Abstract
Purpose
Previous studies investigating the association between EPHX1 polymorphisms (Tyr113His and His139Arg) and cancer risk have yielded inconsistent results. This meta-analysis was performed to derive a more precise estimation of relationship between two EPHX1 polymorphisms and risk of different types of cancer.
Methods
Data were extracted from relevant studies detected by a systematic literature search. Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to assess the strength of the association between EPHX1 polymorphisms and cancer risk.
Results
This meta-analysis carefully collected 99 studies on these two polymorphisms and cancer risk published up to March 2014, consisting of 45 studies (20,091 cases and 27,396 controls) for Tyr113His and 54 studies (19,437 cases and 27,289 controls) for His139Arg. The results in overall population did not show any significant association between these two polymorphisms and cancer risk for all genetic models. However, EPHX1 Tyr113His homozygote individuals have a significantly increased risk of cancer among Asians (homozygote model: OR =1.46, 95% CI=1.05–2.03; recessive model: OR =1.39, 95% CI =1.10–1.76) and mixed population (homozygote model: OR =1.17, 95% CI =1.02–1.34; recessive model: OR =1.17, 95% CI =1.02–1.33), but not Caucasians.
Conclusion
His/His genotype of EPHX1 Tyr113His polymorphism is a risk factor for developing caner for Asian and mixed population, while no evidence was found for the association between the EPHX1 His139Arg polymorphism and increased cancer risk.
Electronic supplementary material
The online version of this article (doi:10.1186/s13046-014-0082-9) contains supplementary material, which is available to authorized users.
Keywords: EPHX1 polymorphisms, Meta-analysis, Cancer risk
Background
Xenobiotic catalytic pathway is an important defense mechanism against carcinogenesis [1]. As a critical biotransformation enzyme of this pathway, microsomal epoxide hydrolase (EPHX1) plays a key role in the detoxification of potential carcinogens from endogenous compounds as well as exogenous chemicals, which ultimately convert them into less toxic metabolites [2-5].
The EPHX1 gene is located on chromosome 1q42 with 9 exons and 8 introns. Functional studies have shown that two common polymorphic sites in the gene affecting EPHX1 enzyme activity. The tyrosine to histidine substitution in exon 3 (Tyr113His, site: T337C, dbSNP: rs1051740) sharply decreases its enzyme activity by nearly 40%, whereas the histidine to arginine substitution in exon 4 (His139Arg, site: A415G, dbSNP: rs2234922) could increase the enzyme activity by approximately 25% [6]. Given the significance of EPHX1 in eliminating carcinogenicity of toxic compounds like epoxides, it could be proposed that these two functional polymorphisms may lead to individual variations of xenobiotic detoxification and further influence susceptibility to chemical carcinogen-induced cancers.
Over the past two decades, a number of studies have been conducted to investigate the relationship between EPHX1 polymorphisms and cancer in different populations. However, the results of these studies are conflicting rather than conclusive. Several previous meta-analyses were flawed in their lack of sufficient data or there were methodological problems. One meta-analysis by Li et al. found that no significant association between EPHX1 polymorphisms (Tyr113His and His139Arg) and increased risk of cancers [7]. However, several studies [8-10] included in this meta-analysis were incorrectly classified according to source of controls, which may lead to an inaccurate result. Some recent studies did not evaluate the deviations from Hardy–Weinberg equilibrium (HWE) in control subjects [11-13], which could bias the estimates of genetic effects in genetic association studies and meta-analysis [14]. Since that date, several more studies have emerged to assess the relationship between the Tyr113His and/or His139Arg polymorphisms of the EPHX1 gene and susceptibility to a variety of cancers. Given the new information, we systematically evaluated the effect of these two polymorphisms on cancer risk in an updated meta-analysis with increased statistical power in order to get a more precise and reliable assessment of the association.
Materials and methods
Search strategy
A comprehensive literature search was performed using PubMed database for relevant articles published (last search: March 14, 2014) with the following terms: ((“epoxide hydrolase 1”) OR EPHX1) AND (((polymorphism) OR (SNP)) OR variant)) AND ((((neoplasm) OR cancer) OR carcinoma) OR leukemia). All the references of retrieved articles and supplementary data were checked when key information relevant to the meta-analysis was missing.
Inclusion criteria
All studies were included if they met the following criteria: (1) case–control study; (2) studies to evaluate the association between EPHX1 gene polymorphisms (Tyr113His and His139Arg) and risk of cancer; (3) sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI); (4) full-text in English available and (5) more than 100 patients. When the same population was included in several publications, only the most complete one was included in this meta-analysis.
Data extraction
Data were carefully evaluated and extracted from the eligible studies by two investigators independently according to the inclusion criteria listed above. The following characters were collected from eligible studies: first author’s name, year of publication, ethnicity (categorized as Asian, Caucasian, African, or mixed), source of control groups (population-based [PB], hospital-based [HB], family-based [FB] or unknown), genotype frequency of cases and controls, and the results of Hardy-Weinberg equilibrium (HWE) test. When it came to discrepancy between two investigators, another investigator was invited to discuss and check the data until a consensus was reached.
Statistical analysis
The departure from the Hardy-Weinberg equilibrium for the control group in each study was assessed with Pearson's goodness-of-fit Chi-square test with 1 degree of freedom by a web-based program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) and the violation of HWE was determined with a threshold of p < 0.05. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of association between the EPHX1 gene polymorphisms and cancer susceptibility. Pooled ORs were performed for dominant model (aa + Aa vs. AA, a was for the minor allele and A was for the major allele), recessive model (aa vs. Aa + AA), homozygote comparison (aa vs. AA), heterozygote comparison (Aa vs. AA), and additive model (a vs. A), respectively. Heterogeneity among pooled studies were evaluated by the Chi-square-based Cochran’s Q test [15] and I2 statistics [16]. To be more conservative, heterogeneity was considered to be present when the Cochran’s Q-test P-value was less than 0.1, then random-effects model (the DerSimonian and Laird method) [17] was utilized, otherwise, fixed-effects model was used (the Mantel-Haenszel method) [18]. In addition, inconsistency across studies was quantified by means of I2 statistic, with I2 < 25%, 25-75%, and >75% considered to represent low, moderate and high degree of heterogeneity, respectively [16]. Stratification analyses were performed to test the effects of cancer types, source of control, ethnicity and smoking status, respectively. To explore the source of heterogeneity among the studies of this meta-analysis, a multivariate meta-regression analysis subjected to 10,000 permutations was undertaken to explore the possible sources of heterogeneity. The following study characteristics were included as covariates in the meta-regression analysis: ethnicity, source of control, cancer types. Sensitivity analysis was carried out through omitting individual study in turn to check the consistency of the results. Publication bias was evaluated by visual inspection of the Begg’s funnel plots [19] and the Egger’s linear regression (P < 0.05 was considered a significant publication bias) [20]. All statistical tests were performed with metafor [21] and meta (http://cran.r-project.org/web/packages/meta/) packages of R (version 3.0.1), using two-sided p-values.
This meta-analysis followed the guidelines of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement [22] (Additional file 1: Table S1).
Results
Study characteristics
The initial literature search through PubMed database yielded 192 published articles. Totally, when reviewed in full-text, 4 were not concerned with Tyr113His or His139Arg polymorphisms in EPHX1 gene, 37 were not cancer risk studies, 1 was not published in English, 1 was not provided in full text, 7 were not case–control studies, 30 were no usable reported data, and 19 were meta-analysis or reviews; all these publications were excluded. Among the remaining 87 articles, studies presented separate OR by different polymorphisms, cancer types or ethnicity, and each of them was considered separately for pooling analysis. Furthermore, 32 studies not in HWE and 39 with less 100 patients were also deleted. Hence, 45 studies [8-10,23-63] for Tyr113His polymorphism (20,091 cases and 27,396 controls) and 54 studies [8-10,24-26,28,29,32,34-45,47-51,53,55-61,63-81] for His139Arg polymorphism (19,437 cases and 27,289 controls) were included eventually. Genotype distributions in the controls of all selected studies are in agreement with HWE. The flow of study selection was shown in Figure 1, and the main characteristics of eligible studies were summarized (Additional file 2: Table S2 and Additional file 3: Table S3).
Figure 1.

The flow diagram of the literature search and the study selection.
Quantitative synthesis
The pooled results of meta-analysis for the association between EPHX1 polymorphisms (Tyr113His and His139Arg) and cancer susceptibility are shown in Tables 1 and 2. Heterogeneity across studies must be considered because it may affect the strengths of the meta-analysis. Significant heterogeneity was observed in some comparisons for both EPHX1 Tyr113His and His139Arg polymorphisms. Thus, random-effect model was used when heterogeneity identified.
Table 1.
Overall and stratified meta-analyses of the association between the EPHX1 Tyr113His polymorphism and cancer risk
| Variables | No a | Case/control | Homozygote comparison | Heterozygote comparison | Dominant model | Recessive model | Additive model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | |||
| Total | 45 | 20091/27396 | 1.05(0.95 ~ 1.16) | 0.37 | 0.00(50.8) | 0.94(0.88 ~ 1.01) | 0.08 | 0.0(54.0) | 0.96(0.90 ~ 1.03) | 0.26 | 0.0(56.8) | 1.08(0.99 ~ 1.18) | 0.08 | 0.0(37.4) | 1.00(0.95 ~ 1.05) | 0.94 | 0.0(57.7) |
| Cancer type | |||||||||||||||||
| Blood | 7 | 2419/2319 | 1.05(0.76 ~ 1.46) | 0.76 | 0.03(56.1) | 0.89(0.73 ~ 1.01) | 0.24 | 0.06(49.8) | 0.92(0.75 ~ 1.13) | 0.42 | 0.02(60.3) | 1.17(0.96 ~ 1.41) | 0.12 | 0.14(37.3) | 0.97(0.82 ~ 1.15) | 0.76 | 0.01(65.9) |
| Prostate | 3 | 1706/1192 | 1.54(0.57 ~ 4.16) | 0.86 | 0.00(88.9) | 1.31(0.82 ~ 2.01) | 0.26 | 0.00(79.7) | 1.39(0.80 ~ 2.41) | 0.24 | 0.00(87.1) | 1.29(0.62 ~ 2.65) | 0.50 | 0.0(82.2) | 1.27(0.81 ~ 1.97) | 0.30 | 0.00(89.7) |
| Esophageal | 3 | 593/1086 | 1.09(0.78 ~ 1.51) | 0.63 | 0.19(40.5) | 1.01(0.52 ~ 1.93) | 0.99 | 0.00(86.0) | 1.08(0.63 ~ 1.84) | 0.79 | 0.00(81.6) | 1.16(0.85 ~ 1.58) | 0.34 | 0.20(38.4) | 1.09(0.84 ~ 1.41) | 0.51 | 0.07(61.9) |
| Colorectal | 9 | 5512/6787 | 0.98(0.86 ~ 1.11) | 0.75 | 0.85(0.0) | 1.00(0.93 ~ 1.08) | 0.98 | 0.79(0.0) | 0.99(0.93 ~ 1.07) | 0.98 | 0.80(0.0) | 0.98(0.87 ~ 1.11) | 0.76 | 0.87(0.0) | 1.00(0.94 ~ 1.05) | 0.87 | 0.84(0.0) |
| Other | 7 | 2425/2872 | 1.20(0.90 ~ 1.59) | 0.22 | 0.06(50.4) | 1.00(0.81 ~ 1.24) | 0.99 | 0.01(66.7) | 1.04(0.84 ~ 1.29) | 0.74 | 0.00(69.4) | 1.18(0.98 ~ 1.42) | 0.08 | 0.40(2.8) | 1.06(0.91 ~ 1.23) | 0.45 | 0.01(65.5) |
| Lung | 9 | 2065/5429 | 0.80(0.57 ~ 1.12) | 0.20 | 0.00(62.5) | 0.80(0.65 ~ 0.98) | 0.03 | 0.00(65.0) | 0.81(0.68 ~ 0.98) | 0.03 | 0.01(62.5) | 0.91(0.66 ~ 1.25) | 0.57 | 0.01(61.7) | 0.87(0.75 ~ 1.01) | 0.08 | 0.0(65.5) |
| Head and neck | 3 | 825/821 | 1.04(0.74 ~ 1.47) | 0.81 | 0.98(0.0) | 0.87(0.71 ~ 1.07) | 0.19 | 0.82(0.0) | 0.90(0.74 ~ 1.10) | 0.30 | 0.85(0.0) | 1.11(0.80 ~ 1.54) | 0.53 | 0.99(0.0) | 0.96(0.83 ~ 1.12) | 0.61 | 0.91(0.0) |
| Breast cancer | 4 | 4546/6890 | 1.02(0.90 ~ 1.17) | 0.73 | 0.15(43.0) | 0.99(0.91 ~ 1.07) | 0.73 | 0.60(0.0) | 0.99(0.92 ~ 1.07) | 0.35 | 0.99(0.0) | 1.14(0.89 ~ 1.46) | 0.31 | 0.05(62.6) | 1.00(0.97 ~ 1.03) | 0.93 | 0.68(0.0) |
| Source of control | |||||||||||||||||
| PB | 31 | 16035/23111 | 1.03(0.93 ~ 1.14) | 0.54 | 0.06(29.7) | 0.97(0.92 ~ 1.03) | 0.28 | 0.10(25.1) | 0.98(0.93 ~ 1.04) | 0.45 | 0.06(30.4) | 1.03(0.96 ~ 1.11) | 0.38 | 0.14(21.9) | 1.00(0.95 ~ 1.04) | 0.91 | 0.03(34.7) |
| HB | 11 | 3517/3627 | 0.94(0.77 ~ 1.15) | 0.56 | 0.08(40.9) | 0.81(0.69 ~ 0.96) | 0.01 | 0.00(59.4) | 0.85(0.74 ~ 0.98) | 0.03 | 0.03(49.4) | 1.05(0.86 ~ 1.27) | 0.66 | 0.05(45.7) | 0.93(0.84 ~ 1.03) | 0.16 | 0.05(45.3) |
| Ethnicity | |||||||||||||||||
| Caucasian | 26 | 11757/18447 | 0.94(0.87 ~ 1.03) | 0.17 | 0.29(11.7) | 0.93(0.87 ~ 1.00) | 0.04 | 0.09(28.7) | 0.93(0.87 ~ 1.00) | 0.04 | 0.06(31.7) | 0.97(0.89 ~ 1.05) | 0.45 | 0.49(0.0) | 0.96(0.91 ~ 1.00) | 0.07 | 0.08(29.6) |
| Mixed | 7 | 5645/5502 | 1.17(1.02 ~ 1.34) | 0.03 | 0.42(0.7) | 1.00(0.93 ~ 1.09) | 0.96 | 0.57(0.0) | 1.03(0.96 ~ 1.11) | 0.44 | 0.53(0.0) | 1.17(1.02 ~ 1.33) | 0.02 | 0.44(0.0) | 1.05(0.99 ~ 1.11) | 0.11 | 0.43(0.0) |
| Asian | 11 | 2534/3205 | 1.46(1.05 ~ 2.03) | 0.04 | 0.00(75.1) | 1.04(0.77 ~ 1.40) | 0.81 | 0.00(81.7) | 1.16(0.88 ~ 1.53) | 0.30 | 0.00(56.8) | 1.39(1.10 ~ 1.76) | 0.01 | 0.01(60.7) | 1.19(0.99 ~ 1.42) | 0.05 | 0.0(79.4) |
| Smoking status | |||||||||||||||||
| Smoker | 9 | 1786/2114 | 0.99(0.80 ~ 1.23) | 0.95 | 0.42(1.4) | 0.85(0.66 ~ 1.10) | 0.21 | 0.03(52.1) | 0.89(0.71-1.11) | 0.30 | 0.05(47.8) | 1.07(0.87 ~ 1.31) | 0.56 | 0.43(0.5) | 0.97(0.88 ~ 1.08) | 0.60 | 0.13(36.7) |
| Non-smoker | 8 | 1357/1947 | 1.45(0.89 ~ 2.37) | 0.14 | 0.00(71.4) | 1.19(0.88 ~ 1.60) | 0.27 | 0.01(63.6) | 1.27(0.92 ~ 1.74) | 0.14 | 0.00(71.7) | 1.29(0.92~1.82) | 0.14 | 0.05(50.0) | 1.23(0.97 ~ 1.57) | 0.09 | 0.00(75.8) |
PB: population based; HB: hospital based. P b: P-values for heterogeneity from Q test; I2 refers to the proportion of total variation owing to between-study heterogeneity.
Bold font marks where fixed effect model used.
Table 2.
Overall and stratified meta-analyses of the association between the EPHX1 His139Arg polymorphism and cancer risk
| Variables | No a | Case/control | Homozygote comparison | Heterozygote comparison | Dominant model | Recessive model | Additive model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | OR (95% CI) | P | P b (I 2 ) | |||
| Total | 54 | 19437/27289 | 1.05(0.93 ~ 1.18) | 0.73 | 0.09(21.3) | 0.96(0.91 ~ 1.01) | 0.15 | 0.02(31.5) | 0.97(0.92 ~ 1.03) | 0.30 | 0.00(40.1) | 1.02(0.93 ~ 1.13) | 0.66 | 0.23(12.0) | 0.99(0.94 ~ 1.04) | 0.57 | 0.00(44.9) |
| Cancer type | |||||||||||||||||
| Other | 11 | 3160/4581 | 0.94(0.75 ~ 1.20) | 0.64 | 0.53(0.0) | 0.97(0.87 ~ 1.10) | 0.51 | 0.56(0.0) | 0.97(0.87 ~ 1.06) | 0.47 | 0.53(0.0) | 0.96(0.76 ~ 1.21) | 0.71 | 0.53(0.0) | 0.97(0.89 ~ 1.05) | 0.50 | 0.46(0.0) |
| Blood | 9 | 2929/3629 | 0.87(0.67 ~ 1.13) | 0.29 | 0.90(0.0) | 0.90(0.81 ~ 1.00) | 0.05 | 0.25(21.4) | 0.90(0.81 ~ 0.99) | 0.04 | 0.47(0.0) | 0.90(0.70 ~ 1.17) | 0.44 | 0.82(0.0) | 0.91(0.83 ~ 0.99) | 0.04 | 0.80(0.0) |
| Esophageal | 3 | 593/1081 | 1.30(0.50 ~ 3.38) | 0.59 | 0.07(61.8) | 1.02(0.65 ~ 1.62) | 0.92 | 0.03(72.6) | 1.06(0.66 ~ 1.73) | 0.80 | 0.01(77.3) | 1.30(0.54 ~ 3.13) | 0.56 | 0.10(56.5) | 1.10(0.71 ~ 1.69) | 0.67 | 0.01(79.2) |
| Colorectal | 10 | 5552/7089 | 1.14(0.85 ~ 1.53) | 0.39 | 0.02(53.5) | 0.92(0.85 ~ 0.99) | 0.03 | 0.97(0.0) | 0.94(0.87 ~ 1.01) | 0.09 | 0.73(0.0) | 1.17(0.87 ~ 1.58) | 0.29 | 0.02(54.0) | 0.97(0.91 ~ 1.03) | 0.28 | 0.11(36.8) |
| Lung | 14 | 4767/8411 | 1.14(0.83 ~ 1.56) | 0.42 | 0.02(49.3) | 1.10(0.94 ~ 1.27) | 0.24 | 0.00(63.4) | 1.10(0.93 ~ 1.29) | 0.27 | 0.00(71.9) | 0.97(0.80 ~ 1.17) | 0.76 | 0.16(27.2) | 1.07(0.93 ~ 1.24) | 0.36 | 0.00(74.1) |
| Head and neck | 4 | 1035/1075 | 1.28(0.81 ~ 2.02) | 0.30 | 0.99(0.0) | 0.84(0.70 ~ 1.01) | 0.06 | 0.69(0.0) | 0.88(0.73 ~ 1.05) | 0.14 | 0.70(0.0) | 1.35(0.85 ~ 2.13) | 0.20 | 1.0(0.0) | 0.94(0.81 ~ 1.01) | 0.41 | 0.78(0.0) |
| Breast cancer | 3 | 1401/1423 | 1.04(0.69 ~ 1.59) | 0.85 | 0.49(0.0) | 0.91(0.77 ~ 1.07) | 0.04 | 0.66(0.0) | 0.92(0.79 ~ 1.08) | 0.30 | 0.67(0.0) | 1.09(0.93 ~ 1.13) | 0.70 | 0.49(0.0) | 0.95(0.83 ~ 1.09) | 0.44 | 0.66(0.0) |
| Source of control | |||||||||||||||||
| HB | 16 | 5010/5638 | 1.14(0.93 ~ 1.41) | 0.21 | 0.99(0.0) | 0.96(0.88 ~ 1.05) | 0.37 | 0.20(22.7) | 0.98(0.90 ~ 1.06) | 0.63 | 0.32(11.4) | 1.17(0.95 ~ 1.44) | 0.15 | 0.99(0.0) | 1.00(0.94 ~ 1.08) | 0.94 | 0.66(0.0) |
| PB | 36 | 14191/21351 | 1.04(0.89 ~ 1.22) | 0.61 | 0.01(39.4) | 0.96(0.90 ~ 1.03) | 0.22 | 0.01(39.4) | 0.97(0.90 ~ 1.04) | 0.40 | 0.00(50.7) | 1.04(0.90 ~ 1.21) | 0.56 | 0.05(30.0) | 0.99(0.92 ~ 1.05) | 0.64 | 0.00(57.2) |
| Ethnicity | |||||||||||||||||
| Asian | 13 | 2957/3770 | 0.97(0.75 ~ 1.27) | 0.84 | 0.51(0.0) | 0.99(0.89 ~ 1.11) | 0.90 | 0.26(18.6) | 0.99(0.89 ~ 1.10) | 0.88 | 0.14(30.6) | 0.98(0.76 ~ 1.28) | 0.90 | 0.59(0.0) | 1.01(0.90 ~ 1.13) | 0.90 | 0.09(37.1) |
| Caucasian | 32 | 12294/17831 | 1.10(0.93 ~ 1.30) | 0.26 | 0.03(34.8) | 0.95(0.88 ~ 1.02) | 0.18 | 0.01(44.0) | 0.97(0.89 ~ 1.04) | 0.39 | 0.00(52.8) | 1.07(0.95 ~ 1.21) | 0.27 | 0.11(24.4) | 0.99(0.92 ~ 1.06) | 0.71 | 0.00(57.3) |
| Mixed | 7 | 3921/5262 | 0.90(0.73 ~ 1.12) | 0.35 | 0.81(0.0) | 0.94(0.86 ~ 1.04) | 0.22 | 0.43(0.0) | 0.94(0.86 ~ 1.03) | 0.16 | 0.69(0.0) | 0.92(0.74 ~ 1.14) | 0.45 | 0.71(0.0) | 0.95(0.88 ~ 1.02) | 0.15 | 0.93(0.0) |
| African | 2 | 265/426 | 0.81(0.22 ~ 2.96) | 0.75 | 0.08(21.3) | 1.09(0.79 ~ 1.50) | 0.60 | 0.78(0.0) | 1.06(0.78 ~ 1.45) | 0.69 | 0.46(0.0) | 0.78(0.23 ~ 2.73) | 0.70 | 0.08(66.6) | 1.03(0.94 ~ 1.01) | 0.84 | 0.22(34.7) |
| Smoking status | |||||||||||||||||
| Smoker | 9 | 2331/2542 | 1.08(0.64 ~ 1.83) | 0.77 | 0.01(61.5) | 1.19(0.88 ~ 1.60) | 0.27 | 0.00(76.6) | 1.16(0.85 ~ 1.58) | 0.35 | 0.00(79.9) | 1.04(0.68~1.60) | 0.86 | 0.06(46.8) | 1.09(0.84 ~ 1.41) | 0.52 | 0.00(80.1) |
| Non-smoker | 8 | 1498/2336 | 1.25(0.90 ~ 1.75) | 0.19 | 0.67(0.0) | 1.05(0.91 ~ 1.21) | 0.49 | 0.78(0.0) | 1.08(0.94 ~ 1.23) | 0.30 | 0.71(0.0) | 1.24(0.89 ~ 1.72) | 0.21 | 0.71(0.0) | 1.08(0.96 ~ 1.22) | 0.19 | 0.60(0.0) |
PB: population based; HB: hospital based. P b: P-values for heterogeneity from Q test; I2 refers to the proportion of total variation owing to between-study heterogeneity.
Bold font marks where fixed effect model used.
For Tyr113His polymorphism, overall, no significantly elevated cancer risk could be observed in all genetic models (Table 1. homozygote model: OR = 1.05, 95% CI = 0.95–1.16; heterozygote model: OR = 0.94, 95% CI = 0.88–1.01; additive model: OR = 1.00, 95% CI = 0.95–1.05; dominant model: OR = 0.96, 95% CI = 0.90–1.03, Figure 2; recessive model: OR = 1.08, 95% CI = 0.99–1.18). When stratified by ethnicity, the significantly increased cancer risks were found among Asian population (homozygote model: OR = 1.46, 95% CI = 1.05–2.03; recessive model: OR = 1.39, 95% CI = 1.10–1.76) and Mixed population (homozygote model: OR = 1.17, 95% CI = 1.02–1.34; recessive model: OR = 1.17, 95% CI = 1.02–1.33). Stratified analyses by cancer types, smoking status and source of controls indicated no evidence of significant association between Tyr113His polymorphism and the cancer risk. Furthermore, individuals carrying Tyr/His or His/His genotype have a significantly reduced risk of lung cancer (heterozygote model: OR = 0.80, 95% CI = 0.65-0.98; dominant model: OR = 0.81, 95% CI = 0.68–0.98).
Figure 2.
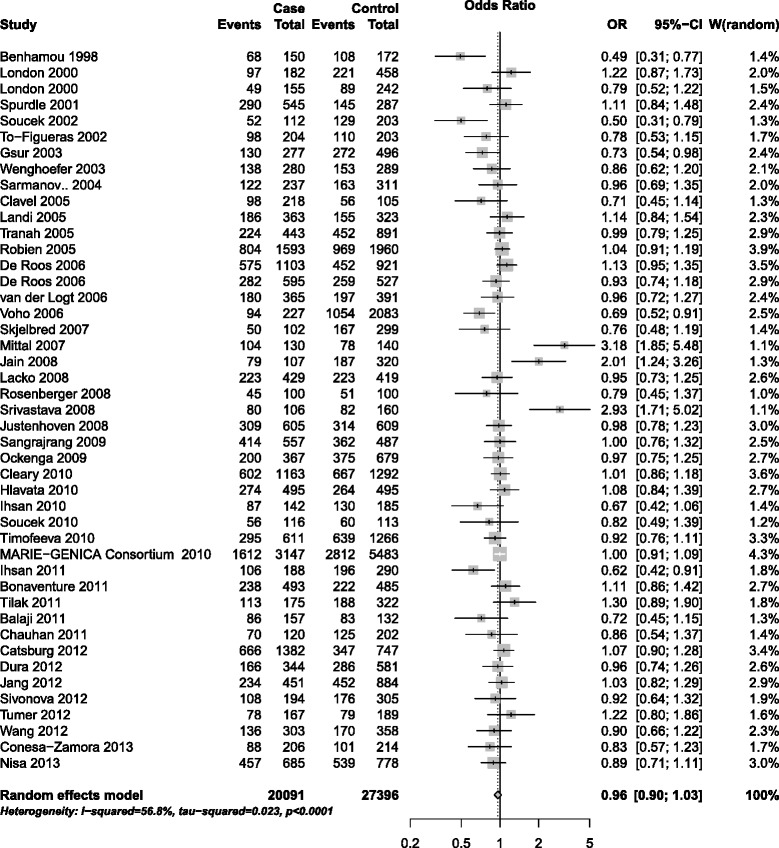
Forest plot for association of EPHX1 Tyr113His polymorphism and cancer risk (dominant model, CT + CC vs. TT).
With respect to His139Arg polymorphism, similarly, the combined results did not show any association with the elevated risk of cancer for all genetic models (homozygote model: OR = 1.05, 95% CI = 0.93–1.18; heterozygote model: OR = 0.96; 95% CI = 0.91–1.01; additive model: OR = 0.99, 95% CI = 0.94–1.04; dominant model: OR = 0.97, 95% CI = 0.92–1.03, Figure 3; recessive model: OR = 1.02, 95% CI = 0.93–1.13). When stratified according to cancer types, no significant association with increased cancer risk was demonstrated in all subgroups for overall population. However, the result suggested a decreased risk for blood cancers (additive model: OR = 0.91, 95% CI = 0.83–0.99; dominant model: OR = 0.90, 95% CI = 0.81–0.99) and colorectal cancer (heterozygote model: OR = 0.92; 95% CI = 0.85–0.99). In the subgroup analysis by source of controls, smoking status and ethnicity, no significant association with cancer risk was observed in all subgroups (Table 2).
Figure 3.
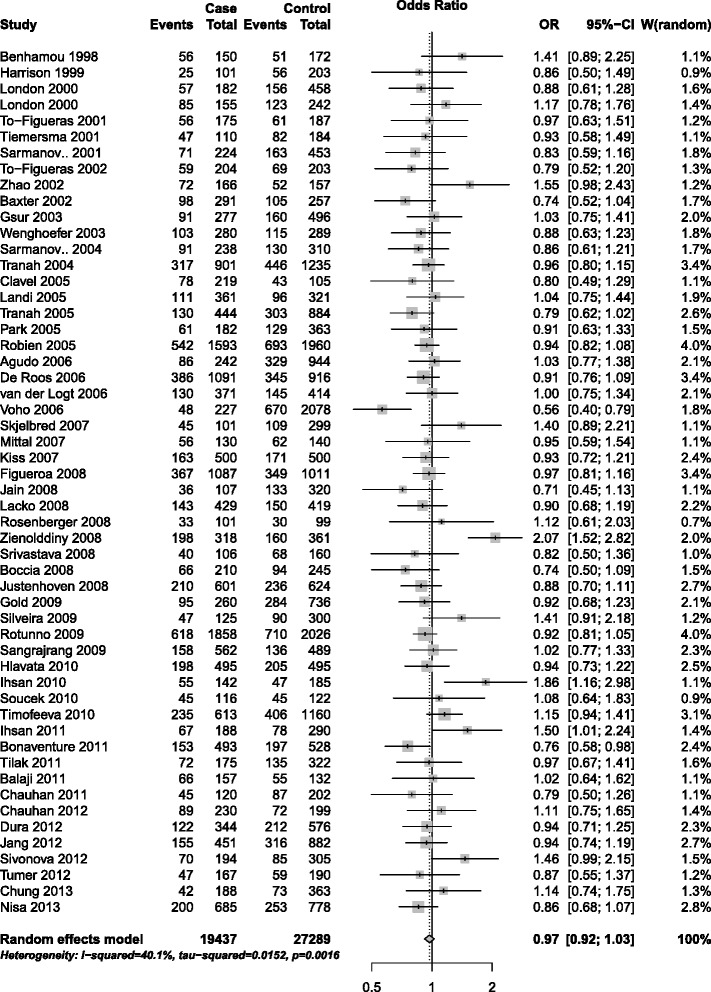
Forest plot for association of EPHX1 His139Arg polymorphism and cancer risk (dominant model, GA + GG vs. AA).
Meta-regression and sensitivity analyses
Heterogeneity is a potential issue that may affect the interpretation of the results. As for Tyr113His polymorphism, the heterogeneity was observed in all genetic models and the detailed data are shown in Table 1. With respect to His139Arg polymorphism (Table 2), the heterogeneity was detected in homozygote comparison, heterozygote model, dominant model and additive model. We therefore explored the source of heterogeneity by cancer type, ethnicity, and source of control by meta-regression in all comparisons with significant heterogeneity. As a result, for Tyr113His polymorphism, source of control may be the major source of heterogeneity in homozygote model (P = 0.024), heterozygote model (P = 0.000), dominant model (P = 0.002) and additive model (P = 0.005), but not recessive model. However, for His139Arg polymorphism, none of these variables showed statistically significant associations in multivariate meta-regression model (P > 0.05), suggesting factors mentioned above could not explain the heterogeneity among studies.
The influences of each individual study on the overall ORs for Tyr113His/His139Arg polymorphisms were evaluated. The results showed the pooled ORs of these two polymorphisms were not materially altered by the omission of any individual study, suggesting credibility for the conclusions (Additional file 4: Figure S1 and Additional file 5: Figure S2).
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures. The shape of funnel plots (Figures 4 and 5) did not reveal any evidence of asymmetry. The statistical results of Egger’s test still did not show publication bias for Tyr113His polymorphism (additive model: P = 0.146; homozygote comparison: P = 0.620; heterozygote model: P = 0.189; dominant model: P = 0.054; recessive model: P = 0.915) and His139Arg polymorphism (additive model: P = 0.125; homozygote comparison: P = 0.847; heterozygote model: P = 0.255; dominant model: P = 0.111; recessive model: P = 0.153).
Figure 4.
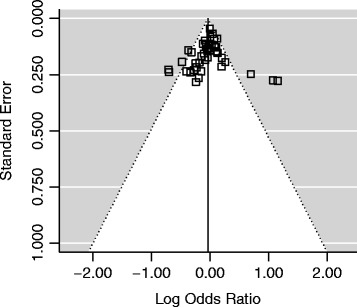
Begg’s funnel plot for publication bias in studies on EPHX1 Tyr113His polymorphism and cancer (dominant model, CT + CC vs. TT).
Figure 5.
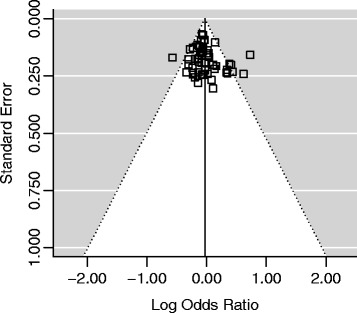
Begg’s funnel plot for publication bias in studies on EPHX1 His139Arg polymorphism and cancer (dominant model, GA + GG vs. AA).
Discussion
With increased knowledge of human gene functions and the architecture of genetic variations, it has become clear that individual variation in genetic backgrounds, such as single nucleotide polymorphism, could substantially influence cancer risk with specific environmental exposure. However, evidences from studies of genetic epidemiology were usually too conflicting to draw conclusions. Meta-analysis shed light on objective and comprehensive assessment of the associations between polymorphisms and cancer risks. Several single nucleotide polymorphisms were identified as cancer risk factors for specific populations by means of meta-analysis [82-84].
The genetic polymorphisms of EPHX1, Tyr113His and His139Arg, may affect enzyme activity involved in general oxidative defenses against a number of environmental substances [6]. Variations in the expression and activity level of EPHX1 as a result of such polymorphisms could cause individual variations of detoxifying capability, then further influence the risk of chemical carcinogen-induced cancers [85]. The findings from some previous studies suggested that genetic polymorphism in EPHX1 has important roles in the development of cancers [55,86-88]. However, others reported no association of EPHX1 polymorphisms with risk of cancers [24,26,34,45]. This inconsistency may be due to tremendous difference in sample size, diverse ethnic background, sampling bias, publication bias, or inadequate statistical power. The benefits of meta-analysis include a larger number of participants, different geographic locations, and the possibility of inclusion of a wider range of population groups, all of which could derive a more precise estimation and further increase the generalizability of the results.
In this meta-analysis, 99 eligible case–control studies including 39,528 cases and 54,685 controls were included to provide a comprehensive assessment of the relationship between EPHX1 polymorphisms and cancer risk. The results revealed that neither EPHX1 Tyr113His polymorphism, nor His139Arg polymorphism have significant association with the cancer susceptibility for all comparing models when all studies were accumulated together. Further stratified analysis according to cancer types, smoking status, or source of controls did not suggest a significantly increased risk. Moreover, His139Arg polymorphism might play a potentially protective role in the development of blood cancer based on dominant model and additive model, and colorectal cancer by heterozygous model. Meanwhile Tyr113His polymorphism showed possible protective effect on risk for lung cancer by heterozygote model and dominant model.
Stratified analysis by ethnicity allowed for assessing the ethnic differences in the association of cancer risk. As for His139Arg polymorphism, no significant associations were found in any genetic model among all populations. However, with respect to the Tyr113His polymorphism, an increased risk of cancer based on homozygote and recessive model could be observed in Asian and mixed population, indicating there is an obvious race-specific effect in the association. It was consistent with the results from two recent meta-analyses for hepatocellular cancer [89] and lung cancer [90]. Further subgroup analysis by the cancer types in Asian and Mixed population was not performed due to the limited data for individual cancer type according to our inclusion criteria.
Heterogeneity between studies should be noted because it may potentially affect the strengths of the meta-analysis. In the current meta-analysis, significance heterogeneity was observed for both EPHX1 Tyr113His and His139Arg polymorphisms. Thus, random-effect models were used if significant heterogeneity was identified. Furthermore, multivariate meta-regression analysis involving covariates, such as source of control, cancer type, ethnicity, was performed to explore the source of heterogeneity. The results from meta-regression emphasized that the heterogeneity of polymorphism Tyr113His was associated with source of control in homozygote model, heterozygote model, dominant model and additive model, but not recessive model. Neither cancer type, nor ethnicity was found to be the source of heterogeneity. However, as for His139Arg, results indicated none of these three covariates could be the main source of the between-study heterogeneity. It suggested that some other confounding factors, such as environmental exposures, gene-gene interaction, and lifestyle might lead to the heterogeneity. Large studies for both polymorphisms with comprehensive classification information are needed to facilitate the subgroup analysis according to these factors, which is unavailable for present meta-analysis because of inadequate information from our original data sources.
Although our result is suggestive, there are still some limitations inherited from the published studies and our analysis strategies. First, the present conclusion was drawn based on unadjusted estimates, while a more precise analysis with the necessary adjustment by other covariates including age, lifestyle, gene–gene interactions and environmental factors should be conducted when more detailed individual data were available. Second, although some results were significant, the p–values were on the borderline, i.e. slightly less than 0.05. Further large and well-designed studies are required for confirmation. Finally, most studies were from Caucasian population, it is critical that larger and well-designed multi-centric studies based on Asians and other racial-ethnic groups should be performed to re-evaluate the association. In spite of these, our meta-analysis also had some advantages. First, the quality of case–control studies included in current meta-analysis was satisfactory and met our inclusion criterion, ensuring the quality of our results. Second, the sensitivity analysis showed that no individual study materially altered the pooled ORs indicating statistical stableness and robustness of the current meta-analysis. In addition, no publication bias for the association between these two polymorphisms and cancer risk could be observed, which further confirmed the credibility.
In conclusion, our investigations suggested that the EPHX1 His139Arg polymorphism might not contribute to the susceptibility of all cancer types for overall population, whereas Tyr113His polymorphism might be associated with increased risk of cancer in the Asian and mixed population. Larger well-designed epidemiological studies with different cancer types, ethnically diverse populations and functional evaluations are warranted to confirm our findings.
Acknowledgments
This work was supported by a Natural Science Foundation of Guangdong Province, China(S2011010004147) and a doctoral startup fund for scientific research at Guangzhou Medical University (2011C06). We thank Lei Deng from the Department of Bioinformatics of Tongji University and Dr. Jie Gu from Department of Biology, Hong Kong Baptist University, for their sincerely assistance in literature searching. We also appreciate Dr. Mian Li and Dr. Chenglin Li from the Institute for Nutritional Sciences, Chinese Academy of Sciences, Shanghai, for their helpful suggestions.
Abbreviations
- HWE
Hardy–Weinberg equilibrium
- EPHX1
Epoxide hydrolase 1
- SNP
Single nucleotide polymorphism
- OR
Odds ratio
- CI
Confidence interval
Additional files
PRISMA Checklist for this meta-analysis.
Principal characteristics of the studies included on EPHX1 Tyr113His polymorphism.
Principal characteristics of the studies included on EPHX1 His139Arg polymorphism.
Sensitivity analysis of the summary OR of the association between EPHX1 Tyr113His polymorphism and cancer susceptibility in dominant model. Results were computed by omitting each study in turn. Random-effects model was used. The two ends of the dotted lines represent the 95% confidence interval.
Sensitivity analysis of the summary OR of the association between EPHX1 His139Arg polymorphism and cancer susceptibility in dominant model. Results were computed by omitting each study in turn. Random-effects model was used. The two ends of the dotted lines represent the 95% confidence interval.
Footnotes
Competing interests
The authors have declared that no competing interests exist.
Authors’ contributions
YXQ and WGP conceived and designed the study, YXQ, WYB and WGP participated in selecting study, extracting data and performing the statistical analysis. YXQ and WGP were involved in drafting and revising the manuscript. All authors read and approved the final manuscript.
Contributor Information
Xiaoqin Yang, Email: sirxqyang@gmail.com.
Yubing Wang, Email: 2.08bing@163.com.
Guiping Wang, Email: docgpwang@163.com.
References
- 1.Gonzalez FJ. The role of carcinogen-metabolizing enzyme polymorphisms in cancer susceptibility. Reprod Toxicol. 1997;11:397–412. doi: 10.1016/S0890-6238(96)00154-2. [DOI] [PubMed] [Google Scholar]
- 2.Decker M, Arand M, Cronin A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch Toxicol. 2009;83:297–318. doi: 10.1007/s00204-009-0416-0. [DOI] [PubMed] [Google Scholar]
- 3.Oesch F. Mammalian epoxide hydrases: inducible enzymes catalysing the inactivation of carcinogenic and cytotoxic metabolites derived from aromatic and olefinic compounds. Xenobiotica. 1973;3:305–340. doi: 10.3109/00498257309151525. [DOI] [PubMed] [Google Scholar]
- 4.Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 5.Arand M, Cronin A, Adamska M, Oesch F. Epoxide hydrolases: structure, function, mechanism, and assay. Methods Enzymol. 2005;400:569–588. doi: 10.1016/S0076-6879(05)00032-7. [DOI] [PubMed] [Google Scholar]
- 6.Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet. 1994;3:421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Hu Z, Qu X, Zhu J, Li L, Ring BZ, Su L. Putative EPHX1 enzyme activity is related with risk of lung and upper aerodigestive tract cancers: a comprehensive meta-analysis. PloS one. 2011;6:e14749. doi: 10.1371/journal.pone.0014749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landi S, Gemignani F, Moreno V, Gioia-Patricola L, Chabrier A, Guino E, Navarro M, de Oca J, Capella G, Canzian F. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenetics Genom. 2005;15:535–546. doi: 10.1097/01.fpc.0000165904.48994.3d. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava DS, Mandhani A, Mittal RD. Genetic polymorphisms of cytochrome P450 CYP1A1 (*2A) and microsomal epoxide hydrolase gene, interactions with tobacco-users, and susceptibility to bladder cancer: a study from North India. Arch Toxicol. 2008;82:633–639. doi: 10.1007/s00204-007-0276-4. [DOI] [PubMed] [Google Scholar]
- 10.Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Laird NM, Khuhaprema T, Brennan P, Boffetta P, Yoshida T. Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. Int J Cancer. 2009;125:837–843. doi: 10.1002/ijc.24434. [DOI] [PubMed] [Google Scholar]
- 11.Tan X, He WW, Wang YY, Shi LJ, Chen MW. EPHX1 Tyr113His and His139Arg polymorphisms in esophageal cancer risk: a meta-analysis. GMR. 2014;13:649–659. doi: 10.4238/2014.January.28.10. [DOI] [PubMed] [Google Scholar]
- 12.Hu JJ, Wang ZT, Li B. Meta-analysis demonstrates lack of an association of microsomal epoxide hydrolase 1 polymorphisms with esophageal cancer risk. GMR. 2013;12:4540–4548. doi: 10.4238/2013.October.15.2. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, Luo J, Cai X. Association between microsomal epoxide hydrolase 1 polymorphisms and susceptibility to esophageal cancer: a meta-analysis. Tumour Biol. 2013;34:2383–2388. doi: 10.1007/s13277-013-0787-y. [DOI] [PubMed] [Google Scholar]
- 14.Zintzaras E. Impact of Hardy-Weinberg equilibrium deviation on allele-based risk effect of genetic association studies and meta-analysis. Eur J Epidemiol. 2010;25:553–560. doi: 10.1007/s10654-010-9467-z. [DOI] [PubMed] [Google Scholar]
- 15.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101–129. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 16.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed) 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DerSimonian R, Laird N. Meta-analysis in clinical trials. Contr Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 18.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 19.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 20.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG, The PG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk Postmenopausal estrogen monotherapy-associated breast cancer risk is modified by CYP17A1_-34_T > C polymorphism. Breast Cancer Res Treat. 2010;120:737–744. doi: 10.1007/s10549-009-0490-2. [DOI] [PubMed] [Google Scholar]
- 24.Balaji L, Lakkakula BV, Krishna BS, Paul SF. Lack of association of EPHX1 genotypes and haplotypes with oral cancer in South Indians. Genet Test Mol Biomarkers. 2011;15:595–599. doi: 10.1089/gtmb.2010.0260. [DOI] [PubMed] [Google Scholar]
- 25.Benhamou S, Reinikainen M, Bouchardy C, Dayer P, Hirvonen A. Association between lung cancer and microsomal epoxide hydrolase genotypes. Cancer Res. 1998;58:5291–5293. [PubMed] [Google Scholar]
- 26.Bonaventure A, Goujon-Bellec S, Rudant J, Orsi L, Leverger G, Baruchel A, Bertrand Y, Nelken B, Pasquet M, Michel G, Sirvent N, Bordigoni P, Ducassou S, Rialland X, Zelenika D, Hemon D, Clavel J. Maternal smoking during pregnancy, genetic polymorphisms of metabolic enzymes, and childhood acute leukemia: the ESCALE study (SFCE) Cancer Causes Control. 2012;23:329–345. doi: 10.1007/s10552-011-9882-9. [DOI] [PubMed] [Google Scholar]
- 27.Catsburg C, Joshi AD, Corral R, Lewinger JP, Koo J, John EM, Ingles SA, Stern MC. Polymorphisms in carcinogen metabolism enzymes, fish intake, and risk of prostate cancer. Carcinogenesis. 2012;33:1352–1359. doi: 10.1093/carcin/bgs175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chauhan PS, Ihsan R, Yadav DS, Mishra AK, Bhushan B, Soni A, Kaushal M, Devi TR, Saluja S, Gupta DK, Mittal V, Saxena S, Kapur S. Association of glutathione S-transferase, EPHX, and p53 codon 72 gene polymorphisms with adult acute myeloid leukemia. DNA Cell Biol. 2011;30:39–46. doi: 10.1089/dna.2010.1092. [DOI] [PubMed] [Google Scholar]
- 29.Clavel J, Bellec S, Rebouissou S, Menegaux F, Feunteun J, Bonaiti-Pellie C, Baruchel A, Kebaili K, Lambilliotte A, Leverger G, Sommelet D, Lescoeur B, Beaune P, Hemon D, Loriot MA. Childhood leukaemia, polymorphisms of metabolism enzyme genes, and interactions with maternal tobacco, coffee and alcohol consumption during pregnancy. Eur J Cancer Prev. 2005;14:531–540. doi: 10.1097/00008469-200512000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Cleary SP, Cotterchio M, Shi E, Gallinger S, Harper P. Cigarette smoking, genetic variants in carcinogen-metabolizing enzymes, and colorectal cancer risk. Am J Epidemiol. 2010;172:1000–1014. doi: 10.1093/aje/kwq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conesa-Zamora P, Ruiz-Cosano J, Torres-Moreno D, Espanol I, Gutierrez-Meca MD, Trujillo-Santos J, Perez-Ceballos E, Gonzalez-Conejero R, Corral J, Vicente V, Perez-Guillermo M. Polymorphisms in xenobiotic metabolizing genes (EPHX1, NQO1 and PON1) in lymphoma susceptibility: a case control study. BMC Cancer. 2013;13:228. doi: 10.1186/1471-2407-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Roos AJ, Gold LS, Wang S, Hartge P, Cerhan JR, Cozen W, Yeager M, Chanock S, Rothman N, Severson RK. Metabolic gene variants and risk of non-Hodgkin's lymphoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1647–1653. doi: 10.1158/1055-9965.EPI-06-0193. [DOI] [PubMed] [Google Scholar]
- 33.De Roos AJ, Rothman N, Brown M, Bell DA, Pittman GS, Shapiro WR, Selker RG, Fine HA, Black PM, Inskip PD. Variation in genes relevant to aromatic hydrocarbon metabolism and the risk of adult brain tumors. Neuro Oncol. 2006;8:145–155. doi: 10.1215/15228517-2005-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dura P, Bregitha CV, Te Morsche RH, Roelofs HM, Kristinsson JO, Wobbes T, Witteman BJ, Tan AC, Drenth JP, Peters WH. EPHX1 polymorphisms do not modify esophageal carcinoma susceptibility in Dutch Caucasians. Oncol Rep. 2012;27:1710–1716. doi: 10.3892/or.2012.1734. [DOI] [PubMed] [Google Scholar]
- 35.Gsur A, Zidek T, Schnattinger K, Feik E, Haidinger G, Hollaus P, Mohn-Staudner A, Armbruster C, Madersbacher S, Schatzl G, Trieb K, Vutuc C, Micksche M. Association of microsomal epoxide hydrolase polymorphisms and lung cancer risk. Br J Canc. 2003;89:702–706. doi: 10.1038/sj.bjc.6601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hlavata I, Vrana D, Smerhovsky Z, Pardini B, Naccarati A, Vodicka P, Novotny J, Mohelnikova-Duchonova B, Soucek P. Association between exposure-relevant polymorphisms in CYP1B1, EPHX1, NQO1, GSTM1, GSTP1 and GSTT1 and risk of colorectal cancer in a Czech population. Oncol Rep. 2010;24:1347–1353. doi: 10.3892/or_00000992. [DOI] [PubMed] [Google Scholar]
- 37.Ihsan R, Chattopadhyay I, Phukan R, Mishra AK, Purkayastha J, Sharma J, Zomawia E, Verma Y, Mahanta J, Saxena S, Kapur S. Role of epoxide hydrolase 1 gene polymorphisms in esophageal cancer in a high-risk area in India. J Gastroenterol Hepatol. 2010;25:1456–1462. doi: 10.1111/j.1440-1746.2010.06354.x. [DOI] [PubMed] [Google Scholar]
- 38.Ihsan R, Chauhan PS, Mishra AK, Yadav DS, Kaushal M, Sharma JD, Zomawia E, Verma Y, Kapur S, Saxena S. Multiple analytical approaches reveal distinct gene-environment interactions in smokers and non smokers in lung cancer. PloS one. 2011;6:e29431. doi: 10.1371/journal.pone.0029431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain M, Tilak AR, Upadhyay R, Kumar A, Mittal B. Microsomal epoxide hydrolase (EPHX1), slow (exon 3, 113His) and fast (exon 4, 139Arg) alleles confer susceptibility to squamous cell esophageal cancer. Toxicol Appl Pharmacol. 2008;230:247–251. doi: 10.1016/j.taap.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 40.Jang JH, Cotterchio M, Borgida A, Gallinger S, Cleary SP. Genetic variants in carcinogen-metabolizing enzymes, cigarette smoking and pancreatic cancer risk. Carcinogenesis. 2012;33:818–827. doi: 10.1093/carcin/bgs028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Justenhoven C, Hamann U, Schubert F, Zapatka M, Pierl CB, Rabstein S, Selinski S, Mueller T, Ickstadt K, Gilbert M, Ko YD, Baisch C, Pesch B, Harth V, Bolt HM, Vollmert C, Illig T, Eils R, Dippon J, Brauch H. Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat. 2008;108:137–149. doi: 10.1007/s10549-007-9586-8. [DOI] [PubMed] [Google Scholar]
- 42.Lacko M, Roelofs HM, Te Morsche RH, Voogd AC, Oude Ophuis MB, Peters WH, Manni JJ. Microsomal epoxide hydrolase genotypes and the risk for head and neck cancer. Head Neck. 2008;30:836–844. doi: 10.1002/hed.20781. [DOI] [PubMed] [Google Scholar]
- 43.London SJ, Smart J, Daly AK. Lung cancer risk in relation to genetic polymorphisms of microsomal epoxide hydrolase among African-Americans and Caucasians in Los Angeles County. Lung Cancer. 2000;28:147–155. doi: 10.1016/S0169-5002(99)00130-0. [DOI] [PubMed] [Google Scholar]
- 44.Mittal RD, Srivastava DL. Cytochrome P4501A1 and microsomal epoxide hydrolase gene polymorphisms: gene-environment interaction and risk of prostate cancer. DNA Cell Biol. 2007;26:791–798. doi: 10.1089/dna.2007.0630. [DOI] [PubMed] [Google Scholar]
- 45.Nisa H, Budhathoki S, Morita M, Toyomura K, Nagano J, Ohnaka K, Kono S, Ueki T, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Maekawa T, Yasunami Y, Takenaka K, Ichimiya H, Terasaka R. Microsomal epoxide hydrolase polymorphisms, cigarette smoking, and risk of colorectal cancer: the Fukuoka Colorectal Cancer Study. Mol Carcinog. 2013;52:619–626. doi: 10.1002/mc.21897. [DOI] [PubMed] [Google Scholar]
- 46.Ockenga J, Strunck S, Post C, Schulz HU, Halangk J, Pfutzer RH, Lohr M, Oettle H, Kage A, Rosendahl J, Keim V, Drenth JP, Jansen JB, Lochs H, Witt H. The role of epoxide hydrolase Y113H gene variant in pancreatic diseases. Pancreas. 2009;38:e97–e101. doi: 10.1097/MPA.0b013e31819feeed. [DOI] [PubMed] [Google Scholar]
- 47.Robien K, Curtin K, Ulrich CM, Bigler J, Samowitz W, Caan B, Potter JD, Slattery ML. Microsomal epoxide hydrolase polymorphisms are not associated with colon cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:1350–1352. doi: 10.1158/1055-9965.EPI-04-0877. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberger A, Illig T, Korb K, Klopp N, Zietemann V, Wolke G, Meese E, Sybrecht G, Kronenberg F, Cebulla M, Degen M, Drings P, Groschel A, Konietzko N, Kreymborg KG, Haussinger K, Hoffken G, Jilge B, Ko YD, Morr H, Schmidt C, Schmidt EW, Tauscher D, Bickeboller H, Wichmann HE. Do genetic factors protect for early onset lung cancer? A case control study before the age of 50 years. BMC Cancer. 2008;8:60. doi: 10.1186/1471-2407-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarmanova J, Susova S, Gut I, Mrhalova M, Kodet R, Adamek J, Roth Z, Soucek P. Breast cancer: role of polymorphisms in biotransformation enzymes. Eur J Hum Genet. 2004;12:848–854. doi: 10.1038/sj.ejhg.5201249. [DOI] [PubMed] [Google Scholar]
- 50.Sivonova MK, Dobrota D, Matakova T, Dusenka R, Grobarcikova S, Habala V, Salagovic J, Tajtakova M, Pidanicova A, Valansky L, Lachvacs L, Kliment JJ, Nagy V, Kliment J. Microsomal epoxide hydrolase polymorphisms, cigarette smoking and prostate cancer risk in the Slovak population. Neoplasma. 2012;59:79–84. doi: 10.4149/neo_2012_010. [DOI] [PubMed] [Google Scholar]
- 51.Skjelbred CF, Saebo M, Hjartaker A, Grotmol T, Hansteen IL, Tveit KM, Hoff G, Kure EH. Meat, vegetables and genetic polymorphisms and the risk of colorectal carcinomas and adenomas. BMC Cancer. 2007;7:228. doi: 10.1186/1471-2407-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soucek P, Sarmanova J, Kristensen VN, Apltauerova M, Gut I. Genetic polymorphisms of biotransformation enzymes in patients with Hodgkin's and non-Hodgkin's lymphomas. Int Arch Occup Environ Health. 2002;75(Suppl):S86–S92. doi: 10.1007/s00420-002-0353-1. [DOI] [PubMed] [Google Scholar]
- 53.Soucek P, Susova S, Mohelnikova-Duchonova B, Gromadzinska J, Moraviec-Sztandera A, Vodicka P, Vodickova L. Polymorphisms in metabolizing enzymes and the risk of head and neck squamous cell carcinoma in the Slavic population of the central Europe. Neoplasma. 2010;57:415–421. doi: 10.4149/neo_2010_05_415. [DOI] [PubMed] [Google Scholar]
- 54.Spurdle AB, Purdie DM, Webb PM, Chen X, Green A, Chenevix-Trench G. The microsomal epoxide hydrolase Tyr113His polymorphism: association with risk of ovarian cancer. Mol Carcinog. 2001;30:71–78. doi: 10.1002/1098-2744(200101)30:1<71::AID-MC1015>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 55.Tilak AR, Kumar S, Jain M, Pant MC, Das BC, Guleria R, Mittal B, Mathur N, Kumar A. Association of functionally important polymorphism of microsomal epoxide hydrolase gene (EPHX1) with lung cancer susceptibility. Canc Investig. 2011;29:411–418. doi: 10.3109/07357907.2011.587070. [DOI] [PubMed] [Google Scholar]
- 56.Timofeeva M, Kropp S, Sauter W, Beckmann L, Rosenberger A, Illig T, Jager B, Mittelstrass K, Dienemann H, Bartsch H, Bickeboller H, Chang-Claude J, Risch A, Wichmann HE. Genetic polymorphisms of MPO, GSTT1, GSTM1, GSTP1, EPHX1 and NQO1 as risk factors of early-onset lung cancer. Int J Cancer. 2010;127:1547–1561. doi: 10.1002/ijc.25175. [DOI] [PubMed] [Google Scholar]
- 57.To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, Caballero M, Cruellas F, Raya A, Dicenta M, Corbella J. Microsomal epoxide hydrolase and glutathione S-transferase polymorphisms in relation to laryngeal carcinoma risk. Cancer Lett. 2002;187:95–101. doi: 10.1016/S0304-3835(02)00406-8. [DOI] [PubMed] [Google Scholar]
- 58.Tranah GJ, Chan AT, Giovannucci E, Ma J, Fuchs C, Hunter DJ. Epoxide hydrolase and CYP2C9 polymorphisms, cigarette smoking, and risk of colorectal carcinoma in the Nurses’ Health Study and the Physicians’ Health Study. Mol Carcinog. 2005;44:21–30. doi: 10.1002/mc.20112. [DOI] [PubMed] [Google Scholar]
- 59.Tumer TB, Sahin G, Arinc E. Association between polymorphisms of EPHX1 and XRCC1 genes and the risk of childhood acute lymphoblastic leukemia. Arch Toxicol. 2012;86:431–439. doi: 10.1007/s00204-011-0760-8. [DOI] [PubMed] [Google Scholar]
- 60.van der Logt EM, Bergevoet SM, Roelofs HM, Te Morsche RH, Dijk Y, Wobbes T, Nagengast FM, Peters WH. Role of epoxide hydrolase, NAD(P)H:quinone oxidoreductase, cytochrome P450 2E1 or alcohol dehydrogenase genotypes in susceptibility to colorectal cancer. Mutat Res. 2006;593:39–49. doi: 10.1016/j.mrfmmm.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 61.Voho A, Metsola K, Anttila S, Impivaara O, Jarvisalo J, Vainio H, Husgafvel-Pursiainen K, Hirvonen A. EPHX1 gene polymorphisms and individual susceptibility to lung cancer. Cancer Lett. 2006;237:102–108. doi: 10.1016/j.canlet.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Joshi AD, Corral R, Siegmund KD, Marchand LL, Martinez ME, Haile RW, Ahnen DJ, Sandler RS, Lance P, Stern MC. Carcinogen metabolism genes, red meat and poultry intake, and colorectal cancer risk. Int J Cancer. 2012;130:1898–1907. doi: 10.1002/ijc.26199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wenghoefer M, Pesch B, Harth V, Broede P, Fronhoffs S, Landt O, Bruning T, Abel J, Bolt HM, Herberhold C, Vetter H, Ko YD. Association between head and neck cancer and microsomal epoxide hydrolase genotypes. Arch Toxicol. 2003;77:37–41. doi: 10.1007/s00204-002-0414-y. [DOI] [PubMed] [Google Scholar]
- 64.Agudo A, Sala N, Pera G, Capella G, Berenguer A, Garcia N, Palli D, Boeing H, Del Giudice G, Saieva C, Carneiro F, Berrino F, Sacerdote C, Tumino R, Panico S, Berglund G, Siman H, Stenling R, Hallmans G, Martinez C, Bilbao R, Barricarte A, Navarro C, Quiros JR, Allen N, Key T, Bingham S, Khaw KT, Linseisen J, Nagel G, et al. Polymorphisms in metabolic genes related to tobacco smoke and the risk of gastric cancer in the European prospective investigation into cancer and nutrition. Cancer Epidemiol Biomarkers Prev. 2006;15:2427–2434. doi: 10.1158/1055-9965.EPI-06-0072. [DOI] [PubMed] [Google Scholar]
- 65.Baxter SW, Choong DY, Campbell IG. Microsomal epoxide hydrolase polymorphism and susceptibility to ovarian cancer. Cancer Lett. 2002;177:75–81. doi: 10.1016/S0304-3835(01)00782-0. [DOI] [PubMed] [Google Scholar]
- 66.Boccia S, Cadoni G, Sayed-Tabatabaei FA, Volante M, Arzani D, De Lauretis A, Cattel C, Almadori G, van Duijn CM, Paludetti G, Ricciardi G. CYP1A1, CYP2E1, GSTM1, GSTT1, EPHX1 exons 3 and 4, and NAT2 polymorphisms, smoking, consumption of alcohol and fruit and vegetables and risk of head and neck cancer. J Cancer Res Clin Oncol. 2008;134:93–100. doi: 10.1007/s00432-007-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chauhan PS, Ihsan R, Mishra AK, Yadav DS, Saluja S, Mittal V, Saxena S, Kapur S. High order interactions of xenobiotic metabolizing genes and P53 codon 72 polymorphisms in acute leukemia. Environ Mol Mutagen. 2012;53:619–630. doi: 10.1002/em.21723. [DOI] [PubMed] [Google Scholar]
- 68.Chung CJ, Huang CY, Pu YS, Shiue HS, Su CT, Hsueh YM. The effect of cigarette smoke and arsenic exposure on urothelial carcinoma risk is modified by glutathione S-transferase M1 gene null genotype. Toxicol Appl Pharmacol. 2013;266:254–259. doi: 10.1016/j.taap.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Figueroa JD, Malats N, Garcia-Closas M, Real FX, Silverman D, Kogevinas M, Chanock S, Welch R, Dosemeci M, Lan Q, Tardon A, Serra C, Carrato A, Garcia-Closas R, Castano-Vinyals G, Rothman N. Bladder cancer risk and genetic variation in AKR1C3 and other metabolizing genes. Carcinogenesis. 2008;29:1955–1962. doi: 10.1093/carcin/bgn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gold LS, De Roos AJ, Brown EE, Lan Q, Milliken K, Davis S, Chanock SJ, Zhang Y, Severson R, Zahm SH, Zheng T, Rothman N, Baris D. Associations of common variants in genes involved in metabolism and response to exogenous chemicals with risk of multiple myeloma. Cancer Epidemiol. 2009;33:276–280. doi: 10.1016/j.canep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harrison DJ, Hubbard AL, MacMillan J, Wyllie AH, Smith CA. Microsomal epoxide hydrolase gene polymorphism and susceptibility to colon cancer. Br J Canc. 1999;79:168–171. doi: 10.1038/sj.bjc.6690028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiss I, Orsos Z, Gombos K, Bogner B, Csejtei A, Tibold A, Varga Z, Pazsit E, Magda I, Zolyomi A, Ember I. Association between allelic polymorphisms of metabolizing enzymes (CYP 1A1, CYP 1A2, CYP 2E1, mEH) and occurrence of colorectal cancer in Hungary. Anticancer Res. 2007;27:2931–2937. [PubMed] [Google Scholar]
- 73.Park JY, Chen L, Elahi A, Lazarus P, Tockman MS. Genetic analysis of microsomal epoxide hydrolase gene and its association with lung cancer risk. Eur J Cancer Prev. 2005;14:223–230. doi: 10.1097/00008469-200506000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rotunno M, Yu K, Lubin JH, Consonni D, Pesatori AC, Goldstein AM, Goldin LR, Wacholder S, Welch R, Burdette L, Chanock SJ, Bertazzi PA, Tucker MA, Caporaso NE, Chatterjee N, Bergen AW, Landi MT. Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PloS one. 2009;4:e5652. doi: 10.1371/journal.pone.0005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarmanova J, Benesova K, Gut I, Nedelcheva-Kristensen V, Tynkova L, Soucek P. Genetic polymorphisms of biotransformation enzymes in patients with Hodgkin's and non-Hodgkin's lymphomas. Hum Mol Genet. 2001;10:1265–1273. doi: 10.1093/hmg/10.12.1265. [DOI] [PubMed] [Google Scholar]
- 76.Silveira Vda S, Canalle R, Scrideli CA, Queiroz RG, Tone LG. Role of the CYP2D6, EPHX1, MPO, and NQO1 genes in the susceptibility to acute lymphoblastic leukemia in Brazilian children. Environ Mol Mutagen. 2010;51:48–56. doi: 10.1002/em.20510. [DOI] [PubMed] [Google Scholar]
- 77.Tiemersma EW, Omer RE, Bunschoten A, van't Veer P, Kok FJ, Idris MO, Kadaru AM, Fedail SS, Kampman E. Role of genetic polymorphism of glutathione-S-transferase T1 and microsomal epoxide hydrolase in aflatoxin-associated hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2001;10:785–791. [PubMed] [Google Scholar]
- 78.To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, Corbella J. Lung cancer susceptibility in relation to combined polymorphisms of microsomal epoxide hydrolase and glutathione S-transferase P1. Cancer Lett. 2001;173:155–162. doi: 10.1016/S0304-3835(01)00626-7. [DOI] [PubMed] [Google Scholar]
- 79.Tranah GJ, Giovannucci E, Ma J, Fuchs C, Hankinson SE, Hunter DJ. Epoxide hydrolase polymorphisms, cigarette smoking and risk of colorectal adenoma in the Nurses’ Health Study and the Health Professionals Follow-up Study. Carcinogenesis. 2004;25:1211–1218. doi: 10.1093/carcin/bgh126. [DOI] [PubMed] [Google Scholar]
- 80.Zhao H, Spitz MR, Gwyn KM, Wu X. Microsomal epoxide hydrolase polymorphisms and lung cancer risk in non-Hispanic whites. Mol Carcinog. 2002;33:99–104. doi: 10.1002/mc.10023. [DOI] [PubMed] [Google Scholar]
- 81.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland LB, Canzian F, Haugen A. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of non-small cell lung cancer in smokers. Carcinogenesis. 2008;29:1164–1169. doi: 10.1093/carcin/bgn020. [DOI] [PubMed] [Google Scholar]
- 82.Peng Q, Mo C, Qin A, Lao X, Chen Z, Sui J, Wu J, Zhai L, Yang S, Qin X, Li S. MDM2 SNP309 polymorphism contributes to endometrial cancer susceptibility: evidence from a meta-analysis. J Exp Clin Cancer Res. 2013;32:85. doi: 10.1186/1756-9966-32-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhuo W, Zhang L, Wang Y, Zhu B, Chen Z. CYP1A1 MspI polymorphism and acute myeloid leukemia risk: meta-analyses based on 5018 subjects. J Exp Clin Cancer Res. 2012;31:62. doi: 10.1186/1756-9966-31-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhan P, Wang Q, Qian Q, Wei SZ, Yu LK. CYP1A1 MspI and exon7 gene polymorphisms and lung cancer risk: an updated meta-analysis and review. J Exp Clin Cancer Res. 2011;30:99. doi: 10.1186/1756-9966-30-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pande M, Amos CI, Osterwisch DR, Chen J, Lynch PM, Broaddus R, Frazier ML. Genetic variation in genes for the xenobiotic-metabolizing enzymes CYP1A1, EPHX1, GSTM1, GSTT1, and GSTP1 and susceptibility to colorectal cancer in Lynch syndrome. Cancer Epidemiol Biomarkers Prev. 2008;17:2393–2401. doi: 10.1158/1055-9965.EPI-08-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sahin O, Arikan S, Oltulu YM, Coskunpinar E, Eren A, Cacina C, Guler E, Yaylim I. Investigation of a possible relationship between EPHX1 gene polymorphisms and colorectal cancer in Turkish society. Genet Test Mol Biomarkers. 2012;16:423–428. doi: 10.1089/gtmb.2011.0223. [DOI] [PubMed] [Google Scholar]
- 87.Erkisi Z, Yaylim-Eraltan I, Turna A, Görmüs U, Camlica H, Isbir T. Polymorphisms in the microsomal epoxide hydrolase gene: role in lung cancer susceptibility and prognosis. Tumori. 2010;96:756–763. doi: 10.1177/030089161009600519. [DOI] [PubMed] [Google Scholar]
- 88.Lee J, Dahl M, Nordestgaard BG. Genetically lowered microsomal epoxide hydrolase activity and tobacco-related cancer in 47,000 individuals. Cancer Epidemiol Biomarkers Prev. 2011;20:1673–1682. doi: 10.1158/1055-9965.EPI-10-1165. [DOI] [PubMed] [Google Scholar]
- 89.Zhong JH, Xiang BD, Ma L, You XM, Li LQ, Xie GS. Meta-analysis of microsomal epoxide hydrolase gene polymorphism and risk of hepatocellular carcinoma. PloS one. 2013;8:e57064. doi: 10.1371/journal.pone.0057064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang S, Zhu J, Zhang R, Wang S, Gu Z. Association between microsomal epoxide hydrolase 1 T113C polymorphism and susceptibility to lung cancer. Tumour Biol. 2013;34:1045–1052. doi: 10.1007/s13277-012-0644-4. [DOI] [PubMed] [Google Scholar]


