum determination of DNA damage at chromosome level (e.g. screening of chromosomal instability, DNA repair capacity, nuclear division rate, mitogenic response and incidence of necrotic and apoptotic cells).6
The genotoxicity tests officially approved as the Organization for Economic Co-operation and Development (OECD) test guidelines include the bacterial reverse mutation test, chromosome aberration test, micronucleus test, and sister chromatid exchange assay. In vivo genotoxicity tests using tissues can be used when obtaining in vitro positive results, that can reflect absorption, excretion, distribution, and metabolism of chemicals but the in vitro test does not.7,8 The in vitro assay has been considered as a genotoxicity test for screening substances (e.g. drug candidates, medicinal plant extract, chemical substances, etc.) and evaluating their initial safety while the in vivo assay provides detailed information of biological and physiological significance. It can determine whether any potential mutagenic effects that have shown in the in vitro step have appeared again in the animal’s whole physiological system.5 Consequently, the in vivo assays have been known as important processes in the verification of in vitro test and risk assessments for humans, indicating that they have more impact than in vitro assays.
The comet assay firstly established by Östling and Johanson has been widely applied for studying DNA strand breaks at the single cell level.9 The in vivo comet assay used to detect the genotoxic potential of chemicals has been recognized as a second in vivo genotoxicity assay by the International Conference on Harmonization (ICH- S2 (R1)) guidance (2012) with in vivo micronucleus (MN) assay. In the field of genotoxicology, the in vivo comet assay has been considered as powerful tools to distinguish between genotoxic carcinogens and nongenotoxic carcinogens as well as to identify carcinogens and mutagens.10 Consequently, the comet assay can be a biomarker for detecting both genetic susceptibility and the DNA damage related to carcinogenesis.11,12 Most carcinomas normally show a greater degree of DNA damage with extensive comet tails than that found in the tissue cells from controls.13 In several researches, CD-1 mouse strain has been commonly used as standard animal model in the in vivo comet assay.
Beside comet assay, the MN assay has been developed for genotoxicity and mutagenicity detection testing of chemicals that induce the formation of small membrane bound DNA fragments in cells (well-known as micronucleus).14–16 In principle, the MN assay is capable of detecting potential genotoxic chemicals that can modify chromosome structure and induce segregation error.17 In number of researches, standard laboratory strains of animals used in the MN assay are F344 rat, SD rat, and CD-1 mouse. Recent studies using the MN assay have shown that increased MN frequency is related with cancer risk, thus supporting the evidence that MN can be a biomarker of carcinogenesis.18,19 As so far application of genotoxicity testing suggests that no single assay can fully detect all genotoxic aspects,20 the ICH guidance on genotoxicity testing have thus proposed combining the in vivo comet assay with the in vivo MN assay.21
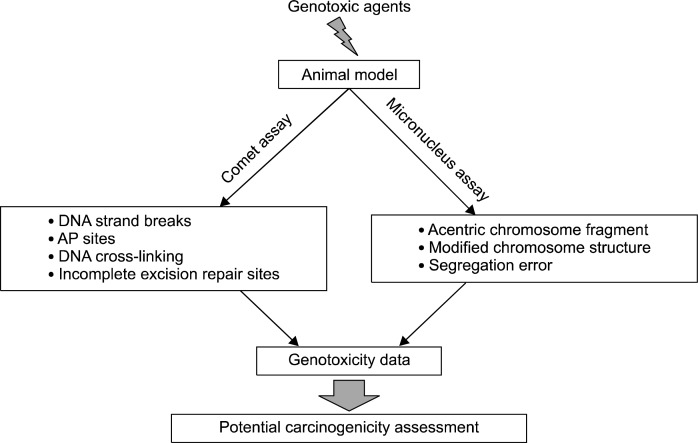
Measurement of DNA damage using genotoxicity assays has been known as a crucial approach for understanding the carcinogenesis and assessing the risk of cancer incidence.22,23 This review will discuss the significance of in vivo comet assay and in vivo MN assay for testing genotoxicity and predicting carcinogenic potential (Fig. 1). By comparing potential carcinogenicity studies using genotoxicity assays with other carcinogenicity databases, prediction of carcinogenic potential will be discussed in the following section and the development of both in vivo assays of comet assay and micronucleus assay will be further warranted.
Fig. 1.
Conceptual scheme representing in vivo micronucleus and comet assay for carcinogenicity study.
PRINCIPLE OF IN VIVO COMET ASSAY
In vivo comet assay has been generally performed to detect DNA strand breaks as the comet tail-like-shapes formed by DNA fragment in cells. In principle, after the lysis and electrophoresis steps of cells, the negatively charged DNA fragments migrate out of the cell toward the anode, and appear in a comet shape visualized under a fluorescence microscope.24 In a testing for genotoxic carcinogen agents, increased migration of the negatively charged DNA fragments toward the anode indicate increased the numbers of DNA strand breaks. Alkaline version of comet assay was introduced for the detection of double strand breaks, single strand breaks, alkali labile sites, DNA cross-linking, and incomplete excision repair sites.10,24 The alkaline-based comet assay has shown increased sensitivity for detecting genotoxic agents because most genotoxic agents may induce more single strand breaks or alkali labile sites than double strand breaks.25 Overall, the advantages of the comet assay in comparison to other genotoxicity assays include: (1) applicability to various tissues and cell types (flexibility), (2) no number of cells requirement, (3) sensitivity to detecting DNA damage, (4) brevity of performance time, and (5) the relatively low cost of the method.10,20,26–29 Prior to the establishment of an OECD test guideline, the in vivo comet assay using tissues was established by JaCVAM validation studies.
METHODOLOGY OF IN VIVO COMET ASSAY
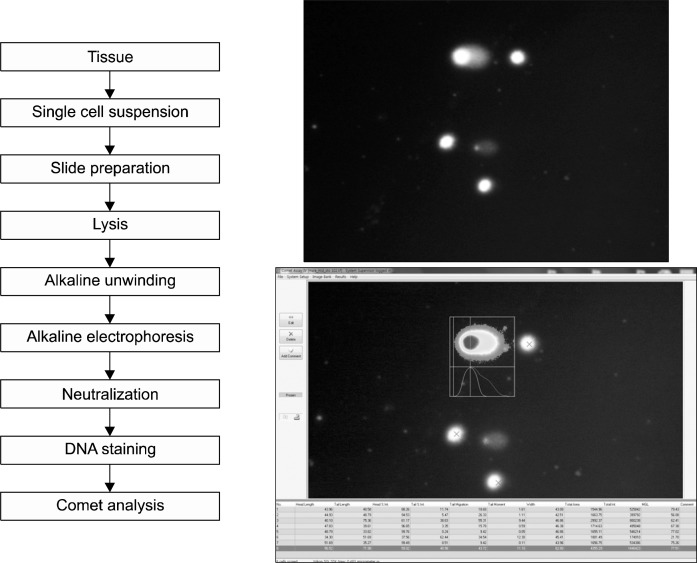
Any type of animal tissue (e.g. liver, stomach, and blood cells) can be applied to the in vivo comet assay, as long as the tissue types can provide high-quality single cell suspension.30 This methodology of comet assay refers to JaCVAM validation studies (Fig. 2).31
Fig. 2.
Scheme illustrating alkaline version of in vivo comet assay.
1. In vivo comet assay using liver tissue
Usually a portion of the left lateral lobe of the liver tissue is removed from a whole liver and then washed sufficiently with an ice-cold appropriate mincing buffer. The washed portion is minced to obtain the single cell suspension. The cell suspension is placed on ice to allow the cluster to settle down. Then, the supernatant can be used to make a comet slide. After preparing the comet slide, the slide is incubated with cold alkaline lysis solution overnight. After the lysis process, the slide is rinsed with deionized water to remove residual detergent and salts. After DNA unwinding using an electrophoresis solution, the slide is electrophoresed. After electrophoresis, the slide is neutralized. Then, the slide is dehydrated by absolute ethanol, air dried at room temperature. The slide is stained with SYBR Gold, and the comet can be visualized under a fluorescence microscope and quantitated via image analyzer system such as Comet IV software (Perspectives, UK). For each sample, one hundred comet cells are subjected for quantitative analysis. Excessively damaged comets showing an appearance of “hedgehog” consisting of very small comet heads and largely-diffused comet tails represent dead cells and should not be analyzed. The parameters used in the comet analysis are as follows: % tail DNA, tail length (a distance between the center of the head mass and the center of the tail mass), and olive tail moment [= tail length × DNA in tail (% tail DNA/100)].31
2. In vivo comet assay using stomach tissue
The whole stomach tissue of a sacrificed animal is initially cut opened and washed until free of food residue using an ice-cold mincing buffer. A portion of the forestomach is removed and then discarded, and the glandular stomach is incubated with the ice-cold mincing buffer. After incubation, the surface epithelial layer is gently scraped. The removed layer is discarded, and the mucosa is rinsed with the ice-cold mincing buffer. Then, the epithelia of the glandular stomach are scraped to obtain cell suspension. The cell suspension is placed on ice to allow cluster to settle down. Then, the cell suspension of the supernatant can be used to make the comet slides. The cell suspension and low-melting-agarose gel are mixed, and then the cell/agar mixture is dispensed onto the comet slide.31 As before, the comet slides follow the process of lysis, DNA unwinding, electrophoresis, neutralization, dehydration, DNA staining, and image analysis.
3. In vivo comet assay using blood sample
Blood sample can be collected by venipuncture. All blood samples must be immediately cooled and processed within 2 hours after collection. The layer made by the blood sample mixed with low-melting-point agarose is placed on the comet slide. As mentioned above, the comet assay is composed of the process of lysis, DNA unwinding, electrophoresis, neutralization, dehydration, staining of comet slide, and image analysis.32
PRINCIPLE OF IN VIVO MICRONUCLEUS ASSAY
The micronucleus (MN) can be generally formed by lagging acentric chromosome fragments, acentric chromatid fragments, or whole chromosome that could not included in the daughter nucleus during mitosis telophase because the whole chromosome did not combine with the spindle during the segregation process of anaphase.15,17,33,34 These chromosome fragments surrounded by nuclear membrane are well known as MN, which is morphologically similar to normal nucleus but smaller in size.35 Compared to other genotoxicity assays, the MN assay is a quick and easy assay at the data analysis step. Moreover, it has no requirement for metaphase cells, and shows identifiable cells under a fluorescence microscope because each cell has only one nuclear division.1,36 The in vivo MN assay was established by the OECD guidelines, and has been continuously being developed through many studies.
METHODOLOGY OF IN VIVO MN ASSAY
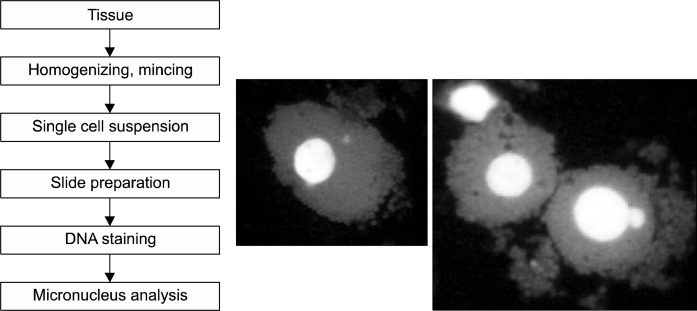
The in vivo MN assay is known as simple and sensitive screening method of clastogenic agents in the target tissues (e.g. blood and lung sample). In addition, many studies of in vivo MN assays using tissues have been so far published (Fig. 3).37
Fig. 3.
Scheme showing main procedure of in vivo micronucleus assay.
1. In vivo MN assay using blood sample
The erythrocyte of a blood sample can be collected by bleeding from a blood vessel (e.g. mouse tail vein), cardiac puncture, or large vessel at animal sacrifice. The blood smears are prepared on glass slides and then subsequently stained for microscopy analysis. Using flow cytometry-based analysis, the sample slides should be fixed and stained. DNA-specific staining using acridine orange can exclude the possibility of any artifacts generated by non-DNA-specific staining.38
2. In vivo MN assay using liver tissue
After liver tissues are removed, it is recommended to immediately perfuse the liver tissues with cold saline solution until the blood is completely removed. The final washing uses a cold homogenizing buffer containing EDTA, NaCl, and DMSO. After weighing the liver sample, the tissues are minced, suspended in cold homogenization buffer, and homogenized maintaining in the cold buffer on ice using a potter-type homogenizator. After centrifugation of the homogenate, the supernatant is removed, and the pellet is resuspended in the homogenization buffer.39 The resuspended pellet needs to be settled down. Then, a drop of the suspension is placed at the end of a pre-cleaned, grease-free, microscopic slide. Subsequently, the drop is spread into a single cell layer without damaging the cell morphology, using a clean cover glass held at 45 degrees.39,40 Next, the prepared slides are air dried and then stained by May-Grunwald stain, followed by a Giemsa solution stain. The stained slides are rinsed with deionized water, air dried, and rinsed with methanol. Then the slides are placed in xylene for clearing. Finally, they are mounted and then analyzed (1000 cells are scored for each sample).39–41
3. In vivo MN assay using lung tissue
After sacrificing animals, cells can be isolated from lung tissue. The inferior vena cava is severed, and then the lung tissue is perfused through the right ventricle with ethy-leneglycoltetraacetic acid (EGTA) solution in Hanks' balanced salt solution (HBSS). After perfusing until it is blanched, the lung tissue is then inflated through the trachea with solution containing trypsin, EDTA, and collagenase. The lung tissue is removed, minced, and incubated with rocking in EDTA containing enzyme solution. The supernatant containing individual cells is collected, and then DNase I is added. After centrifugation of the cells, the pellet is washed with complete medium. Aliquouts of 1×106 cells in 2 ml complete medium are seeded onto square cover glass in a tissue culture dish. To prevent the cells from cytokinesis, an inhibitor of the mitotic spindle namely cytochalasin B (Cyt-B) is added to each culture after 24 hours of the culture, allowing distinguish cells that have completed one nuclear division and consequently become binucleated. After 48 hours of adding Cyt-B, the seeded cells are then fixed with 3:1 methanol:acetic acid or 100% ethanol for staining. The cells are stained with Giemsa solution for calculating the frequency of binucleate cells having MN.42,43
PREDICTION OF CARCINOGENIC POTENTIAL BASED ON COMBINED IN VIVO TEST OF COMET AND MICRONUCLEUS ASSAY
In spite of regulatory directives concerning the reduction of animal use in safety test, modifications to genotoxicity testing guidelines recently offer the utility of two in vivo genotoxicity assays of comet and MN as a follow-up to an in vitro positive.6 Both in vivo assays can be achieved into one informative investigation. Combining these two assays with individual difference in sensitivity, measurable endpoints, and types of parameter analyzed significantly potentiate the current standard capabilities and performance for assessing genotoxicity as well as for evaluating carcinogenesis incidence and safety risk without requirement of additional animals.
Genotoxic events have been regarded as a crucial stage in the initiation of carcinogenesis.44 Genomic instability may enable a cell to accumulate stable genome mutations, and it represents an early step in the carcinogenesis.4 When cells having modified DNA and abnormal genome continually survive, the abnormal cell can be a latent cancer cell or it can give rise to a cancer.45 Most cancers have shown many chromosomal aberrations, and these alterations can be detected in both benign and malignant tumors.2,4 Relying on the standard genotoxicity and/or carcinogenicity tests including in vivo comet and MN assay, the International Agency for Research on Cancer (IARC) has published lists of agents that can cause cancer in humans and has provided reliable data on human carcinogenicity.44 Table 1 presented the results of the in vivo comet and MN assay that have investigated the genotoxicity of potential carcinogens classified by the IARC in carcinogenicity database. Consequently, the genotoxicity results may enable researchers to predict the carcinogenic potential of chemicals (Table 1).
Table 1.
Potential carcinogenicity studies using in vivo comet, MN assays
| Chemical | Species | Tissue | Route | Dose | Sampling time | Result of the Assays | Carcinogenicity Data | Reference |
|---|---|---|---|---|---|---|---|---|
| 1,2-DMH 2HCl | F344 rat | Liver | po | 100, 200 mg/kg | 3, 4, 5 days | Positive at 3, 4, 5 days treatment (MN assay) | IARC: 2A Carcinogenicity: + | [54] |
| 1,4-Dioxane | CD-1 mouse | Liver | po | 1,500, 2,500, 3,500 mg/kg/day for 5 days | 24 h | Positive in 2,500, 3,500 mg/kg (MN assay) | IARC: 2B Carcinogenicity: + | [55] |
| 1,4-Dioxane | CD-1 mouse | Liver | po | 1,000, 2,000, 3,000 mg/kg | 6 days | Positive in 2000mg/kg (MN assay) | IARC: 2B Carcinogenicity: + | [56] |
| 2-Acetylaminofluorene | SD rat | Bone marrow, peripheral blood | po | 125, 500 mg/kg ×2 days | 24 h | Positive in bone marrow, blood at two doses (MN assay) | IARC: nd Carcinogenicity: + | [57] |
| 2-Acetylaminofluorene | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 400 mg/kg | 3, 24 h | Positive in liver and kidney 3 h after treatment (comet assay) | IARC: nd Carcinogenicity: + | [57] |
| 2,4-Diaminotoluene | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 240 mg/kg | 3, 24 h | Positive in liver and kidney 3, 24 h after treatment and in lung 3 h after treatment (comet assay) | IARC: 2B Carcinogenicity: + | [58] |
| 2,4-dinitrotoluene | F344 rat | Liver | po | 200, 400 mg/kg | 3, 4, 5 days | Positive (MN assay) | IARC: 2B Carcinogenicity: + | [54] |
| 2,4-dinitrotoluene | F344 rat | Liver | po | 75, 150, 300 mg/kg (two-dose assay) | 4 days | Positive in 75, 150, 300 mg/kg (MN assay) | IARC: 2B Carcinogenicity: + | [57] |
| 2,6-dinitrotoluene | F344 rat | Liver | po | 50, 100, 200 mg/kg (two-dose assay) | 4 days | Positive in 50, 100, 200 mg/kg (MN assay) | IARC: 2B Carcinogenicity: + | [57] |
| Acrylonitrile | SD rat | Bone marrow, peripheral blood | iv | 124.8, 125 mg/kg × 2 days | 24 h | Positive in bone marrow but not in blood (MN assay) | IARC: 2B Carcinogenicity: + | [57] |
| Arsenic acid solution | CD-1 mouse | Bone marrow | ip | 1, 5, 10, 20 mg/kg/d for 4 days | 24 h | Positive in 10, 20 mg/kg/day (MN assay) | IARC: 1 Carcinogenicity: + | [59] |
| Atrazine | Wistar rat | Liver, blood | po | 300 mg/kg/d for 7, 14, 21 days | Not described | Positive at periods of 7, 14, and 21 days (comet assay, MN assay) | IARC: 3 Carcinogenicity: + | [39] |
| Auramine | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 80 mg/kg | 3, 24 h | Positive in liver, kidney, and lung 3 h after treatment (comet assay) | IARC: 2B Carcinogenicity: + | [58] |
| Benzene | NMRI mice | Blood lymphocytes, bone marrow | po | 40, 200, 450 mg/kg | 6 h | Positive in lymphocytes at 200, 450 mg/kg and in bone marrow at 40, 200, 450 mg/kg (comet assay) | IARC: 1 Carcinogenicity: + | [60] |
| Benzo[α]pyrene | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | po | 250 mg/kg | 3, 24 h | Positive in liver and lung 3 h after treatment (comet assay) | IARC: 2A Carcinogenicity: + | [56] |
| Cadmium chloride | White swiss mouse | Bone marrow | ip | 1.9, 5.7, 7.6 mg/kg | 24 h | Positive in 1.9, 5.7, 7.6 mg/kg (MN assay) | IARC: 1 Carcinogenicity: + | [54] |
| Chloroform | SD rat | Kidney | po | 4 mmol/kg | 2 days | All agents are positive in kidney (MN assay) | IARC: 2B Carcinogenicity: + | [60] |
| Dimethylnitrosamine | F344 rat | Liver | po | 2.5, 5, 10 mg/kg (two-dose assay) | 3, 4, 5 days | Positive in 5, 10 mg/kg (MN assay) | IARC: 2A Carcinogenicity: + | [54] |
| Ethylene thiourea | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 2,000 mg/kg | 3, 24 h | Positive in liver, kidney, lung, and spleen 3, 24 h after treatment (comet assay) | IARC: 3 Carcinogenicity: + | [58] |
| Lambda cyhalothrin | Wistar rat | Bone marrow | po | 0.8, 3.06, 6.12 mg/kg One dose per 48 h for 13 days |
30 h | Positive in all doses (MN assay) | IARC: nd Carcinogenicity: + | [61] |
| Mitomycin C | F344 rat | Liver | ip | 0.5, 1, 2 mg/kg (two-dose assay) | 4 days | Positive in 0.5, 1, 2 mg/kg (MN assay) | IARC: 2B Carcinogenicity: + | [57] |
| N-Methyl-N-nitrosourea | BALB/c mouse | Stomach | po | 100 mg/kg | 3, 4 days | Positive in stomach 3, 4 days after injection (MN assay) | IARC: 2A Carcinogenicity: + | [56] |
| N-Methyl-N-nitrosourea | BALB/c mouse | Peripheral blood | po | 100 mg/kg | 2, 3 days | Positive in peripheral blood 2, 3 days after injection (MN assay) | IARC: 2A Carcinogenicity: + | [56] |
| p-Aminoazobenzene | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 200 mg/kg | 3, 24 h | Positive in liver, kidney, and spleen 3 h after treatment. Positive in all tissues 24 h after treatment (comet assay) | IARC: 2B Carcinogenicity: + | [58] |
| p-Dichlorobenzene | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 2,000 mg/kg | 3, 24 h | Positive in liver and spleen 3 h after treatment (comet assay) | IARC: 2B Carcinogenicity: + | [58] |
| Phenobarbital | SD rat | Liver | po | 30, 90, 120 mg/kg/day for 3 days. 10, 30, 90 mg/kg/day for 29 days |
3 h | Positive at 120 mg/kg/d for 3 days, but not at all doses for 29 days (comet assay) | IARC: 2B Carcinogenicity: + | [62] |
| Potassium chromate(VI) | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 80 mg/kg | 3, 24 h | Positive in liver and lung 3 h after treatment (comet assay) | IARC: 1 Carcinogenicity: + | [4] |
| Quinoline | F344 rat | Liver | po | 30, 60, 90 mg/kg (two-dose assay) | 3, 4, 5 days | Positive in 60, 90 mg/kg (MN assay) | IARC: nd Carcinogenicity: + | [58] |
| Styrene-7,8-oxide | CD-1 mice | Liver, kidney, lung, spleen, bone marrow | ip | 400 mg/kg | 3, 24 h | Positive in all tissues 3 h after treatment (comet assay) | IARC: 2A Carcinogenicity: + | [63] |
| Thioacetamide | C57BL/6 Mouse | Bone marrow | po | 375–1,500 mg/kg | 1, 2 days | Positive (MN assay) | IARC: 2B Carcinogenicity: + | [64] |
| Trichloroethylene | SD rat | Kidney | po | 4 mmol/kg | 2 days | All agents are positive in kidney (MN assay) | IARC: 1 Carcinogenicity: + | [64] |
| Thiabendazole | ddy mouse | Stomach, liver, kidney, bladder, brain, bone marrow, lung | po | 10, 100, 200 mg/kg | 3, 24 h | Positive in all organs at 200 mg/kg, 3 h treatment only positive in stomach at 200 mg/kg, 24 h treatments (comet assay) |
IARC: nd Carcinogenicity: + | [8] |
i Ip, Intraperitoneally; Po, Oral administration; Iv, Intravenous; Nd, not determined; IARC 1, Carcinogenic to humans; IARC 2A, Probably carcinogenic to humans; IARC 2B, Possibly carcinogenic to humans; IARC 3, Not classifiable as to its carcinogenicity to humans; IARC 4, Probably not carcinogenic to humans.
For a more comprehensive investigation of genotoxicity in animal models, the National Toxicology Program (NTP) has performed a combined assay using in vivo comet and MN assay.46 As the comet assay has been known to detect most carcinogens that have been identified as equivocal or negative in the MN assay, the results of both assays are considerably regarded as complementary issue. Therefore, the combined in vivo genotoxicity assay has been recommended to investigate genotoxic potential in several recent studies.46–48 In addition, the combined in vivo genotoxicity assay has been regarded as a useful methodology for evaluation of genetic hazard in safety risk assessment because it offers two distinct genotoxicity endpoints. Based on the advantages of the combined in vivo genotoxicity assay, the NTP has introduced this combined assay as a part of detecting the genotoxicity of substances with public health concern.46 In both in vivo comet and MN assay, mouse strains of BALB/c, C57BL/6, CD-1, ddy, NMRI and White Swiss, and rat strains of F344, SD, and Wistar have been extensively used. In particular, CD-1 mouse strain has been mainly used in the comet assay while strains of F344 rat and SD rat have been frequently used in the MN assay.
The genotoxicity assays have been used for studying the carcinogenesis of chemicals and assessing the potential carcinogenicity of chemicals to humans.44 The findings based on comet assay and MN assay have shown correlations between genotoxicity and preneoplastic/neoplastic changes. The in vivo comet assay studies have been performed to assess genotoxicity in terms of cancer development.7 In addition, correlations between MN induction and cancer development have been reported in several studies.49 Indeed, MN has been known as a manifestation of chromosomal instability that occurs in cancer.35 Numerous studies focused on the application of MN have shown a significant increase of the MN frequency in peripheral blood lymphocytes of cancer patients in comparison with healthy control patients.50 These studies have suggested that increased results of the MN frequency are related with an early step in carcinogenesis.51
Few studies have been performed to investigate the quantitative dose-response relationship between carcinogenesis and in vivo genotoxicity results. Among them, one study was conducted to evaluate their quantitative relation. The quantitation utilized the lowest effective dose (LED) derived from the in vivo genotoxicity tests (MN, comet, sister chromatid exchange, and chromosome aberration test) and the T25 as the chronic daily dose (mg/kg/day that induces tumor in 25% at specific tissue) from rodent carcinogenicity studies. The relation between the LED and T25 was identified as a linear correlation.52,53 In addition, another study utilized the benchmark dose (BMD) to evaluate the quantitative relation among them. The BMD-based approach has been used for correlating equipotent doses of carcinogenicity studies with equipotent doses of in vivo genotoxicity studies. Likewise, positive relation between carcinogenicity and in vivo genotoxicity results were identified using the BMD values. The relation between the lowest BMD10 of MN and the tissue-matched carcinogenicity BMD10 was evaluated, and thus quantitative correlation was observed.52
As mentioned above, the comet and MN assay-based-analysis would enable the prediction of the potential carcinogenicity of diverse substances including chemical and physical agents via carcinogenicity databases. Due to flexibility feature, the in vivo comet assay can be incorporated into most standard testing batteries to provide supplemental data of target tissue without requirement of additional duration or resources in an independent experimentation setting. To further achieve appropriateness of study design and accuracy of comet data interpretation, the recognition of significant discrepancy between the comet assay and the MN assay are necessary for the combination of either the comet assay or the MN assay with standard genotoxicity tests and/or toxicological studies. Herewith, the combined comet and MN assay protocol has proven to be a sensitive and efficient for evaluating within the same animals toward multiple classes of genotoxic agents across a wide range of target tissues. This approach takes advantage of the minimal use of animal number. Taken together, these recommendations provide an effective methodology for combining the in vivo comet and MN assays and for interpreting assay data. This approach would improve safety testing of critical target tissue data with increased sensitivity while minimizing animal use, reducing exposure times and toxicity. Further investigations should be optimized for these two assays to take full advantage of the increased sensitivity, capability, and flexibility.
CONCLUSION
According to several studies and national/international institution guidelines, integrating the in vivo comet and in vivo MN assays has been successfully performed for follow-up testing of positive in vitro results. They may be served as potential tool for the assessment of local gentoxicity, especially for tissues or cell types which cannot be easily measured with other standard testing methods. Several results of these two in vivo genotoxicity assays may indicate that the genotoxicity has positive correlation with the carcinogenicity. Such approach-based findings will provide practical consequences in the risk assessment processes and further development of substances. In many studies, the two in vivo genotoxicity assays have been extensively used to investigate the chemicals classified by IARC, and the genotoxicity assay results indicate that the carcinogenicity of potent substances can be predicted by the in vivo comet and in vivo MN assays. Therefore, the combined in vivo genotoxicity assay may be used to detect the carcinogenic potential of substances for the prevention of cancer. Such approach-based findings will provide practical consequences in the risk assessment processes and further development of substances.
Acknowledgments
This subject is supported by Korea Ministry of Environment as “The Ecoinnovation Project” (412-112-011).
REFERENCES
- 1.Pratheepa Sivasankari N, Kaur S, Reddy KS, Vivekanandam S. Micronucleus Index: an early diagnosis in oral carcinoma. J Anat Soc India. 2008;57:8–13. [Google Scholar]
- 2.Keen-Kim D, Nooraie F, Rao PN. Cytogenetic biomarkers for human cancer. Front Biosci. 2008;13:5928. doi: 10.2741/3127. [DOI] [PubMed] [Google Scholar]
- 3.Miyamae Y, Yamamoto M, Sasaki YF, Kobayashi H, Igarashi-Soga M, Shimoi K, et al. Evaluation of a tissue homogenization technique that isolates nuclei for the in vivo single cell gel electrophoresis (comet) assay: a collaborative study by five laboratories. Mutat Res. 1998;418:131–40. doi: 10.1016/s1383-5718(98)00112-0. [DOI] [PubMed] [Google Scholar]
- 4.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brendler-Schwaab S, Hartmann A, Pfuhler S, Speit G. The in vivo comet assay: use and status in genotoxicity testing. Mutagenesis. 2005;20:245–54. doi: 10.1093/mutage/gei033. [DOI] [PubMed] [Google Scholar]
- 6.Hovhannisyan GG. Fluorescence in situ hybridization in combination with the comet assay and micronucleus test in genetic toxicology. Mol Cytogenet. 2010;3:17. doi: 10.1186/1755-8166-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benigni R, Bossa C, Tcheremenskaia O, Battistelli CL, Crettaz P. The new ISSMIC database on in vivo micronucleus and its role in assessing genotoxicity testing strategies. Mutagenesis. 2012;27:87–92. doi: 10.1093/mutage/ger064. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki YF, Kawaguchi S, Kamaya A, Ohshita M, Kabasawa K, Iwama K, et al. The comet assay with 8 mouse organs: results with 39 currently used food additives. Mutat Res. 2002;519:103–19. doi: 10.1016/s1383-5718(02)00128-6. [DOI] [PubMed] [Google Scholar]
- 9.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123:291–8. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 10.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–91. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 11.Tozan-Beceren A, Omurtag GZ, Yeğen C, Şardaş S. Investigation of DNA damage in patients with colorectal cancer and their first degree relatives. Journal of Marmara University Institute of Health Sciences. 2011;1:155–61. [Google Scholar]
- 12.Zhang H, Buchholz TA, Hancock D, Spitz MR, Wu X. Gamma-radiation-induced single cell DNA damage as a measure of susceptibility to lung cancer: a preliminary report. Int J Oncol. 2000;17:399–803. doi: 10.3892/ijo.17.2.399. [DOI] [PubMed] [Google Scholar]
- 13.Vasavi M, Vedicherala B, Vattam KK, Ahuja YR, Hasan Q. Assessment of genetic damage in inflammatory, precancerous, and cancerous pathologies of the esophagus using the comet assay. Genet Test Mol Biomarkers. 2010;14:477–82. doi: 10.1089/gtmb.2010.0006. [DOI] [PubMed] [Google Scholar]
- 14.Fenech M, Holland N, Chang WP, Zeiger E, Bonassi S. The HUman MicroNucleus Project-An international collaborative study on the use of the micronucleus technique for measuring DNA damage in humans. Mutat Res. 1999;428:271–83. doi: 10.1016/s1383-5742(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 15.Fenech M. Cytokinesis-block micronucleus cytome assay. Nature protocols. 2007;2:1084–104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 16.Fenech M. The in vitro micronucleus technique. Mutat Res. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 17.Savage JR. A comment on the quantitative relationship between micronuclei and chromosomal aberrations. Mutat Res. 1988;207:33–6. doi: 10.1016/0165-7992(88)90008-5. [DOI] [PubMed] [Google Scholar]
- 18.Bonassi S, Znaor A, Ceppi M, Lando C, Chang WP, Holland N, et al. An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogenesis. 2006;28:625–31. doi: 10.1093/carcin/bgl177. [DOI] [PubMed] [Google Scholar]
- 19.Murgia E, Ballardin M, Bonassi S, Rossi AM, Barale R. Validation of micronuclei frequency in peripheral blood lymphocytes as early cancer risk biomarker in a nested case-control study. Mutat Res. 2008;639:27–34. doi: 10.1016/j.mrfmmm.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Lee RF, Steinert S. Use of the single cell gel electrophoresis/comet assay for detecting DNA damage in aquatic (marine and freshwater) animals. Mutat Res. 2003;544:43–64. doi: 10.1016/s1383-5742(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 21.Vasquez MZ. Combining the in vivo comet and micronucleus assays: a practical approach to genotoxicity testing and data interpretation. Mutagenesis. 2010;25:187–99. doi: 10.1093/mutage/gep060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santella RM. DNA damage as an intermediate biomarker in intervention studies. Exp Biol Med. 1997;216:166–71. doi: 10.3181/00379727-216-44166. [DOI] [PubMed] [Google Scholar]
- 23.Mohan V, Ponnala S, Hasan Q, Reddy HM, Jesudasan RA, Sistla R, et al. Chromosome 11 aneusomy in esophageal cancers and precancerous lesions-an early event in neoplastic transformation: Aninterphase fluorescence in situ hybridization study from south India. World J Gastroenterol. 2007;13:503–8. doi: 10.3748/wjg.v13.i4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairbairn DW, Olive PL, O'Neill KL. The comet assay: a comprehensive review. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- 25.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–21. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 26.Collins A, Dušinská M, Franklin M, Somorovská M, Petrovská H, Duthie S, et al. Comet assay in human biomonitoring studies: reliability, validation, and applications. Environ Mol Mutagen. 1997;30:139–46. doi: 10.1002/(sici)1098-2280(1997)30:2<139::aid-em6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 27.Dixon DR, Pruski AM, Dixon LR, Jha AN. Marine invertebrate ecogenotoxicology: a methodological overview. Mutagenesis. 2002;17:495–507. doi: 10.1093/mutage/17.6.495. [DOI] [PubMed] [Google Scholar]
- 28.Jha AN. Genotoxicological studies in aquatic organisms: an overview. Mutat Res. 2004;552:1–17. doi: 10.1016/j.mrfmmm.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann A, Schumacher M, Plappert-Helbig U, Lowe P, Suter W, Mueller L. Use of the alkaline in vivo Comet assay for mechanistic genotoxicity investigations. Mutagenesis. 2004;19:51–9. doi: 10.1093/mutage/geg038. [DOI] [PubMed] [Google Scholar]
- 30.Hartmann A, Agurell E, Beevers C, Brendler-Schwaab S, Burlinson B, Clay P, et al. Recommendations for conducting the in vivo alkaline Comet assay. Mutagenesis. 2003;18:45–51. doi: 10.1093/mutage/18.1.45. [DOI] [PubMed] [Google Scholar]
- 31.Report of the JaCVAM initiative international pre-validation studies of the in vivo rodent alkaline Comet assay for the detection of genotoxic carcinogens. 2013 The comet assay International Validation Management Team. [Google Scholar]
- 32.Garaj-Vrhovac V, Kopjar N. The alkaline Comet assay as biomarker in assessment of DNA damage in medical personnel occupationally exposed to ionizing radiation. Mutagenesis. 2003;18:265–71. doi: 10.1093/mutage/18.3.265. [DOI] [PubMed] [Google Scholar]
- 33.Fenech M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys. 2010;98:234–43. doi: 10.1097/HP.0b013e3181b85044. [DOI] [PubMed] [Google Scholar]
- 34.Mateuca R, Lombaert N, Kirsch-Volders M. Chromosomal changes: induction, detection methods and applicability in human biomonitoring. Biochimie. 2006;88:1515–31. doi: 10.1016/j.biochi.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–32. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 36.Hsu TC, Johnston DA, Cherry LM, Ramkissoon D, Schantz SP, Jessup JM, et al. Sensitivity to genotoxic effects of bleomycin in humans: possible relationship to environmental carcinogenesis. Int J Cancer. 1989;43:403–9. doi: 10.1002/ijc.2910430310. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi M, MacGregor JT, Gatehouse DG, Adler ID, Blakey DH, Dertinger SD, et al. In vivo rodent erythrocyte micronucleus assay. II. Some aspects of protocol design including repeated treatments, integration with toxicity testing, and automated scoring. Environ Mol Mutagen. 2000;35:234–52. [PubMed] [Google Scholar]
- 38.Test No. 474: Mammalian Erythrocyte Micronucleus Test. 2012 OECD. [Google Scholar]
- 39.Singh M, Kaur P, Sandhir R, Kiran R. Protective effects of vitamin E against atrazine-induced genotoxicity in rats. Mutat Res. 2008;654:145–9. doi: 10.1016/j.mrgentox.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Igarashi M, Shimada H. An improved method for the mouse liver micronucleus test. Mutat Res. 1997;391:49–55. doi: 10.1016/s0165-1218(97)00031-1. [DOI] [PubMed] [Google Scholar]
- 41.Schmid W. The micronucleus test. Mutat Res. 1975;31:9–15. doi: 10.1016/0165-1161(75)90058-8. [DOI] [PubMed] [Google Scholar]
- 42.Kligerman AD, Campbell JA, Erexson GL, Allen JW, Shelby MD. Sister chromatid exchange analysis in lung and peripheral blood lymphocytes of mice exposed to methyl iso-cyanate by inhalation. Environ Mutagen. 1987;9:29–36. doi: 10.1002/em.2860090105. [DOI] [PubMed] [Google Scholar]
- 43.Ranaldi R, Bassani B, Villani P, Lombardi CC, Tanzarella C, Pacchierotti F. Measurement and characterization of micronuclei in cultured primary lung cells of mice following inhalation exposure to benzene. Mutagenesis. 1998;13:453–60. doi: 10.1093/mutage/13.5.453. [DOI] [PubMed] [Google Scholar]
- 44.Douglas GR, Blakey DH, Clayson DB. Genotoxicity tests as predictors of carcinogens: an analysis. Mutat Res. 1988;196:83–93. doi: 10.1016/0165-1110(88)90029-2. [DOI] [PubMed] [Google Scholar]
- 45.Weisburger JH, Williams GM. Carcinogen testing: current problems and new approaches. SCIENCE. 1981;214:401–7. doi: 10.1126/science.7291981. [DOI] [PubMed] [Google Scholar]
- 46.Recio L, Hobbs C, Caspary W, Witt KL. Dose-response Assessment of four genotoxic chemicals in a combined mouse and rat micronucleus and Comet assay protocol. J Toxicol Sci. 2010;35:149. doi: 10.2131/jts.35.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirkland D, Speit G. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens: III. Appropriate follow-up testing in vivo. Mutat Res. 2008;654:114–32. doi: 10.1016/j.mrgentox.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Pfuhler S, Albertini S, Fautz R, Herbold B, Madle S, Utesch D, et al. Genetic toxicity assessment: employing the best science for human safety evaluation part IV: Recommendation of a working group of the Gesellschaft fuer Umwelt-Mutationsforschung (GUM) for a simple and straightforward approach to genotoxicity testing. Toxicological sciences. 2007;97:237–40. doi: 10.1093/toxsci/kfm019. [DOI] [PubMed] [Google Scholar]
- 49.Duffaud F, Orsiere T, Digue L, Villani P, Volot F, Favre R, et al. Micronucleated lymphocyte rates from head-and-neck cancer patients. Mutat Res. 1999;439:259–66. doi: 10.1016/s1383-5718(99)00003-0. [DOI] [PubMed] [Google Scholar]
- 50.Iarmarcovai G, Ceppi M, Botta A, Orsiere T, Bonassi S. Micronuclei frequency in peripheral blood lymphocytes of cancer patients: a meta-analysis. Mutat Res. 2008;659:274–83. doi: 10.1016/j.mrrev.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 51.Bonassi S, El-Zein R, Bolognesi C, Fenech M. Micronuclei frequency in peripheral blood lymphocytes and cancer risk: evidence from human studies. Mutagenesis. 2011;26:93–100. doi: 10.1093/mutage/geq075. [DOI] [PubMed] [Google Scholar]
- 52.Hernández LG, Slob W, van Steeg H, van Benthem J. Can carcinogenic potency be predicted from in vivo genotoxicity data? a meta-analysis of historical data. Environ Mol Mutagen. 2011;52:518–28. doi: 10.1002/em.20651. [DOI] [PubMed] [Google Scholar]
- 53.Sanner T, Dybing E. Comparison of carcinogenic and in vivo genotoxic potency estimates. Basic Clin Pharmacol Toxicol. 2005;96:131–9. doi: 10.1111/j.1742-7843.2005.pto960207.x. [DOI] [PubMed] [Google Scholar]
- 54.Fahmy MA, Aly FA. In vivo and in vitro studies on the genotoxicity of cadmium chloride in mice. J Appl Toxicol. 2000;20:231–8. doi: 10.1002/(sici)1099-1263(200005/06)20:3<231::aid-jat653>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 55.Çelik A, Mazmanci B, Çamlica Y, Aşkin A, Çömelekoğlub Ü. Induction of micronuclei by lambda-cyhalothrin in Wistar rat bone marrow and gut epithelial cells. Mutagenesis. 2005;20:125–9. doi: 10.1093/mutage/gei020. [DOI] [PubMed] [Google Scholar]
- 56.Morita T, Asano N, Awogi T, Sasaki YF, Sato SI, Shimada H, et al. Evaluation of the rodent micronucleus assay in the screening of IARC carcinogens (Groups 1, 2A and 2B): The summary report of the 6th collaborative study by CSGMT/JEMSĚMMS. Mutat Res. 1997;389:3–122. doi: 10.1016/s1383-5718(96)00070-8. [DOI] [PubMed] [Google Scholar]
- 57.Wakata A, Miyamae Y, Sato SI, Suzuki T, Morita T, Asano N, et al. Evaluation of the rat micronucleus test with bone marrow and peripheral blood: summary of the 9th collaborative study by CSGMT/JEMS • MMS. Environ Mol Mutagen. 1998;32:84–100. [PubMed] [Google Scholar]
- 58.Sasaki YF, Izumiyama F, Nishidate E, Matsusaka N, Tsuda S. Detection of rodent liver carcinogen genotoxicity by the alkaline single-cell gel electrophoresis (Comet) assay in multiple mouse organs (liver, lung, spleen, kidney, and bone marrow) 1. Mutat Res. 1997;391:201–14. doi: 10.1016/s1383-5718(97)00072-7. [DOI] [PubMed] [Google Scholar]
- 59.Leopardi P, Cordelli E, Villani P, Cremona TP, Conti L, De Luca G, et al. Assessment of in vivo genotoxicity of the rodent carcinogen furan: evaluation of DNA damage and induction of micronuclei in mouse splenocytes. Mutagenesis. 2010;25:57–62. doi: 10.1093/mutage/gep043. [DOI] [PubMed] [Google Scholar]
- 60.Tuo J, Loft S, Thomsen MS, Poulsen HE. Benzene-induced genotoxicity in mice in vivo detected by the alkaline comet assay: reduction by CYP2E1 inhibition. Mutat Res. 1996;368:213–9. doi: 10.1016/s0165-1218(96)90063-4. [DOI] [PubMed] [Google Scholar]
- 61.Okada E, Fujiishi Y, Yasutake N, Ohyama W. Detection of micronucleated cells and gene expression changes in glandular stomach of mice treated with stomach-targeted carcinogens. Mutat Res. 2008;657:39–42. doi: 10.1016/j.mrgentox.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 62.Rothfuss A, O’Donovan M, De Boeck M, Brault D, Czich A, Custer L, et al. Collaborative study on fifteen compounds in the rat-liver Comet assay integrated into 2-and 4-week repeat-dose studies. Mutat Res. 2010;702:40–69. doi: 10.1016/j.mrgentox.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Robbiano L, Mereto E, Migliazzi Morando A, Pastore P, Brambilla G. Increased frequency of micronucleated kidney cells in rats exposed to halogenated anaesthetics. Mutat Res. 1998;413:1–6. doi: 10.1016/s1383-5718(97)00187-3. [DOI] [PubMed] [Google Scholar]
- 64.Mirkova ET. Activities of the rodent carcinogens thio-acetamide and acetamide in the mouse bone marrow micronucleus assay. Mutat Res. 1996;352:23–30. doi: 10.1016/0027-5107(95)00169-7. [DOI] [PubMed] [Google Scholar]