Abstract
Initial bacterial colonization of the gut is a vital component of the development of the gastrointestinal tract, particularly mucosal immune protection, during the neonatal period. Newborn infants in their protected intrauterine environment are suddenly thrust into a highly contaminated extrauterine state. Although mucosal host defenses have developed in utero during fetal maturation due to the stimulation of ingested trophic factors in amniotic fluid, actual active protection only occurs when colonizing bacteria stimulate the gut mucosal barrier. Colonization evolves over a period of about one year and is dependent on the mode of delivery, use of perinatal antibiotics, age at birth and infant feeding. A fully colonized gut consists of 1014 bacteria, establishes a symbiotic relationship with the host and insures normal development and immune homeostasis. Colonizing bacteria can also affect the epithelial mucosal barrier and the innate and adaptive immune systems. Disruption of normal colonization, dysbiosis, is associated with increased expression of disease. Evidence exists that the use of probiotics with dysbiosis may prevent disease expression.
Keywords: intestinal colonization, intestinal defenses, probiotics
INTRODUCTION
Over the last few decades, we have become aware of the importance of initial bacterial colonization in the development of the gastrointestinal tract, particularly mucosal immune protection, during the neonatal period. Appropriate colonization stimulates the maturation of immune defenses to provide protection for the newborn in the extrauterine environment. Normal initial colonization is affected by numerous environmental factors (birth process, use of antibiotics, age of delivery, nature of initial feeding). Under ideal conditions, a full term infant born by vaginal delivery without antibiotics and exclusively breast fed appropriately colonizes the gut in stages over the first year of life. With complete colonization, an individualized, genetically-defined signature of bacteria unique to that individual is established which principally colonizes the distal small intestine and colon with 1014 organisms. This microbiome is made up of balanced phyla (large families of bacteria) and contains about a thousand different species of bacteria. These microbes are said to make up an ancillary body organ which establishes a symbiotic relationship with the human host via expressed molecules on the surface of bacteria or as secreted product of bacteria into the intestinal lumen. These symbiotic bacteria communicate with intestinal epithelial and lymphoid cells via pattern recognition receptors, e.g., toll-like receptors (TLRs), which trigger cellular signaling, activation of transcription factors and translation of cytokines, chemokines, etc. that mediate the immune response, enterocyte development and mucosal barrier function. This review will briefly consider how colonizing bacteria stimulate programming of the intestinal immune response.
SYMBIOSIS AND IMMUNE FUNCTION
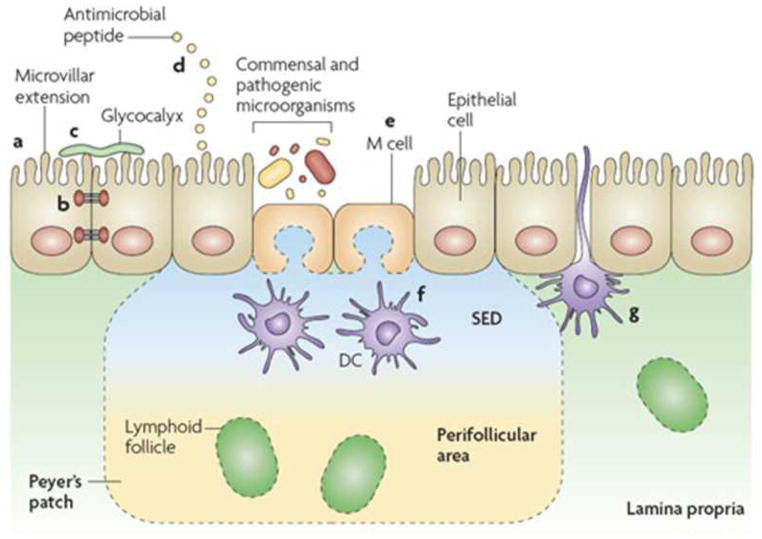
The interaction of symbiotic commensal bacteria with the gut during colonization impacts on development of important mucosal barrier epithelial functions (FIGURE 1) (1). For example, colonizing bacteria can stimulate the release of mucus from goblet cells to coat the intestinal surface as a glycocalyx which contributes an added protective barrier. In addition, commensal bacteria can stimulate the epithelial transcription and translation of tight junction proteins to close the intercellular route for the uptake of large molecules and microbiota. Furthermore, intestinal microbiota can stimulate epithelial cells and Paneth cells to release anti-microbial molecules (defensins) to provide an additional protection against pathologic bacterial penetration.
Figure 1.
The intestinal epithelial-cell barrier. Simple columnar epithelial cells exhibit physical and biochemical adaptations to microbial colonization to maintain barrier integrity including actin-rich microvillar extensions (a), epithelial-cell tight junctions (b), apically attached and secreted mucins that form a glycocalyx (c) and the production of various anti microbial peptides (d). Specialized intestinal epithelial cells known as M (microfold) cells overlie Peyer’s patches and lymphoid follicles to facilitate luminal sampling. M cells exhibit reduced mucin secretion and have modified apical and basolateral surfaces (e) to promote uptake and transport of luminal contents to professional antigen-presenting cells that inhabit the subepithelial dome (SED) of the Peyer’s patches and lymphoid follicles (f). Specialized dendritic cell (CD)subsets can also extend dendrites between the tight junctions of intestinal epithelial cells to sample luminal contents (g). (Reproduced with permission from reference 1.)
In like manner, colonizing bacteria through its symbiotic relationship with gut immune cells can stimulate both innate and adaptive immunity. Intestinal epithelial cells, dendritic cells and lymphocytes express pattern recognition receptors on their cell surface and within their cytoplasm. Molecular patterns expressed on the bacterial cell surface or secreted into the intestinal lumen can stimulate these pattern recognition receptors to evoke a self-limited, acute innate inflammatory response to prevent pathogen penetration of the epithelium (2). The most studied pattern recognition receptors are the toll receptors TLRs. Gram negative bacteria through lypopolysaccarhide (LPS) on their surface can interact with TLR-4 receptors evoking a signaling transduction response leading to a cytoplasmic release of NFκB, a transcription factor, which enters the nucleus and interacts with chromosomes to stimulate the transcription and eventual translation of the chemokine IL-8 which in turn attacks the migration of neutrophils to the site of pathogen interaction. In addition, inflammatory cytokines such as TNF-α are released to activate an inflammatory response to certain pathogens. To create a self-limited inflammatory response and not chronic inflammation, sustained interaction of LPS with TLR-4 activates additional molecules that inhibit the NFκB activation response (LPS tolerance). In like manner peptidoglycans expressed on gram positive organisms can interact with TLR-2 to invoke a self-limited inflammatory response to contain pathogen access to the blood stream (3).
Recent publications (4,5) have shown a close association between the innate and adaptive immune responses mediated by colonizing commensal bacteria. Dendritic cells underlying the intestinal epithelium can separate tight junctions and penetrate its appendage expressing TLRs into the lumen. Interaction of molecular patterns and colonizing bacteria with these expressed TLRs on dendritic cells can invoke a release of cytokines, etc. that create a microenvironment that allows naïve T-helper cells (Th0) to differentiate into a balanced Th sub-class response, e.g., Th1, Th2, Th17 and Tregs (FIGURE 2). This interaction leads to intestinal immune homeostasis and no expression of disease. In the absence of adequate initial colonization, a dysbiosis occurs and there is an increase in immune-mediated diseases (allergy, autoimmune disease including type 1 diabetes).
Figure 2.
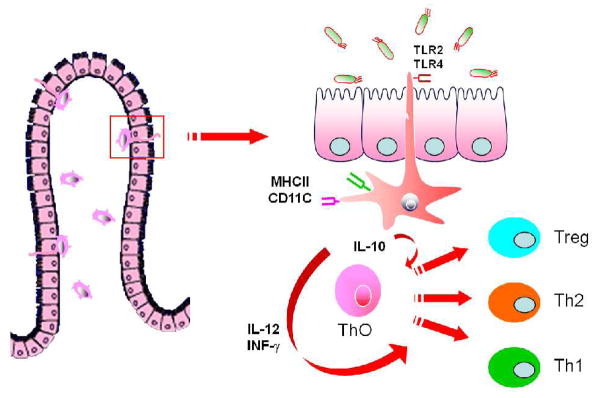
Cartoon depicting sub-epithelial dendritic cells extending their appendages between enterocytes into the intestinal lumen. These appendices, displaying TLRs, are activated by colonizing bacteria, mature and secrete cytokines which activate naïve T cells (TH0) to mature into a balanced T helper cell response (Th1, Th2, Th17 and Treg). (Reproduced with permission from reference 20.)
A major development of mucosal immune homeostasis involves the capacity of T-helper cells to produce tolerance to innocuous antigens and bacteria. Tolerance can not occur in the absence of intestinal colonization (6) and is most effective if colonization occurs in the newborn period (7). Recently it has been shown that breast feeding as the first source of oral feeding for the neonate has an effect on the nature of expression of health promoting bacteria (8). Specific organisms activated by factors in breast milk preferentially stimulate an increase in Bacteroides, Bifidobacteria and Lactobacillus organisms. These organisms have important specific effects on neonatal immune function. For example, a polysaccharide (PSA) expressed on a Bacteroides fragillis species can interact with the TLR-2 receptor on dendritic cells and preferentially create a milieu that stimulates Treg cells (9). Other organisms such as Clostridial species can also provide the same effect. In like manner, Bifidobacteria infantis can preferentially activate B cells to mature into pIgA producing plasma cells (10). These immunoglobulins coat the intestinal surface and help protect against pathogen penetration.
INFANT NUTRITION AND COLONIZATION
An important environmental factor in intestinal colonization is the nature of the diet. We know that specific diets in adults over long periods of time evoke the large families of intestinal bacteria which act as functional units called enterotypes (11). We also know that the composition of intestinal bacteria in African children living in a primitive setting and ingesting a high fiber complex carbohydrate diet compared to age-matched children in Florence, Italy ingesting a typical Westernized diet with processed food and animal fat and protein is strikingly different (12). Since we know that the composition of intestinal bacteria influences the phenotypic expression of disease, this observation may explain the considerable difference between the disease burden in these two child populations.
At no time in the children’s life is nutrition, as an environmental factor for bacterial colonization more important than with initial feeding at birth at the time of the first colonization. More specifically, as mentioned breast fed infants have a different intestinal microbiota than formula fed infants (13). Not only are health-promoting bacteria (Bifidobacteria, Bacteroides, Clostridial and Lactobacillus) present early with breast feeding, but the genes produced by these organisms have a profound effect on bacterial components that are secreted and in turn an effect on the enterocyte’s response with regard to development and protection (14). Several factors in breast milk (non-metabolized oligosaccharide, lysozyme, lactoferrin, antibodies and cytokines) alter the intestinal milieu to stimulate specific indigenous bacteria to proliferate (15).
DYSBIOSIS AND DISEASE
As mentioned before, specific disease states are associated with a dysbiotic microbiome different from their age-matched normal controls (FIGURE 3) (16). Studies transferring the intestinal microbiome from patients with clinical conditions (obesity, diabetes, etc.) or animal models of autoimmune disease to germ-free animals cause these animals to express the disease suggesting that intestinal microbiota at the very least function as an intermediary in disease expression. Although not proven some investigators suggest that microbiota may be the actual cause of disease.
Figure 3.
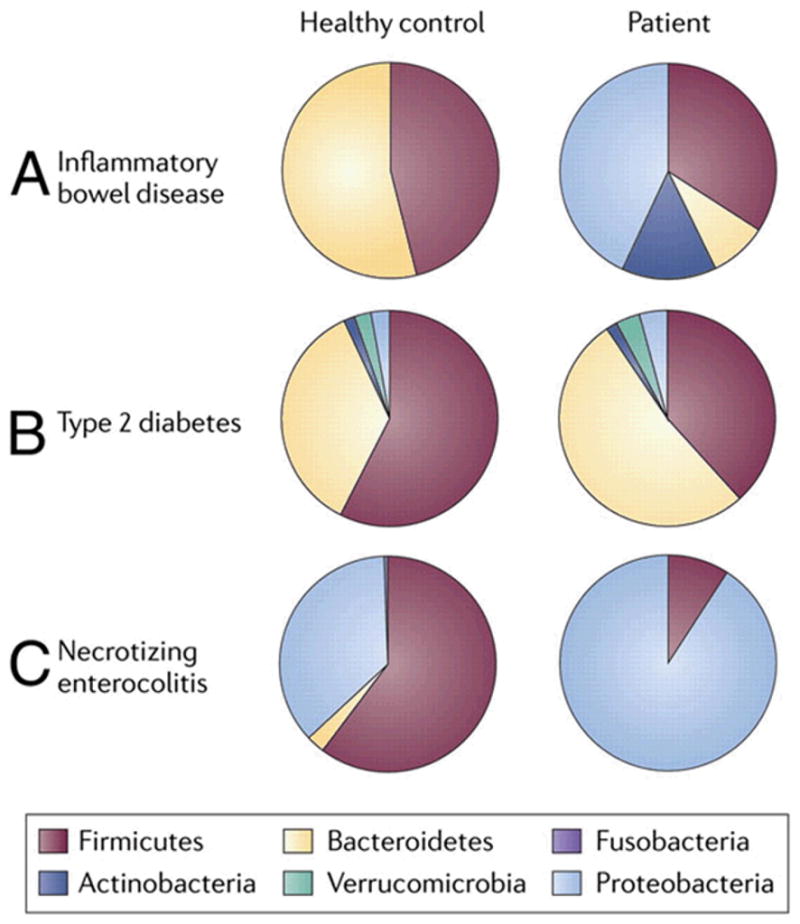
Disease states reveal phylum-level differences compared with healthy controls. Comparisons of the relative abundances of predominant bacterial phyla in IBD, type 2 diabetes, and NEC compared with healthy controls. Fecal samples from infants with NEC and patients with type 2 diabetes were compared with healthy controls revealing a predominance of Proteobacteria in patients with NEC. Cecal samples from patients with IBD were compared with healthy controls, and relative abundances were assessed. (Reproduced with permission from reference 16.)
There are conditions in which the phenotypic expression of disease associated with a dysbiosis may be improved with probiotics. Unfortunately most of the reported conditions have not been subjected to a multi-center, single protocol, double blind study which is necessary and in whose absence weakens the conclusions, even with strong evidence by meta-analysis support. One condition with a known dysbiosis is necrotizing enterocolitis (NEC) which occurs in less than 1500 gms premature at an incidence of between 7 and 12%. These studies have shown that several known probiotics can help to prevent or lessen the severity of NEC and several meta-analysis are supportive. One multi-center study done in Taiwan showed that a combination of Bifidobacteria infantis and Lactobacillus acidophilus given with expressed breast milk from the birth mother was effective (17). We have shown that a 30 kD glycan secreted from Bifidobacteria infantis is anti-inflammatory in human in vitro intestinal models (18) and in vivo in a mouse model of newborn inflammation (19). We plan a clinical trial with a specific protocol in premature newborns after an appropriate safety studies are done.
SUMMARY AND CONCLUSIONS
Extensive studies over the last decade have shown the importance of intestinal colonization in gut function, particularly protective function. The importance of initial colonization is emphasized since the process has a profound effect on the development of mucosal defenses leading to immune homeostasis and the decreased expression of neonatal infectious diseases and immune-mediated disease states later in life. Breast feeding is an important environmental determinant of normal colonization during infancy. Dysbiotic states leading to diseases, e.g. NEC in prematures, may be prevented by aggressive use of probiotics.
Acknowledgments
Supported by National Institute of Health (NIH) grants: National Institute of Digestive, Diabetes and Kidney Diseases P30 DK040561, P01 DK033506; and National Institute of Child Health and Human Development R01 HD012437, R01 HD059126, Bethesda, MD, USA
Footnotes
Disclosure statement: There is no conflict of interest
References
- 1.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian The gut microbiota shapes intestinal responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neish AS. Reviews in basic and clinical gastroenterology: Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Ann Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudo N, Sawamura A, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Imunol. 1997;159:1739–1745. [PubMed] [Google Scholar]
- 7.Karlsson MR, Kahu H, Hanson LA, Telemo E, Dahlgren UIH. Neonatal colonization of rates induces immunological tolerance to bacterial antigens. Eur J Imunol. 1999;29:109–118. doi: 10.1002/(SICI)1521-4141(199901)29:01<109::AID-IMMU109>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Jost T, Lacroix C, Braegger C, Chassard C. New insights into gut microbiota establishment in healthy breast fed neonates. Plos One. 2012;7:e44595–e44603. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 10.Macpherson JS, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 11.Arumugam M, Raes J, Pelletier E, LePaslier, Yamada T, Mande DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFillippo C, Cavalieri D, DiPaola M, Ramazzotti M, Poullet JP, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nature Rev: Gastroenterol & Hepatol. 2010;7:503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandtzaeg P. The mucosal immune system and its integration with the mammary glands. J Pediatr. 2010;156:S8–S15. doi: 10.1016/j.jpeds.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Ballard O, Morrow AL. Human milk composition: Nutrietns and bioavtive factors. Pediatr Clin North Am. 2013;60:49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson CL, Versalovic J. The human microbiome and its potential important to pediatrics. Pediatrics. 2012;129:950–960. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin PW, Nasr TR, Berardinelli AJ, Kumar A, Neish AS. The probiotic Lactobacillus GG may augment intestinal host defense by regulation apoptosis and promoting cytoprotective responses in the developing murine gut. Pediatr Res. 2008;64:511–56. doi: 10.1203/PDR.0b013e3181827c0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguli K, Meng D, Rautava S, Lu L, Walker WA*, Nanthakumar N* Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am J Phys: Gastrointestinal and Liver Physiology. 2013;304:G132–G141. doi: 10.1152/ajpgi.00142.2012. (* shared senior authorship) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng M, Ganguli K, Zhu W, Shi HN, Walker WA. Conditioned media from Bifidobacteria infantis protects against Cronobacter seakzakii-induced intestinal inflammation in newborn mice. Am J Phys- Gastrointestinal and Liver Physiol. 2014 doi: 10.1152/ajpgi.00183.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan JL, Shi H, Walker WA. The role of microbes in developmental immunologic programming: Convergence of the Barker and Hygiene Hypotheses. Pediatr Res. 2011;9(6):465–472. doi: 10.1203/PDR.0b013e318217638a. [DOI] [PubMed] [Google Scholar]
- 21.Walker WA. Bacterial colonization, probiotics and the development of intestinal host defense. Functional Food Reviews. 2009 Spring;1(1):13–19. [Google Scholar]