Abstract
The triggering receptor expressed on myeloid 2 (TREM2) is an immune phagocytic receptor expressed on brain microglia known to trigger phagocytosis and regulate the inflammatory response. Homozygous mutations in TREM2 cause Nasu–Hakola disease, a rare recessive form of dementia. A heterozygous TREM2 variant, p.R47H, was recently shown to increase Alzheimer’'s disease (AD) risk. We hypothesized that if TREM2 is truly an AD risk gene, there would be additional rare variants in TREM2 that substantially affect AD risk. To test this hypothesis, we performed pooled sequencing of TREM2 coding regions in 2082 AD cases and 1648 cognitively normal elderly controls of European American descent. We identified 16 non-synonymous variants, six of which were not identified in previous AD studies. Two variants, p.R47H [P = 9.17 × 10−4, odds ratio (OR) = 2.63 (1.44–4.81)] and p.R62H [P = 2.36 × 10−4, OR = 2.36 (1.47–3.80)] were significantly associated with disease risk in single-variant analyses. Gene-based tests demonstrate variants in TREM2 are genome-wide significantly associated with AD [PSKAT-O = 5.37 × 10−7; OR = 2.55 (1.80–3.67)]. The association of TREM2 variants with AD is still highly significant after excluding p.R47H [PSKAT-O = 7.72 × 10−5; OR = 2.47 (1.62–3.87)], indicating that additional TREM2 variants affect AD risk. Genotyping in available family members of probands suggested that p.R47H (P = 4.65 × 10−2) and p.R62H (P = 6.87 × 10−3) were more frequently seen in AD cases versus controls within these families. Gel electrophoresis analysis confirms that at least three TREM2 transcripts are expressed in human brains, including one encoding a soluble form of TREM2.
INTRODUCTION
Homozygous loss-of-function mutations in the triggering receptor expressed on myeloid cells 2 protein [TREM2 (OMIM 605086)] were initially associated with an autosomal recessive form of early-onset dementia, polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy [PLOSL (MIM 221770)], also known as Nasu–Hakola disease, in Swedish and Norwegian families (1). Subsequently, mutations in TREM2 were found worldwide in PLOSL patients from different countries and ethnic origins (1–4). PLOSL patients carrying different TREM2 variants exhibit a similar clinical phenotype with respect to the neurologic and skeletal abnormalities (1–4). The clinical spectrum of disease associated with TREM2 variants was expanded after the identification of three patients from a Lebanese family carrying mutations in TREM2 that exhibited early-onset dementia without skeletal symptoms (bone cysts) (5). Additional TREM2 variants were also found in three Turkish probands with frontotemporal-like dementia without any bone-associated symptoms (6).
Recently, two independent studies reported that a heterozygous rare variant in TREM2 p.R47H is significantly associated with Alzheimer's disease [AD (MIM 104300)], with an odds ratio (OR) similar to that of an individual carrying one APOE ε4 allele (6,7). Subsequently, the association of p.R47H with AD risk was replicated in Spanish and French populations (8,9). Several studies also found that TREM2 variants are associated with Parkinson's disease, frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (10–20). Thus, TREM2 variants exhibit pleiotropic effects producing a spectrum of disorders that ranges in clinical phenotypes from skeletal abnormalities to neurodegeneration.
Multiple variants in the same gene increase AD risk (e.g. APP, PSEN1, PSEN2, APOE, and PLD3) (21). Interestingly, another APP variant, p.A673T, was reported to reduce risk for AD (22), demonstrating that both protective and risk variants can coexist in the same gene. This notion is further supported by two common variants in APOE (APOE ε2 and APOE ε4), which have strong effects on AD risk but in opposite directions (23,24). To date, the only variant in TREM2 known to significantly affect AD risk is p.R47H (6,7). Recently, a study has sequenced TREM2 coding regions in a Belgian population and found additional coding variants in TREM2 (25). Additionally, an enrichment of TREM2 variants in both AD and FTD patients compared with controls was reported even though none of the rare variants were individually significant (25). Thus, we hypothesize that additional functional variants may be present in TREM2 that significantly increase or reduce AD risk. To test this hypothesis, we performed deep re-sequencing studies in the TREM2 coding regions in samples of European American (EA) descent to identify novel TREM2 variants that are associated with AD risk. Our previous work has shown that the pooled-DNA sequencing strategy can effectively identify novel rare variants in genes of interest associated with AD risk (21,26–30). In this study, we used the same method and demonstrated that TREM2 contains additional rare variants that increase AD risk. Further analysis of the underlying mechanisms by which these variants alter TREM2 function could provide important insights into AD pathogenesis.
RESULTS
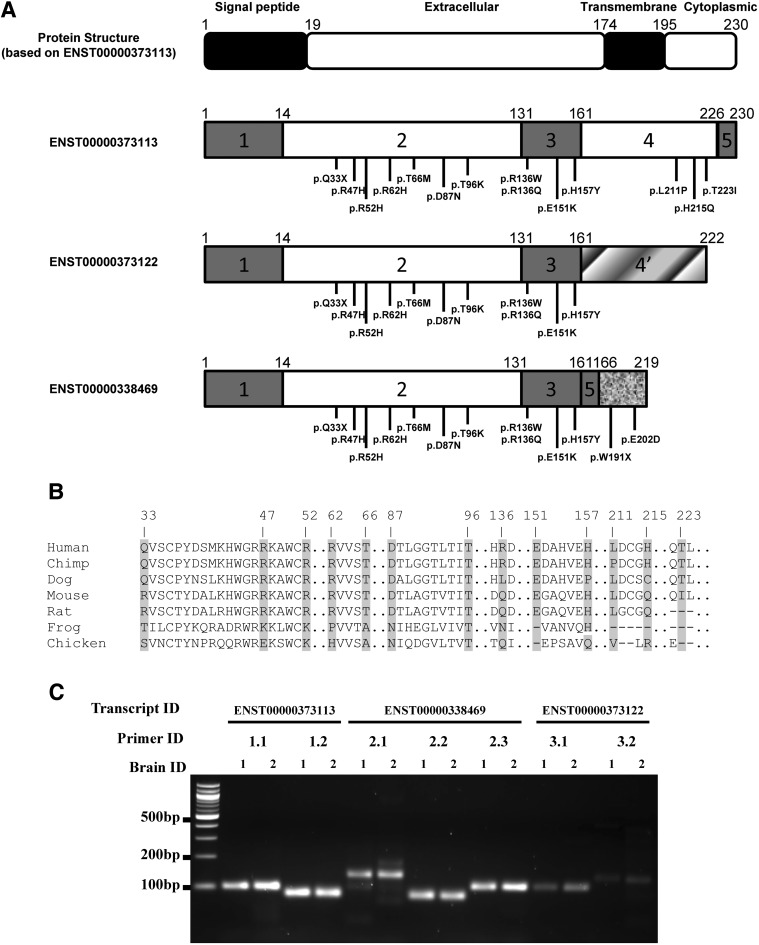
To determine whether additional TREM2 variants contribute to AD risk, AD cases and controls with similar characteristics, including age and gender distributions, were sequenced using a pooled sequencing strategy (Table 1). As expected, the percentage of APOE ε4 allele-positive individuals is significantly higher in cases compared with controls. Pooled sequencing identified 16 rare variants in TREM2, six of which were not identified in the recent studies (6,7,25): p.R52H, p.R136W, p.E151K, p.W191X, p.E202D and p.H215Q (Table 2 and Fig. 1A). Nine variants (p.R52H, p.T66M, p.R136W, p.R136Q, p.H157Y, p.W191X, p.E202D, p.H215Q and p.T223I) were exclusively found in AD cases (a total of 13 carriers; Table 2), three of which were not reported in the exome variant server (EVS) database (p.R136W, p.H215Q and p.E202D). The protein sequence conservation analysis shows that p.R47H, p.R52H, p.R62H, p.T66M, p.D87N, p.T96K, p.E151K, p.H157Y, p.L211P and p.T223I are particularly conserved across species (Fig. 1B). For the single-variant analyses, we replicated the association of p.R47H with AD risk [P = 9.17 × 10−4; OR = 2.63 (1.44–4.81); Table 2]. The minor allele (T) of a second variant, p.R62H was also significantly associated with increased AD risk [P = 2.36 × 10−4; OR = 2.36 (1.47–3.80); Table 2] after multiple-testing correction.
Table 1.
Demographic characteristics of subjects
| Knight-ADRC cases | NIA-LOAD cases | Controls | |
|---|---|---|---|
| N | 1082 | 1000 | 1648 |
| Age ± SD (range) | 72.65 ± 9.17 (44–103) | 71.77 ± 6.98 (48–98) | 76.88 ± 9.00 (50–105) |
| % Female | 57.72 | 64.86 | 60.12 |
| % APOE-ε4 positive | 55.86 | 76.21 | 29.25 |
Sample size (N), mean, standard deviation and range for age in years, percentage of female subjects and percentage of subjects that carry at least one APOE-ε4 allele for the Knight-ADRC AD cases, NIA-LOAD AD cases and cognitively normal elderly controls from both studies.
Table 2.
Rare variant association in sequenced samplesa
| Guerreiro et al.b |
Cuyvers et al.c | AD Cases |
Controls |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA Change | SNP | CHR:BP (hg19) | Pb | OR (95% CI)b | Pc | OR (95% CI)c | No. of Cases | No. of Carriers | No. of Controls | No. of Carriers | Pd | OR (95% CI) | PolyPhen |
| All variants | 5.37 × 10−7e | 2.55 (1.80–3.67) | |||||||||||
| p.Q33X | rs104894002 | 6:41129295 | 0.25 | NA | NA | NA | 2050 | 1 | 1611 | 1 | 1 | 0.81 (0.05–12.97) | NA |
| p.R47H | rs75932628 | 6:41129252 | <0.001 | 4.5 (1.7–11.9) | 0.08 | 3.01 (0.83–10.94) | 2050 | 46 | 1616 | 14 | 9.17 × 10−4 | 2.63 (1.44–4.81) | Damaging |
| p.R52H | rs374851046 | 6:41129237 | NA | NA | NA | NA | 2077 | 1 | 1642 | 0 | 1 | NA | Damaging |
| p.R62H | rs143332484 | 6:41129207 | 0.5 | 0.8 (0.5–1.4) | 0.08 | 1.54 (0.96–2.49) | 2050 | 68 | 1618 | 24 | 2.36 × 10−4 | 2.36 (1.47–3.80) | Benign |
| p.T66M | rs201258663 | 6:41129195 | 0.5 | NA | NA | NA | 2052 | 1 | 1622 | 0 | 1 | NA | Damaging |
| p.D87N | rs142232675 | 6:41129133 | 0.02 | NA | NA | NA | 2051 | 9 | 1619 | 4 | 0.41 | 1.84 (0.56–5.96) | Damaging |
| p.T96K | rs2234253 | 6:41129105 | 0.72 | 1.4 (0.3–6.0) | NA | NA | 2044 | 2 | 1609 | 2 | 1 | 0.81 (0.11–5.77) | Damaging |
| p.R136W | NA | 6:41127606 | NA | NA | NA | NA | 2003 | 2 | 1562 | 0 | 0.51 | NA | NA |
| p.R136Q | rs149622783 | 6:41127605 | 1 | 1.8 (0.1–28.6) | NA | NA | 2047 | 1 | 1623 | 0 | 1 | NA | Benign |
| p.E151K | rs79011726 | 6:41127561 | NA | NA | NA | NA | 2077 | 0 | 1642 | 1 | 0.45 | 0 | Damaging |
| p.H157Y | rs2234255 | 6:41127543 | 0.36 | NA | NA | NA | 2052 | 3 | 1610 | 0 | 1 | NA | Damaging |
| p.W191X | rs2234258 | 6:41126429 | NA | NA | NA | NA | 1816 | 1 | 1440 | 0 | 1 | NA | NA |
| p.E202D | NA | 6:41126395 | NA | NA | NA | NA | 2077 | 1 | 1642 | 0 | 1 | NA | NA |
| p.L211P | rs2234256 | 6:41126655 | 0.56 | 0 | NA | NA | 2043 | 2 | 1605 | 2 | 1 | 0.81 (0.11–5.76) | Benign |
| p.H215Q | NA | 6:41126642 | NA | NA | NA | NA | 2001 | 1 | 1560 | 0 | 1 | NA | NA |
| p.T223I | rs138355759 | 6:41126619 | NA | NA | NA | NA | 2077 | 2 | 1642 | 0 | 0.51 | NA | Benign |
aNA represents not applicable.
dThe Fisher's exact test was used to calculate the P-values using the default commands in PLINK.
eThis P-value summarizes the gene-based association of the identified SNP set and was estimated using the SKAT-O algorithm.
Figure 1.
Schematic representation of protein structure for TREM2 and for the soluble form of TREM2, location of variants, protein conservation of the mutated positions and the results of alternative splicing assays. (A) The top panel shows the protein structure of TREM2 (based on ENST00000373113), a Type I transmembrane receptor that is encoded by a gene containing five exons. The isoform ENST00000373122 encodes a different protein coding sequence after exon 3 (gradient fill rectangle) compared with ENST00000373113. The soluble form of TREM2 (ENST00000338469) lacks exon 4, which encodes the transmembrane domain, and contains a coding region after exon 5 (texture fill rectangle). Figures shown below include the structure of three different TREM2 isoforms, the location of confirmed variants in the most common TREM2 transcript (ENST00000373113), and the location of confirmed variants only in the sTREM2 transcript (ENST00000338469). Most of the variants in the transmembrane form of TREM2 are located in the extracellular domain with three exceptions, located in the cytoplasmic tail. We identified two variants that are located near the C-terminus of the soluble form of TREM2. (B) The protein conservation analysis of confirmed TREM2 variants. Variants are shown with an arrow identifying the corresponding amino acid position. Protein sequences were downloaded from UniProt. The entries used for each species are as follows: Q9NZC2 (human), Q99NH8 (mouse), D3ZZ89 (rat), H2QSZ0 (chimp), F7CW35 (frog), Q2YHU4 (chicken) and E2RP46 (dog). (C) Results of alternative splicing validation. PCR was performed to amplify the cDNA of two AD cases (brain ID = 1 and 2) extracted from autopsy brain tissue obtained from the Knight-ADRC. ENST00000373113, ENST00000338469 and ENST00000373122 were amplified using seven different primer pairs designed to specifically amplify one of the three transcripts (primer ID = 1.1 and 1.2 for ENST00000373113; Primer ID = 2.1, 2.2 and 2.3 for ENST00000338469; Primer ID = 3.1 and 3.2 for ENST00000373122). The amplicon length is 100 bp for 1.1, 84 bp for 1.2, 135 bp for 2.1, 81 bp for 2.2, 104 bp for 2.3, 103 bp for 3.1 and 127 bp for 3.2. The gel electrophoresis analysis clearly shows the presence of three distinct isoforms in the cDNA extracted from brains of two AD cases.
To determine whether TREM2 rare variants collectively contribute to AD risk, we performed a gene-based association test using the optimal SNP-set sequence kernel association test (SKAT-O). Gene-based association testing for TREM2 achieved genome-wide significance [PSKAT-O = 5.37 × 10−7; OR = 2.55 (1.80–3.67); Table 2] and remained highly significant after excluding p.R47H [PSKAT-O = 7.72 × 10−5; OR = 2.47 (1.62–3.87); Supplementary Material, Table S1], the previously confirmed risk factor for AD, in TREM2. This result demonstrates that additional rare variants in TREM2 contribute to AD risk. The cumulative carrier frequency of all TREM2 variants is 6.7% (139 out of 2082) in AD cases and 2.7% (45 out of 1648) in cognitively normal elderly controls.
Next we used the NIA-LOAD family series to test whether TREM2 variants are associated with disease status within families. We found that p.R47H and p.R62H were more frequently found in AD cases than in controls (Fisher's exact P = 4.65 × 10−2 and 6.87 × 10−3 for p.R47H and p.R62H respectively; Table 3) after directly genotyping all sampled individually from 13 (for p.R47H) and 21 (for p.R62H) independent families respectively. Other variants were either too rare or the families were not sufficiently large to provide statistical evidence of association with disease within and across families (Table 3 and Supplementary Material, Table S2). These results strongly support p.R62H as a risk factor for AD in addition to p.R47H.
Table 3.
Segregation of rare variants in available family members
| Affected |
Unaffected |
||||||
|---|---|---|---|---|---|---|---|
| Variant | # of families | Status | Carriers | Non-carriers | Carriers | Non-carriers | P |
| p.Q33X | 1 | Nasu–Hakola variant | 0 | 2 | 1 | 1 | 1 |
| NA | 75 ± 0 | 56 | 58 | ||||
| p.R47H | 13 | Confirmed risk factor | 15 | 4 | 4 | 7 | 4.65 × 10−2* |
| 70.8 ± 7.9 | 72.3 ± 10.7 | 74 ± 4.5 | 78.7 ± 7.7 | ||||
| p.R62H | 21 | Previously identified | 18 | 11 | 11 | 28 | 6.87 × 10−3* |
| 71.8 ± 5.7 | 67.9 ± 7.7 | 71.5 ± 11.4 | 71.4 ± 8.8 | ||||
| p.D87N | 2 | Previously identified | 2 | 0 | 2 | 3 | 4.29 × 10−1 |
| 81.8 ± 1.4 | NA | 71 ± 1.4 | 74.3 ± 9.0 | ||||
| p.H157Y | 1 | Previously identified | 2 | 1 | 0 | 0 | 1 |
| 67 ± 5.7 | 85 | NA | NA | ||||
| p.H215Q | 1 | Novel variant | 1 | 1 | 0 | 0 | 1 |
| 79 | 70 | NA | NA | ||||
| p.T223I | 1 | Novel variant | 1 | 0 | 1 | 0 | 1 |
| 62 | NA | 73 | NA | ||||
Family-based association analysis was performed for variants when samples from family members of the probands were available. The same variant was genotyped to test whether the rare allele is associated with disease status. Variants, number of families performed, variant type, the number of affected carriers, non-carriers and un-affected carriers, non-carriers, the average and standard deviation of age at onset (years) for the affected individuals and the average and standard deviation of age at last assessment (years) for the unaffected individuals were shown. All of the confirmed carriers only carried one rare allele. A two-tailed Fisher's exact test was used to determine evidence of segregation for each variant. * Denotes significant association. NA represents not applicable.
Two of the identified TREM2 (p.W191X and p.E202D, found exclusively in AD cases) variants are located in the coding region of the predicted shortest transcript (ENST00000338469), encoding a soluble form of TREM2 (sTREM2). However, it remains unclear whether sTREM2 results from alternative splicing or sequential cleavage of the transmembrane form of TREM2 molecules. To confirm the existence of this alternative transcript, we performed polymerase chain reaction (PCR) on cDNA from two human brains using transcript-specific primers to amplify each isoform based on the Ensembl database. Gel electrophoresis analysis confirmed that there are at least three distinct TREM2 transcripts: ENST00000373113, ENST00000373122 and ENST00000338469 expressed in the parietal cortex of human brain (Fig. 1C). The variant p.W191X is predicted to result in a premature stop codon in the ENST00000338469 transcript; however, the impact of this variant on AD pathogenesis remains unknown, due to the rarity of the allele (1/1816 cases).
DISCUSSION
TREM2 is a Type I transmembrane receptor protein expressed on myeloid cells including microglia, monocyte-derived dendritic cells, osteoclasts and bone-marrow derived macrophages (31,32). Additionally, protein expression of TREM2 in neurons has been reported (33). TREM2 transduces its intracellular signaling through DAP12 (TYROBP) (31,32). Although the natural ligands of TREM2 remain unknown, upon ligand binding, TREM2 associates with DAP12 to mediate downstream signaling. In the brain, TREM2 is primarily expressed on microglia and has been shown to control two signaling pathways: regulation of phagocytosis and suppression of inflammatory reactivity (34–36). A previous study used microarray and laser microdissection of beta amyloid (Aβ) plaque-associated areas in an animal model of AD and found that TREM2 is differentially expressed in Aβ plaque-associated versus Aβ plaque-free tissue (37). Several studies have shown that homozygous loss-of-function mutations in TREM2 or DAP12 are associated with PLOSL (1–4). Recent studies identified a TREM2 variant p.R47H as a risk factor for LOAD with an OR ∼3 (6,7), which is similar to the increased AD risk associated with carrying one APOE ε4 allele (38). Several additional rare variants were enriched in AD cases; however, these variants failed to reach statistical significance (6,7,25).
To our knowledge, this study is the largest deep re-sequencing study to date which aims to identify novel rare coding variants in TREM2. Sixteen rare coding variants were observed in TREM2, including two variants (p.R47H and p.R62H) that were significantly associated with AD risk and six novel variants that were not found in previous studies (6,7,25). The minor alleles of p.R47H [P = 9.17 × 10−4; OR = 2.63 (1.44–4.81)] and p.R62H [P = 2.36 × 10−4; OR = 2.36 (1.47–3.80)] were associated with increased AD risk after multiple-testing correction. After adjusting for APOE ε2 and ε4 alleles in the logistic regression, the association for p.R47H and p.R62H only changed slightly and remained significant [P = 5.91 × 10−3; OR = 2.48 (1.30–4.75) for p.R47H; P = 8.08 × 10−4; OR = 2.36 (1.43–3.90) for p.R62H; Supplementary Material, Table S3], which suggests that p.R47H and p.R62H affect AD risk independently of APOE ε2 and ε4 alleles. The gene-based test for TREM2 remained highly significant even after dropping p.R47H, suggesting that additional variants in TREM2 influence AD risk. After excluding both p.R47H and p.R62H, the gene-based P-value is 0.09 (Supplementary Material, Table S1), suggesting that most of the statistical significance for the gene-based association comes from these two variants. However, the OR for the gene-based analyses when these two variants were removed was 2.95 (Supplementary Material, Table S1), suggesting that additional very low-frequency variants may have a larger effect size for AD risk than p.R47H and p.R62H. This observation is also supported by the fact that 9 out of 16 TREM2 variants are only identified in 13 AD cases and no controls. The lack of association after excluding p.R47H and p.R62H is likely due to the rarity of the other variants and a lack of statistical power. In order to identify which of the remaining variants affect risk for AD, functional studies will be required. Like p.R47H, most of the identified variants are located in the extracellular domain of TREM2. Our data also support the notion that rare complete or partial loss-of-function mutations in TREM2 affect risk for AD by influencing downstream signaling including mediation of phagocytosis of cell debris and amyloid plaques and suppression of inflammatory reactivity (6,7).
We also evaluated the impact on the analysis of excluding individuals who could not be included in the principal components analysis (PCA) owing to a lack of genome-wide association study (GWAS) data. After removing individuals without GWAS data, a total of 1724 AD cases and 1437 controls were included in the analyses. The single-variant association changed slightly but still surpassed the multiple-testing threshold: p.R47H [P = 2.99 × 10−3; OR = 2.53 (1.35–4.76); Supplementary Material, Table S4] and p.R62H [P = 3.25 × 10−4; OR = 2.54 (1.49–4.35); Supplementary Material, Table S4]. The gene-based association for TREM2 reduced slightly [PSKAT-O = 6.81 × 10−6; OR = 2.56 (1.74–3.83); Supplementary Material, Table S4] but was no longer genome-wide significant (2.5 × 10−6). These results suggest that the SNP-level and gene-level significant associations using data including individuals with/without GWAS data are not false positives due to population substructure.
We also identified two variants, p.W191X and p.E202D, which are predicted to be located in the coding region of the shortest transcript (ENST00000338469), encoding a soluble form of TREM2 (sTREM2) according to Ensembl. A soluble isoform of TREM2 protein has been described as a transcript that encodes a soluble form of TREM2 (39,40). Extracellular TREM2 could be derived from the sTREM2 alternative transcript, a posttranslational cleavage product, or a combination of both. A previous study has described the presence of soluble TREM2 protein in human cerebrospinal fluid (CSF) and serum (41). Furthermore, CSF levels of sTREM2 were found to be elevated in patients with multiple sclerosis (41). Experimental data suggest that soluble TREM1 results from sequential cleavage of the transmembrane form of this related protein (41,42) and so TREM2 may be cleaved in a similar fashion. In this study, we showed the presence of cDNA corresponding to the predicted sTREM2 transcript in the brain tissue from AD cases (Fig. 1C). In summary, this study provides evidence of the presence of sTREM2 mRNA in the human brain. The p.W191X variant introduces a nonsense mutation into this sTREM2 transcript at codon 191. It is unclear whether this would result in a truncated protein or removal of the mutant mRNA by nonsense mediated decay.
In conclusion, we replicated the association for p.R47H with increased risk for AD and uncovered a significant association for p.R62H with increased AD risk. Our family-based association analyses demonstrate that p.R47H and p.R62H are associated with AD, which supports them as risk factors for AD. Our gene-based analyses clearly demonstrate that the combined effects of variants in TREM2 are highly significant even when the previously identified risk variant, p.R47H, is excluded from the analysis. Most of the identified variants are located in the TREM2 ectodomain, presumed to be involved in ligand binding. Additionally, since homozygous p.Q33X variants cause Nasu–Hakola disease due to complete loss of function, we propose that additional variants in the ectodomain of TREM2, including the two significant variants p.R47H and p.R62H, are partial loss of function alleles that affect ligand binding/signal transduction of TREM2.
MATERIALS AND METHODS
Participants and study design
The Institutional Review Board (IRB) at the Washington University School of Medicine in Saint Louis approved the study. A written informed consent was obtained from participants and their family members by the Clinical Core of at the Charles F. and Joanne Knight Alzheimer's Disease Research Center (Knight-ADRC). The approval number for the Knight-ADRC Genetics Core family studies is 93-0006.
Knight-ADRC study
Disease association analyses were initially performed in a total of 1082 AD cases and 706 cognitively normal controls of EA descent confirmed by PCA and matched for age and gender (Table 1). These samples were collected at the Knight-ADRC at Washington University and were evaluated by the Clinical Core of the Knight-ADRC. Cases received a clinical diagnosis of AD dementia in accordance with standard criteria, and dementia severity was determined with the clinical dementia rating (CDR) (43), with higher scores being associated with more severe cognitive decline. Controls underwent the same assessment but were cognitively normal. The Knight-ADRC samples were recruited without enrichment based on family history.
NIA-LOAD study
NIA-LOAD Study case–control series consists of one affected individual from each of 1000 families multiply affected by AD and 942 healthy unrelated controls, with no family history of dementia in first degree relatives, of EA descent (Table 1). All AD cases were diagnosed with dementia of the Alzheimer’'s type (DAT) using criteria equivalent to the National Institute of Neurological and Communication Disorders and Stroke-Alzheimer’'s Disease and Related Disorders Association (NINCDS-ADRDA) for probable AD (44). All NIA-LOAD AD cases had a family history of AD. Probands were required to have a diagnosis of definite or probable AD and a sibling with definite, probable or possible AD with a similar age at onset. A third biologically related family member (first, second or third degree) was also recruited, regardless of cognitive status. Within each pedigree, we screened one individual from each family by selecting the youngest affected family member with the most definitive diagnosis (i.e. individuals with autopsy confirmation were chosen over those with clinical diagnosis only). Written informed consent was obtained from all participants, and the study was approved by local IRB committees. Unrelated controls were cognitively normal and had no family history of dementia.
Human brains and RNA extraction
Parietal lobes from EA autopsy-confirmed AD (N = 2) case brains were acquired from the Knight-ADRC. Subjects selected for validating TREM2 alternative splicing did not carry any TREM2 variants. All subjects signed and provided the hospital autopsy form. If the participant does not provide future consent before death, the DPOA (durable power of attorney) or next of kin provide it after death. The Washington University IRB reviewed the protocol operated by Knight-ADRC Neuropathology Core and determined the study was exempt from approval. RNA was extracted from brain tissue using an RNeasy kit (Qiagen) following the manufacture's protocol. cDNA was synthesized from the extracted RNAs (10 µg) by the PCR using the High-Capacity cDNA Reverse Transcriptase kit (ABI).
TREM2 sequencing
TREM2 was sequenced in 2082 AD cases and 1648 cognitively normal elderly controls of EA descent using pooled-DNA sequencing as previously described (27,45,46). Equimolar amounts of individual DNA samples were pooled together after being measured using Quant-iT™ PicoGreen (Invitrogen) reagent. Pools with 100 ng of DNA from 94 individuals were made. Five exons covering 2090 bp of the target regions of TREM2 were amplified by the PCR using specific primers and Pfu Ultra high-fidelity polymerase (Agilent). An average of 20 diploid genomes (∼0.14 ng DNA) per individual was used as the input. PCR products were cleaned using QIAquick PCR (Qiagen) purification kits, quantified using Quant-iT PicoGreen reagent and ligated in equimolar amounts using T4 Ligase and T4 Polynucleotide Kinase. After ligation, concatenated PCR products were randomly sheared by sonication and prepared for sequencing on an Illumina MiSeq machine according to the manufacturer's specifications. The pCMV6-XL5 amplicon (1908 bp) was included in the reaction as a negative control. The positive controls contained 10 different constructs (p53 gene) with synthetically engineered mutations at an assigned frequency of one mutated copy per 188 normal copies were amplified and pooled with the PCR products. Paired-end reads (150 bp) were aligned to the reference TREM2 sequence using SPLINTER (46). SPLINTER uses the positive controls to estimate sensitivity and specificity for variant calling. The wild-type-to-mutant ratio in the positive control was similar to the relative frequency expected for a single mutation in one pool (1 chromosome mutated in 94 samples = 1/188). SPLINTER uses the negative controls (first 900 bp) to model the error rates across the 150 bp Illumina reads and to create an error model from each sequencing run of the machine. Based on the error model, SPLINTER calculates a P-value for the probability that a predicted variant is a true positive. A P-value at which all mutants in the positive controls were identified was defined as the cutoff to estimate the sensitivity and specificity. All mutants included as part of the amplified positive control vectors were found upon achieving >30-fold coverage at mutated sites (sensitivity = 100%) and only ∼80 sites in the 1908 bp negative control vector were predicted to be polymorphic (specificity = ∼95%). The variants with a P-value below this cutoff value were considered for follow-up genotyping.
SNP genotyping and segregation with disease
All rare missense or splice site variants identified by SPLINTER were validated by directly genotyping all sequenced individuals using Sequenom iPLEX or KASPar genotyping systems as described previously (21,27). To avoid potential batch/plate effects, genotyping was repeated with heterozygous cases or controls that were randomly assigned in the plates. The genotype call rate of these SNPs was >98%. We validated TREM2 variants by genotyping in all available family members to determine whether these variants segregate with disease status.
Population structure
A PCA was conducted to infer genetic structure of individuals who have GWAS data available using the EIGENSTRAT software as previously described (47). Samples were excluded if not located within the EA cluster (Supplementary Material, Fig. S1). Individuals who do not have GWAS data available were included in the study if the self-reported ethnicity was non-Hispanic European.
Statistical analyses
We used the Fisher's exact test to test for association between AD risk and each genetic variant using PLINK (48). For the gene-based association, we tested for association between the confirmed set of variants in TREM2 and AD risk using SKAT-O conducted using R package SKAT (49). The multiple-testing correction cutoff for the single-variant analysis using Bonferroni correction for 16 tests is 3.3 × 10−3 (0.05/16). The gene-level significant threshold is defined by type-I error rate divided by the number of human genes (0.05/20 000 = 2.4 × 10−6). For the family-based association analysis, we used the Fisher's exact test to determine whether any TREM2 variants are associated with disease status within families.
Bioinformatics analysis
The EVS (http://eversusgs.washington.edu/EVS), SeattleSeq Annotation (http://snp.gs.washington.edu/SeattleSeqAnnotation137/) and the Ensembl Genome Database (http://useast.ensembl.org/index.html) were used to annotate the rare variants. Polyphen algorithms were used to predict the functional effect of the identified variants. The Uniprot database (http://www.uniprot.org/) was used to extract and perform alignment of the protein sequences across different species.
Alternative splicing validation
According to Ensembl, TREM2 encodes three alternative transcripts (ENST00000373113, ENST00000373122 and ENST00000338469). To evaluate TREM2 alternative splicing and determine whether these transcripts exist in the human brain, cDNA isolated from parietal lobes of two Alzheimer's disease brains were amplified using PCR with Pfu (Agilent) enzyme. The PrimerQuest Design Tool (Integrated DNA Technology) was used to design primers spanning exon junctions. PCR primers include a forward primer located in the junction between exons 3 and 4 and a reverse primer located in exon 4 of the longest transcript ENST00000373113 (see Supplementary Material, Table S5 and Fig. S2 for primers and the expected amplicon lengths). For the transcript ENST00000373122, we designed a unique forward primer, which only exists in this transcript, located in the exon 3–exon 4 junction and a reverse primer located in exon 4 to amplify the sequence (see Supplementary Material, Table S5 for designed primers and the expected length). For the transcript ENST00000338469, a forward primer located in exons 3–exon 5 junction and a reverse primer located in exon 5 were used to amplify this transcript (see Supplementary Material, Table S5 and Fig. S2 for primers and the expected amplicon lengths). Each PCR reaction contains 7.5 µl of PerfeCTa SYBR Green FastMix (Quanta Biosciences), 720 nm forward and reverse primers, and 15 ng of cDNA in a final volume of 15 µl. Then incubate the reaction mix using a program as follows: (1) 45°C for 2 min; (2) 95°C for 2 min; (3) 95°C for 15 s; (4) 60°C for 1 min; (5) repeat steps 3–4 for 40 cycles. The resulting PCR product was run on a 2% agarose gel and visualized on a Syngene Imaging system.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. A.M.G. has received payment for lectures from Genentech and Amgen, and has served as a consultant for Finnegan LLC. She has received research support from Genentech, Pfizer and Astra Zeneca.
FUNDING
This work was supported by grants from the National Institutes of Health (P30-NS069329, R01-AG044546 and R01-AG035083), and the Alzheimer Association (NIRG-11-200110). This research was conducted while CC was a recipient of a New Investigator Award in Alzheimer's disease from the American Federation for Aging Research.We gratefully thank all the individuals who participated in the study and the scientists, clinicians, technicians and administrative staffs who put efforts into the implementation of this study. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant # UL1TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991 and P01 AG026276. NIALOAD samples were collected under a cooperative agreement grant (U24 AG026395) awarded by the National Institute on Aging. NIALOAD samples were obtained from the National Cell Repository for Alzheimer's Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA).
Supplementary Material
REFERENCES
- 1.Paloneva J., Mandelin J., Kiialainen A., Bohling T., Prudlo J., Hakola P., Haltia M., Konttinen Y.T., Peltonen L. DAP12/TREM2 deficiency results in impaired osteoclast differentiation and osteoporotic features. J. Exp. Med. 2003;198:669–675. doi: 10.1084/jem.20030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klunemann H.H., Ridha B.H., Magy L., Wherrett J.R., Hemelsoet D.M., Keen R.W., De Bleecker J.L., Rossor M.N., Marienhagen J., Klein H.E., et al. The genetic causes of basal ganglia calcification, dementia, and bone cysts: DAP12 and TREM2. Neurology. 2005;64:1502–1507. doi: 10.1212/01.WNL.0000160304.00003.CA. [DOI] [PubMed] [Google Scholar]
- 3.Soragna D., Papi L., Ratti M.T., Sestini R., Tupler R., Montalbetti L. An Italian family affected by Nasu–Hakola disease with a novel genetic mutation in the TREM2 gene. J. Neurol. Neurosurg. Psychiatry. 2003;74:825–826. doi: 10.1136/jnnp.74.6.825-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Numasawa Y., Yamaura C., Ishihara S., Shintani S., Yamazaki M., Tabunoki H., Satoh J.I. Nasu–Hakola disease with a splicing mutation of TREM2 in a Japanese family. Eur. J. Neurol. 2011;18:1179–1183. doi: 10.1111/j.1468-1331.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 5.Chouery E., Delague V., Bergougnoux A., Koussa S., Serre J.L., Megarbane A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum. Mutat. 2008;29:E194–E204. doi: 10.1002/humu.20836. [DOI] [PubMed] [Google Scholar]
- 6.Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. TREM2 variants in Alzheimer's disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benitez B.A., Cooper B., Pastor P., Jin S.C., Lorenzo E., Cervantes S., Cruchaga C. TREM2 is associated with the risk of Alzheimer's disease in Spanish population. Neurobiol. Aging. 2013;34:1711. doi: 10.1016/j.neurobiolaging.2012.12.018. e1715–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pottier C., Wallon D., Rousseau S., Rovelet-Lecrux A., Richard A.C., Rollin-Sillaire A., Frebourg T., Campion D., Hannequin D. TREM2 R47H variant as a risk factor for early-onset Alzheimer's disease. J. Alzheimers Dis. 2013;35:45–49. doi: 10.3233/JAD-122311. [DOI] [PubMed] [Google Scholar]
- 10.Lattante S., Le Ber I., Camuzat A., Dayan S., Godard C., Van Bortel I., De Septenville A., Ciura S., Brice A., Kabashi E., et al. TREM2 mutations are rare in a French cohort of patients with frontotemporal dementia. Neurobiol. Aging. 2013;34:2443. doi: 10.1016/j.neurobiolaging.2013.04.030. e2441–2442. [DOI] [PubMed] [Google Scholar]
- 11.Guerreiro R.J., Lohmann E., Bras J.M., Gibbs J.R., Rohrer J.D., Gurunlian N., Dursun B., Bilgic B., Hanagasi H., Gurvit H., et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 2013;70:78–84. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayaprolu S., Mullen B., Baker M., Lynch T., Finger E., Seeley W.W., Hatanpaa K.J., Lomen-Hoerth C., Kertesz A., Bigio E.H., et al. TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson's disease. Mol. Neurodegener. 2013;8:19. doi: 10.1186/1750-1326-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerreiro R., Hardy J. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1569–1570. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 14.Bird T.D. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1568. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 15.Rajagopalan P., Hibar D.P., Thompson P.M. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1565–1567. doi: 10.1056/NEJMc1306509#SA3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertram L., Parrado A.R., Tanzi R.E. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1565. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 17.Reitz C., Mayeux R., Alzheimer's Disease Genetics C. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1564–1565. doi: 10.1056/NEJMc1306509#SA1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonsson T., Stefansson K. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1568–1569. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 19.Benitez B.A., Cruchaga C., United States-Spain Parkinson's Disease Research G. TREM2 and neurodegenerative disease. N. Engl. J. Med. 2013;369:1567–1568. doi: 10.1056/NEJMc1306509#SA4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cady J., Koval E.D., Benitez B.A., Zaidman C., Jockel-Balsarotti J., Allred P., Baloh R.H., Ravits J., Simpson E., Appel S.H., et al. TREM2 Variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:449–453. doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruchaga C., Chakraverty S., Mayo K., Vallania F.L., Mitra R.D., Faber K., Williamson J., Bird T., Diaz-Arrastia R., Foroud T.M., et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS One. 2012;7:e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonsson T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 23.Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., Roses A.D. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corder E.H., Saunders A.M., Risch N.J., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Jr, Rimmler J.B., Locke P.A., Conneally P.M., Schmader K.E., et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 25.Cuyvers E., Bettens K., Philtjens S., Van Langenhove T., Gijselinck I., van der Zee J., Engelborghs S., Vandenbulcke M., Van Dongen J., Geerts N., et al. Investigating the role of rare heterozygous TREM2 variants in Alzheimer's disease and frontotemporal dementia. Neurobiol. Aging. 2014;35:726. doi: 10.1016/j.neurobiolaging.2013.09.009. e711–729. [DOI] [PubMed] [Google Scholar]
- 26.Haller G., Druley T., Vallania F.L., Mitra R.D., Li P., Akk G., Steinbach J.H., Breslau N., Johnson E., Hatsukami D., et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum. Mol. Genet. 2012;21:647–655. doi: 10.1093/hmg/ddr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin S.C., Pastor P., Cooper B., Cervantes S., Benitez B.A., Razquin C., Goate A., Cruchaga C. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer's disease Ibero-American cohort. Alzheimers Res. Ther. 2012;4:34. doi: 10.1186/alzrt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benitez B.A., Karch C.M., Cai Y., Jin S.C., Cooper B., Carrell D., Bertelsen S., Chibnik L., Schneider J.A., Bennett D.A., et al. The PSEN1, p.E318G variant increases the risk of Alzheimer's disease in APOE-epsilon4 carriers. PLoS Genet. 2013;9:e1003685. doi: 10.1371/journal.pgen.1003685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haller G., Kapoor M., Budde J., Xuei X., Edenberg H., Nurnberger J., Kramer J., Brooks A., Tischfield J., Almasy L., et al. Rare missense variants in CHRNB3 and CHRNA3 are associated with risk of alcohol and cocaine dependence. Hum. Mol. Genet. 2014;23:810–819. doi: 10.1093/hmg/ddt463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruchaga C., Karch C.M., Jin S.C., Benitez B.A., Cai Y., Guerreiro R., Harari O., Norton J., Budde J., Bertelsen S., et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouchon A., Dietrich J., Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 2000;164:4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 32.Bouchon A., Hernandez-Munain C., Cella M., Colonna M. A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J. Exp. Med. 2001;194:1111–1122. doi: 10.1084/jem.194.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sessa G., Podini P., Mariani M., Meroni A., Spreafico R., Sinigaglia F., Colonna M., Panina P., Meldolesi J. Distribution and signaling of TREM2/DAP12, the receptor system mutated in human polycystic lipomembraneous osteodysplasia with sclerosing leukoencephalopathy dementia. Eur. J. Neurosci. 2004;20:2617–2628. doi: 10.1111/j.1460-9568.2004.03729.x. [DOI] [PubMed] [Google Scholar]
- 34.Neumann H., Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J. Neuroimmunol. 2007;184:92–99. doi: 10.1016/j.jneuroim.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K., Prinz M., Stagi M., Chechneva O., Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4:e124. doi: 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbull I.R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., Colonna M. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 37.Frank S., Burbach G.J., Bonin M., Walter M., Streit W., Bechmann I., Deller T. TREM2 is upregulated in amyloid plaque-associated microglia in aged APP23 transgenic mice. Glia. 2008;56:1438–1447. doi: 10.1002/glia.20710. [DOI] [PubMed] [Google Scholar]
- 38.Saunders A.M., Strittmatter W.J., Schmechel D., George-Hyslop P.H., Pericak-Vance M.A., Joo S.H., Rosi B.L., Gusella J.F., Crapper-MacLachlan D.R., Alberts M.J., et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 39.Schmid C.D., Sautkulis L.N., Danielson P.E., Cooper J., Hasel K.W., Hilbush B.S., Sutcliffe J.G., Carson M.J. Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J. Neurochem. 2002;83:1309–1320. doi: 10.1046/j.1471-4159.2002.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gattis J.L., Washington A.V., Chisholm M.M., Quigley L., Szyk A., McVicar D.W., Lubkowski J. The structure of the extracellular domain of triggering receptor expressed on myeloid cells like transcript-1 and evidence for a naturally occurring soluble fragment. J. Biol. Chem. 2006;281:13396–13403. doi: 10.1074/jbc.M600489200. [DOI] [PubMed] [Google Scholar]
- 41.Piccio L., Buonsanti C., Cella M., Tassi I., Schmidt R.E., Fenoglio C., Rinker J., 2nd, Naismith R.T., Panina-Bordignon P., Passini N., et al. Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain. 2008;131:3081–3091. doi: 10.1093/brain/awn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez-Pina V., Soares-Schanoski A., Rodriguez-Rojas A., Del Fresno C., Garcia F., Vallejo-Cremades M.T., Fernandez-Ruiz I., Arnalich F., Fuentes-Prior P., Lopez-Collazo E. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J. Immunol. 2007;179:4065–4073. doi: 10.4049/jimmunol.179.6.4065. [DOI] [PubMed] [Google Scholar]
- 43.Morris J.C. The clinical dementia rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 44.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 45.Cruchaga C., Kauwe J.S., Nowotny P., Bales K., Pickering E.H., Mayo K., Bertelsen S., Hinrichs A., Fagan A.M., Holtzman D.M., et al. Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer's disease. Hum. Mol. Genet. 2012;21:4558–4571. doi: 10.1093/hmg/dds296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallania F.L., Druley T.E., Ramos E., Wang J., Borecki I., Province M., Mitra R.D. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20:1711–1718. doi: 10.1101/gr.109157.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.