Abstract
Redox metabolism plays critical roles in multiple biological processes and diseases. Until recently, knowledge of specific, key redox processes in living systems was limited by the lack of adequate methodology. Reduced nicotinamide adenine dinucleotide (NADH) and its oxidized form (NAD+) is the most important small molecule in the redox metabolism of mammalian cells. We previously reported a series of genetically encoded fluorescent sensors for intracellular NADH detection. Here, we present an accounting of experimental components and considerations, such as protein expression and purification, fluorescence titration, transfections, and confocal imaging, necessary to perform a standardized NADH assay experiment with these probes. In addition, we outline initial experiments used to derive basic principles of NADH/NAD+ redox biology in vitro. Finally, we describe a protocol for a steady-state kinetics experiment, and the processing of experimental data to measure intracellular NADH levels.
Keywords: Redox, glycolysis, mitochondria, fluorescence, metabolism
1. INTRODUCTION
1.1 Cellular redox systems
Oxidation-reduction reactions exist in all cells and are critical for cell homeostasis and signaling, including energy metabolism, gene expression, cell cycle regulation, immune response, cell growth, and cell apoptosis (Balaban et al., 2005; Delaunay et al., 2002; Jones, 2008; Menon and Goswami, 2007; Ying, 2008). The intracellular redox states are mainly determined by pyridine nucleotide redox systems and thiol/disulfide redox systems. Pyridine nucleotide redox system comprises reduced and oxidized nicotinamide adenine dinucleotide (NADH/NAD+) and reduced and oxidized nicotinamide adenine dinucleotide phosphate (NADPH/NADP+). NADH/NAD+ are the most important cofactors involved in energy metabolism, receiving electrons during the oxidation of various nutrients and then oxidized via mitochondrial respiration. In addition, this redox couple also function in circadian rhythms, apoptosis, aging, development, and carcinogenesis (Chen et al., 2009; Dumollard et al., 2007; Lin et al., 2004; Nakahata et al., 2009; Ramsey et al., 2009; Zhang et al., 2006). By contrast, NADPH provides the reducing equivalents for biosynthetic reactions of anabolic pathways and antioxidant defense against oxidative stress, and is also responsible for generating free radicals, such as superoxide and nitric oxide. Thiol/disulfide redox system comprises the glutathione redox system and the thioredoxin redox system. Both reduced glutathione and thioredoxin are regenerated by reductases that consume NADPH (Figure 1).
Figure 1. Simplified scheme for intracellular redox systems.
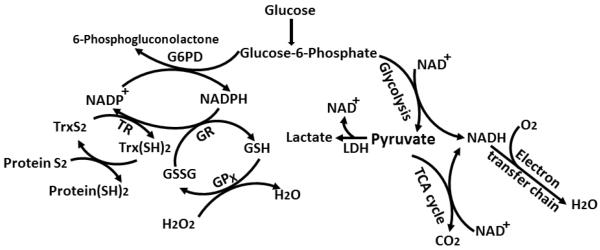
NADH produced during glycolysis and the tricarboxylic acid (TCA) cycle is consumed by the electron transfer chain. NADPH generated by the catalysis of glucose-6-phosphate dehydrogenase (G6PD) is utilized to reduce oxidized glutathione (GSSG) by glutathione reductase (GR) or oxidized thioredoxin (Trx) by thioredoxin reductase (TR).
1.2 Genetically-encoded fluorescent sensors for NADH
For many years, researchers relied on weak autofluorescence to measure the NADH levels in living cells (Kasischke et al., 2004; Mayevsky and Rogatsky, 2007; Patterson et al., 2000; Pellerin and Magistretti, 2004). However, this method has the drawbacks of a weak signal, low sensitivity and specificity or cell injury resulting from the ultraviolet irradiation required to induce the autofluorescent signal. To probe dynamic cellular events in living cells with high spatial and temporal resolution, researchers have developed a diverse set of genetically encoded biosensors, which generally involve the fusion of a fluorescent protein with a specific sensing protein domain. In bacteria, the Rex family proteins are especially sensitive to NADH (Brekasis and Paget, 2003; Gyan et al., 2006; Sickmier et al., 2005; Wang et al., 2008). Crystallographic studies have shown that NADH binding induces dramatic confor-mational changes in the Rex dimer, which shifts from an open to a closed form (McLaughlin et al., 2010). By fusing the Rex protein and circularly permuted fluorescent proteins(cpFPs), Yellen's group and our group independently developed genetically encoded NADH sensors that allow specific monitoring of dynamic changes in NADH levels as affected by NADH transport, glucose metabolism, mitochondrial electron transport, and redox environment (Hung et al., 2011; Zhao et al., 2011). The Frex sensors (Zhao et al., 2011) (Figure 2A) specifically detect NADH levels with large dynamic range and different affinities; however, its fluorescence is pH sensitive. Peredox sensors (Hung et al., 2011) (Figure 2B) are much more resistant to local pH and partially reflect the more physiologically relevant NADH:NAD+ ratio; however its dynamic range of fluorescence is much smaller than that of Frex sensors (150% versus 800% upon NADH binding). The affinity of Peredox for NADH is quite high, which makes these sensors largely saturated under normal physiological conditions (Hung et al., 2011) and, therefore, difficult to reflect redox fluctuations.
Figure 2. Schematic representations of Frex and Peredox.
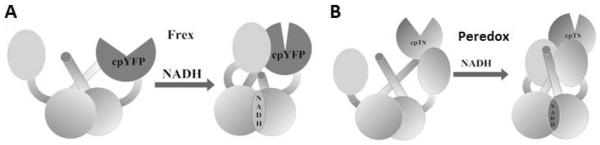
(A) Design of Frex, which is a fusion protein of a complete Rex monomer, cpYFP, and the NADH-binding domain of a second Rex molecule. (B) Design of Peredox, which is a fusion protein of two complete Rex monomers connected by a circular permuted T-Sapphire molecule.
Genetically encoded sensors have also been used to detect disulfides (Cannon and James Remington, 2009; Dooley et al., 2004; Hanson et al., 2004; Meyer and Dick, 2010; Ostergaard et al., 2001), oxidized glutathione (Bjornberg et al., 2006; Gutscher et al., 2008), and hydrogen peroxide (Belousov et al., 2006). Here, we present a detailed protocol for live cell imaging and real time measurement of intracellular NADH level using the Frex series of genetically encoded sensors.
2. EXPERIMENTAL COMPONENTS AND CONSIDERATIONS
2.1 Expression and purification of recombinant Frex sensors
Sufficient quantities of purified Frex sensors are desirable to ensure consistent and reproducible results from in vitro titration. Although there are several options for protein purification, we recommend immobilized metal affinity chromatography and Ni2+ resin for the purification of His-tagged proteins. Protein purity is assessed by SDS-PAGE analysis. The aliquots of purified Frex sensors are stored at −20 °C until experimental use. The steps for the sensor protein preparation are as follows:
E.coli BL21 (DE3) pLys cells carrying the pRSETb-Frex expression plasmid are grown in 10 ml LB media containing 34 μg/ml chloramphenicol and 100 μg/ml ampicillin. Cells are incubated with shaking at 37 °C until OD 600 nm reaches 0.6~0.8.
Add IPTG from a 100 mM stock to a final concentration of 0.1 mM and incubate with shaking at 18 °C overnight.
Harvest the cells by centrifugation at 4,000 ×g for 30 min at 4 °C.
Resuspend the cell pellets in 1 ml 50 mM potassium phosphate buffer (Kpi buffer), pH7.4, containing 0.5 M sodium chloride and 20 mM imidazole (Buffer A), and lyse cells by ultrasonication.
Purify His6-tagged proteins using His MultiTrp 96-well filter plates. First, wash the plate with 2 column volumes of wash buffer (Buffer A contain 50 mM imidazole). Second, elute the protein from the resin using 50 mM Kpi buffer, pH 7.4, containing 0.5 M sodium chloride and 300 mM imidazole (Buffer B).
Desalt and exchange the protein preparations into 100 mM potassium phosphate buffer (pH 7.4) before assay.
Measure the protein concentration by the Lowry assay and store the aliquotted samples at −20 °C.
2.2 Frex sensor expressing in mammalian cells
Frex sensors were expressed in different subcellular compartments by tagging these proteins with organellespecific signal peptides and observing the cells 24–30 h post-transfection, as follows:
The coding sequences of Frex were subcloned into pcDNA3.1 Hygro (+) (Invitrogen) upstream of a Kozak sequence for mammalian expression. For nuclear targeting of Frex, the three-fold nuclear localization signal (3×NLS) DPKKKRKVDPKKKRKVDPKKKRKV was added to the C-terminus. For mitochondrial targeting, the mitochondrial localization signal MRKMLAAVSRVLSGASQKPASRVLVASRNFANDATF was inserted at the N-terminus. 293FT cells (Invitrogen, USA) were maintained in DMEM (high glucose) supplemented with 10% FBS, 0.1 mM MEM non-essential amino acids (Invitrogen), 6 mM L-glutamine, and 1 mM sodium pyruvate (Invitrogen) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Cells were plated in antibiotic-free high glucose-DMEM supplemented with 10% FBS 16 hours before transfection. We typically used 0.8 μg endotoxin-free plasmids with 3.2 μl Lipofectamine 2000 (Invitrogen) for each well of a 12-well plate according to the manufacturer's protocol.
2.3 Fluorescent microplate reading
For high-throughput detection of intracellular NADH levels, a Synergy 2 Multi-Mode Microplate Reader (Biotek) was used for measuring the fluorescence of suspended cells expressing NADH sensors in 96-well or 384-well black flat bottom microplates. In general, 293FT cells were harvested 24–48 h after transfection, counted, washed, and suspended in phosphate-buffered saline preheated at 37 °C, and aliquots of cells were incubated at 37 °C with compounds of interest during the measurement. Dual-excitation ratios were obtained using a Synergy MX Multi-Mode Microplate Reader (Biotek) at 410 nm and 500 nm excitation, and 528 nm emission for both excitation wavelengths. Filters used were 528 BP 20 nm for emission, and 410 BP 20 nm or 485 BP 20 nm for excitation.
2.4 Laser scanning confocal microscopy
For real-time imaging of intracellular NADH, we utilized the Zeiss Laser Scanning Confocal Microscopy (LSCM) system on a Zeiss Axio Observer Z1 inverted microscope equipped with a Plan Apo 63×1.4 NA oil immersion objective. 293FT cells expressing the NADH sensor were maintained at 37 °C in a humidified atmosphere using a CO2 incubator (PECON). We excited NADH sensors sequentially line by line with the 405 nm and 488 nm laser line and with the emission detection set to 500–550 nm. We strongly recommended that microscopes provided with “line mode” scanning ability be used to measure the fluorescence ratio of each pixel accurately by minimizing subcellular particle movements and the effects of vibration. Scanning was performed using the “line mode,” 1024×1024 format, 12 bit depth, 2×line average, and 3.0 pinhole.
2.5 Automatic wide field fluorescence microscopy
To assess Frex and C3L194K sensor occupancy by NADH in mitochondria, a high performance fluorescence microscopy system equipped with a Nikon Eclipse Ti-E automatic microscope, Plan Apo VC 60x1.2 NA water immersion objective, Evolve 512 EMCCD (Photometrics), and the highly stable Lambda XL light source (Sutter Instruments) was used for ratiometric fluorescence imaging of living cells and for calibration with purified recombinant sensor proteins. 410 BP 20 nm and 480 BP 30 nm band-pass excitation filters, and a 535/40 emission filter were switched by a Lambda 10-XL filter wheel (Sutter Instruments) for dual-excitation ratio imaging. Images were captured using a 512×512 format, 16 bit depth, and 100 ms exposure for both channels.
3. INITIAL CHARACTERIZATION EXPERIMENTS
Initial experiments are necessary to determine the fluorescent properties of Frex sensors for each preparation (Kd for NADH and dynamic range), sensor concentration, instrument parameters, and pH fluctuation for the NADH detection. This approach will ensure that both valid and relevant results are achieved in the steady-state & kinetics experiments.
3.1 In vitro titration of Frex sensor by NADH
Frex sensor had an apparent Kd for NADH and NADH analogs of 3.7 μM and >>400 μM, respectively (Zhao et al., 2011). For titration experiments in vitro, when removing samples from −20 °C storage, we recommend that the aliquot of Frex sensors be rapidly defrosted by hand and then placed immediately on ice. Use one aliquot per set of experiments as multiple freeze-thaw cycles may denature and inactivate Frex sensors. Ideally, all experiments should be conducted with Frex sensors from the same purification experiment. A sample protocol is as follows:
Prepare the NADH stock solution by dissolving 7.09 mg NADH ml−1 in 100 mM Kpi buffer, pH 7.4. Dilute 20 μl of the stock solution with 980 μl of 100 mM Kpi buffer to prepare the maximal standard concentration (200 μM) and then make serial dilutions of NADH to concentration of: 200 μM, 100 μM, 50 μM, 20 μM, 10 μM, 5 μM, 2 μM, 1 μM, 0.5 μM, 0.2 μM, and 0.1 μM.
Add 50 μl of different concentrations of NADH to 96-well black flat-bottom microplates.
Dilute Frex protein into 100 mM Kpi buffer (pH 7.4) with 0.1% bovine serum albumin to a final concentration of 1 μM, and then add 50 μl of Frex protein to 96-well black flat-bottom microplates using a dispenser.
Mix immediately and then measure the dual-excitation ratios of the Frex sensor using a synergy 2 Multi-Mode Microplate Reader with 420 nm and 485 nm excitation and 528 nm emission for both excitation wavelengths.
Fluorescence values were background-corrected by subtracting the intensity of different concentrations of NADH solution.
Note: For calibration of mitochondrial NADH levels, prepare NADH stock solutions in 100 mM Kpi buffer, pH 8.0. Dilute Frex and C3L194K protein into 100 mM KPi buffer with 0.1% bovine serum albumin (pH 8.0) to final concentrations of 0.1 μM and 0.3 μM, respectively. For in vitro titration of the Frex sensor and the C3L194K sensor, a series of NADH solutions with concentration (0–200 μM) and (0–1 mM) were used as above.
3.2 Correction of pH effects with cpYFP
As with many other genetically encoded sensors based on cpYFP, Frex sensors are also sensitive to pH. The fluorescence of the cpYFP moiety excited at 485 nm increased significantly with an increase of pH, while the fluorescence excited at 420 nm was much less affected (Figure 3A). Fortunately, the Frex sensor is one of the most responsive genetic encoded sensors currently available, exhibiting an 8-fold increase and 3-fold decrease of fluorescence with excitation at 490 nm and 420 nm, respectively, upon NADH binding, whereas there is less than a 1-fold increase in fluorescence with excitation at 490 nm when the pH increased from 7.0 to 7.4, the range of physiological pH in the cytosol. Thus, pH variations complicated modestly the quantification of free intracellular NADH levels. In order to identify pH fluctuations during the study of biological processes or effects of specific treatments, we suggest that one always measure the fluorescence of cpYFP alone in cells, or purified cpYFP in solution, before or in parallel with measurements in Frex-expressing cells. Under many circumstances, however, the pH variation in the cytosol is too small to induce significant changes in Frex fluorescence (Zhao et al., 2011). pH effects were not obvious in the studies of other cpYFP-based sensors (Belousov et al., 2006; Berg et al., 2009; Nakai et al., 2001). By contrast, the effects of pH variations are more obvious in studies of mitochondrial function, which heavily depend on electron transfer and respiration. Under such circumstances, the Frex fluorescence signal should be corrected by cpYFP fluorescence measured in parallel experiments, owing to the similar pH dependency of Frex and cpYFP (Figure 3B). For example, we found that there was a two-phase increase of mitochondrial-targeted Frex fluorescence during the 1 hr time course upon glucose supplementation in glucose-starved cells (Figure 3C). It turned out that the slow phase was an artifact caused by the increase in pH in the mitochondria matrix as shown by the cpYFP fluorescence. To correct this artifact, Frex-Mit fluorescence was normalized by cpYFP-Mit fluorescence. This corrected Frex-Mit fluorescence reached its maximum over 10 minutes after glucose supplementation, consistent with reports in the literature using other cells (Eto et al., 1999).
Figure 3. pH effect correction of Frex fluorescence.
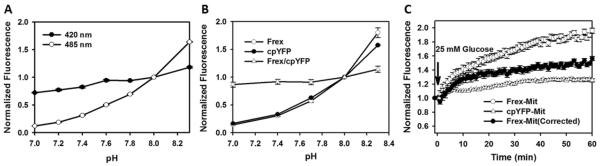
(A) pH dependency of cpYFP fluorescence excited at 420 nm and 485 nm.(B) pH dependency of Frex and cpYFP fluorescence excited at 485 nm. (C) Kinetics of fluorescence response of Frex-Mit and cpYFP-Mit after glucose supplementation. The solid symbol represents the fluorescence response of Frex-Mit corrected for pH effects.
In fluorescence microscopy studies of live cells when cells are imaged with an on-line CO2 incubation system, significant changes in pH may occur when the cells are moved from or placed into a CO2 incubator, as the pH of cell culture medium buffered by bicarbonate salts and is highly dependent on the dissolved CO2 level. Such pH variations may be monitored using cpYFP-expressing cells. The following precautions should be taken under these conditions to control pH:
Adjust the pH of different stock solutions to pH 7.4 to maintain the pH of the buffers and medium for the cells.
Add 100 mM HEPES (pH 7.4) to phenol red-free complete medium to increase the pH buffering capacity of the medium.
Keep cells in the incubator for 10 min before imaging to balance the dissolved CO2.
4. STEADY-STATE & KINETICS EXPERIMENTS
After determination of the critical experimental parameters and precautions from the initial characterization experiments, a full steady-state kinetics experiment can be conducted.
4.1 Quantification of NADH in subcellular compartments
Measuring NADH concentrations in living cells is vital for the understanding of fluctuation in metabolic states. For many years, researchers relied on endogenous NAD(P)H fluorescence for measuring mitochondrial NADH levels(Mayevsky, 2009). The total NAD+–NADH pool in the matrix varies from 300 μM to 3 mM depending on the cell type (Joubert et al., 2004; Yang et al., 2007). Previous reports showed that NAD+/NADH ratio varies from 2 to 16 in the mitochondrial matrix (Henley and Laughrey, 1970; Kasimova et al., 2006; Williamson et al., 1967); however, the ratio of free/bound NADH varies significantly from almost 0:1 (Wakita et al., 1995), 1.5:1 (Blinova et al., 2005) to 1:4 (Yu and Heikal, 2009) as measured by time-resolved fluorescence, fluorescence anisotropy, and fluorescence spectral decomposition analysis. These data may reflect different metabolic conditions in the mitochondria of different organisms or the limitations of the techniques used. Unfortunately, the traditional NAD(P)H fluorescence method is awkward and challenging for measuring free NADH levels in the cytosol or nucleus either because of very low concentrations of NADH in these organelles or the NADPH pool bearing the majority of cytosolic reduced pyridine dinucleotide (Jones, 1981; Shigemori et al., 1996).
NADH sensors are intrinsically ratiometric with two excitation wavelengths (Figure 4A) and allow quantification of free intracellular NADH concentrations in different subcellular compartments (Figure 4B). A standard protocol is as follows:
Figure 4. Subcellular distribution of NADH in mammalian cells.
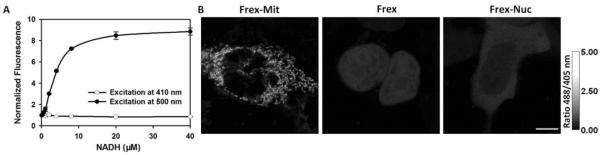
(A) Fluorescence intensities with excitation at 410 nm or 500 nm normalized to the initial value; emission at 528 nm. (B) Ratiometric fluorescence images of cells expressing Frex in mitochondria, the nucleus, and cytosol. The pixel-by-pixel ratios of the 488 nm excitation image by the 405 nm excitation image of the same cell were used to gray the images. Scale bar, 10 μm.
For the digitonin permeabilization assay:
Prepare digitonin stock solution by dissolving 30 mg digitonin in 1 ml PBS.
For digitonin permeabilization assay of Frex-expressing cells, resuspend cells with PBS buffer (pH 7.4) containing 0.001% digitonin.
For digitonin permeabilization assay of Frex-Mit-expressing cells, resuspend cells with PBS and then lyse them with 100 mM KPi buffer (pH 8.0) containing 0.3% digitonin.
Measure the fluorescence (excited at 485 nm) of cells or cell lysates in the presence or absence of 100 μM NADH.
For ratiometric fluorescence imaging:
Measure the ratiometric fluorescence of purified recombinant sensor proteins (0.1 μM Frex or 0.3 μM C3L194K) with or without 100 μM NADH in living cells using a high-performance fluorescence microscopy system equipped with a stable light source and high-performance EMCCD camera.
Assess Frex and C3L194K sensor occupancy by NADH in the mitochondria and then calculate mitochondrial NADH concentration.
For the FrexH fluorescence assay:
Measure the ratiometric fluorescence of purified recombinant FrexH sensor proteins (0.1 μM FrexH) with or without 10 μM NADH and FrexH-expressing cells using a fluorescence plate reader.
Assess the FrexH sensor occupancy by NADH in the cytosol and then calculate cytosolic NADH concentration.
Considering the highly specific affinity of sensors toward NADH compared to other pyridine and adenine nucleotides (Zhao et al., 2011). Frex (Kd ~3.7 μM at pH 7.4) and FrexH (Kd ~40 nM at pH 7.4) sensors are recommended for NADH detection in the cytosol and nucleus, where the free NADH concentration is extremely low. Frex (Kd ~11 μM at pH 8.0) and C3L194K (Kd ~50 μM at pH 8.0) sensors are recommended for NADH detection in the mitochondrial, where the free NADH concentration is extremely comparatively high. We measured subcellular NADH levels in 293FT cells through Frex and FrexH using a microplate reader and found extremely low cytosolic NADH levels ranging from 120 nM to 130 nM (Table 1), in agreement with previous estimations in the nucleus (Zhang et al., 2002). Ratiometric fluorescence imaging and digitonin permeabilization assays showed that the Frex-Mit sensors were ~70% to 75% saturated by NADH in the mitochondrial matrix of 293FT cells, suggesting that the mitochondrial free NADH concentration is 33±9 μM and 26 ± 7 μM, respectively (Table 1). The free NADH concentration in the mitochondrial matrix was also determined to be 27±5 μM using a low-affinity sensor, C3L194K (Table 1).
Table 1.
Subcellular NADH concentrations in 293FT cells
| Sensors | Kdfor NADH (μM) | Subcellular location | Detection methods | Sensor occupancy (%) | [NADH] (μM) | Reference (μM) |
|---|---|---|---|---|---|---|
| FrexH-Cyt | 0.04 | cytosol | Microplate | ~76 | ~0.13 | ~0.13*2 |
| Frex-Cyt | 3.7 | cytosol | Microplate | ~3.3 | ~0.12 | |
| Frex-Mit | 11*1 | Mitochondria | Imaging | ~70%–75% | ~33±9 | ~60–75*3 |
| Frex-Mit | 11*1 | Mitochondria | Microplate | ~70%–75% | ~26±7 | |
| C3L194K | 50*1 | Mitochondria | Imaging | ~35% | ~27 ± 5 |
Measured at pH 8.0
The concentration of free NADH in the nucleus is ~130 nM (Zhang et al, 2002).
The concentration of NADH in mitochondria of intact pancreatic islets at resting glucose levels (4–5 mM) is ~ 60–75 μM (Patterson et al., 2000).
Exogenous NADH binding protein expression, i.e., the Frex sensor expression, may contribute to the buffering systems for NADH. However, the fluorescence of Frex expressed in the cytosol responds immediately to exogenous NADH, lactate, and glucose, suggesting that such temporal buffering effects of the Frex sensor are minimal. Cells have several “buffering” systems for NADH, including several NADH binding proteins in the cytosol and mitochondria. Furthermore, NADH and NAD+ are buffered by coupled reaction pairs, pyruvate and lactate via lactate dehydrogenase in the cytosol and oxoglutarate and glutamate via β-hydroxybutyrate dehydrogenase in the mitochondria. The concentrations of these metabolites are in the millimolar range. The Frex protein concentration expressed in the cytosol ranges from 4 μM to 13 μM, depending on cell type and transfection efficiency. Frex protein concentration expressed in the mitochondria was estimated to be within 10 μM to 30 μM, although only 70% was bound with NADH. We estimated that the Frex-Mit-bound NADH is less than 12% of the total mitochondrial NADH in resting cells. According to these calculations, Frex sensors are unlikely to have a major impact on the total NADH concentration and the metabolic status of the cells in which they are expressed (Zhao and Yang, 2012).
4.2 Dynamic monitoring of NADH in single cells
Frex sensors are brighter and more sensitive than endogenous NAD(P)H fluorescence. Thus, they are more suitable for dynamic monitoring of NADH in single cells, particularly in the cytosol and nucleus where the endogenous NAD(P)H fluorescence is extremely weak and derived mainly from NADPH. For single cell imaging, a sample protocol is as follows:
Plate 293 FT cells or cells of interest onto a 35-mm glass-bottom dish with phenol red-free fresh growth medium containing 10% FBS, 0.1 mM MEM nonessential amino acids, 6 mM L-glutamine, and 1 mM sodium pyruvate.
Transfect the cells with 0.8 μg per dish of cDNA (Frex, Frex-Nuc or Frex-Mit) using Lipofectamine 2000 according to the manufactures's instruction, and incubate the cells in a humidified atmosphere at 37 °C under 5% CO2 for 24–30 hr.
Replace the medium with fresh growth medium containing 100 mM HEPES (pH 7.4), and keep cells in a humidified atmosphere using a CO2 incubator (PeCon) for 10 min to balance the dissolved CO2 before imaging.
For single cell imaging, choose moderately bright cells in which the fluorescence is well distributed in the cytosol (Frex), in the nucleus (Frex-Nuc), or in the mitochondria (Frex-Mit), and use the Zeiss 710 laser scanning confocal microscopy (LSCM) system on a Zeiss Axio Observer Z1 inverted microscope equipped with a Plan Apo 63×1.4 NA oil immersion objective for green fluorescence of the indicator (Frex).
Switch on the lasers, controller, scanning head, and computer. Note: The lasers need 30 min to warm up and stabilize.
Start the scanning process using the “line mode” and excite the indicator (Frex) sequentially line by line with the 405 nm and 488 nm laser line and with emission detection set to 500–550 nm.
For fluorescence images, adjust the 405 nm and 488 nm laser intensity to the minimum level that allows easy identification of single cells, adjust the size of the the pinhole for acceptable depth of the image, and adjust the sensitivity of the photomultiplier tube (PMT) for optimal signal-to-noise ratio.
Acquire images every 15 s for 5 min to 10 min.
During the image acquisition, add fresh growth medium containing reagents of interest.
Determine the image ratio by dividing the image intensities acquired with excitation at 488 nm by those acquired at 405 nm.
4.3 High throughput measurement of NADH by microplate reader
Frex sensors are highly specific, have a large dynamic range of fluorescence upon NADH binding, and can be targeted to different subcellular compartments. These advantages make Frex compatible with high throughput measurement of NADH by microplate reader. A sample protocol is as follows:
Plate 293 FT cells into 12-well plates with fresh growth medium containing 10% FBS, 0.1 mM MEM nonessential amino acids, 6 mM L-glutamine, and 1 mM sodium pyruvate.
Transfect the cells with 0.8 μg per dish of cDNA (Frex-Mit or Frex-cpYFP) using 3.2 μl Lipofectamine 2000 according to the manufacturer's instruction, and incubate the cells in a humidified atmosphere at 37 °C under 5% CO2 for 24–30 hr.
Harvest 293FT cells expressing Frex-Mit and cpYFP-Mit from 12-well plates by trypsinization, and then centrifuge at 100 ×g for 5 minutes.
Carefully remove the supernant with a 1-ml pipette tip or an aspirator.
Gently add PBS preheated to 37 °C along the wall of 1.5 ml Eppendorf tube, and carefully remove the supernant.
Maintain the cells in PBS without glucose for 1 h to consume intracellular glucose.
Prepare glucose stock solution by dissolving 90 mg glucose ml−1 in PBS with (500 mM glucose), dilute 100 μl of the stock solution with 900 μl of PBS to make the maximal standard concentration (50 mM), and then make serial dilutions for a series of glucose solutions with concentration: 50 mM, 20 mM, 10 mM, 5 mM, 2 mM, 1 mM, 0.5 mM, 0.2 mM, 0.1 mM, 0.05 mM, and 0.02 mM.
Add 50 μl of different concentrations of glucose to 96-well black flat-bottom microplates.
Count cells with a hemacytometer, and then add 50 μl of cell suspensions to 96-well black flat-bottom microplates using a dispenser.
Mix immediately and then measure the dual-excitation ratios of the Frex sensor using a synergy 2 Multi-Mode Microplate Reader with 420 nm and 485 nm excitation and 528 nm emission for both excitation wavelengths.
Correct the fluorescence values by subtracting the intensity of 293FT cell samples not expressing Frex.
Correct the Frex-Mit fluorescence signal by cpYFP fluorescence measured in parallel experiments to eliminate the effects of pH variation.
5. DATA HANDLING/PROCESSING
For microplate reader assays, samples containing equal numbers of Frex expressing cells or untransfected (control) cells were both measured. Fluorescence values were background-corrected by subtracting the intensity of 293FT cell samples not expressing Frex. Unlike endogenous NAD(P)H fluorescence, it is possible that the free NADH concentration in different cells and different organelles can be quantified and compared after calibration of Frex fluorescence in live cells. For the calibration of cytosolic NADH levels, Frex expressing cells were re-suspended in PBS buffer, pH 7.4, containing 0.001% digitonin. The fluorescence (excited at 485 nm) of cells or cell lysates in the presence or absence of 100 μM NADH was then measured.
For imaging, raw data were exported to Image J software as 12-bit TIF for analysis. The pixel-by-pixel ratio of the 488 nm excitation image by the 405 nm excitation image of the same cell were used to gray the images in HSB gray space. Simply, the RGB value (255, 0, 255) represents the lowest ratio, and the red (255, 0, 0) represents the highest ratio, while the gray brightness is proportional to the fluorescence signals in both channels. In order to assess the Frex sensor occupancy by NADH in the mitochondria matrix, a calibration standard of sensor protein with or without NADH should be obtained using the same microscopy setting. For the calibration of mitochondrial NADH level, Frex protein was diluted into 100 mM KPi buffer with 0.1% bovine serum albumin (pH 8.0) to a final concentration of 0.1 μM.
6. SUMMARY
This chapter presents methods for intracellular NADH detection using genetically encoded fluorescence sensors. This protocol is adaptable to determine NADH levels using Frex sensors by a variety of instruments (microplate reader, fluorescence spectrofluorimeter, flow cytometers, laser scanning confocal microscope, or fluorescence microscope). Compared to endogenous NAD(P)H fluorescence, Frex sensors are highly specific, manifest a large change in fluorescence upon NADH binding, and can be targeted to different subcellular compartments. These advantages allow for real time imaging of cytosolic and nuclear NADH concentrations, which are very useful for better understanding of cellular metabolism and signaling involving NADH.
Acknowledgements
We thank Stephanie Tribuna for expert secretarial assistance. Work supported by the 973 Program (2013CB531200), National Natural Science Foundation of China (grants 31225008, 31170815 and 31071260), Key Project of Shanghai Innovation of Science and Technology Plan (NO 12JC1402900), Dawn Program of Shanghai Education Commission (grant 11SG31), Doctoral Fund of Ministry of Education of China, and by NIH grants HL 061795, HL 108630, HL 070819, and HL 048743 (to JL).
REFERENCES
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornberg O, Ostergaard H, Winther JR. Mechanistic insight provided by glutaredoxin within a fusion to redox-sensitive yellow fluorescent protein. Biochemistry. 2006;45:2362–2371. doi: 10.1021/bi0522495. [DOI] [PubMed] [Google Scholar]
- Blinova K, Carroll S, Bose S, Smirnov AV, Harvey JJ, Knutson JR, Balaban RS. Distribution of mitochondrial NADH fluorescence lifetimes: steady-state kinetics of matrix NADH interactions. Biochemistry. 2005;44:2585–2594. doi: 10.1021/bi0485124. [DOI] [PubMed] [Google Scholar]
- Brekasis D, Paget MS. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2) EMBO J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MB, James Remington S. Redox-sensitive green fluorescent protein: probes for dynamic intracellular redox responses. A review. Methods Mol Biol. 2009;476:50–64. doi: 10.1007/978-1-59745-129-1_4. [DOI] [PubMed] [Google Scholar]
- Chen S, Whetstine JR, Ghosh S, Hanover JA, Gali RR, Grosu P, Shi Y. The conserved NAD(H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc Natl Acad Sci U S A. 2009;106:1496–1501. doi: 10.1073/pnas.0802674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Ward Z, Carroll J, Duchen MR. Regulation of redox metabolism in the mouse oocyte and embryo. Development. 2007;134:455–465. doi: 10.1242/dev.02744. [DOI] [PubMed] [Google Scholar]
- Eto K, Tsubamoto Y, Terauchi Y, Sugiyama T, Kishimoto T, Takahashi N, Yamauchi N, Kubota N, Murayama S, Aizawa T, et al. Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science. 1999;283:981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- Gyan S, Shiohira Y, Sato I, Takeuchi M, Sato T. Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J Bacteriol. 2006;188:7062–7071. doi: 10.1128/JB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- Henley KS, Laughrey EG. The redox state of the mitochondrial NAD system in cirrhosis of the liver and in chronic quantitative undernutrition in the rat. Biochim Biophys Acta. 1970;201:9–12. doi: 10.1016/0304-4165(70)90003-6. [DOI] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging Cytosolic NADH-NAD(+) Redox State with a Genetically Encoded Fluorescent Biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP. Determination of pyridine dinucleotides in cell extracts by high-performance liquid chromatography. J Chromatogr. 1981;225:446–449. doi: 10.1016/s0378-4347(00)80293-5. [DOI] [PubMed] [Google Scholar]
- Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert F, Fales HM, Wen H, Combs CA, Balaban RS. NADH enzyme-dependent fluorescence recovery after photobleaching (ED-FRAP): applications to enzyme and mitochondrial reaction kinetics, in vitro. Biophys J. 2004;86:629–645. doi: 10.1016/S0006-3495(04)74141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimova MR, Grigiene J, Krab K, Hagedorn PH, Flyvbjerg H, Andersen PE, Moller IM. The free NADH concentration is kept constant in plant mitochondria under different metabolic conditions. Plant Cell. 2006;18:688–698. doi: 10.1105/tpc.105.039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayevsky A. Mitochondrial function and energy metabolism in cancer cells: past overview and future perspectives. Mitochondrion. 2009;9:165–179. doi: 10.1016/j.mito.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Mayevsky A, Rogatsky GG. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Physiol Cell Physiol. 2007;292:C615–640. doi: 10.1152/ajpcell.00249.2006. [DOI] [PubMed] [Google Scholar]
- McLaughlin KJ, Strain-Damerell CM, Xie K, Brekasis D, Soares AS, Paget MS, Kielkopf CL. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol Cell. 2010;38:563–575. doi: 10.1016/j.molcel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Menon SG, Goswami PC. A redox cycle within the cell cycle: ring in the old with the new. Oncogene. 2007;26:1101–1109. doi: 10.1038/sj.onc.1209895. [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Knobel SM, Arkhammar P, Thastrup O, Piston DW. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci U S A. 2000;97:5203–5207. doi: 10.1073/pnas.090098797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Neuroscience. Let there be (NADH) light. Science. 2004;305:50–52. doi: 10.1126/science.1100428. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemori K, Ishizaki T, Matsukawa S, Sakai A, Nakai T, Miyabo S. Adenine nucleotides via activation of ATP-sensitive K+ channels modulate hypoxic response in rat pulmonary artery. Am J Physiol. 1996;270:L803–809. doi: 10.1152/ajplung.1996.270.5.L803. [DOI] [PubMed] [Google Scholar]
- Sickmier EA, Brekasis D, Paranawithana S, Bonanno JB, Paget MS, Burley SK, Kielkopf CL. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Wakita M, Nishimura G, Tamura M. Some characteristics of the fluorescence lifetime of reduced pyridine nucleotides in isolated mitochondria, isolated hepatocytes, and perfused rat liver in situ. J Biochem. 1995;118:1151–1160. doi: 10.1093/oxfordjournals.jbchem.a125001. [DOI] [PubMed] [Google Scholar]
- Wang E, Bauer MC, Rogstam A, Linse S, Logan DT, von Wachenfeldt C. Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol Microbiol. 2008;69:466–478. doi: 10.1111/j.1365-2958.2008.06295.x. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- Yu Q, Heikal AA. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B. 2009;95:46–57. doi: 10.1016/j.jphotobiol.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, Xu L, Wang X, Yang Y, Loscalzo J. Genetically Encoded Fluorescent Sensors for Intracellular NADH Detection. Cell Metab. 2011;14:555–566. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang Y. Frex and FrexH: Indicators of metabolic states in living cells. Bioeng Bugs. 2012;3:181–188. doi: 10.4161/bbug.19769. [DOI] [PMC free article] [PubMed] [Google Scholar]


