Abstract
Background
Cerebral ischemic stroke (CIS) is a major cause of morbidity and mortality. Its main pathological basis is atherosclerosis (AS); in turn, the main risk factor in AS is dyslipidemia. Human proprotein convertase subtilisin/kexin9 (PCSK9) plays a key role in regulating plasma low-density lipoprotein (LDL) cholesterol levels. We sought to assess the association between PCSK9 and CIS in Chinese Han and Uygur populations.
Material/Methods
We selected 408 CIS patients and 348 control subjects and used a single-base terminal extension (SNaPshot) method to detect the genotypes of the 20 single-nucleotide polymorphisms (SNPs) in PCSK9.
Results
Distribution of SNP8 (rs529787) genotypes showed a significant difference between CIS and control participants (P=0.049). However, when analyzing Han and Uygur populations separately, we found that only Han subjects showed distribution of SNP1 (rs1711503), SNP2 (rs2479408), and SNP8 (rs529787) alleles that was significantly different between CIS and control participants (P=0.028, P=0.013, P=0.006, respectively), and distribution of SNP2 (rs2479408) in the dominant model (CC vs. CG + GG) was significantly different between CIS and control participants (P=0.013), even after adjustment for covariates (OR: 75.262, 95% confidence interval [CI]: 7.232–783.278, P<0.001). Distribution of the 2 haplotypes (A-C and G-C) (rs1711503 and rs2479408) was significantly different between CIS and control participants (both, P=0.011).
Conclusions
Both rs1711503 and rs2479408 of PCSK9 genes were associated with CIS in the Han population of China. A-C haplotype may be a genetic marker of CIS risk in this population.
MeSH Keywords: Atherosclerosis, Ethnic Groups, Ischemic Attack, Transient, Proprotein Convertases
Background
Cerebral ischemic stroke (CIS) is a major cause of morbidity and mortality, and is expected to remain so until at least 2030 [1]. CIS and coronary heart disease (CHD) are major manifestations of atherosclerotic processes. High plasma levels of low-density lipoprotein cholesterol (LDL-C) have consistently been shown to be a risk factor for the development of atherosclerosis [2]. Plasma concentrations of LDL-C are determined primarily by the activity of the LDL receptor (LDLR) in the liver. Proprotein convertase subtilisin-like kexin type 9 (PCSK9) was recently discovered to be a major factor in cholesterol homeostasis through enhanced degradation of LDLR [3–6] and possibly in neural development. However, both rare mutations and common variants in the coding regions of PCSK9 can affect LDL cholesterol levels and stroke risk. Recent studies identified several PCSK9 variants influencing circulating LDL-C levels [7,8]. Since the first identification mutation of PCSK9 was implicated in autosomal dominant hypercholesterolemia by Abifadel [9], more than 53 missense variants have been identified. A common SNP, E670G (rs505151) in exon 12 of PCSK9, results in the substitution of glutamate for a glycine residue at position 670 in the protein [10] Carriers of 670 Gln in the general population presented increased plasma TC, LDL-C, and ApoB levels. Another study suggested a key role played by the E670G polymorphism in determining plasma LDL-C levels and the severity of coronary atherosclerosis in the United States [11]. More recently, the presence of the 670G allele was significantly associated with an increased risk of large-vessel atherosclerosis (LVA) stroke [12] and intimal media thickness (IMT) [13]. However, these studies were inconsistent with previous studies [14–16], which were conducted in Caucasian and African populations and failed to find this association. Furthermore, the carriers of 670G showed significantly increased LDL in men but not in women in a European population [17]. In addition, the rs72555377 insertion polymorphism in exon 1 of PCSK9 is associated with lower LDL-C in Caucasian populations [18], while the L11 allele, with insertion of 2 Leucines, is associated with higher LDL-C [11], and rs562556 (Ile474Val) in exon9 of the PCSK9 gene is associated with approximately 7% lower LDL cholesterol levels in carriers in a Japanese population [19].
In our study, we used a single-base terminal extension (SNaPshot) method to detect the genotypes of the 20 single-nucleotide polymorphisms (SNP) in the PCSK9 gene to assess the association between the human PCSK9 gene polymorphism and CIS in members of the Han and Uygur populations of China.
Material and Methods
Ethics approval of the study protocol
Written informed consent was obtained from all participants. All participants explicitly provided permission for DNA analyses as well as collection of relevant clinical data. This study was approved by the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University, Urumqi, China (NO. 20120510). It was conducted according to the standards of the Declaration of Helsinki.
Subjects
Subjects were from a Han population and a Uygur population who lived in the Xinjiang Uygur Autonomous Region of China. We recruited the CIS group from the First Affiliated Hospital of Xinjiang Medical University Neurology Department between since October 2011 and May 2012, and the control group came from the same hospital in the same period.
In the CIS group, there were 408 CIS patients (158 Uygur, 250 Han), mean age 61.97±11.80 years. Inclusion criteria were: (1) diagnosed in accordance with the standards set at 10 international classifications of diseases (ICD10); (2) confirmed by MRI. Exclusion criteria were: (1) patients with CHD; (2) hemorrhagic cerebrovascular disease confirmed by CT or MRI; (3) refused to participate in trials.
In the Control group there were 348 of healthy controls (149 Uygur, 199 Han), mean age 61.84±11.65 years. Inclusion criteria were: (1) aged >40; (2) no known family history of cerebrovascular disease; (3) the cardiopulmonary physical examination and nervous system examination did not find abnormalities; (4) MRI negative except for cerebrovascular disease. Exclusion criteria: acute or chronic infection, malignant tumor, autoimmune diseases.
Clinical characteristics of the study participants
All patients completed the standard test registration form, and disclosed the following data: (1) General information: age, sex, race. (2) Personal history: smoking history (daily average smoking, smoking an average of ≥1 day or more, time >1 year, defined as smoking), (drinking alcohol an average of ≥3 times per week, more than 50 g each time >1 year, defined as drinking), hypertension, diabetes, hyperlipidemia, transient ischemic attack (TIA), atrial fibrillation (AF), heart valve disease, heart valve replacement, peripheral vascular disease. Hypertension: the Seventh World Health Organization /International Society of Hypertension League Conference defined the new standard for the diagnosis of hypertension; in our study, the diagnosis of hypertension was established if patients were treated with antihypertensive medication or if the mean of 3 measurements of systolic blood pressure (SBP) >140 mm Hg or diastolic blood pressure (DBP) >90 mm Hg, respectively. Diabetes mellitus was diagnosed according to the criteria of the American Diabetes Association [20]. Individuals with daytime random blood glucose ≥11.1 mmol/l or after fasting glucose ≥7.0mmol/l or glucose in line 2 h ≥11.1mmol/l or with a history of diabetes or treatment with insulin were considered diabetic. (3) Medical history prior to admission: treatment with antihypertensive drugs, antiplatelet drugs and anticoagulants, diabetes, lipid drug, anti-seizure medication, birth control pills, hormones. (4) Family history: whether grandparents, parents, siblings, and children had hypertension, diabetes, cerebral hemorrhage, cerebral infarction, myocardial infarction, coronary heart disease, or arrhythmia incidence. (5) Physical examination: height, weight, blood pressure, pulse, temperature. (6) Special tests: electrocardiogram, chest X-ray, heart neck ultrasound, blood routine, blood glucose, blood lipids.
Biochemical analysis
Serum concentrations of total cholesterol (TC), triglyceride (TG), glucose (Glu), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C), apolipo protein A1 (ApoA1), apolipo protein B (ApoB), and apolipo lipoprotein a (ApoLpa) were measured using standard methods in the Central Laboratory of First Affiliated Hospital of Xinjiang Medical University.
Blood collection and DNA extraction
Fasting blood samples (5 mL) drawn by venipuncture were taken from all participants early in the morning. The blood samples were drawn into a 5-mL ethylene diamine tetraacetic acid (EDTA) tube and centrifuged at 4000×g for 5 min to separate the plasma content. Genomic DNA was extracted from the peripheral leukocytes using standard phenol-chloroform method. The DNA samples were stored at −80°C until use, then diluted to 50 ng/μL concentration.
SNaPshot Reactions
We selected the genotypes of the 20 SNPs in the PCSK9 gene using the Haploview 4.2 software and the HapMap phrase II database by using minor allele frequency (MAF) ≤0.1 and linkage disequilibrium patterns with r2 ≥0.5 as a cut-off. The position of the 20 SNPs was by order of increasing distance from the gene PCSK9 5′ end (Table 1). We used single-base terminal extension (SNaPshot) method to genotype. SNaPshot reactions were performed as described by the manufacturer (Applied Biosystems, Warrington, UK). Briefly, 4.0-μl of PCR product was incubated at 37°C for 60 min with 2-U shrimp alkaline phosphatase (SAP) and 2-U Exonuclease I (ExoI). Following a 15-min incubation to inactivate the enzymes, 1 ul of digested PCR product was mixed with 5 ul of ready reaction premix, 1 ul of 1.0- UM primer (Table 1), and 3 ul of dH2O. This mixture was placed in the thermal cycler and underwent 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. When completed, 0.5-U SAP was added and the reaction mixture was incubated for 60 min. Prior to loading onto the PRISM 310, 10 ul of formamide was added to 1 ul of reaction mixture and samples were heated to 95°C for 5 min.
Table 1.
Genotype and allele distributions of the twenty SNPs in patients with CIS and control subjects.
| SNP | Chr. 1 postion | Function | dbSNP allele | MAF | Total | Han | Uygur | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIS | Control | P value | CIS | Control | P value | CIS | Control | P value | |||||||||
| 1 | rs17111503 | 55503448 | 5′ near gene | Upstream variant 2KB | A/G | 0.3375 | Genotype | AA | 81 | 59 | 0.223 | 42 | 22 | 0.094 | 39 | 37 | 0.757 |
| SNP1 | AG | 197 | 158 | 115 | 86 | 82 | 72 | ||||||||||
| GG | 130 | 131 | 93 | 91 | 37 | 40 | |||||||||||
| Allele | A | 359 | 276 | 0.088 | 199 | 130 | 0.028* | 160 | 146 | 0.684 | |||||||
| G | 457 | 420 | 301 | 268 | 156 | 152 | |||||||||||
| 2 | rs2479408 | 55504118 | 5′ near gene | Upstream variant 2KB | C/G | 0.1708 | Genotype | CC | 385 | 314 | 0.064 | 249 | 192 | 0.013* | 136 | 122 | 0.446 |
| SNP2 | CG | 21 | 33 | 1 | 7 | 20 | 36 | ||||||||||
| GG | 2 | 1 | 0 | 0 | 2 | 1 | |||||||||||
| Allele | C | 791 | 661 | 0.050 | 499 | 391 | 0.013* | 292 | 270 | 0.423 | |||||||
| G | 25 | 35 | 1 | 7 | 24 | 28 | |||||||||||
| rs2479409 | 55504650 | 5′ near gene | Upstream variant 2KB | A/G | 0.4362 | Genotype | AA | 75 | 58 | 0.789 | 34 | 24 | 0.864 | 41 | 34 | 0.715 | |
| 3 | SNP3 | AG | 190 | 162 | 110 | 87 | 80 | 75 | |||||||||
| GG | 143 | 128 | 106 | 88 | 37 | 40 | |||||||||||
| Allele | A | 340 | 278 | 0.496 | 178 | 135 | 0.599 | 162 | 143 | 0.416 | |||||||
| G | 476 | 418 | 322 | 263 | 154 | 155 | |||||||||||
| 4 | rs11583680 | 55505668 | Exon 1 | Missense (V-A) | T/C | 0.0905 | Genotype | CC | 328 | 280 | 0.237 | 194 | 159 | 0.099 | 134 | 121 | 0.673 |
| SNP4 | CT | 78 | 62 | 56 | 37 | 22 | 25 | ||||||||||
| TT | 2 | 6 | 0 | 3 | 2 | 3 | |||||||||||
| Allele | C | 734 | 622 | 0.710 | 444 | 355 | 0.850 | 290 | 267 | 0.350 | |||||||
| T | 82 | 74 | 56 | 43 | 26 | 31 | |||||||||||
| 5 | rs10888896 | 555509213 | Intron 1 | Intron variant | C/G | 0.2374 | Genotype | CC | 326 | 280 | 0.797 | 203 | 169 | 0.360 | 123 | 111 | 0.782 |
| SNP5 | CG | 76 | 61 | 45 | 27 | 31 | 34 | ||||||||||
| GG | 6 | 7 | 2 | 3 | 4 | 4 | |||||||||||
| Allele | C | 728 | 621 | 0.995 | 451 | 365 | 0.436 | 277 | 256 | 0.521 | |||||||
| G | 88 | 75 | 49 | 33 | 39 | 42 | |||||||||||
| 6 | rs4927193 | 55509872 | Intron 2 | Intron variant | C/T | 0.1377 | Genotype | CC | 2 | 5 | 0.347 | 0 | 3 | 0.097 | 2 | 2 | 0.863 |
| SNP6 | CT | 82 | 64 | 59 | 39 | 23 | 25 | ||||||||||
| TT | 324 | 279 | 191 | 157 | 133 | 122 | |||||||||||
| Allele | C | 86 | 74 | 0.953 | 59 | 45 | 0.818 | 27 | 29 | 0.609 | |||||||
| T | 730 | 622 | 441 | 353 | 289 | 269 | |||||||||||
| 7 | rs499718 | 55512549 | Intron 3 | Intron variant | C/T | 0.247 | Genotype | CC | 276 | 240 | 0.829 | 154 | 130 | 0.564 | 122 | 110 | 0.778 |
| SNP7 | CT | 116 | 97 | 86 | 64 | 30 | 33 | ||||||||||
| TT | 16 | 11 | 10 | 5 | 6 | 6 | |||||||||||
| Allele | C | 668 | 577 | 0.570 | 394 | 324 | 0.332 | 274 | 253 | 0.520 | |||||||
| T | 148 | 119 | 106 | 74 | 42 | 45 | |||||||||||
| 8 | rs529787 | 55513521 | Intron 3 | Intron variant | C/G | 0.1166 | Genotype | CC | 384 | 312 | 0.049* | 249 | 191 | 0.006* | 135 | 121 | 0.452 |
| SNP8 | CG | 22 | 35 | 1 | 8 | 21 | 27 | ||||||||||
| GG | 2 | 1 | 0 | 0 | 2 | 1 | |||||||||||
| Allele | C | 790 | 659 | 0.039* | 499 | 390 | 0.006* | 291 | 269 | 0.426 | |||||||
| G | 26 | 37 | 1 | 8 | 25 | 29 | |||||||||||
| 9 | rs11206514 | 55516004 | Intron 3 | Intron variant | A/C | 0.4096 | Genotype | AA | 248 | 212 | 0.670 | 152 | 124 | 0.899 | 96 | 88 | 0.727 |
| SNP9 | AC | 141 | 115 | 89 | 67 | 52 | 48 | ||||||||||
| CC | 19 | 21 | 9 | 8 | 10 | 13 | |||||||||||
| Allele | A | 637 | 539 | 0.772 | 393 | 315 | 0.842 | 244 | 224 | 0.551 | |||||||
| C | 179 | 157 | 107 | 83 | 72 | 74 | |||||||||||
| 10 | rs572512 | 55517344 | Intron 3 | Intron variant | C/T | 0.4596 | Genotype | CC | 54 | 40 | 0.711 | 26 | 14 | 0.365 | 28 | 26 | 0.993 |
| SNP10 | CT | 171 | 144 | 102 | 78 | 69 | 66 | ||||||||||
| TT | 183 | 164 | 122 | 107 | 61 | 57 | |||||||||||
| Allele | C | 279 | 224 | 0.469 | 154 | 106 | 0.171 | 125 | 118 | 0.991 | |||||||
| T | 537 | 472 | 346 | 292 | 191 | 180 | |||||||||||
| 11 | rs2479413 | 55518682 | Intron 5 | Intron variant | C/T | 0.3191 | Genotype | CC | 225 | 193 | 0.090 | 141 | 124 | 0.183 | 84 | 69 | 0.054 |
| SNP11 | CT | 150 | 140 | 95 | 70 | 55 | 70 | ||||||||||
| TT | 33 | 15 | 14 | 5 | 19 | 10 | |||||||||||
| Allele | C | 600 | 526 | 0.363 | 377 | 318 | 0.109 | 223 | 208 | 0.834 | |||||||
| T | 216 | 170 | 123 | 80 | 93 | 90 | |||||||||||
| 12 | rs7552841 | 55518752 | Intron 5 | Intron variant | C/T | 0.284 | Genotype | CC | 264 | 235 | 0.602 | 176 | 143 | 0.886 | 88 | 92 | 0.502 |
| SNP12 | CT | 126 | 96 | 65 | 48 | 61 | 48 | ||||||||||
| TT | 18 | 17 | 9 | 8 | 9 | 9 | |||||||||||
| Allele | C | 654 | 566 | 0.564 | 417 | 334 | 0.834 | 237 | 232 | 0.406 | |||||||
| T | 162 | 130 | 83 | 64 | 79 | 66 | |||||||||||
| 13 | rs557435 | 55520864 | Intron 5 | Intron variant | A/G | 0.1662 | Genotype | AA | 5 | 1 | 0.242 | 1 | 0 | 0.668 | 4 | 1 | 0.260 |
| SNP13 | AG | 56 | 56 | 32 | 25 | 24 | 31 | ||||||||||
| GG | 347 | 291 | 217 | 174 | 130 | 117 | |||||||||||
| Allele | A | 66 | 58 | 0.862 | 34 | 25 | 0.755 | 32 | 33 | 0.703 | |||||||
| G | 750 | 638 | 466 | 373 | 284 | 265 | |||||||||||
| 14 | rs693668 | 55521109 | Intron 5 | Intron variant | A/G | 0.3912 | Genotype | AA | 212 | 189 | 0.772 | 131 | 116 | 0.441 | 81 | 73 | 0.908 |
| SNP14 | AG | 164 | 135 | 100 | 71 | 64 | 64 | ||||||||||
| GG | 32 | 24 | 19 | 12 | 13 | 12 | |||||||||||
| Allele | A | 588 | 513 | 0.472 | 362 | 303 | 0.205 | 226 | 210 | 0.774 | |||||||
| G | 228 | 183 | 138 | 95 | 90 | 88 | |||||||||||
| 15 | R434W | 5552339? | Exon 8 | Missense (R-W) | C/T | / | Genotype | CC | 408 | 348 | 250 | 199 | 158 | 149 | |||
| SNP15 | CT | ||||||||||||||||
| TT | |||||||||||||||||
| Allele | C | 816 | 696 | 500 | 199 | 316 | 298 | ||||||||||
| T | |||||||||||||||||
| 16 | rs540796 | 55524197 | Exon 9 | Synonymous codon (V-V) | G/A | 0.1354 | Genotype | AA | 1 | 2 | 0.585 | 0 | 0 | 0.716 | 1 | 2 | 0.340 |
| SNP16 | AG | 29 | 20 | 5 | 5 | 24 | 25 | ||||||||||
| GG | 378 | 326 | 245 | 194 | 133 | 132 | |||||||||||
| Allele | A | 31 | 24 | 0.716 | 5 | 5 | 0.714 | 26 | 19 | 0.378 | |||||||
| G | 785 | 672 | 495 | 393 | 290 | 279 | |||||||||||
| 17 | rs149311926 | 55525315 | Exon 10 | Missense (E-Q) | G/C | 0.0005 | Genotype | CC | 408 | 348 | 250 | 199 | 158 | 149 | |||
| SNP17 | CG | ||||||||||||||||
| GG | |||||||||||||||||
| Allele | C | 816 | 696 | 500 | 398 | 316 | 298 | ||||||||||
| G | |||||||||||||||||
| 18 | rs483462 | 55525400 | Intron 10 | Intron variant | A/G | 0.3223 | Genotype | AA | 279 | 234 | 0.939 | 170 | 135 | 0.907 | 109 | 99 | 0.837 |
| SNP18 | AG | 116 | 102 | 73 | 57 | 43 | 45 | ||||||||||
| GG | 13 | 12 | 7 | 7 | 6 | 5 | |||||||||||
| Allele | A | 674 | 570 | 0.721 | 413 | 327 | 0.863 | 261 | 243 | 0.734 | |||||||
| G | 142 | 126 | 87 | 71 | 55 | 55 | |||||||||||
| 19 | rs10465832 | 55528807 | Intron 11 | Intron variant | C/G | 0.1483 | Genotype | CC | 2 | 3 | 0.778 | 1 | 2 | 0.654 | 1 | 1 | 0.419 |
| SNP19 | CG | 75 | 67 | 54 | 39 | 21 | 28 | ||||||||||
| GG | 331 | 278 | 195 | 158 | 136 | 120 | |||||||||||
| Allele | C | 79 | 73 | 0.603 | 56 | 43 | 0.850 | 23 | 30 | 0.218 | |||||||
| G | 737 | 623 | 444 | 355 | 293 | 268 | |||||||||||
| 20 | rs505151 | 55529187 | Exon 12 | Missense (E-G) | A/G | 0.0983 | Genotype | AA | 365 | 310 | 0.878 | 219 | 179 | 0.537 | 146 | 131 | 0.399 |
| SNP20 | AG | 41 | 37 | 30 | 20 | 11 | 17 | ||||||||||
| GG | 2 | 1 | 1 | 0 | 1 | 1 | |||||||||||
| Allele | A | 771 | 657 | 0.940 | 468 | 378 | 0.380 | 303 | 279 | 0.207 | |||||||
| G | 45 | 39 | 32 | 20 | 13 | 19 | |||||||||||
Statistical analysis
All continuous variables (e.g., age, BMI, pulse, and cholesterol levels) are presented as means ± standard deviation (S.D.). The difference between the CIS and control groups was analyzed using an independent-sample T-test. The differences in the frequencies of sex, hypertension, diabetes mellitus, smoking, drinking, and genotypes were analyzed using chi-square test or Fisher’s exact test, as appropriate. Hardy-Weinberg equilibrium was assessed by chi-square analysis. Logistic regression analyses with effect ratios (odds ratio [OR] and 95% CI) were used to assess the contribution of the major risk factors. All statistical analyses were performed using SPSS 17.0 for Windows (SPSS Institute, Chicago, USA). Haplotypes were estimated using the SHEsis platform [21,22]. P-values of less than 0.05 were considered to be statistically significant.
Results
Table 2 showed the clinical characteristics of the CIS patients (n=408) and control participants (n=348). For all Han and Uygur subjects, there were no significant differences in age and sex between CIS patients and control subjects, indicating the study was an age- and sex-matched case-control study. We observed several differences between the groups of patients. As expected, several common risk factors for CIS were significantly different between the 2 subgroups: Glu, low HDL-C, high LDL-C, EH, and DM. Other CIS risk factors, such as high TC, TG levels, and cigarette smoking and drinking, were not significantly different.
Table 2.
Characteristics of study participants.
| Total | Han | Uygur | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Stroke patients | Control subjects | p Value | Stroke patients | Control subjects | p Value | Stroke patients | Control subjects | p Value | |
| Number (n) | 408 | 348 | 250 | 199 | 158 | 149 | |||
| Sex(M/W) | 242/166 | 183/165 | 0.063 | 144/106 | 102/97 | 0.183 | 98/60 | 81/68 | 0.203 |
| Age (years) | 61.97±11.80 | 61.84±11.65 | 0.885 | 63.56±11.37 | 62.35±11.79 | 0.269 | 59.44±12.01 | 61.17±11.45 | 0.198 |
| BMI (kg/m2) | 24.67±3.36 | 24.51±2.93 | 0.508 | 24.30±3.30 | 24.20±3.13 | 0.728 | 25.23±3.37 | 24.93±2.60 | 0.386 |
| Glu (mmol/L) | 6.90±3.30 | 5.45±2.68 | <0.001* | 6.86±3.12 | 5.24±1.44 | <0.001* | 6.98±3.55 | 5.74±3.73 | <0.003* |
| TG (mmol/L) | 1.90±1.12 | 2.04±1.30 | 0.122 | 1.81±1.08 | 1.90±1.21 | 0.406 | 2.03±1.17 | 2.22±1.40 | 0.221 |
| TC (mmol/L) | 4.38±0.96 | 4.27±1.24 | 0.182 | 4.35±0.95 | 4.43±1.24 | 0.444 | 4.42±0.98 | 4.06±1.22 | 0.004* |
| HDL (mmol/L) | 1.05±0.35 | 1.36±0.90 | <0.001* | 1.07±0.26 | 1.35±0.84 | <0.001* | 1.02±0.44 | 1.37±0.98 | <0.001* |
| LDL (mmol/L) | 2.76±0.88 | 2.52±0.78 | <0.001* | 2.68±0.86 | 2.51±0.78 | 0.038* | 2.87±0.89 | 2.52±0.79 | <0.001* |
| ApoA1 (mmol/L) | 1.25±0.27 | 1.22±0.35 | 0.216 | 1.27±0.22 | 1.24±0.30 | 0.310 | 1.21±0.32 | 1.18±0.40 | 0.538 |
| ApoB (mmol/L) | 0.89±0.76 | 0.89±0.61 | 0.909 | 0.90±0.75 | 0.90±0.79 | 0.986 | 0.87±0.24 | 0.88±0.23 | 0.612 |
| ApL(a) (mmol/L) | 195.27±146.14 | 172.94±113.84 | 0.019* | 199.72±146.08 | 192.68±136.62 | 0.602 | 188.20±146.42 | 146.57±64.73 | 0.001* |
| EH (Y/N) | 284/118 | 98/246 | <0.001* | 175/72 | 54/143 | <0.001* | 129/46 | 44/103 | <0.001* |
| DM (Y/N) | 125/269 | 65/272 | <0.001* | 78/164 | 31/168 | <0.001* | 47/105 | 34/104 | 0.242 |
| Smoke (Y/N) | 117/279 | 86/250 | 0.234 | 79/169 | 51/144 | 0.208 | 38/110 | 35/106 | 0.893 |
| Drinking(Y/N) | 0.204 | 44/290 | 0.161 | 43/204 | 26/167 | 0.292 | 23/120 | 18/123 | 0.500 |
BMI – body mass index; BUN – blood urea nitrogen; Glu – glucose; TG – triglyceride; TC – total cholesterol; HDL – high density lipoprotein; LDL – low density lipoprotein; EH – essential hypertension; DM – diabetes mellitus. Continuous variable were expressed as mean ± standard deviation. P value of continuous variables was calculated by independent T-T test. The P value of categorical variable was calculated by Fisher’s exact test.
P<0.05.
Table 1 shows the basic information and the distribution of genotypes and alleles of the 20 SNPs for the PCSK9 gene. The position of the 20 SNPs was by order of increasing distance from the gene PCSK9 5′ end. We observed that the distribution of genotypes and alleles of 3 SNPs (SNP1, SNP2, and SNP8) were significantly different between CIS group and control participants. All SNPs were consistent with Hardy-Weinberg expectations (data not shown).
The 3 SNPs among the 3 groups (Total, Han, and Uyghur) were examined by Hardy-Weinberg equilibrium test and no significant differences were found in these 3 groups (data not shown).
In the study, we confirmed the distribution of genotypes and alleles of the 3 SNPs (SNP1, SNP2, and SNP8) for the PCSK9 gene. For SNP1 (rs17111503), the distribution of alleles showed a significant difference between CIS and control participants (P=0.028) in the Han group, but not in the total group and Uygur group. For SNP2 (rs2479408), the distribution of alleles, the dominant model (CC vs. CG + GG), and the additive model (CG vs. CC + GG) showed a significant difference between CIS and control participants in total and Han groups, but not in the Uygur group. C allele of rs2479408 was significantly higher in CIS patients than in control participants (total: 96.94% vs. 94.97%; Han: 99.80% vs. 98.24%). For SNP3 (rs529787), the distribution of alleles, the dominant model (CC vs. CG + GG) and the additive model (CG vs. CC + GG) showed a significant difference between CIS and control participants in the total and Han groups, but not in the Uygur group. C allele of rs529787 was significantly higher in CIS patients than in control participants (Total: 96.81% vs. 94.68%; Han: 99.80% vs. 97.99%) (data no shown).
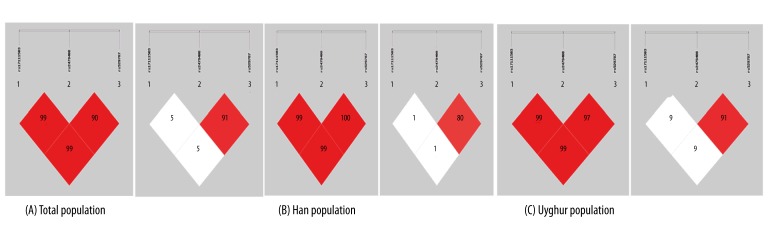
Table 3 and Figure 1 show patterns of linkage disequilibrium in the PCSK9 gene, with their |D′| and r2 values. |D′| values from 0.7 to 1 indicate strong LD between a pair of SNPs. |D′| values from 0.25 to 0.7 indicate moderate LD and |D′| values of 0–0.25 indicate low LD. In the study, 3 strong LD patterns were observed between SNP1 and SNP2 (|D′|=0.999), SNP2 and SNP8 (|D′|=0.983), and SNP1 and SNP8 (|D′|=0.999). We consider that all 3 SNPs were located in 1 haplotype block. The r2 value of SNP2–SNP8 >0.5 means the SNP2 and SNP8 can replace each other [11] and they cannot construct haplotypes simultaneously. Therefore, given that the position of SNP1 and SNP2 are both in 2KB upstream of PCSK9 gene and the position of SNP8 is in intron3, we constructed the haplotypes using SNP1 and SNP2.
Table 3.
Pairwise linkage disequilibrium (| D’| above diagonal and r2 below diagonal) for the three SNPs.
| Total | Han | Uygur | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |D′| | |D′| | |D′| | ||||||||||
| SNP | SNP1 | SNP2 | SNP8 | SNP | SNP1 | SNP2 | SNP8 | SNP | SNP1 | SNP2 | SNP8 | |
| r2 | SNP1 | 0.999 | 0.999 | SNP1 | 1.000 | 0.988 | SNP1 | 0.999 | 0.999 | |||
| SNP2 | 0.057 | 0.983 | SNP2 | 0.016 | 1.000 | SNP2 | 0.093 | 0.979 | ||||
| SNP8 | 0.060 | 0.918 | SNP3 | 0.017 | 0.888 | SNP8 | 0.097 | 0.919 | ||||
Figure 1.
Pairwise estimates of linkage disequilibrium (LD) between each PCSK9 polymorphism were plotted using SHEsis platform. Each polymorphism is numbered according to its position in the PCSK9 gene as presented (left shows |D′| and right shows r2).
Table 4 shows the distribution of haplotypes in CIS patient and control participants. There were 4 haplotypes established in all subjects. The overall distribution of the haplotypes were significantly different between the CIS patients and the control subjects (all P<0.05). The most frequent haplotype in this study was A-C haplotype. For Han, the frequency of A-C was significantly higher in the CIS patients than in the control subjects (P=0.0011). In addition, the frequency of the G-C haplotype was lower in the CIS patients than in the control subjects (P=0.0011).
Table 4.
Haplotype analysis of the two SNPs (rs17111503 and rs2479408).
| Haplotype | Case (freq) | Control (freq) | Odds Ratio [95% CI] | P | |
|---|---|---|---|---|---|
| Total | AC | 334.02 (0.409) | 241.01 (0.346) | 1.308 [1.061–1.613] | 0.012* |
| AG | 24.98 (0.031) | 34.99 (0.050) | 0.597 [0.353–1.007] | 0.051 | |
| GC | 456.98 (0.560) | 419.99 (0.603) | 0.837 [0.681–1.027] | 0.088 | |
| GG | 0.02 (0.000) | 0.01 (0.000) | |||
| Han | AC | 198.05 (0.396) | 123.00 (0.309) | 1.434 [1.085–1.895] | 0.011* |
| AG | 0.95 (0.002) | 7.00 (0.018) | |||
| GC | 300.95 (0.602) | 268.00 (0.673) | 0.697 [0.528–0.922] | 0.011* | |
| GG | 0.05 (0.000) | 0.00 (0.000) | |||
| Uygur | AC | 136.01 (0.430) | 118.01 (0.396) | 1.153 [0.836–1.590] | 0.387 |
| AG | 23.99 (0.076) | 27.99 (0.094) | 0.792 [0.448–1.401] | 0.423 | |
| GC | 155.99 (0.494) | 151.99 (0.510) | 0.936 [0.682–1.285] | 0.685 | |
| GG | 0.01 (0.000) | 0.01 (0.000) |
All those frequency<0.03 will be ignored in analysis.
Table 5 showed that multiple logistic regression analyses were performed with age, sex, BMI, HDL-C, LDL-C, TC, TG, ApoA1 ApoB, ApoLpa, EH, DM, and smoking and drinking, because these variables were the major confounding factors for CIS. The significant difference of the dominant model (CC vs. CG + GG) of rs2479408 was retained after adjustment for covariates in the Han, but not in the Uygur group (OR: 75.262, 95% confidence interval [CI]: 7.232–783.278, P<0.001).
Table 5.
Multiple logistic regression analysis for stoke patients and control subjects.
| Total | Han | Uygur | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | ||||
| Lower | Upper | Lower | Upper | Lower | Upper | |||||||
| rs2479408 (CC/CG+GG) | 10.544 | 3.336 | 33.328 | 0.000* | 75.262 | 7.232 | 783.278 | 0.000* | 2.229 | 0.449 | 11.060 | 0.327 |
| sex | 10.544 | 3.336 | 33.328 | 0.613 | 1.147 | 0.651 | 2.019 | 0.635 | 1.045 | 0.558 | 1.956 | 0.891 |
| age | 1.001 | 0.986 | 1.016 | 0.901 | 0.997 | 0.976 | 1.018 | 0.762 | 1.017 | 0.991 | 1.043 | 0.196 |
| BMI | 0.981 | 0.924 | 1.041 | 0.522 | 0.983 | 0.905 | 1.068 | 0.686 | 0.987 | 0.897 | 1.086 | 0.789 |
| TG | 1.109 | 0.953 | 1.291 | 0.181 | 1.228 | 0.981 | 1.537 | 0.073 | 1.118 | 0.882 | 1.418 | 0.356 |
| TC | 1.031 | 0.851 | 1.250 | 0.756 | 1.239 | 0.933 | 1.646 | 0.139 | 0.715 | 0.508 | 1.008 | 0.055 |
| HDL-C | 1.783 | 1.288 | 2.468 | 0.000* | 2.568 | 1.413 | 4.666 | 0.002* | 1.297 | 0.854 | 1.970 | 0.223 |
| LDL-C | 0.685 | 0.528 | 0.889 | 0.004* | 0.660 | 0.453 | 0.961 | 0.030* | 0.752 | 0.483 | 1.169 | 0.205 |
| APOA1 | 0.990 | 0.556 | 1.762 | 0.974 | 0.744 | 0.269 | 2.061 | 0.570 | 1.348 | 0.617 | 2.945 | 0.453 |
| APOB | 1.103 | 0.889 | 1.370 | 0.373 | 1.114 | 0.873 | 1.421 | 0.388 | 1.292 | 0.380 | 4.392 | 0.681 |
| APL (a) | 0.999 | 0.997 | 1.000 | 0.061 | 0.999 | 0.997 | 1.001 | 0.225 | 0.996 | 0.993 | 1.000 | 0.031 |
| EH | 5.308 | 3.700 | 7.615 | 0.000* | 6.366 | 3.877 | 10.453 | 0.000* | 5.112 | 2.836 | 9.215 | 0.000 |
| DM | 2.407 | 1.546 | 3.746 | 0.000* | 4.746 | 2.403 | 9.376 | 0.000* | 1.379 | 0.717 | 2.655 | 0.336 |
| Smoking | 1.137 | 0.656 | 1.972 | 0.647 | 1.133 | 0.542 | 2.370 | 0.739 | 0.956 | 0.376 | 2.433 | 0.925 |
| Drinking | 8.645 | 3.174 | 23.549 | 0.000* | 52.408 | 5.808 | 472.912 | 0.000* | 1.883 | 0.495 | 7.165 | 0.353 |
Discussion
PCSK9, also known as neural apoptosis-regulated convertase 1 (NARC1), is the ninth member of the proprotein convertase (PC) family [23]. The human PCSK9 gene is located on chromosome 1p32.3; it encompasses 12 exons and encodes a 692 amino acid glycoprotein. PCSK9 is synthesized as an inactive zymogen, pro-PCSK9 (73 kDa) and contains a signal peptide, a prodomain (residues 31–152) and a catalytic domain (residues 153–451) followed by a C-terminal domain (residues 452–692) [24]. PCSK9 acts as a serine protease and molecular chaperone that reduces both hepatic and extrahepatic low-density lipoprotein receptor levels through an endosomal/lysosomal pathway and increases plasma LDL cholesterol [4,25]. PCSK9 may also regulate apolipoprotein B-containing lipoprotein production and apoB secretion [26,27].
Recent advances revealed a large number of genetic variants of PCSK9 that may modulate plasma cholesterol levels either positively or negatively. “Gain of function” missense mutations in PCSK9 were associated with autosomal-dominant hypercholesterolemia (ADH), a rare form of familial hypercholesterolemia (FH) in which neither the LDLR nor the ligand binding domain of apolipoprotein (apo) B100 are mutated [28,29]. “Loss of function” nonsense mutations in PCSK9 were associated with low plasma LDL-C levels and a reduced incidence of cardiovascular disease [30,31]. Later, many in vitro and in vivo overexpression and knockout/knockdown studies confirmed that PCSK9 targets the LDLR for degradation [32–34]. Studies have confirmed that both rare mutations and common variants in the coding regions of PCSK9 affect LDL cholesterol levels and stroke risk. In this study, we selected 20 SNPs of PCSK9 and used case-control analyses to assess the association between the human PCSK9 gene polymorphism and CIS in the Han and Uygur populations.
Our findings showed the distribution of SNP8 (rs529787) genotypes were significantly different between CIS and control participants (P=0.049). However, when analyzing Han and Uygur groups separately, we found that only in the Han population was the distribution of SNP1 (rs1711503), SNP2 (rs2479408), and SNP8 (rs529787) alleles significantly different between CIS and control participants (P=0.028, P=0.013, P=0.006, respectively). For SNP1 (rs17111503), the frequency of A allele was higher in CIS than in control participants (P=0.028, 39.80% vs. 32.66%) in the Han group, indicating that the risk of CIS was increased with the A allele of rs17111503. For SNP2 (rs2479408), the distribution of alleles, the dominant model (CC vs. CG + GG), and the additive model (CG vs. CC + GG) showed a significant difference between CIS and control participants in total and Han groups, but not in the Uygur group. C allele of rs2479408 was significantly higher in CIS patients than in control participants (total: 96.94% vs. 94.97%; Han: 99.80% vs. 98.24%). Moreover, the significant difference of the dominant model (CC vs. CG + GG) of rs2479408 was retained after adjustment for covariates: age, sex, BMI, HDL-C, LDL-C, TC, TG, ApoA1 ApoB, ApoLpa, EH, DM, and smoking and drinking in the Han group (OR: 75.262, 95% confidence interval [CI]: 7.232–783.278, P<0.001), indicating that the risk of CIS was increased with the C allele of rs2479408. For SNP3 (rs529787), the distribution of alleles, the dominant model (CC vs. CG + GG), and the additive model (CG vs. CC + GG) showed a significant difference between CIS and control participants in total and Han groups, but not in Uygurs. C allele of rs529787 was significantly higher in CIS patients than in control participants (total: 96.81% vs. 94.68%; Han: 99.80% vs. 97.99%). When we constructed the haplotypes using SNP1 and SNP2, we found that the most frequent haplotype in this study was A-C haplotype. For Han, the frequency of A-C was significantly higher in the CIS patients than in the control subjects (P=0.0011), but the frequency of the G-C haplotype was lower in the CIS patients than in the control subjects (P=0.0011). This fully showed that A allele of rs17111503 and C allele of rs2479408 may be the risk factor of CIS, and G allele of rs17111503 and G allele of rs2479408 may be the protective factor of CIS.
SNP20 (rs505151) was observed in the exon12 of the PCSK9 gene and the polymorphisms caused the substitution of glutamate for a glycine residue at position 670 in the protein. The studies about the association between rs505151 of PCSK9 gene polymorphisms (E670G) and the cardiovascular risk have provided inconsistent results, as the introduction of description. Our study was consistent with previous studies [14–16] showing no significant association between the polymorphism of PCSK9 (rs505151) and CIS. By comparison, we found the age of our control subjects was higher than the other studies [11,12] and the study by Afef Slimani [35]. In our study, there were no significant difference in age between CIS patients (age: 63.56±11.37) and control subjects (age: 62.35±11.79) (P=0.269), but in the study by Afef Slimani, there were significant difference in age between CIS patients (age: 66/54.5–76.50) and control subjects (age: 49/45–55) (P<0.0001). Age is a risk factor for stroke, and this may be why our conclusions were not consistent with their conclusions. In addition, there may be differences in populations and geographical factors that explain some differences.
Conclusions
We found that both rs1711503 and 2479408 of PCSK9 were associated with CIS in the Han population of China. A-C haplotype may be a risk genetic marker of CIS in Han in China. A allele of rs17111503 and C allele of rs2479408 may be the risk marker of CIS. Studies with statistically significant numbers of clinical samples are needed for further research in China.
Footnotes
Source of support: This work was supported by grants from the National Natural Science Foundation of China (81160145, 81260180), the proprotein convertase Hay bacteriolysin 9 gene polymorphism studies in patients with ischemic stroke with carotid atherosclerosis in Xinjiang Han and Uyghur populations (81160145), and the apoplexy folic acid metabolism pathway gene of rare and common variants of high-throughput sequencing studies in patients with ischemic stroke with carotid atherosclerosis in Xinjiang Han and Uyghur populations (81260180)
References
- 1.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS medicine. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobbs HH, Russell DW, Brown MS, Goldstein JL. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu Rev Genet. 1990;24:133–70. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 3.Park SW, Moon YA, Horton JD. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J Biol Chem. 2004;279:50630–38. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 4.Benjannet S, Rhainds D, Essalmani R, et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol chem. 2004;279:48865–75. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 5.Peterson AS, Fong LG, Young SG. PCSK9 function and physiology. J Lipid Res. 2008;49:1595–99. doi: 10.1194/jlr.CX00001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton JD, Cohen JC, Hobbs HH. PCSK9: a convertase that coordinates LDL catabolism. J Lipid Re. 2000;50(Suppl):S172–77. doi: 10.1194/jlr.R800091-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folsom AR, Peacock JM, Boerwinkle E Atherosclerosis Risk in Communities Study I. Variation in PCSK9, low LDL cholesterol, and risk of peripheral arterial disease. Atherosclerosis. 2009;202:211–15. doi: 10.1016/j.atherosclerosis.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abifadel M, Varret M, Rabes JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–56. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 10.Aung LH, Yin RX, Miao L, et al. The proprotein convertase subtilisin/kexin type 9 gene E670G polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2011;10:5. doi: 10.1186/1476-511X-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SN, Ballantyne CM, Gotto AM, Jr, et al. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am Coll Cardiol. 2005;45:1611–19. doi: 10.1016/j.jacc.2005.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abboud S, Karhunen PJ, Lutjohann D, et al. Proprotein convertase subtilisin/kexin type 9 (PCSK9) gene is a risk factor of large-vessel atherosclerosis stroke. PloS one. 2007;2:e1043. doi: 10.1371/journal.pone.0001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norata GD, Garlaschelli K, Grigore L, et al. Effects of PCSK9 variants on common carotid artery intima media thickness and relation to ApoE alleles. Atherosclerosis. 2010;208:177–82. doi: 10.1016/j.atherosclerosis.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Kotowski IK, Pertsemlidis A, Luke A, et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–22. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polisecki E, Peter I, Robertson M, et al. Genetic variation at the PCSK9 locus moderately lowers low-density lipoprotein cholesterol levels, but does not significantly lower vascular disease risk in an elderly population. Atherosclerosis. 2008;200:95–101. doi: 10.1016/j.atherosclerosis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scartezini M, Hubbart C, Whittall RA, et al. The PCSK9 gene R46L variant is associated with lower plasma lipid levels and cardiovascular risk in healthy U.K. men. Clin Sci. 2007;113:435–41. doi: 10.1042/CS20070150. [DOI] [PubMed] [Google Scholar]
- 17.Evans D, Beil FU. The E670G SNP in the PCSK9 gene is associated with polygenic hypercholesterolemia in men but not in women. BMC Med Genet. 2006;7:66. doi: 10.1186/1471-2350-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue P, Averna M, Lin X, Schonfeld G. The c.43_44insCTG variation in PCSK9 is associated with low plasma LDL-cholesterol in a Caucasian population. Hum Mutat. 2006;27:460–66. doi: 10.1002/humu.20316. [DOI] [PubMed] [Google Scholar]
- 19.Miyake Y, Kimura R, Kokubo Y, et al. Genetic variants in PCSK9 in the Japanese population: rare genetic variants in PCSK9 might collectively contribute to plasma LDL cholesterol levels in the general population. Atherosclerosis. 2008;196:29–36. doi: 10.1016/j.atherosclerosis.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 20.American Diabetes Association. clinical practice recommendations 1997. Diabetes Care. 1997;20:S1–S70. [PubMed] [Google Scholar]
- 21.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis ( http://analysis.bio-x.cn) Cell Res. 2009;19:519–23. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- 23.Seidah NG, Benjannet S, Wickham L, et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci USA. 2003;100:928–33. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert G, Charlton F, Rye KA, Piper DE. Molecular basis of PCSK9 function. Atherosclerosis. 2009;203:1–7. doi: 10.1016/j.atherosclerosis.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt RJ, Beyer TP, Bensch WR, et al. Secreted proprotein convertase subtilisin/kexin type 9 reduces both hepatic and extrahepatic low-density lipoprotein receptors in vivo. Biochem Biophys Res Commun. 2008;370:634–40. doi: 10.1016/j.bbrc.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Ouguerram K, Chetiveaux M, Zair Y, et al. Apolipoprotein B100 metabolism in autosomal-dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler Thromb Vasc Biol. 2004;24:1448–53. doi: 10.1161/01.ATV.0000133684.77013.88. [DOI] [PubMed] [Google Scholar]
- 27.Sun XM, Eden ER, Tosi I, et al. Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet. 2005;14:1161–69. doi: 10.1093/hmg/ddi128. [DOI] [PubMed] [Google Scholar]
- 28.Allard D, Amsellem S, Abifadel M, et al. Novel mutations of the PCSK9 gene cause variable phenotype of autosomal dominant hypercholesterolemia. Hum Mutat. 2005;26:497. doi: 10.1002/humu.9383. [DOI] [PubMed] [Google Scholar]
- 29.Leren TP. Mutations in the PCSK9 gene in Norwegian subjects with autosomal dominant hypercholesterolemia. Clin Genet. 2004;65:419–22. doi: 10.1111/j.0009-9163.2004.0238.x. [DOI] [PubMed] [Google Scholar]
- 30.Hallman DM, Srinivasan SR, Chen W, et al. Relation of PCSK9 mutations to serum low-density lipoprotein cholesterol in childhood and adulthood (from The Bogalusa Heart Study) Am J Cardiol. 2007;100:69–72. doi: 10.1016/j.amjcard.2007.02.057. [DOI] [PubMed] [Google Scholar]
- 31.Hooper AJ, Marais AD, Tanyanyiwa DM, Burnett JR. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 2007;193:445–48. doi: 10.1016/j.atherosclerosis.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 32.Benjannet S, Rhainds D, Hamelin J, et al. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J Biol Chem. 2006;281:30561–72. doi: 10.1074/jbc.M606495200. [DOI] [PubMed] [Google Scholar]
- 33.Lagace TA, Curtis DE, Garuti R, et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert G, Jarnoux AL, Pineau T, et al. Fasting induces hyperlipidemia in mice overexpressing proprotein convertase subtilisin kexin type 9: lack of modulation of very-low-density lipoprotein hepatic output by the low-density lipoprotein receptor. Endocrinology. 2006;147:4985–95. doi: 10.1210/en.2006-0098. [DOI] [PubMed] [Google Scholar]
- 35.Slimani A, Harira Y, Trabelsi I, et al. Effect of E670G Polymorphism in PCSK9 Gene on the Risk and Severity of Coronary Heart Disease and Ischemic Stroke in a Tunisian Cohort. J Mol Neurosci. 2014;53(2):150–57. doi: 10.1007/s12031-014-0238-2. [DOI] [PubMed] [Google Scholar]