Abstract
Fungal diseases pose constant threats to the global economy and food safety. As the largest group of plant fungal pathogens, necrotrophic fungi cause heavy crop losses worldwide. The molecular mechanisms of the interaction between necrotrophic fungi and plants are complex and involve sophisticated recognition and signaling networks. Here, we review recent findings on the roles of phytotoxin and proteinaceous effectors, pathogen-associated molecular patterns (PAMPs), and small RNAs from necrotrophic fungi. We also consider the functions of damage-associated molecular patterns (DAMPs), the receptor-like protein kinase BIK1, and epigenetic regulation in plant immunity to necrotrophic fungi.
Keywords: effectors, necrotrophic fungi, innate immunity, defense response, PRR and epigenetic modification
Introduction
Plant fungal diseases and biotrophic, hemibiotrophic, and necrotrophic pathogens
Plant fungal diseases represent a worldwide threat to food security and ecosystem health.1 Based on their lifestyle, plant-pathogenic fungi have been classified as biotrophs, hemi-biotrophs, and necrotrophs. Biotrophic pathogens must obtain nutrients from living host cells and tissues and often secrete limited amounts of cell wall-degrading enzymes and effectors to suppress the host immune system.2 In contrast, necrotrophic pathogens thrive on the dead host tissues that they kill before or during colonization; to induce cell necrosis, they often secrete phytotoxic secondary metabolites (SMs) and peptides, and produce reactive oxygen species (ROS).3 Hemi-biotrophic pathogens display a biotrophic phase early during infection and display a necrotrophic phase only later; these pathogens produce toxins only at late stages in order to kill the host cells and complete their life cycle on dead tissues.3
Necrotrophs can be further divided into host-specific and broad host-range species. Host-specific necrotrophs produce host-specific toxins (HSTs) that are toxic only to host plants of the fungus. Host-specific necrotrophs include Cochliobolus carbonum (causal agent of northern corn leaf spot), C. heterostrophus (causal agent of southern corn leaf blight), C. victoriae (causal agent of Victoria blight of oats), Parastagonospora nodorum (previously Stagonospora nodorum,4 causal agent of Stagonospora nodorum blotch of wheat), and Pyrenophora tritici-repentis (causal agent of tan spot of wheat). The archetypical broad host-range fungal necrotrophs are Botrytis cinerea, Alternaria brassicicola, Plectosphaerella cucumerina, and Sclerotinia sclerotiorum.
Necrotrophic pathogens cause severe economic losses in agriculture
The economic impact of necrotrophic pathogens on agriculture was highlighted by a recent survey.5 The report indicated that the losses in wheat and barley in Australia resulting from tan spot and Stagonospora nodorum blotch, both of which are caused by necrotrophic pathogens, significantly exceeded losses resulting from wheat rusts and mildews, which are caused by biotrophic pathogens. In addition, the necrotroph B. cinerea infects almost all vegetable and fruit crops and annually results in worldwide losses of $10 to $100 billion. It is clearly important to develop effective methods to control plant diseases caused by necrotrophic fungi. Knowledge on the mechanism of pathogen virulence and host immune responses is most relevant to future management of necrotrophic pathogens.
Plant innate immunity
Because they lack somatic adaptive immune systems, plants depend solely on innate immunity to cope with pathogens.6-8 Regardless of the lifestyle of the attacking pathogen, the plant innate immune system has two layers: pathogen-associated molecular pattern (PAMP)-triggered immunity or PTI, and effector-triggered immunity or ETI. PTI is the first layer of innate immunity and is initiated in plants when PAMPs are recognized by pattern recognition receptors (PRRs); such recognition triggers a relatively weak but broad-spectrum immune response to pathogen infection. In contrast, ETI (the second layer of innate immunity) is induced by direct or indirect recognition of highly variable pathogen avirulence effectors by host disease-resistance (R) proteins; the recognition in this case leads to a rapid and robust response that is often referred to as a hypersensitive reaction (HR).
Plant immune responses to necrotrophs may be similar to or different from plant immune responses to biotrophs depending on the pathogen species and the primary determinant of virulence. In the case of necrotrophs, plant immune systems are very complex and reflect the multiplicity of necrotroph virulence mechanisms targeting diverse host cellular processes. Plants have evolved effective immune responses to counter the “pro-death” virulence strategies of necrotrophic fungi. Recent research has increased our understanding of the recognition events and defense signaling processes in necrotroph–host interactions. In this review, we highlight the recent advances in elucidating the roles of immune-related molecules from both necrotrophic fungi and plants in various plant pathosystems (Fig. 1).
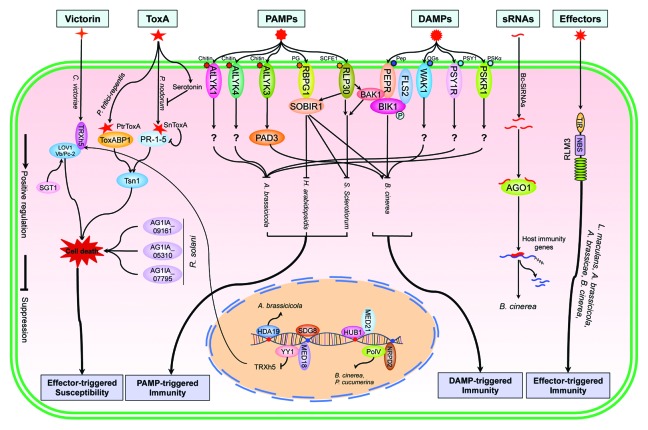
Figure 1. The major immune signaling pathways in the interaction between necrotrophic fungi and plants. The effector victorin binds to the host virulence target Trx-h5, which activates the NBS-LRR protein LOV1-mediated susceptibility to Cochliobolus victoriae. The transcription of Trx-h5 is regulated by the transcription factor YY1 through the interaction with the mediator MED18. In addition, the chaperone SGT1 is required for victorin-mediated cell death by affecting the accumulation of LOV1.ToxA-triggered susceptibility to necrotrophic pathogens is governed by an R-like protein Tsn1. PtrToxA targets to a chloroplastic protein ToxABP1 and this interaction may trigger ToxA-mediated cell death. Moreover, a pathogenesis-related protein PR-1–5 is a potential target of SnToxA and the interaction between PR-1–5 and ToxA may mediate ToxA-induced necrosis in sensitive wheat. Three effectors, AG1IA_09161, AG1IA_05310 and AG1IA_07795, secreted by Rhizoctonia solani are delivered into rice cells and induce cell death in rice. Two major LysM-containing receptor-like kinases AtLYK1 and AtLYK4 perceive the PAMP chitin to induce a PTI response. AtLYK1 and AtLYK4 positively regulate Arabidopsis resistance to necrotrophic fungi. However, AtLYK3 as a negative regulator in Arabidopsis modulates the resistance to the necrotrophic fungi depending on PAD3. The PAMPs PG and SCFE1 are recognized by the Arabidopsis LRR-RLP RBPG1 and RLP30, respectively, to trigger an Arabidopsis PTI response. SOBIR1 is required for the activation of RBPG1- and RLP30-mediated immune response. SCFE1-triggered immune responses also require the LRR-RLK BAK1. The DAMPs OGs, PEP1, PSKα and PSY1 are perceived by the PRRs WAK1, PEPR1/PEPR2, PSKR1 and PSY1R, respectively. DAMP and PRR recognition can trigger immune responses and may overlap with PAMP-triggered immunity. In particular, the Arabidopsis peptide Pep1 triggers immunity through the receptor kinases PEPR1 and PEPR2. PEPR1 directly phosphorylates BIK1 and BAK1 to activate downstream signaling. Some small RNAs delivered by Botryits cinerea into host cells can bind to the Arabidopsis RNA interference machinery and suppress host immune responses. The TIR-NB RLM3 protein shows a gene-for-gene resistance relationship to the semibiotrophic fungus Leptosphaeria maculans and three necrotrophic fungi B. cinerea, A. brassicicola, and A. brassicae although the cognate effector needs to be determined.
Effector-Triggered Immunity to Necrotrophic Fungi
Gene-for-gene resistance to necrotrophic fungi is rare in plants. The semibiotrophic fungus Leptosphaeria maculans displays a gene-for-gene relationship with both Brassica napus and Arabidopsis.9 Using a microarray-based cloning strategy, researchers identified the RLM3 locus on chromosome 4 of Arabidopsis.10 RLM3 encodes a putative Toll interleukin-1 receptor-nucleotide binding (TIR-NB) class protein. Interestingly, the rlm3 mutant not only loses resistance to L. maculans but also exhibits enhanced susceptibility to the necrotrophic fungi B. cinerea, A. brassicicola, and A. brassicae. A 3:1 segregation of resistance against A. brassicicola, A. brassiciae, and B. cinerea was observed in the F2 population, indicating that RLM3 is a single dominant gene that governs resistance to these necrotrophic pathogens. The effector of RLM3 in the necrotrophic pathogens and the function of the RLM3 gene in response to these pathogens are unknown.
Effector-Triggered Susceptibility to Necrotrophic Fungi
In a broad sense, effectors are defined as any pathogen proteins and small molecules that can alter the structure and function of host cells.11 Effectors of necrotrophic pathogens include phytotoxins and traditional proteinaceous effectors (Table 1). Phytotoxins can be either non-HSTs that affect a broad range of plant species or HSTs that affect only a particular plant species or more often genotypes of that species.53,54 Based on their chemical structure, phytotoxins are commonly classified as polyketides, nonribosomal peptides, alkaloids, terpenes, or metabolites of mixed biosynthetic origin.55 So far, the protein effectors found in necrotrophs don't have a conserved domain like the RXLR motif in oomycetes.56
Table 1. Overview of effectors of necrotrophic fungi.
| Pathogen | Effectors | Structure | Plant target | R gene | References |
|---|---|---|---|---|---|
| Parastagonospora nodorum | SnToxA | Protein | Chloroplasts, ToxABP1 | Tsn1 | 12 and 13 |
| P. nodorum | SnTox1 | Protein | Probably chloroplasts | Snn1 | 14 and 15 |
| P. nodorum | SnTox2 | Protein | Probably chloroplasts | Snn2 | 16 |
| P. nodorum | SnTox3 | Protein | Unknown | Snn3 | 17 |
| P. nodorum | SnTox4 | Protein | Probably chloroplasts | Snn4 | 18 |
| P. nodorum | SnTox5 | Protein | Unknown | Snn5 | 19 |
| Pyrenophora tritici-repentis | PtrToxA | Protein | Chloroplasts, ToxABP1 | Tsn1 | 12, 13, 20, and 21 |
| P. tritici-repentis | PtrToxB | Protein | Probably chloroplasts | Tsc2 | 22 |
| Alternaria alternata | AM-toxin | Cyclic depsipeptide | Plasma membrane and chloroplasts | Unknown | 23 and 24 |
| A. alternata | AAL-toxin | Aminopentol ester | Asc | Unknown | 25 and 26 |
| A. alternata | AT-toxin | Unknown | Unknown | Unknown | 27 and 28 |
| A. alternata | AF-toxin | Epoxy-decatrienoic acid | Unknown | Unknown | 29 and 30 |
| A. alternata | AK-toxin | Epoxy-decatrienoic acid | Unknown | Unknown | 27 and 31 |
| A. alternata | ACT-toxin | Epoxy-decatrienoic acid | Unknown | Unknown | 32 |
| A. alternata | ACR-toxin | Polyketide | ACRS | Unknown | 33–35 |
| Cochliobolus heterostrophus | T-toxin | Linear polyketide | URF13 | Unknown | 36 and 37 |
| C. carbonum | HC-toxin | Cyclic tetrapeptide | HDACs | Unknown | 38 |
| C. victoriae | victorin | Cyclic chlorinated pentapeptide | Unknown | LOV1 | 39 and 40 |
| Rhynchosporium commune | NIP1 | A phytotoxic protein | Plasma membrane H+-ATPase | Rrs1 | 41–43 |
| R. commune | NIP2 | Protein | Unknown | Unknown | 41 |
| R. commune | NIP3 | A glycoprotein | Plasma membrane H+-ATPase | Unknown | 41 and 42 |
| Botrytis cinerea | NEP1-like | Protein | Cell membranes and nuclear envelope | Unknown | 44 |
| Mycosphaerella zeae-maydis | PM-toxin | Linear polyketide | URF13 13-KDa mitochondrial protein | Unknown | 45 |
| Periconia circinata | PC toxin | Unknown | Unknown | PC | 46 |
| Sclerotinia sclerotiorum | SSITL | Integrin protein | Unknown | Unknown | 47 |
| S. sclerotiorum | Ss-caf1 | Protein with a putative Ca2+-binding EF-hand motif | Unknown | Unknown | 48 |
| S. sclerotiorum | SSV263 | A hypothetical secreted novel protein | Unknown | Unknown | 49 |
| S. sclerotiorum | Sspg1d | Endopolygalacturonases | IPG-1 | Unknown | 50 and 51 |
| R. solani | AG1IA_09161 | Protein with a glycosyltransferase GT family 2 domain | Unknown | Unknown | 52 |
| R. solani | AG1IA_05310 | Protein with a cytochrome C oxidase assembly protein CtaG/cox11 domain | Unknown | Unknown | 52 |
| R. solani | AG1IA_07795 | Protein with a peptidase inhibitor I9 domain | Unknown | Unknown | 52 |
ETI is triggered by the recognition of pathogen effectors by plant R proteins, a recognition that leads to a HR and localized host cell death.57 As a consequence, biotrophic pathogens, which require living host tissues, fail to survive and infect. Interestingly, HSTs secreted by necrotrophic fungi activate R protein-mediated ETI to cause HR cell death, which leads to effector-triggered susceptibility (ETS), as is the case for the cyclic peptide HST victorin that is produced and secreted by C. victoriae. Pathogenesis by C. victoriae, the causal agent of Victoria blight of oats, is determined by its production of victorin.58,59 Victoria blight exclusively appeared on oat plants carrying the R gene Pc2, which is associated with disease resistance to the biotrophic fungus Puccinia coronate.60 HST victorin sensitivity is dominated by the Vb R gene in oats. The results of many genetic and mutagenic efforts to separate Pc2 and Vb suggest that Pc2 and Vb are the same gene.61-64 Susceptibility of Arabidopsis to C. victoriae is mediated by the NBS-LRR R protein LOCUS ORCHESTRATING VICTORIN EFFECTS1 (LOV1).39
TRX-h5, a defense-associated thioredoxin, is required for victorin sensitivity mediated by LOV1 in Arabidopsis, and the trx-h5 mutant is insensitive to victorin.65 In LOV1’s absence, victorin inhibits TRX-h5, resulting in compromised defense but no disease symptoms after C. victoriae infection. In LOV1’s presence, the binding of victorin to TRX-h5 activates LOV1 and elicits a HR-like response that confers susceptibility to C. victoriae. Recently, Lai et al.66 confirmed that the transcription of the TRX-h5 gene is repressed by the transcription factor YIN YANG1 through the interaction with mediator MED18. The chaperone SGT1 (SUPPRESSOR OF G2 ALLELE OF SKP1), which affects the accumulation of R proteins, is also involved in victorin-mediated cell death resulting from the reduced accumulation of LOV1 protein.67 In addition, the silencing of six genes that encode different metabolic enzymes in Nicotiana benthamiana suppresses the LOV1-mediated, victorin-induced cell death. Simultaneously, cell death induced by a closely related RPP8 R gene is also suppressed due to silencing of these six genes.67
The proteinaceous HST ToxA is produced by two fungal pathogens of wheat, P. nodorum and P. tritici-repentis.12,20,68 The ToxA gene was originated in P. nodorum and was only recently transferred to P. tritici-repentis through interspecies hybridization69,70 The dominant Tsn1 allele in wheat confers susceptibility to ToxA.12,71 In P. tritici-repentis, PtrToxA activates host responses that are typically observed in resistance responses to biotrophic pathogens, thereby providing additional evidence that necrotrophic pathogens such as P. tritici-repentis subvert host resistance mechanisms to cause disease.72-74 Recently, Du et al.75 demonstrate the induction of the monoamine serotonin in wheat after SnToxA infiltration. As a phytoalexin, serotonin can inhibit sporulation of P. nodorum by interfering with spore formation and maturation within pycnidial structures.75 PtrToxA targets to the chloroplastic protein ToxABP1 and this interaction may induce alterations in photosystems I and II leading to a light-dependent accumulation of ROS in chloroplasts that disrupts their photosynthetic capacity and triggers PCD.13,14,73 In addition, PR-1–5, a dimeric PR-1-type pathogenesis-related protein, is a potential target of ToxA, and the site-specific interaction between PR-1–5 and ToxA may mediate ToxA-induced necrosis in sensitive wheat.76 How the ToxABP1 and PR-1–5 mediate ToxA-induced necrosis remain to be investigated.
PAMP-Triggered Immunity to Necrotrophic Fungi
PAMPs are conserved microbe-specific molecules or signatures that activate the plant defense response in a manner analogous to the way in which molecules trigger an immune response in animals.77 PAMPs are often structural components of the pathogen cell wall or other conserved macromolecules.78 As noted earlier, PAMPs are perceived by PRRs, which are currently divided into receptor-like kinase proteins (RLKs) and receptor-like proteins (RLPs). RLKs have a cytoplasmic kinase domain that participates in intracellular signal transduction and an extracellular domain that is potentially responsible for ligand perception. RLPs have structures similar to RLKs but lack the cytoplasmic kinase domain. Plant resistance against necrotrophic pathogens with a broad host range is considered to be complex early in the research. Recent studies showed that the plant resistance to necrotrophs also involves PRR perception of PAMPs (Table 2). The following examples demonstrate the relevance of PTI in resistance against necrotrophic fungi.
Table 2. Overview of PAMPs/DAMPs of necrotrophic fungi and plant pattern recognition receptors (PRRs).
| Molecule | PAMP/DAMP | Structure | PRR | PRR structure | Target | References |
|---|---|---|---|---|---|---|
| Chitin | PAMP | A polymer of N-acetyl-D-glucosamine | AtCERK1/ AtLYK4 | LysM receptor kinase | ? | 79–81 |
| PGs | PAMP | Enzyme | RBPG1 | Leucine-rich repeat receptor-like protein | SOBIR1 | 82 |
| SCFE1 | PAMP | Peptide | RLP30 | Receptor like protein | BAK1/SOBIR1 | 83 |
| Pep1/Pep2 | DAMP | Peptide | PEPR1/ PEPR2 | Leucine-rich repeat protein kinase | ? | 84 and 85 |
| OGs | DAMP | A polymer of 1,4-linked α-D-galacturonic acid | WAK1 | Wall-associated kinase1 | ? | 86 |
| PSKα and PSY1 | DAMP | The tyrosine-sulfated peptides | PSKR1 | Phytosulfokine (PSK) receptor | ? | 110 and 111 |
Chitin
Chitin perception and signaling has been well characterized in Arabidopsis. Chitin perception depends on the lysin motif (LysM)-containing receptor-like kinases such as LysM RLK1/CHITIN ELICITOR RECEPTOR KINASE 1 (AtLYK1/AtCERK1). Arabidopsis resistance against an incompatible fungus, A. brassicicola, was partly impaired in the AtLYK1/AtCERK1 mutant.79,80 The lysM domain directly binds to chitin, and the intracellular kinase domain is responsible for the activation of the downstream signaling.79,80 The binding of chitin to the LysM motif induces the dimerization of AtCERK1, which is essential for the downstream response signaling.87 In addition, AtLYK4 binds to chitin and is required for full induction of chitin signaling; the lyk4 mutant confers enhanced susceptibility to the necrotrophic fungus A. brassicicola.81
There are five LYK genes (AtLYK1/AtCERK1, AtLYK2, AtLYK3, AtLYK4, and AtLYK5) in Arabidopsis. In contrast to the other members of the Arabidopsis LYK gene family, AtLYK3 is transcriptionally repressed in response to B. cinerea infection and to treatments with different elicitors, including chitin.88-90 The atlyk3 mutant shows increased expression of basal levels of defense-related genes and enhanced resistance to B. cinerea. Furthermore, the enhanced resistance of atlyk3 to B. cinerea depends on the increased expression of PHYTOALEXIN DEFICIENT 3 (PAD3), a defense-related gene that is regulated by fungal infection and elicitors independently of SA-, JA-, and ethylene-mediated pathways.88 In addition, AtLYK3 is a positive regulator of ABA signaling, which is involved in the plant immune response to necrotrophic fungi.91 These results demonstrate that AtLYK3 regulates the cross talk between immunity and ABA responses.91
Fungal endopolygalacturonases
Fungal endopolygalacturonases (PGs) act as PAMPs that are recognized by the Arabidopsis LRR-RLP RBPG1 (RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1). Infiltration of B. cinerea PGs into the Arabidopsis accession Columbia induced necrotic cell death. Coimmunoprecipitation experiments demonstrated that RBPG1 and PG form a complex in Nicotiana benthamiana, which also involves the Arabidopsis leucine-rich repeat receptor-like protein SOBIR1 (for SUPPRESSOR OF BIR1).82 PGs do not induce necrosis in sobir1 mutant plants, and PG-induced resistance to Hyaloperonospora arabidopsidis is compromised in such plants.82
SCLEROTINIA CULTURE FILTRATE ELICITOR1 (SCFE1)
Recently, the novel proteinaceous elicitor SCFE1 was identified in the necrotrophic fungal pathogen S. sclerotiorum.83 Its corresponding receptor in Arabidopsis is the RECEPTOR LIKE PROTEIN30 (RLP30). The rlp30 mutant is more susceptible than the wild type to both S. sclerotiorum and B. cinerea. In addition, SCFE1-mediated immunity is dependent of the receptor-like kinase BRASSINOSTEROID (BR) INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1, BAK1, and SOBIR1/EVR. Double mutants of bak1 and sobir1 are more susceptible to S. sclerotiorum and the related fungus B. cinerea than the wild type.83
Three new PAMP effectors identified from Rhizoctonia solani
The fungus Rhizoctonia solani is an important soil-borne, necrotrophic pathogen with a broad host range.1 R. solani is the causal agent of the sheath blight of rice, a disease that causes severe yield losses in many rice-growing areas. There is little effective resistance against R. solani in rice or other crop plants. A large and diverse set of secreted proteins, enzymes of primary and secondary metabolism, carbohydrate-active enzymes, and transporters were identified from the draft sequence of the AG1 IA strain of R. solani.52 Among the 25 candidate pathogen effectors, three secreted effectors, AG1IA_09161 (glycosyltransferase GT family 2 domain), AG1IA_05310 (cytochrome C oxidase assembly protein CtaG/cox11 domain), and AG1IA_07795 (peptidase inhibitor I9 domain) caused cell death phenotypes after inoculation in rice. In addition, these effectors show host-specific toxin characteristics for different hosts (rice, maize, and soybean). These results demonstrate that the three effectors are delivered into rice cells and might play a role in inducing cell death in rice during infection. How these effectors are delivered into rice cells and the identity of their host targets, however, remain unknown.
Damage-Associated Molecular Pattern (DAMP)-Triggered Immunity
DAMPs are endogenous molecules with elicitor activity and are released from host cellular components during pathogen attack or abiotic stress. Well-characterized DAMPs include oligogalacturonides (OGs), peptides, and cutin monomers (Table 2). The responses triggered by DAMPs largely overlap with those activated by PAMPs. For instance, transcript profiling of seedlings treated with OGs and flg22 revealed an extensive overlap of responses at least during the early stages after treatments.89
Oligogalacturonides
Researchers have speculated that OGs are derived from the degradation of a major component of pectin in plant cell walls by microbial polygalacturonases during infections or by the action of endogenous polygalacturonases that are induced by mechanical damage.92,93 OGs elicit a wide range of defense responses, including an oxidative burst,94 accumulation of phytoalexins,95 an increase of glucanase, and chitinase activity,96,97 deposition of callose,98 increased hormone biosynthesis,89 and enhanced resistance to B. cinerea.88 Cell wall-associated receptor kinases have historically been considered potential receptors of OGs because of their ability to bind to OGs in vitro.99 WAK1, a member of the wall-associated kinase family, acts as a receptor of OGs as revealed by a receptor domain-exchange approach.86 The EF-Tu receptor (EFR) is a LRR receptor kinase for the bacterial PAMP elf18.96 The chimeric receptor by fusing the extracellular domain of WAK1 with the kinase portion of EFR is able to activate the kinase domain in response to OGs.86,100 On the other hand, after the treatment with the cognate ligand elf18, the EFR ectodomain activates the WAK1 kinase that triggers defense responses similar with those activated by OGs and effective against fungal and bacterial pathogens. In addition, WAK1 overexpression in Arabidopsis increases resistance to B. cinerea.86,101
Peptides
In Arabidopsis, Peps, the small peptides derived from PROPEP genes, act as DAMPs. Six members of the PROPEP family are transcriptionally induced by pathogen infection and by PAMPs like elf18 and flg22.102,103 By binding to the promoter of the W-boxes, WRKY-type transcription factors are the major regulators of PAMP-induced PROPEP2 and PROPEP3 expression.104 Perception of Peps by the LRR receptor kinases PEPR1 and PEPR2 amplifies the immune response.84,85 The importance of the Pep immune signaling pathway is indicated by the finding that PEPR can activate the PTI response.105 The importance of the Pep immune signaling pathway has been further demonstrated that PEPR1 specifically interacts with BIK1 and PBS1-like 1 (PBL1) to mediate Pep1-induced defenses and both PEPR1 and PEPR2 directly phosphorylate BIK1 in response to Pep1 treatment. In addition, PEPR1, and PEPR2 were found to interact with BAK1.106 The pepr1/pepr2 double-mutant has a reduced sensitivity to ethylene (ET) and confers susceptibility to B. cinerea, demonstrating an important role of PEPRs in the ET-mediated defense response to necrotrophic pathogens.107 In addition, a recent study indicated that PEPRs are required to connect local immunity to systemic immunity by reinforcing the separate defense signaling pathways.108
Phytosulfokine receptor
Phytosulfokines (PSKs) are secreted sulfated pentapeptides that have been purified from plant cell culture media.109 PSKα, a bisulfated five-amino acid peptide, and PSY1, an 18-amino acid sulfated and glycosylated peptide, are perceived by LRR-RLK receptors PSKR1 (phytosulfokine receptor) and PSY1R, which are involved in plant growth and development.110,111 The Arabidopsis pskr1 plants showed enhanced PTI against the bacterial pathogen Pseudomonas syringae, suggesting that PSKRs are involved in PAMP responses.112 Similarly, Mosher et al.111 determined that PSKα and PSY1 are involved in the PAMP-mediated defense. pskr1 and psyr1 mutants exhibit enhanced defense gene expression and heightened resistance to the biotrophic pathogen P. syringae. Conversely, pskr1 mutants showed enhanced susceptibility to the necrotrophic fungus A. brassicicola. Molecular analysis revealed that the mutants accumulate elevated levels of salicylate, enhance the transcription of salicylic acid (SA) responsive or inducible pathogenesis-related genes, but repress the expression of jasmonic acid (JA)-responsive genes. These findings are consistent with the antagonistic effect of SA and JA on biotrophic and necrotrophic pathogen resistance. Integration of PSKα and PSY1 signaling in plant development and defense may involve the interactions between different phytohormones. Further investigation about hormone crosstalk in PSKα/PSY1 signaling is needed to determine the causal relationships in this complex network.111
Fungal Small RNAs as a Novel Type of Effector for Suppression of Plant Immunity
In most eukaryotic organisms, small RNAs regulate many biological processes, including development, stress responses, metabolism, and maintenance of genome integrity. Accumulating evidence has revealed that small RNAs play critical roles in plant–microbe interactions.113,114 A recent, surprising finding is that pathogen small RNAs and the host RNA silencing machinery are important in the B. cinerea–plant interaction.115 The authors found that some B. cinerea small RNAs (Bc-sRNAs) can bind to the Arabidopsis Argonaute 1 (AGO1), a component of the RNA interference machinery, and selectively silence host immunity genes.115 The ago1 mutant confers reduced susceptibility to B. cinerea while the B. cinerea mutant that could not produce these Bc-sRNAs had reduced pathogenicity on Arabidopsis and tomato. These results demonstrate that the necrotrophic pathogen B. cinerea can deliver “virulent” small RNAs into host cells and that such small RNAs function as effectors that suppress plant immunity.
The Functions of the Receptor-Like Kinase BIK1 in PTI to Necrotrophs
BIK1, a receptor-like cytoplasmic kinase in Arabidopsis, is induced early during infection by B. cinerea and plays an essential role in mediating plant resistance to necrotrophic pathogens.116 BIK1 is also a positive regulator in plant immunity as a component of the FLS2-BAK1 immune receptor complex and is directly phosphorylated by BAK1.117,118 The ligand-binding to FLS2 recruits BAK1, forming active receptor complexes.117 The activation of these receptors results in a rapid phosphorylation of BIK1, which then dissociates from the receptor complexes to activate downstream signaling. BIK1 also associates with several RLKs including BRI1, elongation factor-Tu receptor (EFR), and the LysM-RK CERK1, DAMP peptide 1 receptor (AtPEPR1).107,118 Lin et al.119 show that BIK1 acts as a negative regulator in BR signaling because the bik1 mutant displays various BR hypersensitive phenotypes. A recent study shows that BIK1 directly interacts with and phosphorylates RBOHD upon PAMP perception, which is critical for the PAMP-induced ROS burst and antibacterial immunity.120 These results indicate that BIK1 acts as a central regulator of defense signals in that it integrates PAMP, DAMP, ET, and BR signals from multiple surface-localized receptors. However, it is not known how signals from distinct receptors are integrated to activate an overlapping set of downstream defense responses.
A recent study indicates that BIK1 is a dual-specific kinase and that both tyrosine and serine/threonine kinase activity are essential for its function in Arabidopsis innate immunity.121 The difference in BIK1 phosphorylation by BAK1 and BRI1 may account for the distinct functions of BIK1 in different signaling pathways. It will be interesting to determine whether tyrosine phosphorylation activity is required for BIK1 function in BR and ET signaling. In addition, more experiments are needed to determine whether the multiple functions of BIK1 are achieved through different phosphorylation sites mediated by different receptors or co-receptors.
Epigenetic Regulation of Plant Innate Immunity to Necrotrophic Fungi
A growing body of evidence shows that epigenetic mechanisms, including DNA methylation and histone modifications, play important roles in plant defense responses.122-125 RNA-directed DNA methylation (RdDM) is a small interfering RNA-mediated epigenetic modification that induces de novo methylation of cytosines at the target genomic regions and leads to transcriptional gene silencing.122,126 Lopez et al. identified NRPD2, which encodes the subunit of the plant-specific RNA Polymerases IV and V (Pol IV and Pol V); these polymerases are important components of the RdDM pathway and have been implicated in immune responses.123 The nrpd2 mutants are more susceptible than the wild type to the necrotrophic pathogens B. cinerea and P. cucumerina. Studies on other defective mutants related to the RdDM pathway suggest that Pol V is required for plant immunity to necrotrophs.123
Histone modifications are also involved in the regulation of defense genes.127 For example, HISTONE DEACETYLASE19 (HDA19) modulates the resistance to A. brassicicola by mediating the JA and ET signaling pathways.124 Interestingly, some toxins produced by necrotrophic pathogens can inhibit histone deacetylases and thereby suppress immune responses and facilitate infection.128-130 For example, the necrotrophic pathogen C. carbonum produces HC toxins that induce histone hyperacetylation in maize.129 A. brassicicola produces the toxin depudecin, which can also inhibit histone deacetylase. In addition, the histone methyltransferase SET DOMAIN GROUP8 (SDG8)-mediated histone H3 lysine 36 methylation is required for basal and R protein-mediated resistance to necrotrophs and biotrophs.126,131
Furthermore, HISTONE MONOUBIQUITINATION1 (HUB1), a RING E3 ligase that ubiquitinates histone H2B, has been proved to regulate plant resistance to necrotrophic fungi.125 HUB1 can interact with MED21, which is a subunit of the Arabidopsis mediator and an evolutionarily conserved transcriptional cofactor complex in all eukaryotes. MED21 is involved not only in immune responses to necrotrophs but also in embryo development.125 Several mediator subunits in Arabidopsis (MED8, MED15, MED16, MED18, and MED25) are also implicated in resistance to necrotrophs and/or biotrophs.66,132-134 It remains unclear, however, how these mediator components and chromatin modification regulate resistance.
Summary Points
1. Toxin effectors from necrotrophic fungi can target a host’s central signal regulator to trigger R gene-mediated resistance and to thereby increase host susceptibility to attack by necrotrophic fungi.
2. Chitin, PGs, SCFE1, and other PAMP effectors secreted by necrotrophic fungi can be recognized by RLPs or RLKs, and such recognition triggers a series of PTI responses.
3. Although necrotrophic fungi can secrete enzymes that degrade the host cell wall, some of the degradation products, i.e., DAMPs, act as elicitors that trigger host immune responses.
4. By binding to the host RNAi machinery, small RNAs delivered by necrotrophic fungi into host cells can act as virulence effectors that suppress host immune responses.
5. Although PAMPs/DAMPs are initially recognized by distinct upstream PRRs, the immune signaling pathways triggered by those PRRs may converge on a central regulator like BIK1 and SOBIR1.
6. By regulating the expression of defense genes, epigenetic modifications, including DNA methylation and histone modifications, play important roles in plant immunity to necrotrophic fungi.
Future Issues
1. To date, RLM3, a TIR domain-encoding gene in Arabidopsis, is the only cloned R gene that is involved in broad-range immunity to necrotrophic fungal pathogens. Identifying additional R genes in host plants and their corresponding avirulence effector genes from necrotrophic fungi will provide new insights into plant immunity against this group of important fungal pathogens.
2. Although the mechanisms by which effectors are translocated into host cells during infection have been elucidated for biotrophic/hemibiotrophic plant pathogens, further research is required to determine how necrotrophic fungal effectors enter host cells.
3. Although a few PRRs targeted by effectors have been characterized, additional effector targets should be identified and their functions in plant innate immunity to necrotrophic fungi should be determined.
4. Small RNAs from B. cinerea have been identified as a new type of effector that suppresses the host innate immunity. As additional necrotrophic fungi are sequenced in the next few years, new small RNAs with similar effector functions will be identified.
5. BIK1 is a central regulator connecting plant development to immune responses through its function in ET signaling. It will be interesting to determine how diverse biological processes are integrated in a way that increases plant fitness in dynamic environments that provide only limited resources.
6. So far, only a few gene promoters that are targeted by epigenetic regulators have been described. Genome-level binding studies will be required to identify gene promoters that are targeted by epigenetic regulators during infection. Similarly, it will be important to determine the specific epigenetic modifications that occur after the recognition of PAMP or DAMP effectors and to establish the direct relationship to the plant immune response.
7. Although recent advances in the understanding of necrotrophic fungal and plant interactions have been substantial, few breakthroughs have been exploited for practical application. It is imperative that we begin to use this new understanding of pathogen effectors and R genes for the development of sustainable resistance to necrotrophic fungi.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from the National Transgenic Project (Sheath Blight Transgenic Project 2012ZX08009001), the National Natural Science Foundation of China (31101404) and the Program for Innovative Research Team in University (IRT1239) to Hunan Agricultural University. We thank Dr. Bruce Jaffee for English editing.
References
- 1.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–30. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stergiopoulos I, de Wit PJ. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–63. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 3.Horbach R, Navarro-Quesada AR, Knogge W, Deising HB. When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J Plant Physiol. 2011;168:51–62. doi: 10.1016/j.jplph.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Quaedvlieg W, Verkley GJ, Shin H-D, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW. Sizing up Septoria. Stud Mycol. 2013;75:307–90. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray GM, Brennan JP. Estimating disease losses to the Australian wheat industry. Australas Plant Pathol. 2009;38:558–70. doi: 10.1071/AP09053. [DOI] [Google Scholar]
- 6.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–33. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–9. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 8.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–14. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Staal J, Kaliff M, Bohman S, Dixelius C. Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 2006;46:218–30. doi: 10.1111/j.1365-313X.2006.02688.x. [DOI] [PubMed] [Google Scholar]
- 10.Staal J, Kaliff M, Dewaele E, Persson M, Dixelius C. RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 2008;55:188–200. doi: 10.1111/j.1365-313X.2008.03503.x. [DOI] [PubMed] [Google Scholar]
- 11.Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009;22:115–22. doi: 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Friesen TL, Ling H, Meinhardt SW, Oliver RP, Rasmussen JB, Faris JD. The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome. 2006;49:1265–73. doi: 10.1139/g06-088. [DOI] [PubMed] [Google Scholar]
- 13.Manning VA, Hardison LK, Ciuffetti LM. Ptr ToxA interacts with a chloroplast-localized protein. Mol Plant Microbe Interact. 2007;20:168–77. doi: 10.1094/MPMI-20-2-0168. [DOI] [PubMed] [Google Scholar]
- 14.Stergiopoulos I, Collemare J, Mehrabi R, De Wit PJ. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 2013;37:67–93. doi: 10.1111/j.1574-6976.2012.00349.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu ZH, Faris JD, Meinhardt SW, Ali S, Rasmussen JB, Friesen TL. Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology. 2004;94:1056–60. doi: 10.1094/PHYTO.2004.94.10.1056. [DOI] [PubMed] [Google Scholar]
- 16.Friesen TL, Meinhardt SW, Faris JD. The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. Plant J. 2007;51:681–92. doi: 10.1111/j.1365-313X.2007.03166.x. [DOI] [PubMed] [Google Scholar]
- 17.Friesen TL, Zhang Z, Solomon PS, Oliver RP, Faris JD. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 2008;146:682–93. doi: 10.1104/pp.107.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abeysekara NS, Friesen TL, Keller B, Faris JD. Identification and characterization of a novel host-toxin interaction in the wheat-Stagonospora nodorum pathosystem. Theor Appl Genet. 2009;120:117–26. doi: 10.1007/s00122-009-1163-6. [DOI] [PubMed] [Google Scholar]
- 19.Friesen TL, Chu C, Xu SS, Faris JD. SnTox5-Snn5: a novel Stagonospora nodorum effector-wheat gene interaction and its relationship with the SnToxA-Tsn1 and SnTox3-Snn3-B1 interactions. Mol Plant Pathol. 2012;13:1101–9. doi: 10.1111/j.1364-3703.2012.00819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuori RP, Wolpert TJ, Ciuffetti LM. Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici-repentis. Mol Plant Microbe Interact. 1995;8:41–8. doi: 10.1094/MPMI-8-0041. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Friesen TL, Ling H, Meinhardt SW, Oliver RP, Rasmussen JB, Faris JD. The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome. 2006;49:1265–73. doi: 10.1139/g06-088. [DOI] [PubMed] [Google Scholar]
- 22.Friesen TL, Faris JD. Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor Appl Genet. 2004;109:464–71. doi: 10.1007/s00122-004-1678-9. [DOI] [PubMed] [Google Scholar]
- 23.Park P, Nishimura S, Kohmoto K, Otani H, Tsujimoto K. Two action sites of AM-toxin I produced by apple pathotype of Alternaria alternata in host cells: an ultrastructural study. Can J Bot. 1981;59:301–10. doi: 10.1139/b81-043. [DOI] [Google Scholar]
- 24.Saito A, Nakazawa N, Suzuki M. Selection of mutants resistant to Alternaria blotch from in vitro-cultured apple shoots irradiated with X-and γ-rays. J Plant Physiol. 2001;158:391–400. doi: 10.1078/0176-1617-00235. [DOI] [Google Scholar]
- 25.Gilchrist D, Grogan R. Production and nature of a host-specific toxin from Alternaria alternata f. sp. lycopersici. Phytopathology. 1976;66:165–71. doi: 10.1094/Phyto-66-165. [DOI] [Google Scholar]
- 26.Brandwagt BF, Mesbah LA, Takken FL, Laurent PL, Kneppers TJ, Hille J, Nijkamp HJJ. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc Natl Acad Sci U S A. 2000;97:4961–6. doi: 10.1073/pnas.97.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima T, Ueno T, Fukami H, Taga T, Masuda H, Osaki K, Otani H, Kohmoto K, Nishimura S. Isolation and structures of AK-toxin I and II, host-specific phytotoxic metabolites produced by Alternaria alternata Japanese pear pathotype. Agric Biol Chem. 1985;49:807–15. doi: 10.1271/bbb1961.49.807. [DOI] [Google Scholar]
- 28.Kodama M, Suzuki T, Otani H, Kohmoto K, Nishimura S. Purification and bioassay of host-selective AT-toxin from Alternaria alternata causing brown spot of tobacco [Nicotiana tabacum] Ann Phytopathological Soc Jpn. 1990;59 [Google Scholar]
- 29.Maekawa N, Kohmoto K, Kuwada M, Yamamoto M, Nishimura S, Watanabe Y. Studies on host-specific AF-toxins produced by Alternaria alternata strawberry pathotype causing Alternaria black spot of strawberry, 1: Production of host-specific toxins and their biological activities. Ann Phytopathological Soc Jpn . 1984 doi: 10.3186/jjphytopath.50.600. [DOI] [Google Scholar]
- 30.Nakatsuka S-i, Ueda K, Goto T, Yamamoto M, Nishimura S, Kohmoto K. Structure of AF-toxin II, one of the host-specific toxins produced by alternaria alternata strawberry pathotype. Tetrahedron Lett. 1986;27:2753–6. doi: 10.1016/S0040-4039(00)84635-3. [DOI] [Google Scholar]
- 31.Tanaka A, Tsuge T. Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata. Mol Plant Microbe Interact. 2000;13:975–86. doi: 10.1094/MPMI.2000.13.9.975. [DOI] [PubMed] [Google Scholar]
- 32.Kohmoto K, Itoh Y, Shimomura N, Kondoh Y, Otani H, Kodama M, Nishimura S, Nakatsuka S. Isolation and biological activities of two host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology. 1993;83:495–502. doi: 10.1094/Phyto-83-495. [DOI] [Google Scholar]
- 33.Kohmoto K, Kohguchi T, Kondoh Y, Otani H, Nishimura S, Nakatsuka S-i. The mitochondrion: The prime site for a host-selective toxin (ACR-toxin I) produced by alternaria alternata pathogenic to rough lemon. Proc Jpn Acad, Ser B, Phys Biol Sci. 1985;61:269–72. doi: 10.2183/pjab.61.269. [DOI] [Google Scholar]
- 34.Ohtani K, Yamamoto H, Akimitsu K. Sensitivity to Alternaria alternata toxin in citrus because of altered mitochondrial RNA processing. Proc Natl Acad Sci U S A. 2002;99:2439–44. doi: 10.1073/pnas.042448499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izumi Y, Ohtani K, Miyamoto Y, Masunaka A, Fukumoto T, Gomi K, Tada Y, Ichimura K, Peever TL, Akimitsu K. A polyketide synthase gene, ACRTS2, is responsible for biosynthesis of host-selective ACR-toxin in the rough lemon pathotype of Alternaria alternata. Mol Plant Microbe Interact. 2012;25:1419–29. doi: 10.1094/MPMI-06-12-0155-R. [DOI] [PubMed] [Google Scholar]
- 36.Wise RP, Fliss AE, Pring DR, Gengenbach BG. urf13-T of T cytoplasm maize mitochondria encodes a 13 kD polypeptide. Plant Mol Biol. 1987;9:121–6. doi: 10.1007/BF00015644. [DOI] [PubMed] [Google Scholar]
- 37.Yang G, Rose MS, Turgeon BG, Yoder OC. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell. 1996;8:2139–50. doi: 10.1105/tpc.8.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walton JD. HC-toxin. Phytochemistry. 2006;67:1406–13. doi: 10.1016/j.phytochem.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 39.Lorang JM, Sweat TA, Wolpert TJ. Plant disease susceptibility conferred by a “resistance” gene. Proc Natl Acad Sci U S A. 2007;104:14861–6. doi: 10.1073/pnas.0702572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarre DA, Wolpert TJ. Inhibition of the glycine decarboxylase multienzyme complex by the host-selective toxin victorin. Plant Cell. 1995;7:463–71. doi: 10.1105/tpc.7.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wevelsiep L, Kogel K-H, Knogge W. Purification and characterization of peptides from Rhynchosporium secalis inducing necrosis in barley. Physiol Mol Plant Pathol. 1991;39:471–82. doi: 10.1016/0885-5765(91)90013-8. [DOI] [Google Scholar]
- 42.Wevelsiep L, Rupping E, Knogge W. Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol. 1993;101:297–301. doi: 10.1104/pp.101.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hahn M, Jüngling S, Knogge W. Cultivar-specific elicitation of barley defense reactions by the phytotoxic peptide NIP1 from Rhynchosporium secalis. Mol Plant Microbe Interact. 1993;6:745–54. doi: 10.1094/MPMI-6-745. [DOI] [PubMed] [Google Scholar]
- 44.Schouten A, van Baarlen P, van Kan JA. Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associate with membranes and the nucleus of plant cells. New Phytol. 2008;177:493–505. doi: 10.1111/j.1469-8137.2007.02274.x. [DOI] [PubMed] [Google Scholar]
- 45.Yun S-H, Turgeon B, Yoder O. REMI-induced mutants of Mycosphaerella zeae-maydis lacking the polyketide PM-toxin are deficient in pathogenesis to corn. Physiol Mol Plant Pathol. 1998;52:53–66. doi: 10.1006/pmpp.1997.0134. [DOI] [Google Scholar]
- 46.Nagy ED, Lee T-C, Ramakrishna W, Xu Z, Klein PE, SanMiguel P, Cheng C-P, Li J, Devos KM, Schertz K, et al. Fine mapping of the Pc locus of Sorghum bicolor, a gene controlling the reaction to a fungal pathogen and its host-selective toxin. Theor Appl Genet. 2007;114:961–70. doi: 10.1007/s00122-006-0481-1. [DOI] [PubMed] [Google Scholar]
- 47.Zhu W, Wei W, Fu Y, Cheng J, Xie J, Li G, Yi X, Kang Z, Dickman MB, Jiang D. A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS One. 2013;8:e53901. doi: 10.1371/journal.pone.0053901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao X, Xie J, Cheng J, Li G, Yi X, Jiang D, Fu Y. Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol Plant Microbe Interact. 2014;27:40–55. doi: 10.1094/MPMI-05-13-0145-R. [DOI] [PubMed] [Google Scholar]
- 49.Liang Y, Yajima W, Davis MR, Kav NN, Strelkov SE. Disruption of a gene encoding a hypothetical secreted protein from Sclerotinia sclerotiorum reduces its virulence on canola (Brassica napus) Can J Plant Pathol. 2013;35:46–55. doi: 10.1080/07060661.2012.745904. [DOI] [Google Scholar]
- 50.Zuppini A, Navazio L, Sella L, Castiglioni C, Favaron F, Mariani P. An endopolygalacturonase from Sclerotinia sclerotiorum induces calcium-mediated signaling and programmed cell death in soybean cells. Mol Plant Microbe Interact. 2005;18:849–55. doi: 10.1094/MPMI-18-0849. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Li Q, Niu X, Chen H, Xu L, Qi C. Characterization of a canola C2 domain gene that interacts with PG, an effector of the necrotrophic fungus Sclerotinia sclerotiorum. J Exp Bot. 2009;60:2613–20. doi: 10.1093/jxb/erp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng A, Lin R, Zhang D, Qin P, Xu L, Ai P, Ding L, Wang Y, Chen Y, Liu Y, et al. The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat Commun. 2013;4:1424. doi: 10.1038/ncomms2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolpert TJ, Dunkle LD, Ciuffetti LM. Host-selective toxins and avirulence determinants: what’s in a name? Annu Rev Phytopathol. 2002;40:251–85. doi: 10.1146/annurev.phyto.40.011402.114210. [DOI] [PubMed] [Google Scholar]
- 54.Berestetskiy A. A review of fungal phytotoxins: from basic studies to practical use. Appl Biochem Microbiol. 2008;44:453–65. doi: 10.1134/S0003683808050013. [DOI] [PubMed] [Google Scholar]
- 55.Collemare J, Lebrun MH. Fungal secondary metabolites: ancient toxins and novel effectors in plant–microbe interactions. Effectors in Plant-Microbe Interactions 2011:377-400. [Google Scholar]
- 56.Win J, Krasileva KV, Kamoun S, Shirasu K, Staskawicz BJ, Banfield MJ. Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 2012;8:e1002400. doi: 10.1371/journal.ppat.1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 58.Navarre DA, Wolpert TJ. Victorin induction of an apoptotic/senescence-like response in oats. Plant Cell. 1999;11:237–49. doi: 10.1105/tpc.11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorang JM, Carkaci-Salli N, Wolpert TJ. Identification and characterization of victorin sensitivity in Arabidopsis thaliana. Mol Plant Microbe Interact. 2004;17:577–82. doi: 10.1094/MPMI.2004.17.6.577. [DOI] [PubMed] [Google Scholar]
- 60.Litzenberger S. Nature of susceptibility to Helminthosporium victoriae and resistance to Puccinia coronata in Victoria oats. Phytopathology. 1949;39:300–18. [Google Scholar]
- 61.Luke H, Murphy H, Petr F. INHERITANCE OF SPONTANEOUS MUTATIONS OF VICTORIA LOCUS IN OATS. Phytopathology. 1966;56:210. [Google Scholar]
- 62.Mayama S, Bordin A, Morikawa T, Tanpo H, Kato H. Association of avenalumin accumulation with co-segregation of victorin sensitivity and crown rust resistance in oat lines carrying the Pc-2 gene. Physiol Mol Plant Pathol. 1995;46:263–74. doi: 10.1006/pmpp.1995.1021. [DOI] [Google Scholar]
- 63.Rines HW, Luke HH. Selection and regeneration of toxin-insensitive plants from tissue cultures of oats (Avena sativa) susceptible to Helminthosporium victoriae. Theor Appl Genet. 1985;71:16–21. doi: 10.1007/BF00278247. [DOI] [PubMed] [Google Scholar]
- 64.Welsh J, Peturson B, Machacek J. Associated inheritance of reaction to races of crown rust, Puccinia coronata avenae Erikss., and to Victoria blight, Helminthosporium victoriae M. and M., in oats. Can J Bot. 1954;32:55–68. doi: 10.1139/b54-008. [DOI] [Google Scholar]
- 65.Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng S-C, Maier CS, Wolpert TJ. Tricking the guard: exploiting plant defense for disease susceptibility. Science. 2012;338:659–62. doi: 10.1126/science.1226743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun D-J, Mengiste T. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat Commun. 2014;5:3064. doi: 10.1038/ncomms4064. [DOI] [PubMed] [Google Scholar]
- 67.Gilbert BM, Wolpert TJ. Characterization of the LOV1-mediated, victorin-induced, cell-death response with virus-induced gene silencing. Mol Plant Microbe Interact. 2013;26:903–17. doi: 10.1094/MPMI-01-13-0014-R. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H-F, Francl LJ, Jordahl JG, Meinhardt SW. Structural and physical properties of a necrosis-inducing toxin from Pyrenophora tritici-repentis. Phytopathology. 1997;87:154–60. doi: 10.1094/PHYTO.1997.87.2.154. [DOI] [PubMed] [Google Scholar]
- 69.Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, Rasmussen JB, Solomon PS, McDonald BA, Oliver RP. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet. 2006;38:953–6. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- 70.McDonald MC, Oliver RP, Friesen TL, Brunner PC, McDonald BA. Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 2013;199:241–51. doi: 10.1111/nph.12257. [DOI] [PubMed] [Google Scholar]
- 71.Manning VA, Ciuffetti LM. Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell. 2005;17:3203–12. doi: 10.1105/tpc.105.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adhikari TB, Bai J, Meinhardt SW, Gurung S, Myrfield M, Patel J, Ali S, Gudmestad NC, Rasmussen JB. Tsn1-mediated host responses to ToxA from Pyrenophora tritici-repentis. Mol Plant Microbe Interact. 2009;22:1056–68. doi: 10.1094/MPMI-22-9-1056. [DOI] [PubMed] [Google Scholar]
- 73.Pandelova I, Betts MF, Manning VA, Wilhelm LJ, Mockler TC, Ciuffetti LM. Analysis of transcriptome changes induced by Ptr ToxA in wheat provides insights into the mechanisms of plant susceptibility. Mol Plant. 2009;2:1067–83. doi: 10.1093/mp/ssp045. [DOI] [PubMed] [Google Scholar]
- 74.Pandelova I, Figueroa M, Wilhelm LJ, Manning VA, Mankaney AN, Mockler TC, Ciuffetti LM. Host-selective toxins of Pyrenophora tritici-repentis induce common responses associated with host susceptibility. PLoS One. 2012;7:e40240. doi: 10.1371/journal.pone.0040240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du Fall LA, Solomon PS. The necrotrophic effector SnToxA induces the synthesis of a novel phytoalexin in wheat. New Phytol. 2013;200:185–200. doi: 10.1111/nph.12356. [DOI] [PubMed] [Google Scholar]
- 76.Lu S, Faris JD, Sherwood R, Friesen TL, Edwards MC. A dimeric PR-1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA-induced necrosis in sensitive wheat. Mol Plant Pathol. 2014 doi: 10.1111/mpp.12122. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mackey D, McFall AJ. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol. 2006;61:1365–71. doi: 10.1111/j.1365-2958.2006.05311.x. [DOI] [PubMed] [Google Scholar]
- 78.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 79.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:19613–8. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wan J, Zhang X-C, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–81. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan J, Tanaka K, Zhang X-C, Son GH, Brechenmacher L, Nguyen THN, Stacey G. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012;160:396–406. doi: 10.1104/pp.112.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang L, Kars I, Essenstam B, Liebrand TW, Wagemakers L, Elberse J, Tagkalaki P, Tjoitang D, van den Ackerveken G, van Kan JA. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 2014;164:352–64. doi: 10.1104/pp.113.230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FF, Tör M, Zipfel C, Gust AA, Brunner F. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell. 2013;25:4227–41. doi: 10.1105/tpc.113.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2010;285:13471–9. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell. 2010;22:508–22. doi: 10.1105/tpc.109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci U S A. 2010;107:9452–7. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336:1160–4. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 88.Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007;144:367–79. doi: 10.1104/pp.107.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant. 2008;1:423–45. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–32. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paparella C, Savatin DV, Marti L, De Lorenzo G, Ferrari S. The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 2014;165:262–76. doi: 10.1104/pp.113.233759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cervone F, Hahn MG, De Lorenzo G, Darvill A, Albersheim P. Host-Pathogen Interactions : XXXIII. A Plant Protein Converts a Fungal Pathogenesis Factor into an Elicitor of Plant Defense Responses. Plant Physiol. 1989;90:542–8. doi: 10.1104/pp.90.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orozco-Cardenas M, Ryan CA. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci U S A. 1999;96:6553–7. doi: 10.1073/pnas.96.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galletti R, Denoux C, Gambetta S, Dewdney J, Ausubel FM, De Lorenzo G, Ferrari S. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 2008;148:1695–706. doi: 10.1104/pp.108.127845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davis KR, Darvill AG, Albersheim P, Dell A. Host-Pathogen Interactions : XXIX. Oligogalacturonides Released from Sodium Polypectate by Endopolygalacturonic Acid Lyase Are Elicitors of Phytoalexins in Soybean. Plant Physiol. 1986;80:568–77. doi: 10.1104/pp.80.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davis KR, Hahlbrock K. Induction of defense responses in cultured parsley cells by plant cell wall fragments. Plant Physiol. 1987;84:1286–90. doi: 10.1104/pp.84.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Broekaert WF, Peumans WJ. Pectic polysaccharides elicit chitinase accumulation in tobacco. Physiol Plant. 1988;74:740–4. doi: 10.1111/j.1399-3054.1988.tb02046.x. [DOI] [Google Scholar]
- 98.Bellincampi D, Dipierro N, Salvi G, Cervone F, De Lorenzo G. Extracellular H(2)O(2) induced by oligogalacturonides is not involved in the inhibition of the auxin-regulated rolB gene expression in tobacco leaf explants. Plant Physiol. 2000;122:1379–85. doi: 10.1104/pp.122.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Decreux A, Thomas A, Spies B, Brasseur R, Van Cutsem P, Messiaen J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry. 2006;67:1068–79. doi: 10.1016/j.phytochem.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 100.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–60. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 101.Li H, Zhou S-Y, Zhao W-S, Su S-C, Peng Y-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol. 2009;69:337–46. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- 102.Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci U S A. 2006;103:10098–103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huffaker A, Ryan CA. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci U S A. 2007;104:10732–6. doi: 10.1073/pnas.0703343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Logemann E, Birkenbihl RP, Rawat V, Schneeberger K, Schmelzer E, Somssich IE. Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol. 2013;198:1165–77. doi: 10.1111/nph.12233. [DOI] [PubMed] [Google Scholar]
- 105.Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nürnberger T, Tsuda K, Saijo Y. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci U S A. 2013;110:6211–6. doi: 10.1073/pnas.1216780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Postel S, Küfner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, Halter T, Kemmerling B, Nürnberger T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89:169–74. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 107.Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou J-M. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A. 2013;110:6205–10. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ross A, Yamada K, Hiruma K, Yamashita-Yamada M, Lu X, Takano Y, Tsuda K, Saijo Y. The Arabidopsis PEPR pathway couples local and systemic plant immunity. EMBO J. 2014;33:62–75. doi: 10.1002/embj.201284303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci U S A. 1996;93:7623–7. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–2. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- 111.Mosher S, Seybold H, Rodriguez P, Stahl M, Davies KA, Dayaratne S, Morillo SA, Wierzba M, Favery B, Keller H, et al. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 2013;73:469–82. doi: 10.1111/tpj.12050. [DOI] [PubMed] [Google Scholar]
- 112.Igarashi D, Tsuda K, Katagiri F. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J. 2012;71:194–204. doi: 10.1111/j.1365-313X.2012.04950.x. [DOI] [PubMed] [Google Scholar]
- 113.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010;48:225–46. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seo J-K, Wu J, Lii Y, Li Y, Jin H. Contribution of small RNA pathway components in plant immunity. Mol Plant Microbe Interact. 2013;26:617–25. doi: 10.1094/MPMI-10-12-0255-IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weiberg A, Wang M, Lin F-M, Zhao H, Zhang Z, Kaloshian I, Huang H-D, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–23. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18:257–73. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 119.Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci U S A. 2013;110:12114–9. doi: 10.1073/pnas.1302154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 121.Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci U S A. 2014;111:3632–7. doi: 10.1073/pnas.1318817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 123.López A, Ramírez V, García-Andrade J, Flors V, Vera P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 2011;7:e1002434. doi: 10.1371/journal.pgen.1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dhawan R, Luo H, Foerster AM, Abuqamar S, Du H-N, Briggs SD, Mittelsten Scheid O, Mengiste T. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell. 2009;21:1000–19. doi: 10.1105/tpc.108.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berr A, McCallum EJ, Alioua A, Heintz D, Heitz T, Shen W-H. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 2010;154:1403–14. doi: 10.1104/pp.110.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berr A, Shafiq S, Shen W-H. Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta. 2011;1809:567–76. doi: 10.1016/j.bbagrm.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 128.Brosch G, Ransom R, Lechner T, Walton JD, Loidl P. Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell. 1995;7:1941–50. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ransom RF, Walton JD. Histone hyperacetylation in maize in response to treatment with HC-toxin or infection by the filamentous fungus Cochliobolus carbonum. Plant Physiol. 1997;115:1021–7. doi: 10.1104/pp.115.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Privalsky ML. Depudecin makes a debut. Proc Natl Acad Sci U S A. 1998;95:3335–7. doi: 10.1073/pnas.95.7.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P, Hofius D, Petersen M, Mundy J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010;6:e1001137. doi: 10.1371/journal.ppat.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell. 2009;21:2237–52. doi: 10.1105/tpc.109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Canet JV, Dobón A, Tornero P. Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell. 2012;24:4220–35. doi: 10.1105/tpc.112.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR. The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 2003;34:395–406. doi: 10.1046/j.1365-313X.2003.01734.x. [DOI] [PubMed] [Google Scholar]