Abstract
The most common cystic fibrosis (CF) mutation, ΔF508, causes protein misfolding, leading to proteosomal degradation. We recently showed that expression of miR-138 enhances CF transmembrane conductance regulator (CFTR) biogenesis and partially rescues ΔF508-CFTR function in CF airway epithelia. We hypothesized that a genomic signature approach can be used to identify new bioactive small molecules affecting ΔF508-CFTR rescue. The Connectivity Map was used to identify 27 small molecules with potential to restore ΔF508-CFTR function in airway epithelia. The molecules were screened in vitro for efficacy in improving ΔF508-CFTR trafficking, maturation, and chloride current. We identified four small molecules that partially restore ΔF508-CFTR function in primary CF airway epithelia. Of these, pyridostigmine showed cooperativity with corrector compound 18 in improving ΔF508-CFTR function. There are few CF therapies based on new molecular insights. Querying the Connectivity Map with relevant genomic signatures offers a method to identify new candidates for rescuing ΔF508-CFTR function.
Keywords: cystic fibrosis, small molecule compound, Connectivity Map, airway epithelia, corrector compound 18
Clinical Relevance
Mutations in the cystic fibrosis (CF) transmembrane conductance regulator gene cause CF; the most common mutation, ΔF508, results in protein misfolding and degradation. Restoring function to the misfolded protein could prevent or slow disease. Here we use a genomics signature approach to provide proof of principle for a new paradigm for drug discovery in CF.
Cystic fibrosis (CF), an autosomal recessive disease caused by mutations in the CF transmembrane conductance regulator (CFTR) gene, is the most common lethal genetic disease among Caucasians. The CFTR mutation, ΔF508, is present on approximately 70% of mutant alleles and causes protein misfolding, leading to proteosomal degradation. If ΔF508-CFTR trafficks to the cell membrane, as occurs with low temperature (1) or chemical chaperone treatment (2), the mutant protein retains channel function, although its residency and open-state probability are reduced (3, 4). Because approximately 90% of people with CF have at least one ΔF508 allele, a major focus of current CF research and therapeutics development is understanding factors regulating CFTR expression and biosynthesis, the cellular mechanisms underlying ΔF508-CFTR degradation, and exploiting this knowledge to identify drugs or small molecules that might restore ΔF508-CFTR function (5–8).
We recently reported that transfection of polarized primary cultures of human airway epithelia with a miR-138 mimic reduced, and that of a miR-138 anti-miR increased, SIN3A mRNA and protein levels (9). Furthermore, we showed, using polarized primary non-CF human airway epithelia, that transfection with a miR-138 mimic or a Dicer-substrate small interfering RNA (DsiRNA) against SIN3A increased, and that of a miR-138 anti-miR reduced, CFTR mRNA and protein levels (9). Treatment with the miR-138 mimic and the SIN3A DsiRNA increased cAMP-stimulated conductance (Gt), whereas the miR-138 anti-miR reduced Gt responses to cAMP-dependent stimulation. The reciprocal effects of the miR-138 anti-miR in decreasing CFTR messenger RNA (mRNA), protein, and transepithelial Cl− permeability emphasized the role of miR-138 in regulating CFTR expression (9). Furthermore, we demonstrated, in polarized primary CF human airway epithelia, that manipulating the miR-138/SIN3A network with a miR-138 mimic or a DsiRNA against SIN3A rescued ΔF508-CFTR–mediated Cl− transport in CF primary airway epithelial cells. Both interventions increased the abundance of fully glycosylated band C and restored ΔF508-CFTR–mediated Cl− transport (9). Significant restoration of ΔF508-CFTR–mediated Cl− transport was observed to varying levels in primary CF epithelia from multiple human donors, and similar results were obtained in a cell line homozygous for the ΔF508 mutation (CF bronchial epithelial [CFBE] cells) (9).
Global mRNA transcript profiling in Calu-3 epithelia treated with the miR-138 mimic or SIN3A DsiRNA identified a common set of 773 genes whose expression changed in response to these interventions (9). Pathway and gene ontology analysis of the 773 differentially expressed genes revealed a significant enrichment of gene sets in pathways including chaperones, unfolded protein response, protein ubiquitination and proteosomal catabolic processes, negative regulation of apoptosis, and heat shock (9). These results support the conclusion that miR-138 enhances CFTR biogenesis and influences the expression of genes at multiple steps along its biosynthetic pathway.
The Connectivity Map (CMAP) (10), introduced in 2006, is a reference collection of gene-expression profiles from cultured human cells treated with bioactive small molecules, together with a pattern-matching software to mine these data. CMAP (build 02) includes 7,000 expression profiles representing 1,309 compounds used in 3,442 individual treatments (multiple doses, durations) and matched vehicle control pairs applied to freely cycling human cell lines (10). Since the development of CMAP, several labs have independently used this tool to successfully investigate breast (11), colon (12), and prostate cancers (13–15) and other disease targets (16–18). Adams and colleagues used this resource and mRNA expression signatures of human skeletal muscle atrophy to discover a natural compound that increases muscle mass and demonstrated in vivo efficacy (19). This variety of reports provides confidence in the broad utility of CMAP to link cell biology, gene expression, disease, and therapeutics.
The observation that manipulating the miR-138/SIN3A network partially restored ΔF508-CFTR protein function suggests that we might exploit this interaction for therapeutic purposes (9). This previously unrecognized gene network provides a new understanding of how CFTR expression and biosynthesis is regulated and suggests therapeutic targets for rescuing the function of the mutant CFTR protein most commonly associated with CF. We hypothesized that a genomic signature approach can be used to identify bioactive small molecules effecting ΔF508-CFTR rescue. Using previously generated gene expression signatures (9) to query CMAP, we identified 27 small molecules that mimicked the SIN3A DsiRNA and miR-138 treatments. We screened these small molecules at different doses and in combination with the CFTR corrector compound 18 (C18) to test for efficacy in improving ΔF508-CFTR maturation and function.
Materials and Methods
Detailed descriptions of materials and methods are available in the online supplement.
Results
CMAP-Based Identification of Novel Bioactive Small Molecules
Global mRNA transcript profiling in Calu-3 epithelia treated with the miR-138 mimic or SIN3A DsiRNA (9) generated a list of 1,608 and 1,723 differentially expressed genes, respectively, with a P value < 0.05 and a fold change > 1.2. Intersection of these two data sets identified a common set of 773 genes (Set 1; see Figure E1A in the online supplement) whose expression changed in response to both interventions (9). On increasing the stringency to a P value < 0.01 and a fold change > 1.5, we generated a list of 284 differentially expressed genes for the miR-138 treatment (Set 2; Figure E1A) and 239 differentially expressed genes for the SIN3A DsiRNA treatment (Set 3; Figure E1A). We used these three gene sets to query the CMAP 2.0 database for small molecules that mimic the query signature.
For each query a unique small molecule enrichment profile was generated by CMAP. To prioritize small molecules for our screen, we mined the profiles using three distinct strategies (Figure E1B). First, we selected small molecules in each profile based on “connectivity scores” closest to +1 and picked candidate small molecules from the intersection of the three profiles. The connectivity score provides the relative strength of a drug signature to the query (20). We sought the best positive correlation, thus a connectivity score closest to +1. Second, in each profile, we prioritized small molecules based on “up” and “down” scores (Figure E1B). The “up” score is the absolute enrichment of up-regulated genes in the query for a given drug signature (20). The opposite is true for the “down” score. We selected small molecules (common to the three profiles) with the highest “up” score (closest to +1) or the lowest “down” score (closest to −1). Finally, for each query, we separated the profile by cell line (HL60, MCF7, PC3) and repeated the first strategy, prioritizing by connectivity score (Figure E1B). The connectivity score for a small molecule comprises it enrichment score across all three cell lines (20). A small molecule may only work in one or two of the cell lines, resulting in a low score for a specific query. We counteracted this by analyzing the data for each cell line separately. Using these strategies, we selected 27 small molecules for initial experiments.
Valproic Acid, Pizotifen, Biperiden, and Pyridostigmine Improve ΔF508-CFTR Biosynthesis
We screened the 27 candidate small molecules for efficacy in rescuing ΔF508-CFTR biosynthesis by three different methods: (1) surface display of ΔF508-CFTR in HeLa cells stably expressing the HA-tagged ΔF508-CFTR (21) (HeLa-ΔF508-CFTR-HA), (2) rescue of ΔF508-CFTR maturation in CFBE cells (grown submerged) (22) measured as the formation of fully glycosylated CFTR (band C), and (3) functional rescue of ΔF508-CFTR in well-differentiated passage 2 (P2) CF airway epithelial cultures measured as cAMP-activated Cl−, GlyH-101–inhibited current.
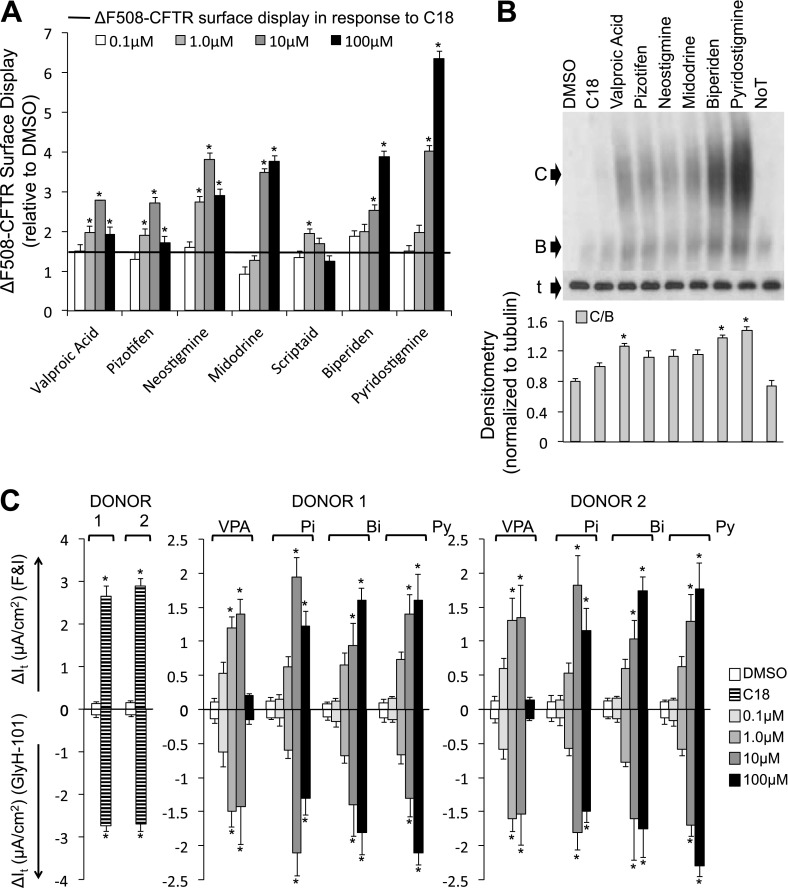
In HeLa cells, we screened each small molecule at four doses (0.1, 1.0, 10, and 100 μM) and measured ΔF508-CFTR trafficking 24 hours after treatment. We identified seven small molecules that rescued ΔF508-CFTR surface display greater than the C18 control (> 1.5-fold) at two or more doses (Figures 1A and E2). We performed a similar experiment in CFBE cells. We harvested protein 24 hours after treatment and identified six small molecules that variably improved ΔF508-CFTR maturation. These include valproic acid, pizotifen, neostigmine, midodrine, biperiden, and pyridostigmine (Figures 1B and E3). Next, we measured functional improvement of ΔF508-CFTR in well-differentiated P2 CF airway epithelial cultures exposed to the small molecules basolaterally for 24 hours. Valproic acid, pizotifen, biperiden, and pyridostigmine partially restored ΔF508-CFTR–mediated Cl− current (Figures 1C and E4). However, these changes were significantly less than that achieved with C18 alone (note differences in Y-axis scales).
Figure 1.
Four small molecules rescue ΔF508–cystic fibrosis transmembrane conductance regulator (CFTR) function in primary cystic fibrosis airway cultures. (A) Average surface display of ΔF508-CFTR in HeLa cells measured by cell-surface ELISA 24 hours after the indicated treatments. Fold increase relative to DMSO treatment. C18 (represented as black horizontal line) was added for 24 hours at 6 μM concentration (n = 24). Each small molecule (including all doses) was controlled by independent DMSO treatments. Results from full screen described in Figure E2. (B) Representative immunoblot depicting ΔF508-CFTR expression in cystic fibrosis bronchial epithelial (CFBE) cells (grown submerged). B, CFTR band B; C, CFTR band C; noT, no treatment; t, α-tubulin. Protein was harvested 24 hours after treatment. Densitometry (normalized to α-tubulin) is presented as the average ratio of band C to band B (C/B) in CFBE cells (n = 4). Significance was calculated against DMSO treatment. Results from full screen are presented in Figure E3. (C) Average change in transepithelial current (It) in response to forskolin and 3-isobutyl-1-methylxanthine (FandI) and GlyH-101 measured in well-differentiated human (ΔF508/ΔF508) passage 2 airway epithelial cultures from two donors (n = 6 per donor). Small molecules were administered basolaterally 24 hours before the electrophysiology study. Each small molecule (including all doses) was controlled by independent DMSO treatments. Results from full screen are described in Figure E5. Concentration of small molecules is noted in key. C18 was administered at 6 μM concentration; note the difference in Y-axis scale for C18. Bi, biperiden; Pi, pizotifen; Py, pyridostigmine; VPA, valproic acid. All panels: DMSO administered at 1/1,000 concentration; error bars indicate standard error; statistical significance was determined by the Holm-Bonferroni method (*P < 0.05).
Pyridostigmine and C18 Synergistically Rescue ΔF508-CFTR Function
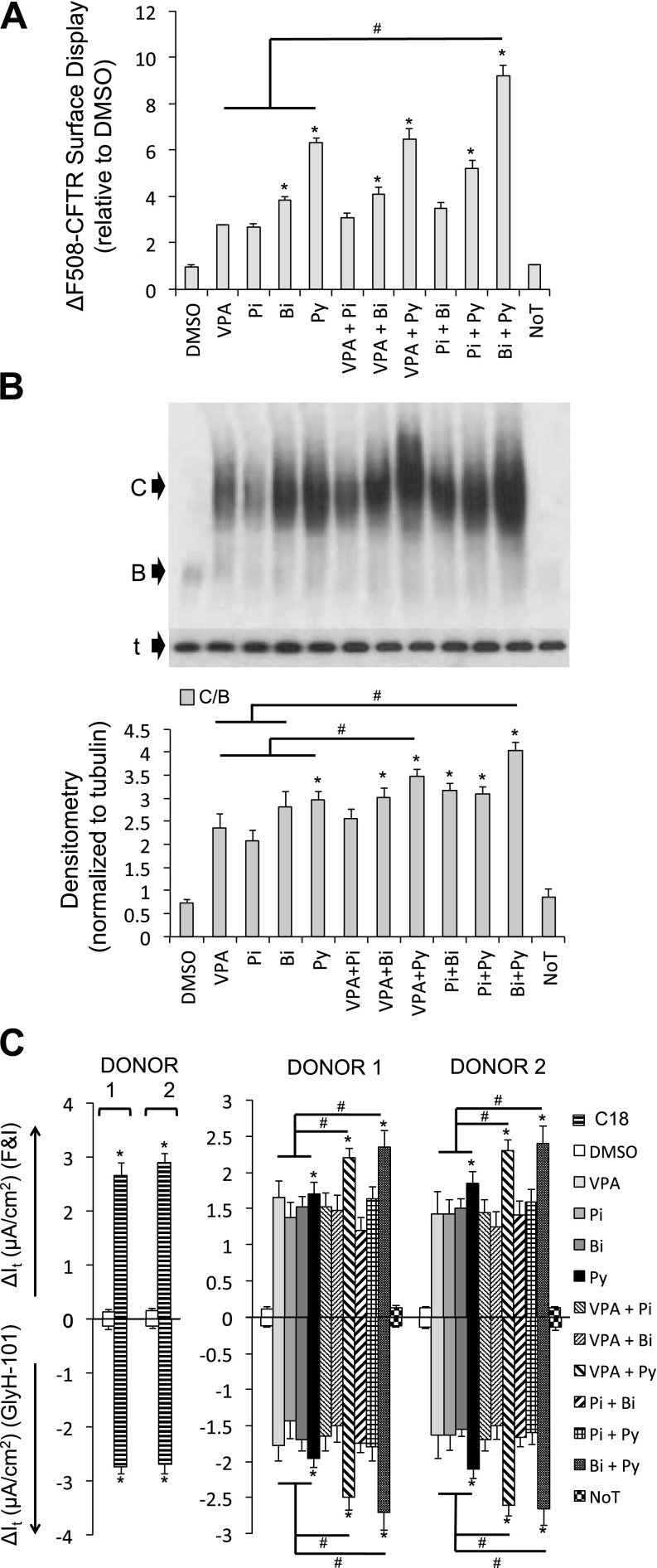
Early studies by Lim and colleagues reported the cooperative actions of small-molecule compounds in restoring protein trafficking and ΔF508-CFTR–mediated Cl− current in IB3–1 cells (23). Studies such as this were among the first to suggest the possibility of combination therapy in restoring function of ΔF508-CFTR. As understanding of the complexity of the defect caused by the ΔF508-CFTR mutation has increased and as the inefficiencies of individual corrector compounds in restoring ΔF508-CFTR–mediated Cl− current are recognized, multiple groups have suggested that combination therapies may be required (24–28). For these reasons, we asked whether combining the four candidate small molecules with each other or with C18 (6.0 μM) resulted in cooperative interactions. We selected the concentration with the best functional rescue (Figure 1C) and cotreated cells for 24 hours. The combination of pyridostigmine and biperiden cooperatively improved ΔF508-CFTR surface display (HeLa cells; Figure 2A) and maturation (CFBE cells; Figure 2B). Upon treating well-differentiated P2 CF airway epithelial cells, the combination of pyridostigmine with valproic acid or biperiden cooperatively improved ΔF508-CFTR–mediated Cl− current. However, these changes were still significantly lower than that achieved with the C18 treatment alone (Figure 2C).
Figure 2.
Combination of small molecules fails to rescue ΔF508-CFTR function greater than C18. (A) Average surface display of ΔF508-CFTR in HeLa cells measured by cell-surface ELISA 24 hours after the indicated treatments. Fold increase relative to DMSO treatment (n = 24). Combinatorial treatments were applied simultaneously. (B) Representative immunoblot depicting ΔF508-CFTR expression in CFBE cells (grown submerged). B, CFTR band B; C, CFTR band C; noT, no treatment; t, α-tubulin. Protein was harvested 24 hours after treatment. Densitometry (normalized to α-tubulin) is presented as the average ratio of band C to band B (C/B) in CFBE cells (n = 4). (C) Average change in transepithelial current (It) in response to FandI and GlyH-101 measured in well-differentiated human (ΔF508/ΔF508) passage 2 airway epithelial cultures from two donors (n = 6 per donor). Small molecules were administered basolaterally 24 hours before the electrophysiology study. C18 was administered at 6 μM concentration; note the difference in Y-axis scale for C18. All panels: Bi, biperiden (100 μM); Pi, pizotifen (10 μM); Py, pyridostigmine (100 μM); VPA, valproic acid (10 μM). Error bars indicate standard error; experiments were controlled by independent DMSO (1/1,000) treatments; statistical significance was determined by the Holm-Bonferroni method (*P < 0.05; #P < 0.05).
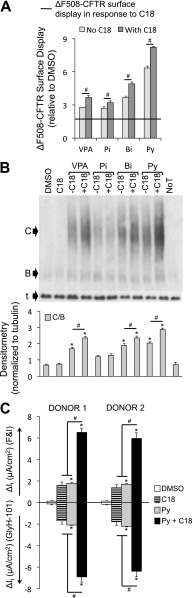
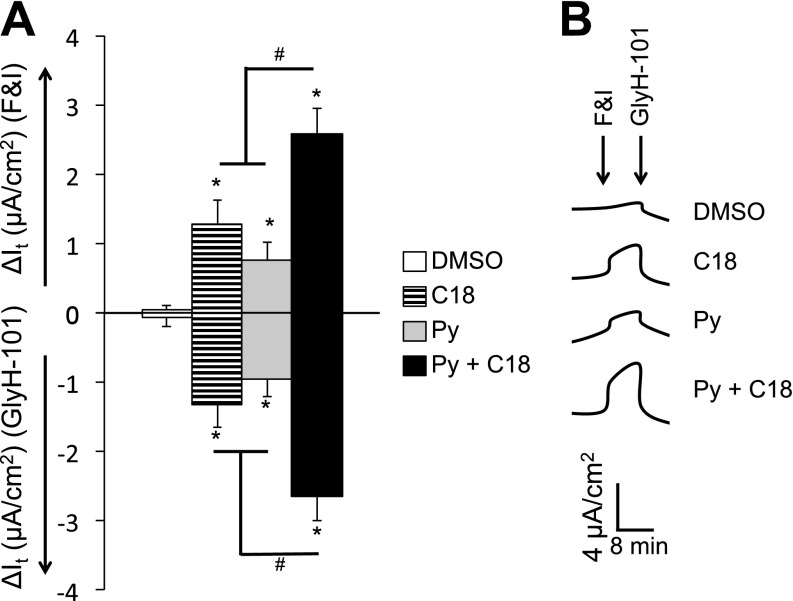
On combining the candidate small molecules with C18, the combination of pyridostigmine and C18 gave the greatest increase in ΔF508-CFTR surface display in HeLa cells (Figure 3A) and maturation in CFBE cells (Figure 3B). Furthermore, upon treatment of well-differentiated P2 cultures of CF airway epithelia with drug combinations, the combined use of pyridostigmine and C18 significantly improved ΔF508-CFTR–mediated Cl− current more than either treatment alone (Figures 3C and E5). We next treated well-differentiated P0 primary cultures of CF airway epithelia with pyridostigmine and C18 alone or in combination. Although pyridostigmine and C18 partially restored ΔF508-CFTR–mediated Cl− current, the combination of pyridostigmine and C18 conferred significantly greater CFTR anion channel function (Figure 4).
Figure 3.
Pyridostigmine and C18 cooperatively rescue ΔF508-CFTR biosynthesis. (A) Average surface display of ΔF508-CFTR in HeLa cells measured by cell-surface ELISA 24 hours after the indicated treatments. Fold increase relative to DMSO treatment (n = 24). Combinatorial treatments were applied simultaneously. C18, represented as a black horizontal line, was added for 24 hours at 6 μM concentration (n = 24). (B) Representative immunoblot depicting ΔF508-CFTR expression in CFBE cells (grown submerged). B, CFTR band B; C, CFTR band C; noT, no treatment; t, α-tubulin. Protein was harvested 24 hours after treatment. Densitometry (normalized to α-tubulin) is presented as the average ratio of band C to band B (C/B) in CFBE cells (n = 4). (C) Average change in transepithelial current (It) in response to FandI and GlyH-101 measured in well-differentiated human (ΔF508/ΔF508) passage 2 airway epithelial cultures from two donors (n = 6 per donor). Small molecules were administered basolaterally 24 hours before the electrophysiology study. All panels: Bi, biperiden (100 μM); Pi, pizotifen (10 μM); Py, pyridostigmine (100 μM); VPA, valproic acid (10 μM). C18 was administered at 6 μM concentration. Error bars indicate standard error; experiments were controlled by independent DMSO (1/1,000) treatments; statistical significance was determined by the Holm-Bonferroni method (*P < 0.05; #P < 0.05).
Figure 4.
Pyridostigmine and C18 partially rescue ΔF508-CFTR function in primary airway epithelial cultures. (A) Average change in transepithelial current (It) in response to FandI and GlyH-101 measured in well-differentiated human (ΔF508/ΔF508) primary passage 0 airway epithelial cultures from four donors (n = 6 per donor). (B) Representative It tracings of responses to FandI followed by GlyH-101 treatment. Small molecules were administered basolaterally 24 hours before the electrophysiology study. Py, pyridostigmine (100 μM). C18 was administered at 6 μM concentration. Experiments were controlled by DMSO (1/1,000) treatment. Error bars indicate standard error; statistical significance was determined by the Holm-Bonferroni method (*P < 0.05; #P < 0.05).
Discussion
Although knowledge of CFTR function has advanced greatly since the discovery of the gene in 1989, treatments for the disease remain suboptimal, and CF remains progressive and fatal. For rare CFTR conductance mutants such as G551D, a class of agents termed “potentiators” is under development. The first of these to reach clinical trials, VX-770, showed efficacy and is now FDA approved (8). High throughput screens have identified several promising correctors of ΔF508 (5–7, 28–33), but the first agent in clinical trials, VX-809, showed no efficacy (34), and better agents are needed.
Although transient, partial, and airway-specific delivery of a miR-138 mimic or anti-SIN3A small interfering RNA (siRNA) might be therapeutic (9), the delivery of siRNA or miR mimics to airway epithelia is inefficient with current technologies (35, 36). Further advancements in this field may lead to new clinical applications. An alternative strategy is to focus on the downstream targets of SIN3A that directly mediate the observed ΔF508-CFTR rescue. Drug screens concentrated on the identification of small molecules or pharmaceuticals that inhibit SIN3A, or a core subset of gene products responsible for ΔF508-CFTR rescue, represent a promising therapeutic approach (37). We identified genomic signatures for miR-138 mimic expression and SIN3A inhibition and used these gene sets in iterative CMAP database mining to identify candidate drugs with similar effects on gene expression. Treatment of epithelia with a subset of these candidates showed evidence of ΔF508-CFTR rescue.
Seven small molecules improved ΔF508-CFTR trafficking greater than C18 at two or more doses, and four small molecules consistently improved ΔF508-CFTR maturation in CFBE cells. A more detailed analysis of these four molecules—pyridostigmine, pizotifen, biperiden, and valproic acid—revealed evidence of cooperation between C18 and pyridostigmine in improving ΔF508-CFTR biosynthesis. Combined treatment with pyridostigmine and C18 further enhanced surface presentation of ΔF508-CFTR in HeLa cells, increased band C expression in CFBE cells, and enhanced cAMP-activated Cl− current in P0 and P2 primary CF airway epithelia from multiple human donors.
Variations in the response of different cell types to small molecule treatments are expected. Typically, there are more positive hits in simple cell models, and the number of compounds that work in primary CF cells is considerably less. HeLa cells are a heterologous cell line stably expressing the ΔF508-CFTR transgene, and CFBEs are an immortalized CF airway epithelial cell line also stably expressing the ΔF508-CFTR transgene. Both of these cell types are inherently different from each other and the CF primary (P0 and P2) airway epithelia in terms of CFTR expression and other cellular properties, such as quality control checkpoints. Our aim was to identify small molecules that rescued aspects of ΔF508-CFTR biosynthesis—trafficking, maturation, and function—across the four cell types used. With functional rescue in well-differentiated P2 cultures of CF airway epithelia as perhaps the most stringent or informative readout for a compounds’ capacity to rescue ΔF508-CFTR function, we used those results (Figures 1C and E4) as the deciding factor in selecting pyridostigmine for experiments in the precious P0 primary airway epithelial cultures.
Pyridostigmine, biperiden, pizotifen, and valproic acid are FDA-approved drugs, and all improved ΔF508-CFTR maturation and function to varying degrees. These small molecules belong to different classes of drug with unrelated modes of action, and their mechanisms for improving ΔF508-CFTR maturation and function are not known. Pyridostigmine is a reversible cholinesterase inhibitor used to treat muscle weakness in myasthenia gravis, and biperiden is an anticholinergic drug used to treat Parkinsonism. Pizotifen is a serotonin antagonist used for migraine prevention, and valproic acid is a histone deacetylase inhibitor with clinical applications as an anticonvulsant and mood stabilizer. Histone deacetylase inhibitors have been reported to improve ΔF508-CFTR processing (29). The discovery that pyridostigmine in combination with biperiden or valproic acid enhanced ΔF508-CFTR band C formation and function in epithelial cells supports a strategy whereby bioactive small molecules that mimic SIN3A knockdown or miR-138 treatments can be identified using a genomic signature approach. Although the clinical utility of the molecules tested here requires further investigation, the discovery of their effect on ΔF508-CFTR biosynthesis highlights the utility of a genomic signatures approach and CMAP in drug discovery and, more importantly, provides a path for related studies to be conducted.
To our knowledge, this is the first example of using the CMAP to identify drugs that improve ΔF508-CFTR processing and function. Other groups have used genomics-based approaches to investigate interventions that restore ΔF508-CFTR activity. Wright and colleagues used gene expression analysis to determine that 4-phenylbutyrate rescued ΔF508-CFTR trafficking by modulating heat-shock protein expression (38). Galeitta and coworkers evaluated the gene expression profiles from low-temperature and corrector treatments in CFBE41o− cells to study the genetic mechanisms underlying improved ΔF508-CFTR function (5). Zhang and colleagues used genomic signatures from CF epithelia treated with ouabain, low temperature, VX-325, and VX-809 to query CMAP and search for commonality among possible mechanisms of action (33). This approach revealed that low-temperature and ouabain treatments shared similarities in gene expression profiles. Using a large-scale, proteomics-based approach, Wang and colleagues identified proteins that associate with CFTR, the “CFTR-interactome,” and investigated how the disease state changes such interactions (32). This approach has helped characterize the chaperone folding environment and is being exploited to identify candidates contributing to CF pathophysiology and to identify new therapeutic strategies.
It is possible that a single corrector drug may be insufficient to restore ΔF508-CFTR maturation and function to a clinically relevant extent (24–28). This is largely due to the complexity of the CFTR defect caused by the ΔF508 mutation and the redundancy of quality control mechanisms that exist to detect and degrade the mutant protein (24–28). Thibodeau and colleagues showed that second-site suppressor mutations, located within the second transmembrane domain, rescued ΔF508-CFTR domain–domain assembly. This result suggests that ΔF508-CFTR correction requires stabilization of nucleotide-binding domain 1 and the interface between membrane-spanning domains and nucleotide-binding domain 1 (25). Okiyoneda and colleagues proposed and investigated the mechanisms of action for three different classes of correctors (27). They demonstrated that corrector compound VX-809 can be potentiated by coadministration with other classes of correctors (27). Similarly, Wang and colleagues demonstrated that ΔF508-CFTR steady-state maturation efficiency doubled when corrector compound VRT-325 was combined with corrector-4a or corrector-2b (39). These results highlight the limited efficacy of a single drug approach to rescue ΔF508-CFTR function and suggest that a combination of correctors may overcome their individual modest effects. This concept led to a clinical trial evaluating the combination of VX-770 and VX-809 (NCT01225211, clinicaltrials.gov) (38).
C18 is an analog of corrector VX-809 made available by Cystic Fibrosis Foundation Therapeutics, Inc. (Bethesda, MD). C18 not only affects biosynthetic processing but may also induce conformational changes that improve the gating defect (40, 41). Thus, C18 and related compounds are excellent candidates for codelivery with another drug that rescues ΔF508-CFTR biosynthesis by a different mechanism. The observed cooperativity between pyridostigmine and C18 in ΔF508-CFTR rescue supports the potential of such a strategy. There may be further opportunities to repurpose FDA-approved drugs for this application (37, 42, 43). These findings also suggest that congeners of drugs identified with activity might be candidates for further derivatization.
As the fields of genomics and pharmacogenomics advance, the availability of gene expression data from healthy and diseased tissues and cells and of tissues and cells treated with a variety of agents continues to increase. The CMAP (10) introduced a new paradigm by which bioactive small molecules can be identified solely on the basis of the genomic signature of a disease or phenotype. CMAP has now merged with the larger NIH LINCS Program (LINCS: library of integrated network-based cellular signatures). The expanded LINCS database currently archives approximately 1,000,000 gene expression profiles, representing the responses of 20 cell lines to approximately 10,000 different interventions. These perturbations include 3,000 gain- and loss-of-function treatments, many more FDA approved drugs than previously available in CMAP, and 4,000 small molecule treatments. In addition to CMAP, Butte and colleagues developed ProfileChaser (http://profilechaser.stanford.edu) (44) and GeneChaser (http://genechaser.stanford.edu/) (45) as open access tools to mine microarray data in public repositories and query gene expression levels from test cells and tissues in response to biological conditions including disease states, drug treatments, and individuals (46, 47). These tools further enable the search for bioactive compounds with therapeutic relevance. This greatly increases the opportunities for discovering novel therapeutic agents.
Acknowledgments
Acknowledgments
The authors thank Patrick Sinn, John Engelhardt, and Lynda Ostedgaard for critically reviewing the manuscript.
Footnotes
This work was supported by National Institutes of Health grants R21 HL-91808 and RO1 HL-118000 (P.B.M.), by the Roy J. Carver Charitable Trust, and by the Cystic Fibrosis Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI:10.1165/rcmb.2014-0007OC on March 26, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown CR, Hong-Brown LQ, Biwersi J, Verkman AS, Welch WJ. Chemical chaperones correct the mutant phenotype of the delta F508 cystic fibrosis transmembrane conductance regulator protein. Cell Stress Chaperones. 1996;1:117–125. doi: 10.1379/1466-1268(1996)001<0117:ccctmp>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott CJ, Bischof JM, Unti KM, Gillen AE, Leir SH, Harris A. Nucleosome occupancy reveals regulatory elements of the CFTR promoter. Nucleic Acids Res. 2012;40:625–637. doi: 10.1093/nar/gkr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedemonte N, Lukacs GL, Du K, Caci E, Zegarra-Moran O, Galietta LJ, Verkman AS. Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J Clin Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J, Olson ER, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sondo E, Tomati V, Caci E, Esposito AI, Pfeffer U, Pedemonte N, Galietta LJ. Rescue of the mutant CFTR chloride channel by pharmacological correctors and low temperature analyzed by gene expression profiling. Am J Physiol Cell Physiol. 2011;301:C872–C885. doi: 10.1152/ajpcell.00507.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Accurso FJ, Rowe SM, Clancy JP, Boyle MP, Dunitz JM, Durie PR, Sagel SD, Hornick DB, Konstan MW, Donaldson SH, et al. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N Engl J Med. 2010;363:1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramachandran S, Karp PH, Jiang P, Ostedgaard LS, Walz AE, Fisher JT, Keshavjee S, Lennox KA, Jacobi AM, Rose SD, et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci USA. 2012;109:13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 11.Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Lee AV. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garman KS, Acharya CR, Edelman E, Grade M, Gaedcke J, Sud S, Barry W, Diehl AM, Provenzale D, Ginsburg GS, et al. A genomic approach to colon cancer risk stratification yields biologic insights into therapeutic opportunities. Proc Natl Acad Sci USA. 2008;105:19432–19437. doi: 10.1073/pnas.0806674105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andren O, Johnson LA, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, Opferman JT, Sallan SE, den Boer ML, Pieters R, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Chen MH, Lin KJ, Yang WL, Kao YW, Chen TW, Chao SC, Chang PM, Liu CY, Tzeng CH, Chao Y, et al. Gene expression-based chemical genomics identifies heat-shock protein 90 inhibitors as potential therapeutic drugs in cholangiocarcinoma. Cancer. 2013;119:293–303. doi: 10.1002/cncr.27743. [DOI] [PubMed] [Google Scholar]
- 17.Edris B, Fletcher JA, West RB, van de Rijn M, Beck AH.Comparative gene expression profiling of benign and malignant lesions reveals candidate therapeutic compounds for leiomyosarcoma. Sarcoma2012. 2012:805614 [DOI] [PMC free article] [PubMed]
- 18.Campbell JD, McDonough JE, Zeskind JE, Hackett TL, Pechkovsky DV, Brandsma CA, Suzuki M, Gosselink JV, Liu G, Alekseyev YO, et al. A gene expression signature of emphysema-related lung destruction and its reversal by the tripeptide GHK. Genome Med. 2012;4:67. doi: 10.1186/gm367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel SD, Suneja M, Ebert SM, Bongers KS, Fox DK, Malmberg SE, Alipour F, Shields RK, Adams CM. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 2011;13:627–638. doi: 10.1016/j.cmet.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb J, Lu X, Peck D, Wrobel M, Subramanian S, Blat I, Modell J, Lerner J, Liu E, Crawford E, et al. the MSigDB curation team, the Genetic Analysis Platform. The Broad Institute. Cambridge, MA: The Broad Institute of MIT and Harvard; 2006. Available from: http://www.broadinstitute.org/cmap/
- 21.Sharma M, Benharouga M, Hu W, Lukacs GL. Conformational and temperature-sensitive stability defects of the delta F508 cystic fibrosis transmembrane conductance regulator in post-endoplasmic reticulum compartments. J Biol Chem. 2001;276:8942–8950. doi: 10.1074/jbc.M009172200. [DOI] [PubMed] [Google Scholar]
- 22.Kunzelmann K, Schwiebert EM, Zeitlin PL, Kuo WL, Stanton BA, Gruenert DC. An immortalized cystic fibrosis tracheal epithelial cell line homozygous for the delta F508 CFTR mutation. Am J Respir Cell Mol Biol. 1993;8:522–529. doi: 10.1165/ajrcmb/8.5.522. [DOI] [PubMed] [Google Scholar]
- 23.Lim M, McKenzie K, Floyd AD, Kwon E, Zeitlin PL. Modulation of deltaF508 cystic fibrosis transmembrane regulator trafficking and function with 4-phenylbutyrate and flavonoids. Am J Respir Cell Mol Biol. 2004;31:351–357. doi: 10.1165/rcmb.2002-0086OC. [DOI] [PubMed] [Google Scholar]
- 24.Pedemonte N, Galietta LJ. Pharmacological correctors of mutant CFTR mistrafficking. Front Pharmacol. 2012;3:175. doi: 10.3389/fphar.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thibodeau PH, Richardson JM, III, Wang W, Millen L, Watson J, Mendoza JL, Du K, Fischman S, Senderowitz H, Lukacs GL, et al. The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem. 2010;285:35825–35835. doi: 10.1074/jbc.M110.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galietta LJ. Managing the underlying cause of cystic fibrosis: a future role for potentiators and correctors. Paediatr Drugs. 2013;15:393–402. doi: 10.1007/s40272-013-0035-3. [DOI] [PubMed] [Google Scholar]
- 27.Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, Roldan A, Verkman AS, Kurth M, Simon A, et al. Mechanism-based corrector combination restores DeltaF508-CFTR folding and function. Nat Chem Biol. 2013;9:444–454. doi: 10.1038/nchembio.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalid O, Mense M, Fischman S, Shitrit A, Bihler H, Ben-Zeev E, Schutz N, Pedemonte N, Thomas PJ, Bridges RJ, et al. Small molecule correctors of F508del-CFTR discovered by structure-based virtual screening. J Comput Aided Mol Des. 2010;24:971–991. doi: 10.1007/s10822-010-9390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutt DM, Herman D, Rodrigues AP, Noel S, Pilewski JM, Matteson J, Hoch B, Kellner W, Kelly JW, Schmidt A, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holleran JP, Glover ML, Peters KW, Bertrand CA, Watkins SC, Jarvik JW, Frizzell RA. Pharmacological rescue of mutant CFTR detected using a novel fluorescence platform. Mol Med. 2012;18:685–696. doi: 10.2119/molmed.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phuan PW, Yang B, Knapp JM, Wood AB, Lukacs GL, Kurth MJ, Verkman AS. Cyanoquinolines with independent corrector and potentiator activities restore DeltaPhe508-cystic fibrosis transmembrane conductance regulator chloride channel function in cystic fibrosis. Mol Pharmacol. 2011;80:683–693. doi: 10.1124/mol.111.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Ciciriello F, Anjos SM, Carissimo A, Liao J, Carlile GW, Balghi H, Robert R, Luini A, Hanrahan JW, et al. Ouabain mimics low temperature rescue of F508del-CFTR in cystic fibrosis epithelial cells. Front Pharmacol. 2012;3:176. doi: 10.3389/fphar.2012.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clancy JP, Rowe SM, Accurso FJ, Aitken ML, Amin RS, Ashlock MA, Ballmann M, Boyle MP, Bronsveld I, Campbell PW, et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012;67:12–18. doi: 10.1136/thoraxjnl-2011-200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moschos SA, Frick M, Taylor B, Turnpenny P, Graves H, Spink KG, Brady K, Lamb D, Collins D, Rockel TD, et al. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 38.ClinicalTrials.gov results database. Study of VX-809 alone and in combination with VX-770 in cystic fibrosis (CF) patients homozygous or heterozygous for the F508del-CFTR mutation. ClinicalTrials.gov Identifier: NCT01225211. National Library of Medicine (NLM) at the National Institutes of Health (NIH); 2010 [updated 2014 Feb 27]. Available from: http://www.clinicaltrials.gov/
- 39.Wright JM, Zeitlin PL, Cebotaru L, Guggino SE, Guggino WB. Gene expression profile analysis of 4-phenylbutyrate treatment of IB3–1 bronchial epithelial cell line demonstrates a major influence on heat-shock proteins. Physiol Genomics. 2004;16:204–211. doi: 10.1152/physiolgenomics.00160.2003. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Loo TW, Bartlett MC, Clarke DM. Additive effect of multiple pharmacological chaperones on maturation of CFTR processing mutants. Biochem J. 2007;406:257–263. doi: 10.1042/BJ20070478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He L, Kota P, Aleksandrov AA, Cui L, Jensen T, Dokholyan NV, Riordan JR. Correctors of DeltaF508 CFTR restore global conformational maturation without thermally stabilizing the mutant protein. FASEB J. 2013;27:536–545. doi: 10.1096/fj.12-216119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plewczynski D, Rychlewski L. Meta-basic estimates the size of druggable human genome. J Mol Model. 2009;15:695–699. doi: 10.1007/s00894-008-0353-5. [DOI] [PubMed] [Google Scholar]
- 44.Engreitz JM, Chen R, Morgan AA, Dudley JT, Mallelwar R, Butte AJ. ProfileChaser: searching microarray repositories based on genome-wide patterns of differential expression. Bioinformatics. 2011;27:3317–3318. doi: 10.1093/bioinformatics/btr548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen R, Mallelwar R, Thosar A, Venkatasubrahmanyam S, Butte AJ. GeneChaser: identifying all biological and clinical conditions in which genes of interest are differentially expressed. BMC Bioinformatics. 2008;9:548. doi: 10.1186/1471-2105-9-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kodama K, Horikoshi M, Toda K, Yamada S, Hara K, Irie J, Sirota M, Morgan AA, Chen R, Ohtsu H, et al. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proc Natl Acad Sci USA. 2012;109:7049–7054. doi: 10.1073/pnas.1114513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;17:93–96. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]