Abstract
Several hallmarks of cancer cells are their display of metabolic changes and enhanced proliferation. Highly proliferating cells utilize glutamine as a source of nitrogen, and therefore, one of the commonly seen metabolic changes is increased glutaminolysis, or glutamine catabolism. In addition, glutamine is an important anaplerotic source by which cells support the pools of TCA cycle intermediates in Myc-expressing cancer cells. Glutamine is converted to aspartate, which forms oxaloacetate, malate, and pyruvate. These conversions increase the NADPH/NADP+ ratio and maintain redox balance, which supports proliferation in K-ras-expressing cells. Therefore, glutamine is important for cancer cell proliferation and survival. On the other hand, glutamine stimulates the activation of the tumor suppressor p53, which induces apoptosis and tumor regression. The tumor suppressor SIRT4 inhibits glutamate dehydrogenase, which converts glutamic acid to α-ketoglutarate, an intermediate in the TCA cycle. Overall, the expression levels of oncogenes and tumor suppressors are critical to determine whether glutamine supports or suppresses proliferation and survival of cancer cells.
Keywords: Oncogenes, Tumor suppressors, Glutamine metabolism, Cancer cells
INTRODUCTION
The global cancer incidence has been steadily increasing, along with its correlated morbidity and mortality. Therefore, there has been rising interest in cancer prevention in order to decrease or delay the initial carcinogenesis and development of premalignant cells.1 One focus is on chemoprevention, which involves the use of natural, synthetic, or biological agents.2,3 Nutrients are also recognized as chemopreventive factors that significantly influence cancer development and prevention. The metabolism of nutrients such as glucose and glutamine, and the activity of plant-derived phytochemicals are associated with cancer suppression or prevention.4–6
Glutamine plays several important roles in the cell. It serves as a constituent of protein, carbon source for energy production, and a precursor of antioxidant glutathione (GSH). Glutamine prevents inflammation by improving immune function7 and is involved in signal transduction.8 Although glutamine is traditionally considered to be a non-essential amino acid, it is conditionally essential during catabolic stress such as severe injury, trauma, and sepsis.9 Most importantly, glutamine metabolism supports cell proliferation. Tumor cells display changes in metabolism induced by oncogenes, which also increase proliferation.10 As a result of the energy demands in tumor cells, glutaminolysis, or glutamine catabolism, is heightened, as is aerobic glycolysis.11 Specifically, glutamine supports the viability and proliferation of cancer cells by providing pools of TCA cycle intermediates and by the biosynthesis of proteins, lipids, and nucleotides.12 In addition, glutaminolysis is required to suppress oxidative stress in cancer cells because glutamine participates in GSH synthesis. Glutamine also contributes to restoring GSH to its reduced form by increasing the production of NADPH through malic enzyme regulation.13 In this review, the role of glutamine in cancer cells and how oncogenes and tumor suppressors control glutamine metabolism shall be discussed.
p53
p53 is a well-known protein which is involved in many cellular functions including cell cycle arrest, apoptosis, senescence, and differentiation. As a tumor suppressive transcription factor that regulates its target genes both positively and negatively, p53 selectively destroys abnormal or stressed cells to prevent transformation into cancer cells.14 In fact, mutational inactivation of p53 is strongly associated with increased cancer risks due to lack of p53-mediated apoptosis.15,16 p53 mediates the major apoptotic pathways in the cell by stimulating death receptor signaling and mitochondrial perturbations.17,18
One way p53 may mediate tumor suppression is through its induction of the expression of glutaminase 2 (GLS2). GLS is an enzyme that converts glutamate from glutamine, which is how GLS regulates GSH biosynthesis19 and mitochondrial oxidative phosphorylation. There are two phosphate-activated GLS isoforms: GLS1 and GLS2.20,21 GLS1 is involved in cell proliferation, whereas GLS2 is associated with the cell resting or quiescent state.21 GLS2 is induced in response to ROS in a p53-dependent manner, and overexpression of GLS2 results in a reduction of tumor cell growth. GLS2 levels were significantly reduced in liver tumors, and these results provide evidence that glutamine metabolism may contribute to the tumor suppressive role of p53.
Other experiments also show a relationship between p53 and glutamine in regards to tumor suppression. Oral glutamine supplementation increases phosphorylation of p53 and apoptosis in DMBA-induced mammary tumors.5,22 In this experiment, the levels of phosphorylated p53 in tumors of rats given glutamine supplementation were higher than those without glutamine supplementation. In addition, the expression of p21Waf1/Cip1, PTEN, mdm2, and IGF-IR, which are target proteins of p53, was regulated by glutamine supplementation. Because p53 is required for improvement of tumor regression and apoptosis in response to chemotherapy,23,24 glutamine supplementation may be a beneficial approach for suppression of cancer cells in a p53-dependent manner.
Conversely, under low glutamine conditions, p53 is required for cellular survival and adaptation to glutamine deprivation. Reid et al. demonstrated that glutamine deprivation activates p53 by regulating protein phosphatase 2A (PP2A).25 In this experiment, B55α, one of the regulatory subunit of PP2A, was specifically induced during glutamine starvation in a ROS-dependent manner. Additionally, B55α activates p53 through direct binding and dephosphorylation of EDD, an inhibitor of p53. Glutamine is converted to glutamic acid, which is composed of GSH,26 and during glutamine deprivation, GSH is reduced and ROS levels are increased in the cells. Increased ROS levels induce the expression of B55α and subsequently, the activation of p53. In other words, p53 is activated upon glutamine deprivation. Since there is controversy surrounding p53 activation and glutamine status (supplementation or deprivation), carefully designed studies should be performed to explore these complex relationships during different cell states.
Myc
Myc is a transcription factor that becomes deregulated in tumors. Myc is a proto-oncogene that promotes cell growth and regulates cell proliferation,27,28 and it is highly involved in glycolytic metabolism.29 Myc expression is tightly controlled by external signals and cell cycle arrest signals. Although its levels are low in the resting state, once the cell cycle starts, Myc levels increase significantly.30 As a result, abnormal regulation of Myc expression causes uncontrolled cell proliferation with aberrant apoptotic pathways.31
Several studies have revealed that Myc regulates glutamine metabolism and uptake.32,33 In these experiments, glutamine addiction was induced by increasing Myc expression, and glutamine deprivation resulted in apoptosis. In the experiment by Wise et al., the Myc transformation increased cellular dependence on glutamine in order to support TCA cycle anaplerosis.33 In addition, Myc increased the expression of GLS1, which is involved in cell proliferation.32 The experiment by Le et al. also found that glutamine consumption was increased in Myc-expressing cells, but the glutamine was preferentially utilized in GSH synthesis instead of TCA anaplerosis. By participating in GSH synthesis, glutamine plays a pivotal role in cell survival in these Myc-expressing cells. This study demonstrates the antioxidant role of glutamine to be essential for cell survival in Myc-expressing cells.34
SIRT4
Sirtuins are NAD+-dependent deacetylases, targeting a wide variety of proteins in the nucleus, cytosol, and mitochondria.35 Since sirtuins respond to cellular stress and DNA damage, they have been extensively studied as therapeutic targets for human diseases, such as cancer.36,37 The sirtuin (SIRT) family consists of seven members (SIRT1-7), and each of the members differs in its activity and function. SIRT4 and SIRT6 catalyze ADP-ribosylations, whereas SIRT1, SIRT2, SIRT3, SIRT5, and SIRT7 catalyze deacetylation reactions.38 Importantly, it has been reported that SIRT4 expression is reduced in cancer cells, and overexpression of SIRT4 inhibits cell proliferation and tumor development.35,39,40
Previous studies have shown that SIRT4 inhibits glutamine dehydrogenase (GDH),41 and it is well established that glutamine is an important source for TCA anaplerosis in proliferating cells.42 Glutamate is produced from glutamine by glutaminase (GLS), and then glutamate is convertted to α-ketoglutarate by GDH. The α-ketoglutarate produced participates in the TCA cycle as an intermediate. Jeong et al. demonstrated that SIRT4 inhibits glutamine uptake and anaplerosis of cells with DNA damage, resulting in cell cycle arrest.43 These findings suggest that SIRT4-mediated inhibition of glutamine anaplerosis may function as a tumor suppressor mechanism.
Another way in which the relationship between SIRT4 and glutamine may uncover information about potential cancer therapy targets was reported by Csibi et al. In this study, the activation of mammalian target of rapamycin complex 1 (mTORC1) was associated with increased glutamine metabolism.44 Specifically, mTORC1 increased glutamine anaplerosis by activating GDH. Furthermore, mTORC1 mechanically inhibited SIRT4. This transcriptional repression of SIRT4 is essential for glutamine metabolism, and targeting glutamine metabolism in cancers with more active mTORC1 pathways may be useful for cancer therapy.
K-ras
Activated Ras regulates a wide range of effectors such as Raf kinase and PI-3 kinase, and activated Ras produces pleiotropic cellular effects. Since Ras is important in oncogenesis, Ras signaling in cell growth and survival has been extensively studied.45 K-ras has been accepted to be one of the most frequently mutated oncogenes, and K-ras mutations have been frequently found in a number of tumors, suggesting K-ras as an effective target for cancer therapy.46 One of K-ras’s main functions is to mediate metabolic transformation during tumorigenesis. The transformed cells that express high levels of K-ras protein show heightened glucose sensitivity, reduced mitochondrial function, and reduced proliferation in response to glutamine deprivation.47,48 In addition, K-ras transformation increases glycolytic flux, reduces oxidative TCA cycle flux, and enhances the utilization of glutamine in mouse fibro-blasts.49
As mentioned, K-ras-transformed cells utilize glutamine differently than non-transformed cells. Specifically, Son et al. suggests that oncogenic K-ras mediates the reprogramming of glutamine utilization in human pancreatic ductal adenocarcinoma (PDAC) cells,50 resulting in cells that are more sensitive to low glutamine levels. [KR4]K-ras induces the cytosolic conversion of aspartate into oxaloacetate, malate, and pyruvate. These conversions increase the NADPH/NADP+ ratio by which cells are able to maintain redox balance and support proliferation. Unlike most cells, which utilize glutamate dehydrogenase (GDH) in the mitochondria to convert glutamine-derived glutamate into α-ketoglutarate for the TCA cycle, these PDAC cells inhibit GDH and activate aspartate transaminases (GOT1) through this K-ras-regulated metabolic pathway.
CONCLUSION
As illustrated, oncogenes and tumor suppressors play a major role in regulating glutamine metabolism. In general, glutamine contributes to cancer cell proliferation and survival by maintaining the pool of TCA cycle intermediates or by regulating redox homeostasis. However, many oncogenes can significantly alter the function of glutamine or its metabolites. For example, Myc transformations change GSH synthesis, K-ras reprograms glutamine utilization under low glutamine levels, and the p53 level is altered by glutamine starvation. Glutamine is converted to aspartate, which forms oxaloacetate, malate, and pyruvate. These conversions increase the NADPH/NADP+ ratio and maintain redox balance, which supports proliferation in K-ras-expressing cells. Therefore, glutamine is an important player in cancer cell proliferation and survival.
On the other hand, glutamine stimulates the activation of the tumor suppressor p53, which induces apoptosis and tumor regression. p53 induces the expression of GLS2, which is associated with the slowing of tumor growth. In addition, the tumor suppressor SIRT4 inhibits GDH, which converts glutamic acid to the TCA cycle intermediate, α-ketoglutarate. Overall, the availability or levels of glutamine influence the signal transduction pathways of the oncogenes myc and K-ras and the tumor suppressors p53 and SIRT4.
Beyond its traditional role as the most abundant amino acid in the human body, recent studies illustrate gluta-mine’s role in cancer and proliferating cells. Glutamine’s relationship with cancer is complicated since it acts through several oncogenes and tumor suppressors. Although the literature suggests that the expression levels of oncogenes and tumor suppressors may affect glutamine metabolism in cancer cells, the metabolism of glutamine in cancer is not well understood, especially when compared to our current knowledge of glucose metabolism in cancer. With better understanding of glutamine metabolism in cancer and highly proliferating cells, we expect the use of glutamine for chemoprevention and cancer therapy to be a viable option in the future.
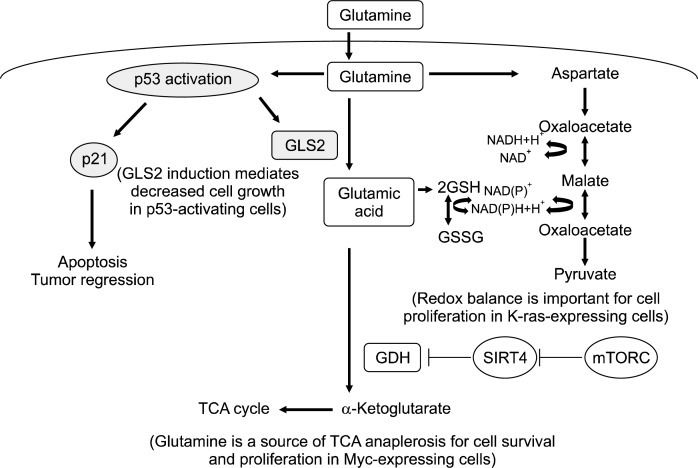
Fig. 1.
Schematic overview of glutamine metabolism, which is regulated by oncogenes and tumor suppressors in cancer cells. Glutamine stimulates the activation of tumor suppressor p53, which induces apoptosis and tumor regression through the expression of p21. Also, p53 induces the expression of GLS2, which is associated with reductions in tumor growth (left panel). Glutamine is an important anaplerotic source that support pools of TCA cycle intermediates in Myc-expressing cancer cells. Glutamine is converted to aspartate, which forms oxaloacetate, malate, and pyruvate. These conversions increase the NADPH/NADP+ ratio and maintain redox balance, supporting proliferation in K-ras-expressing cells. The tumor suppressor SIRT4 inhibits GDH, which converts glutamic acid to the TCA cycle intermediate, α-ketoglutarate. mTORC1 mechanically inhibits SIRT4 and activates GDH (right panel). Overall, the expression levels of oncogenes and tumor suppressors are critical to determine whether glutamine supports or suppresses the proliferation and survival of cancer cells. GLS, glutaminase; GDH, glutamine dehydrogenase; SIRT4, Sirtuin4; mTORC1, mammalian target of rapamycin complex 1.
Acknowledgments
This work was supported by the NRF of Korea grant, which is funded by the Korean government (MSIP) (2007-0056092).
REFERENCES
- 1.Sidorenko Iu S, Shaposhnikov AV. Long- and short-range strategies in cancer prevention. Vopr Onkol. 2009;55:671–8. [PubMed] [Google Scholar]
- 2.Sporn MB. Approaches to prevention of epithelial cancer during the preneoplastic period. Cancer Res. 1976;36:2699–702. [PubMed] [Google Scholar]
- 3.Steward WP, Brown K. Cancer chemoprevention: a rapidly evolving field. Br J Cancer. 2013 doi: 10.1038/bjc.2013.280. e-pub ahead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biscaro F, Parisotto EB, Zanette VC, Gunther TM, Ferreira EA, Gris EF, et al. Anticancer activity of flavonol and flavan-3-ol rich extracts from Croton celtidifolius latex. Pharm Biol. 2013;51:737–43. doi: 10.3109/13880209.2013.764331. [DOI] [PubMed] [Google Scholar]
- 5.Lim V, Korourian S, Todorova VK, Kaufmann Y, Klimberg VS. Glutamine prevents DMBA-induced squamous cell cancer. Oral Oncol. 2009;45:148–55. doi: 10.1016/j.oraloncology.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Sak K. Chemotherapy and dietary phytochemical agents. Chemother Res Pract. 2012;2012:282570. doi: 10.1155/2012/282570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H. Glutamine as an immunonutrient. Yonsei Med J. 2011;52:892–7. doi: 10.3349/ymj.2011.52.6.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liboni KC, Li N, Scumpia PO, Neu J. Glutamine modulates LPS-induced IL-8 production through IkappaB/NF-kappaB in human fetal and adult intestinal epithelium. J Nutr. 2005;135:245–51. doi: 10.1093/jn/135.2.245. [DOI] [PubMed] [Google Scholar]
- 9.Windmueller HG, Spaeth AE. The Journal of Biological Chemistry, Volume 249, 1974: Uptake and metabolism of plasma glutamine by the small intestine. Nutr Rev. 1990;48:310–2. doi: 10.1111/j.1753-4887.1990.tb02968.x. [DOI] [PubMed] [Google Scholar]
- 10.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–9. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 12.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–33. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vousden KH. p53: death star. Cell. 2000;103:691–4. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y. p53 and its downstream proteins as molecular targets of cancer. Mol Carcinog. 2006;45:409–15. doi: 10.1002/mc.20231. [DOI] [PubMed] [Google Scholar]
- 16.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 17.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 18.Green DR. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695–8. doi: 10.1016/s0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 19.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kvamme E, Nissen-Meyer LS, Roberg BA, Torgner IA. Novel form of phosphate activated glutaminase in cultured astrocytes and human neuroblastoma cells, PAG in brain pathology and localization in the mitochondria. Neurochem Res. 2008;33:1341–5. doi: 10.1007/s11064-008-9589-9. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Gomez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sanchez P, et al. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–42. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todorova VK, Kaufmann Y, Luo S, Klimberg VS. Modulation of p53 and c-myc in DMBA-induced mammary tumors by oral glutamine. Nutr Cancer. 2006;54:263–73. doi: 10.1207/s15327914nc5402_13. [DOI] [PubMed] [Google Scholar]
- 23.Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–10. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 24.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–67. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 25.Reid MA, Wang WI, Rosales KR, Welliver MX, Pan M, Kong M. The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol Cell. 2013;50:200–11. doi: 10.1016/j.molcel.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Carretero J, Obrador E, Pellicer JA, Pascual A, Estrela JM. Mitochondrial glutathione depletion by glutamine in growing tumor cells. Free Radic Biol Med. 2000;29:913–23. doi: 10.1016/s0891-5849(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 27.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang CV. Rethinking the Warburg effect with Myc micro-managing glutamine metabolism. Cancer Res. 2010;70:859–62. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:d250–68. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 31.Cole MD, Nikiforov MA. Transcriptional activation by the Myc oncoprotein. Curr Top Microbiol Immunol. 2006;302:33–50.s. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- 32.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Front Pharmacol. 2012;3:4. doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2013 doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebastian C, Satterstrom FK, Haigis MC, Mostoslavsky R. From sirtuin biology to human diseases: an update. J Biol Chem. 2012;287:42444–52. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez-Marcos PJ, Serrano M. Sirt4: the glutamine gate-keeper. Cancer Cell. 2013;23:427–8. doi: 10.1016/j.ccr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 41.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 42.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–54. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Mancera PA, Tuveson DA. Physiological analysis of oncogenic K-ras. Methods Enzymol. 2006;407:676–90. doi: 10.1016/S0076-6879(05)07053-9. [DOI] [PubMed] [Google Scholar]
- 46.Adjei AA. K-ras as a target for lung cancer therapy. J Thorac Oncol. 2008;3:S160–3. doi: 10.1097/JTO.0b013e318174dbf9. [DOI] [PubMed] [Google Scholar]
- 47.Baracca A, Chiaradonna F, Sgarbi G, Solaini G, Alberghina L, Lenaz G. Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim Biophys Acta. 2010;1797:314–23. doi: 10.1016/j.bbabio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Chiaradonna F, Gaglio D, Vanoni M, Alberghina L. Expression of transforming K-Ras oncogene affects mitochondrial function and morphology in mouse fibroblasts. Biochim Biophys Acta. 2006;1757:1338–56. doi: 10.1016/j.bbabio.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]