Abstract
Fibroblast growth factor receptor 2 (FGFR2) promotes osteoprogenitor proliferation and differentiation during bone development, yet how the receptor elicits these distinct cellular responses remains unclear. Analysis of the FGFR2-skeletal disorder bent bone dysplasia syndrome (BBDS) demonstrates that FGFR2, in addition to its canonical signaling activities at the plasma membrane, regulates bone formation from within the nucleolus. Previously, we showed that the unique FGFR2 mutations that cause BBDS reduce receptor levels at the plasma membrane and diminish responsiveness to extracellular FGF2. In this study, we find that these mutations, despite reducing canonical signaling, enhance nucleolar occupancy of FGFR2 at the ribosomal DNA (rDNA) promoter. Nucleolar FGFR2 activates rDNA transcription via interactions with FGF2 and UBF1 by de-repressing RUNX2. An increase in the nucleolar activity of FGFR2 in BBDS elevates levels of ribosomal RNA in the developing bone, consequently promoting osteoprogenitor cell proliferation and decreasing differentiation. Identifying FGFR2 as a transcriptional regulator of rDNA in bone unexpectedly reveals a nucleolar route for FGF signaling that allows for independent regulation of osteoprogenitor cell proliferation and differentiation.
INTRODUCTION
Congenital defects in skeletal development affect a significant proportion of the population, with an incidence rate of 1 case per 3000 live births (1). Many of these skeletal disorders arise from mutations in genes that define the size and shape of bones during embryonic development. Fibroblast growth factor receptor 2 (FGFR2) (MIM 176 943) is one such gene. FGFR2 mutations that enhance receptor activity cause abnormal fusions of the bones within the skull and limbs in Apert, Crouzon, Jackson-Weiss and Pfeiffer syndromes (2). In contrast, FGFR2 mutations that reduce receptor activity cause bone hypoplasia within the skull and limb in Lacrimo-auriculo-dento-digital (LADD) syndrome (3). Genetic studies in mouse show that Fgfr2 mutations disrupt skeletal development by altering the ability of the receptor to regulate osteoprogenitor cell proliferation and differentiation: increased Fgfr2 function enhances proliferation and differentiation (4,5), whereas diminished Fgfr2 function decreases proliferation and differentiation (6,7). While Fgfr2 dually promotes osteoprogenitor cell proliferation and differentiation in the developing skeleton, the mechanisms through which the receptor elicits these seemingly opposed cellular responses remain unclear.
Evidence suggests that the assortment of FGF signaling components functioning either upstream or downstream of FGFR2 do not correlate in a simple way with either proliferation or differentiation in osteoprogenitor cells. For example, different FGF ligands, such as FGF2 and FGF18, promote both osteoprogenitor cell proliferation and differentiation (8,9). Similarly, FGFR2-mediated activation of distinct downstream signaling cascades, such as MAPK or PLCγ, in osteoprogenitor cells promotes both proliferation and differentiation (10–12). The dual role for FGFR2 in osteoprogenitor cells could be explained by differences in the transcriptional profile initiated upon receptor activation. Nevertheless, the mechanism through which FGF signaling elicits a primary transcriptional response is not clearly delineated because the transcriptional regulatory proteins targeted by the pathway are ubiquitously expressed and employed by multiple signaling pathways (13). Intriguingly, however, the FGF signaling pathway has a direct presence in the nucleus. FGF2 produces five protein isoforms through alternative translation, all of which contain a nuclear localization signal (NLS) (14). The low-molecular-weight FGF2 is initially secreted and undergoes nuclear translocation upon endocytosis, while the high-molecular-weight isoforms remain intracellular and move to the nucleus after synthesis to regulate proliferation (15). The NLS of FGF2 has also been shown to facilitate nuclear import of FGFR1 via importin-β (16). Colocalization of nuclear FGFR1–FGF2 to active nuclear speckles is correlated with transcriptional activation (17); however, the direct gene targets occupied by nuclear FGFR1–FGF2 remain unclear.
In this study, our analysis of the unique FGFR2 mutations in bent bone dysplasia syndrome (BBDS; MIM 614 592) reveals a direct gene target for nuclear FGF signaling and, at the same time, resolves the dual functions of FGFR2 in bone (18). BBDS, distinct from other FGFR2 disorders, presents with deficient ossification in the craniofacial and appendicular skeleton despite a rich supply of osteoprogenitor cells (18). This phenotype suggests that the FGFR2 missense mutations exclusive to this disorder uncouple the dual functions of FGFR2 osteoprogenitor cell proliferation and differentiation. The dominant FGFR2 mutations in BBDS, p.Met391Arg or p.Tyr381Asp, are located in the transmembrane domain, selectively reduce plasma membrane levels of FGFR2 and consequently diminish receptor responsiveness to extracellular FGFs (18). While aspects of the BBDS phenotype likely result from decreased canonical FGFR2 signaling, there remains compelling evidence that this disorder does not result from a strict loss of FGFR2 function: the skeletal defects in BBDS do not overlap with those in LADD syndrome or mice with a conditional knockout of Fgfr2 in the skeletogenic mesenchyme (3,6). Here, we provide strong evidence that enhanced nuclear FGFR2 activity can explain the pathogenesis of the skeletal defects in BBDS. We demonstrate that in skeletal precursors nucleolar FGFR2 directly activates ribosomal DNA (rDNA) transcription by interacting at the rDNA promoter with FGF2 and UBF1 and limiting transcriptional repression by RUNX2. Furthermore, we show that the FGFR2 mutations in BBDS increase nucleolar FGFR2 activity, consequently elevating levels of ribosomal RNA (rRNA), promoting osteoprogenitor cell proliferation and decreasing osteoblast differentiation. By identifying FGFR2 as a transcriptional regulator of rDNA in bone, this work unexpectedly reveals a nucleolar route for FGF signaling that allows for independent regulation of osteoprogenitor cell proliferation and differentiation.
RESULTS
Nuclear localization of FGFR2 and FGF2 is altered in BBDS
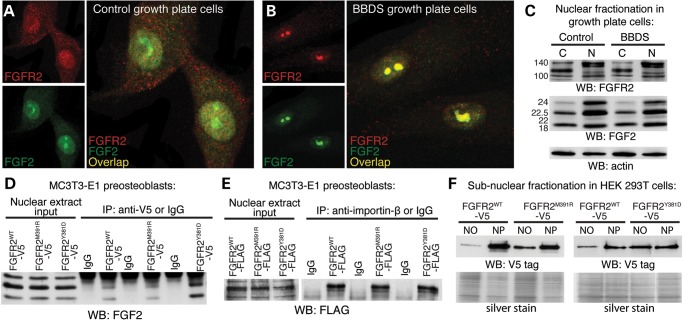
Our previous work identified FGFR2 within the nucleus of both control and BBDS femoral growth plate cells (18). Since the NLS within FGF2 has been shown to facilitate nuclear import of FGFR1 by importin-β (16), we examined a nuclear connection between FGFR2 and FGF2 in human growth plate cells. Immunofluorescence detected colocalization of FGFR2 and FGF2 in the nuclear matrix and discrete subnuclear domains of control femoral growth plate cells (Fig. 1A). In BBDS growth plate cells, nuclear colocalization of FGFR2 and FGF2 was enhanced to distinct subnuclear domains (Fig. 1B). Protein levels of nuclear FGFR2 and FGF2 were unaffected in BBDS indicating altered localization was due to nuclear redistribution rather than a change in the levels of expression and/or nuclear import (Fig. 1C). This suggests that the FGFR2 mutations in BBDS alter nuclear localization of FGF2 through direct interactions. Immunoprecipitation of epitope-tagged FGFR2WT, FGFR2M391R and FGFR2Y381D from MC3T3-E1 mouse calvarial preosteoblast nuclear extracts confirmed that both the wild-type and mutant receptors interact with nuclear Fgf2 (Fig. 1D). Furthermore, immunoprecipitation of nuclear FGFR2WT, FGFR2M391R and FGFR2Y381D identified interactions between the receptors and importin-β, the transport protein responsible for nuclear import of Fgf2 (Fig. 1E). Thus, interactions between FGFR2 and FGF2 could explain nuclear transport of FGFR2 and altered subnuclear localization of FGFR2 and FGF2 in BBDS.
Figure 1.
Nuclear colocalization of FGFR2 and FGF2 is altered in BBDS. (A) Immunofluorescent analysis showed that FGFR2 (red) and FGF2 (green) were colocalized in the nucleus of control human growth plate cells. (B) Immunofluorescent analysis showed nuclear colocalization of FGFR2 (red) and FGF2 (green) was enriched to subnuclear domains in BBDS growth plate cells. (C) Western blot showed that levels of endogenous FGFR2 and FGF2 isoforms in the cytosol (C) and nucleus (N) of control and BBDS growth plate cells were equivalent. (D) Immunoprecipitation of nuclear V5-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D from MC3T3-E1 preosteoblasts identified interactions between the receptors and nuclear isoforms of FGF2 (n = 3). (E) Immunoprecipitation of nuclear FLAG-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D from MC3T3-E1 preosteoblasts identified interactions between the receptors and importin-β (n = 3). (F) Western blot of nucleolar (NO) fractions in HEK 293T cells showed 4.3- and 3.9-fold increases in the nucleolar levels of V5-tagged FGFR2M391R and FGFR2Y381D, respectively, compared with FGFR2WT (n = 3). Nucleoplasmic (NP) fractions showed equivalent levels of mutant and wild-type FGFR2. Equal protein loading was confirmed by silver stain.
FGFR2 mutations in BBDS enhance nucleolar localization of FGFR2 with regulators of rDNA transcription
FGF2 has been previously shown to occupy the nucleolus of cells (19,20). To investigate the possibility that the nuclear regions with FGFR2 and FGF2 enrichment in BBDS growth plate cells were nucleoli, we performed subnuclear fractionation in HEK 293T cells expressing epitope-tagged FGFR2WT, FGFR2M391R and FGFR2Y381D. While nucleoplasmic levels of FGFR2M391R and FGFR2Y381D were equal to that of FGFR2WT, nucleolar levels of FGFR2M391R and FGFR2Y381D were increased 4.3- and 3.9-fold, respectively, compared with FGFR2WT (Fig. 1F). These data reveal that the FGFR2 mutations in BBDS specifically enhance nucleolar localization of FGFR2.
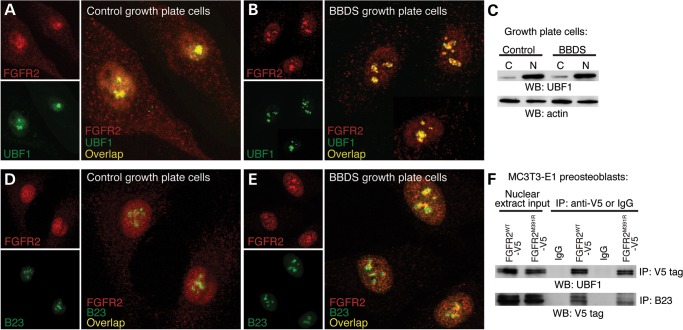
Nucleoli are non-membrane bound structures that organize around the chromosomal locales of 200 rDNA tandem repeats to regulate rRNA expression and ribosome assembly. In the nucleolus, FGF2 interacts with UBF1, a transcription factor for RNA Polymerase I (20). The nucleolar interaction of FGF2 with UBF1 at rDNA is presumed to be receptor independent; however, our data suggested that this might not be the case. Immunofluorescent detection of FGFR2 and UBF1 showed that they are colocalized in the nucleus and nucleoli of control growth plate cells (Fig. 2A). In BBDS growth plate cells, UBF1 was more focally localized to the nucleoli in regions overlapping FGFR2 (Fig. 2B). Protein levels of UBF1 were unaffected in BBDS indicating that altered localization was not due to changes in expression (Fig. 2C). This suggests that the FGFR2 mutations in BBDS alter nuclear localization of UBF1 via protein interactions. Immunoprecipitation of epitope-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D from nuclear extracts of MC3T3-E1 preosteoblasts showed that both the wild-type and mutant receptors interact with Ubf1 (Fig. 2F). This suggests that FGFR2, by forming a complex with UBF1, can enhance nucleolar localization of UBF1 in BBDS.
Figure 2.
Nucleolar localization of FGFR2 with UBF1 and B23 is enriched in BBDS. (A) Immunofluorescent analysis showed that FGFR2 (red) and UBF1 (green) were colocalized to the nucleoli of control human growth plate cells. (B) Immunofluorescent analysis showed that colocalization of FGFR2 (red) and UBF1 (green) was enriched in the nucleoli of BBDS growth plate cells. (C) Western blot showed that levels of UBF1 in the nucleus (N) and cytoplasm (C) were equivalent between control and BBDS growth plate cells. (D) Immunofluorescent analysis showed colocalization of FGFR2 (red) and B23 (green) to the nucleoli of control human growth plate cells. (E) Immunofluorescent analysis showed that FGFR2 (red) and B23 (green) colocalization was enriched in the nucleoli of BBDS growth plate cells. (F) Immunoprecipitation of nuclear V5-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D from MC3T3-E1 preosteoblasts identified receptor interactions with both UBF1 and B23.
To further assess the functional significance of FGFR2 interactions with UBF1 and FGF2, we asked whether FGFR2 is colocalized with B23, a histone chaperone recruited by UBF1 to rDNA to promote transcription (21). Immunofluorescence detected nucleolar overlap of FGFR2 and B23 in control human growth plate cells (Fig. 2D). In BBDS growth plate cells, FGFR2 showed enrichment to B23-positive domains, supporting the idea that there is enhanced localization of FGFR2 to regions of active rDNA transcription within the nucleoli (Fig. 2E). Immunoprecipitation of nuclear extracts from MC3T3-E1 preosteoblasts expressing epitope-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D showed that both the wild-type and mutant receptors interact with B23 (Fig. 2F). This interaction between FGFR2 and B23 further supports the hypothesis that FGFR2 functions together with UBF1 and FGF2 in the nucleolus at rDNA.
Enhanced nucleolar localization of FGFR2 augments activation of the ERK–MAPK pathway
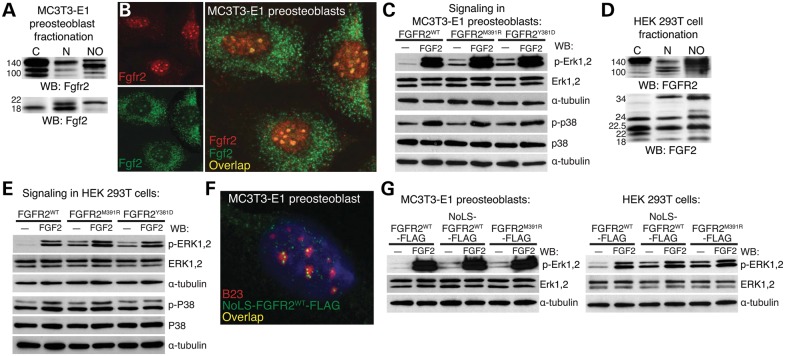
Enrichment of FGFR2 and FGF2 in nucleoli of BBDS growth plate cells suggested that the BBDS mutation in FGFR2 augment normal nuclear signaling, as nuclear FGFs activate the ERK–MAPK pathway (22,23). We assayed the signaling potential of the mutant receptors in calvarial MC3T3-E1 preosteoblasts, which express endogenous nuclear Fgf2 and allow for nuclear import of Fgfr2 (Fig. 3A and B). In serum-starved conditions, where the receptor is only exposed to endogenous nuclear Fgf2, FGFR2M391R and FGFR2Y381D exhibited higher levels p-Erk1,2 compared with FGFR2WT (Fig. 3C). These findings suggest that responsiveness of FGFR2M391R and FGFR2Y381D to nuclear Fgf2 is enhanced. Serum starvation increases cell surface localization of wild-type FGFRs. The ability of the mutant receptors to enhance p-Erk1,2 activation under these conditions reflects increased accessibility of the mutant receptor to nuclear Fgf2. Increased activation of the MAPK pathway was specific to Erk1,2, as there was no change in the levels of p-p38. Upon stimulation with exogenous FGF2, preosteoblasts expressing FGFR2WT, FGFR2M391R and FGFR2Y381D display similar levels of p-Erk1,2 and p-p38, suggesting that the mutant receptors do not block activation of endogenous wild-type Fgfr2 expressed in MC3T3-E1 preosteoblasts (Fig. 3C).
Figure 3.
Nucleolar FGFR2 activates the ERK–MAPK pathway through intracellular FGF2. (A) Western blot showed MC3T3-E1 preosteoblasts have endogenous Fgfr2 and Fgf2 isoforms in the cytosol (C), nucleus (N) and nucleolus (NO). (B) Immunofluorescent analysis showed that Fgfr2 (red) and Fgf2 (green) were colocalized in the nucleoli of MC3T3-E1 preosteoblasts. (C) Western blots of MC3T3-E1 preosteoblasts expressing FGFR2M391R and FGFR2Y381D showed a 5.3- and 5.5-fold increase, respectively, in p-Erk1,2 activation compared with FGFR2WT under serum-starved conditions (n = 4). No difference was found in p-p38 activation (n = 4). (D) Western blot showed HEK 293T cells have endogenous FGFR2 and FGF2 isoforms in the cytosol (C), nucleus (N) and nucleoli (NO). (E) Western blots of HEK 293T cells expressing FGFR2M391R and FGFR2Y381D showed a 5.7- and 5.4-fold increase in p-ERK1,2 activation, respectively, compared with FGFR2WT under serum-starved conditions (n = 4). No difference was found in p-P38 activation (n = 4). (F) Immunofluorescent analysis for B23 (red) and anti-FLAG (green) showed targeting of NoLS-FGFR2WT-FLAG to the nucleoli of MC3T3-E1 preosteoblasts. (G) Western blots of MC3T3-E1 preosteoblasts and HEK 293T cells expressing NoLS-FGFR2WT-FLAG showed a 3.1- and 5.4-fold increase in p-Erk1,2 activation, respectively, compared with FGFR2WT-FLAG under serum-starved conditions (n = 3).
FGFR2 also activates the PI3K, JAK/STAT and PLCγ signaling pathways. Since MC3T3-E1 preosteoblasts expressed the components of these pathways at very low levels, we further tested the ability of the mutant receptor to signal in HEK 293T cells, which have endogenous nuclear and nucleolar FGF2 and FGFR2 (Fig. 3D). Consistent with our results in MC3T3-E1 preosteoblasts, serum-starved HEK 293T cells expressing FGFR2M391R and FGFR2Y381D showed increased p-ERK1,2 levels in response to endogenous nuclear FGF2 compared with cells expressing FGFR2WT (Fig. 3E). FGFR2M391R and FGFR2Y381D did not alter the activation of the P38-MAPK, PI3K, JAK/STAT or PLCγ1 pathways (Fig. 3E; Supplementary Material, Fig. S1A). Together, these data show that BBDS mutations in FGFR2 enhance the ability of the receptor to signal through nuclear FGF2, which specifically activates the ERK–MAPK pathway.
To test the idea that enhanced nucleolar localization of FGFR2 leads to increased activation of the ERK pathway, we fused FGFR2WT to a nucleolar localization signal (NoLS) (24) and challenged it to respond to nuclear FGF2 (Fig. 3F). In MC3T3-E1 preosteoblasts and HEK 293T cells, under serum-starved conditions, expression of NoLS-FGFR2WT increased p-ERK1,2 levels in response to nuclear FGF2 compared with FGFR2WT expression (Fig. 3G). Activation of the P38-MAPK, PI3K, JAK/STAT and PLCγ pathways remained unaltered by NoLS-FGFR2WT (Supplementary Material, Fig. S1B). Together, these data show that nucleolar localization of FGFR2 with FGF2 supports activation of the ERK-MAPK pathway and that the FGFR2 mutations that cause BBDS further enhance this nucleolar signaling.
FGFR2 mutations in BBDS promote the mitogenic activity of FGFR2 by increasing rDNA transcription
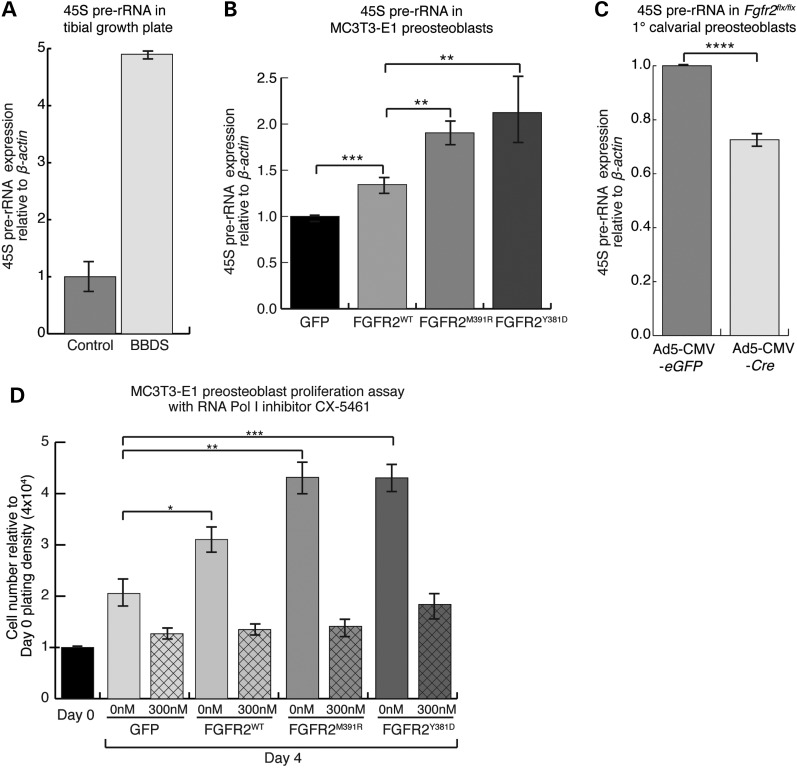
Enhanced nucleolar colocalization of FGFR2 with FGF2 and UBF1 in BBDS growth plate cells suggested that the disease process might affect rDNA transcription. Changes in active rDNA transcription in the human tibial growth plate were detected by qPCR of 45S pre-rRNA, the primary transcript that is subsequently processed to form the 18S, 5.8S and 28S rRNAs. Levels of the 45S pre-rRNA were increased nearly 5-fold in the BBDS tibial growth plate compared with a stage-match control (Fig. 4A). This suggests that enhanced nucleolar localization of FGFR2 with FGF2 and UBF1 can lead to increased rDNA transcription in individuals affected by BBDS.
Figure 4.
FGFR2 mutations in BBDS enhance rDNA transcription and cell proliferation. (A) Levels of 45S precursor rRNA, detected by qPCR relative to β-actin, were elevated nearly 5-fold in the BBDS tibial growth plate compared with control. (B) Levels of 45S precursor rRNA, detected by qPCR relative to β-actin, were elevated 1.9-fold, and 2.2-fold in MC3T3-E1 cells expressing FGFR2M391R and FGFR2Y381D compared with GFP. MC3T3-E1 cells expressing FGFR2WT showed a 1.3-fold increase, compared with GFP (n = 3). (C) Levels of 45S precursor rRNA, detected by qPCR relative to β-actin, were reduced 27% in Fgfr2flx/flx primary calvarial preosteoblasts transduced with Ad5-CMV-Cre compared with those preosteoblasts transduced with Ad5-CMV-eGFP (n = 3). (D) MC3T3-E1 preosteoblasts expressing FGFR2WT showed a 1.5-fold increase in proliferation, whereas those expressing FGFR2M391R or FGFR2Y381D showed a 2-fold increase in proliferation compared with GFP. CX-5461 treatment decreased proliferation of MC3T3-E1 preosteoblasts expressing FGFR2WT by 56%, FGFR2M391R by 67%, and FGFR2Y381D by 58% compared with a 35% reduction for GFP (hatched bars, n = 4). Cell numbers counted on Day 4 were made relative to Day 0 plating density at 4 × 104 cells (solid bars, n = 4).
In mouse calvarial preosteoblasts, we found that levels of 45S pre-rRNA are linked to nucleolar FGFR2 activity. Expression of FGFR2WT in MC3T3-E1 preosteoblasts increased levels of the 45S pre-rRNA by 1.3-fold, whereas expression of FGFR2M391R and FGFR2Y381D increased levels 1.9- and 2.2-fold, respectively, compared with a GFP control (Fig. 4B). We also identified similar changes in 45S pre-rRNA in HEK 293T cells expressing the wild-type and mutant receptors (Supplementary Material, Fig. S2A). This supports the hypothesis that the mutations in FGFR2 that cause BBDS are directly responsible for the observed increase in rDNA transcription. Correspondingly, when we transduced primary calvarial preosteoblasts from Fgfr2flx/flx mice with a Cre-expressing adenovirus to generate Fgfr2Δ/Δ preosteoblasts, we detected a 27% drop in the levels of 45S rRNA compared with Fgfr2flx/flx calvarial preosteoblasts transduced with an eGFP-expressing adenovirus (Fig. 4C). While these differences appear modest, a relatively small fold difference in rRNA transcripts can have a large biological impact, as rRNA accounts for up to 80% of the total RNA in eukaryotic cells (25). Together, these data show that FGFR2 promotes rDNA transcription and that the FGFR2 mutations that cause BBDS enhance this activity.
The proliferative potential of a cell relies heavily on ribosome biogenesis, as a high translational capacity allows cells to gain the mass necessary for division (26). The rate-limiting step in ribosome production is rDNA transcription (27). Thus, proteins that directly regulate rDNA transcription through UBF1 influence proliferation (28). Expression of FGFR2WT in MC3T3-E1 preosteoblasts increased proliferation 1.5-fold compared with GFP, while that of FGFR2M391R and FGFR2Y381D increased cell proliferation over 2-fold (Fig. 4D). Treatment of these MC3T3-E1 preosteoblasts with the RNA Polymerase I inhibitor CX-5461 considerably reduced proliferation, with the greatest reduction occurring in preosteoblasts that expressed FGFR2M391R and FGFR2Y381D (Fig. 4D). Together, these data show that FGFR2 promotes proliferation by activating rDNA transcription (Fig. 4B) and that the mutations in FGFR2 that cause BBDS increase this mitogenic activity. That FGFR2M391R and FGFR2Y381D increase the proliferation of preosteoblasts through rDNA transcription offers an explanation for the increased numbers of osteoprogenitor cells observed in the bones of individuals with BBDS.
FGFR2 mutations in BBDS enrich FGFR2 at UBF1-binding sites in the rDNA repeat
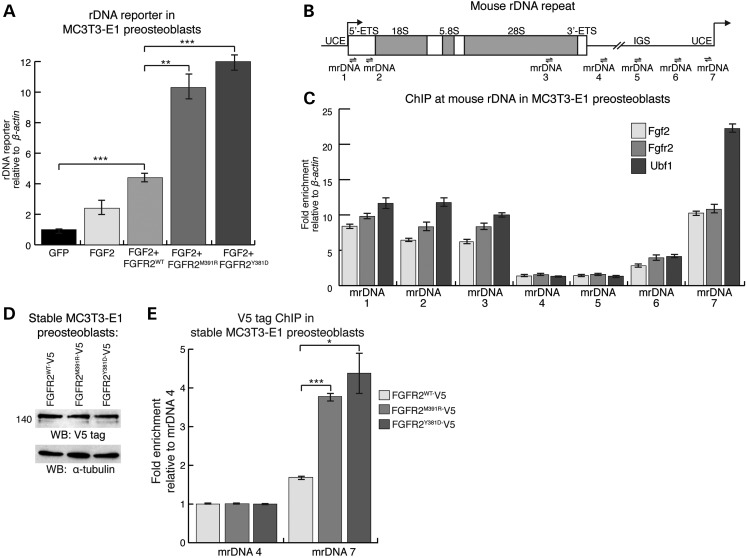
To determine whether FGFR2 promotes rDNA transcription at the level of the rDNA promoter, we utilized the pMr1930-BH rDNA minigene reporter, which contains the proximal rDNA promoter with the Upstream Core Enhancer (UCE) fused to a pUC9 sequence and terminator elements (29). In MC3T3-E1 preosteoblasts, FGFR2WT expression increased activation of the rDNA reporter 4.4-fold compared with GFP, whereas FGFR2M391R and FGFR2Y381D expression increased activation by 10.3- and 12.0-fold, respectively (Fig. 5A). These findings demonstrate that the FGFR2 mutations that cause BBDS enhance the ability of FGFR2 to activate rDNA transcription through the rDNA promoter.
Figure 5.
FGFR2 occupies the rDNA promoter with FGF2 and UBF1. (A) Activity of a rDNA minigene reporter, measured by qPCR of a unique pUC9 sequence within the minigene, was activated in MC3T3-E1 preosteoblasts expressing FGFR2WT by 4.4-fold, FGFR2M391R by 10.3-fold and FGFR2Y381D by 12-fold compared with GFP (n = 3). (B) Schematic representation of the mouse rDNA repeat with the upstream control element (UCE), external transcribed spacer (ETS) and IGS. Arrows denote the approximate locations of the qPCR primer sets mrDNA1-mrDNA7 used for ChIP. (C) ChIP-qPCR at the mouse rDNA repeat in MC3T3-E1 preosteoblasts showed co-enrichment of endogenous Fgfr2 and Fgf2 at known Ubf1-binding sites in the UCE. The qPCR data were normalized to a non-target region in β-actin (n = 3). (D) Western blot of MC3T3-E1 preosteoblasts stably expressing FGFR2WT-V5, FGFR2M391R-V5 or FGFR2Y381D-V5 showed equivalent levels of FGFR2 expression. (E) ChIP-qPCR in stably expressing MC3T3-E1 preosteoblasts showed that FGFR2M391R-V5 and FGFR2Y381D-V5 increase FGFR2 occupancy 2.3- and 2.6-fold, respectively, at the UCE (identified by primer set mrDNA7) compared FGFR2WT. A negative control region in rDNA (identified by primer set mrDNA4) did not show any change in receptor occupancy (n = 3). Error bars represent SEM.
UBF1 interacts along the rDNA repeat to promote rDNA transcription, and thus FGFR2 could activate transcription through a direct interaction with the rDNA promoter. Chromatin immunoprecipitation (ChIP)-qPCR for endogenous Fgfr2 and Fgf2 in MC3T3-E1 preosteoblasts showed enrichment of Fgfr2 with Fgf2 at known Ubf1-binding sites within rDNA including the UCE, the 5′ external transcribed sequence (5′-ETS), and the 28S rRNA coding sequence (Fig. 5B and C) (30). Regions relatively free of Ubf1 within the intergenic spacer (IGS) region of rDNA were free of Fgfr2 and Fgf2. The specificity of the anti-Fgfr2 antibody was confirmed by western blot and ChIP-qPCR in primary calvarial preosteoblasts from Fgfr2flx/flx mice that were transduced with either eGFP- or Cre-expressing adenovirus (Supplementary Material, Fig. S2B and C). ChIP-qPCR experiments in HEK 293T cells further corroborated FGFR2 and FGF2 occupancy at UBF1-binding sites within the UCE of human rDNA (Supplementary Material, Fig. S2D and E). Given that the UCE is critical for Ubf1-mediated transcriptional activation (26), these data suggest that Fgfr2, together with Fgf2, promotes rDNA transcription by directly influencing Ubf1. These unexpected findings demonstrate that nucleolar FGFR2 is part of the complex that directly regulates rDNA transcription.
To test how the mutations in FGFR2 that cause BBDS alter FGFR2 occupancy at the UCE of rDNA, we performed ChIP-qPCR in MC3T3-E1 preosteoblasts stably expressing epitope-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D. These stable lines express equivalent levels of the receptors when induced with doxycycline (Fig. 5D). ChIP-qPCR showed that FGFR2M391R and FGFR2Y381D were enriched 2.3- and 2.6-fold at the UCE, respectively, compared with FGFR2WT (Fig. 5E). This demonstrates that the FGFR2 mutations that cause BBDS increase receptor occupancy at a UBF1 interacting region known to be critical for rDNA transcription.
FGFR2 mutations in BBDS reduce osteoblast differentiation
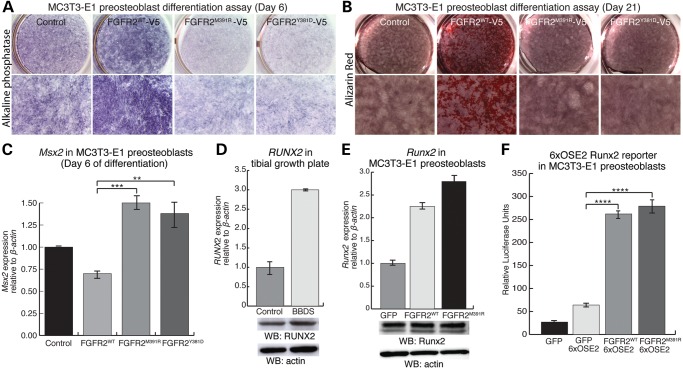
Despite an abundance of osteoprogenitor cells, the bones of BBDS patients are deficient in osteoblasts. To determine the effects of the FGFR2 mutations in BBDS on osteoblast differentiation, we performed differentiation assays in MC3T3-E1 calvarial preosteoblasts. During early stages of osteoblast differentiation, preosteoblasts stably expressing FGFR2WT exhibited increased cellular staining for the early osteoblast marker alkaline phosphatase (Fig. 6A). This is consistent with previous reports showing that FGFR2 promotes osteoblast differentiation (11). Preosteoblasts stably expressing FGFR2M391R and FGFR2Y381D showed decreased alkaline phosphatase staining compared with control, showing that the BBDS mutations reduce differentiation (Fig. 6A). During osteoblast maturation, preosteoblasts expressing FGFR2WT showed enhanced production of mineralized bone matrix compared with control, as detected by alizarin red staining, whereas those expressing FGFR2M391R and FGFR2Y381D did not (Fig. 6B). Msx2 is expressed in the calvarial mesenchyme and maintains early osteoprogenitor cell proliferation (31–33). We found that after 6 days of differentiation, calvarial preosteoblasts expressing FGFR2M391R and FGFR2Y381D exhibited a 2-fold increase in the levels of Msx2 compared with those cells expressing FGFR2WT (Fig. 6C). This provides further evidence that the FGFR2 mutations in BBDS maintain these cells in an osteoprogenitor-like state even under osteogenic conditions. Together, these data show that the FGFR2 mutations that cause BBDS, which enhance FGFR2-mediated activation of rDNA transcription, abrogate the ability of FGFR2 to promote osteoblast differentiation and instead reduce differentiation.
Figure 6.
FGFR2 mutations in BBDS reduce osteoblast differentiation without altering the levels or transactivation potential of RUNX2. (A) Alkaline phosphatase staining was increased in MC3T3-E1 preosteoblasts expressing FGFR2WT, whereas alkaline phosphatase staining was decreased in MC3T3-E1 preosteoblasts expressing FGFR2M391R and FGFR2Y381D, compared with control after 6 days in osteoblast differentiation medium (n = 4). (B) Alizarin red staining was increased in MC3T3-E1 preosteoblasts expressing FGFR2WT, whereas alizarin red staining in preosteoblasts expressing FGFR2M391R and FGFR2Y381D remained similar to the control after 21 days in osteoblast differentiation medium (n = 4). (C) Following 6 days of differentiation, levels of Msx2 detected by qPCR relative to β-actin were elevated at least 2.0-fold in MC3T3-E1 cells expressing FGFR2M391R and FGFR2Y381D compared with those cells expressing FGFR2WT (n = 3). (D) While levels of RUNX2 transcript, detected by qPCR, were increased in BBDS compared with control, levels of RUNX2 protein, detected by western blot, remained equivalent. (E) Levels of RUNX2 transcript, detected by qPCR, were increased in MC3T3-E1 preosteoblasts expressing FGFR2M391R compared with FGFR2WT; however, Runx2 protein levels, detected by Western blot, remained equivalent (n = 3). (F) The Runx2 6XOSE2-luciferase reporter was equivalently activated in MC3T3-E1 preosteoblasts expressing FGFR2WT and FGFR2M391R (n = 3 for each). Error bars represent SEM.
Increased nucleolar FGFR2 activity limits RUNX2-mediated inhibition of rDNA transcription
Interestingly, abnormalities resulting from reduced osteoblast differentiation in BBDS, including deficient skull ossification and hypoplastic clavicles, are also characteristic of cleidocranial dysplasia, a disorder resulting from RUNX2 haploinsufficiency (34). RUNX2 is a transcription factor essential for osteoblast differentiation (35), and this phenotypic overlap suggests that RUNX2 function may be reduced in BBDS. Canonical FGF signaling promotes Runx2 expression in preosteoblasts (36); however, we did not find evidence that RUNX2 levels are altered in BBDS. While RUNX2 transcripts were increased nearly 3-fold in the BBDS tibial growth plate compared with control, RUNX2 protein levels remained comparable to control (Fig. 6D). Expression of FGFR2M391R in MC3T3-E1 preosteoblasts increased levels of Runx2 slightly over cells expressing FGFR2WT, yet Runx2 protein levels remained unchanged (Fig. 6E). This data supports that RUNX2 protein levels are not grossly altered by the FGFR2 mutations that cause BBDS.
We next tested the effects of the FGFR2 mutations that cause BBDS on RUNX2 activity. During osteoblast differentiation, RUNX2 activates transcription of genes that facilitate development of the bone cell phenotype. As canonical FGF signaling promotes this function for RUNX2 (12,37), we examined the effects of the FGFR2 mutations in BBDS on RUNX2 activity at its bone-specific target genes. We found that expression of VEGF and MMP-13, which are RUNX2 targets in developing long bones, were not altered in the BBDS tibial growth plate (Supplementary Material, Fig. S3A). Furthermore, expression of FGFR2M391R in MC3T3-E1 preosteoblasts did not change Runx2-mediated activation of osteocalcin, a Runx2 target in developing calvaria, and the Runx2 reporter 6XOSE2-luciferase compared with FGFR2WT (38) (Supplementary Material, Fig. S3B) (Fig. 6F). Thus, the ability of RUNX2 to activate bone-specific genes in the long bone and calvaria remained unaffected by the FGFR2 mutations that cause BBDS.
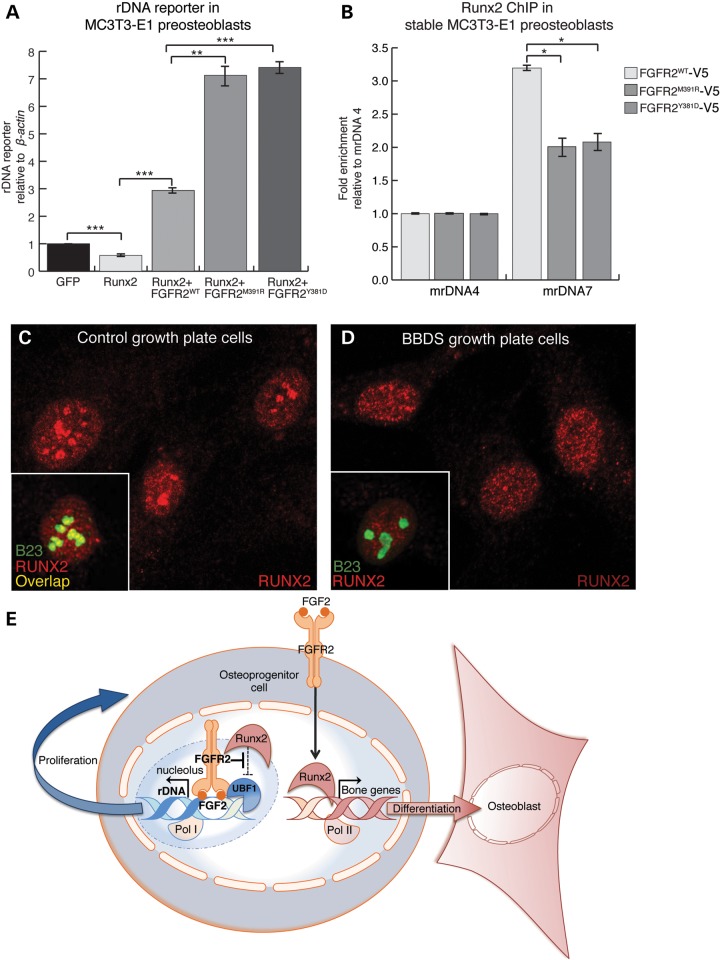
Runx2 function is not restricted to activation of bone-specific genes. Runx2 also promotes osteoblast differentiation by attenuating preosteoblast cell proliferation through repression of rDNA transcription (30). Correspondingly, expression of Runx2 with the mouse rDNA minigene reporter in MC3T3-E1 preosteoblasts reduced reporter activation by 42% compared with reporter alone (Fig. 7A). Expression of FGFR2WT with Runx2 resulted in a 3-fold activation of the rDNA reporter, whereas expression of FGFR2M391R or FGFR2Y381D with Runx2 further increased this activation to over 7-fold. This suggests that the mutations in FGFR2 that cause BBDS enhance the ability of FGFR2 to de-repress Runx2 at the rDNA promoter.
Figure 7.
FGFR2 limits nucleolar occupancy and activity of RUNX2 at rDNA. (A) Activation of the rDNA minigene reporter, detected by qPCR, was reduced 42% in MC3T3-E1 preosteoblasts expressing Runx2. On the other hand, activation of the rDNA minigene reporter was increased 3-fold in preosteoblasts co-expressing Runx2 with FGFR2WT and over 7-fold in preosteoblasts co-expressing Runx2 with either FGFR2M391R or FGFR2Y381D (n = 3). (B) ChIP-qPCR showed that stable expression of FGFR2M391R or FGFR2Y381D in MC3T3-E1 preosteoblasts reduced Runx2 occupancy at the UCE in the rDNA promoter by 37 and 35%, respectively (detected by the mrDNA7 primer set), compared with those preosteoblasts expressing FGFR2WT (n = 3). Error bars represent SEM. (C) Immunofluorescent analysis showed that RUNX2 (red) is localized to the nucleus and nucleoli, as detected by B23 (green), in control growth plate cells. (D) Immunofluorescent analysis showed RUNX2 (red), while localized to the nucleus, is absent from the nucleoli of BBDS growth plate cells. (E) Model for the dual role of FGFR2 signaling in osteoprogenitor cells.
Runx2 interacts with Ubf1 at the UCE to inhibit rDNA transcription (30). Since Fgfr2 occupies this same region and blocks Runx2-mediated repression of the rDNA minigene reporter, we examined endogenous Runx2 occupancy at rDNA in MC3T3-E1 preosteoblasts expressing the BBDS mutations. ChIP-qPCR experiments showed that expression of FGFR2M391R and FGFR2Y381D reduced Runx2 occupancy 37 and 35%, respectively, compared with FGFR2WT (Fig. 7B). Thus, the FGFR2 mutations in BBDS limit Runx2 occupancy at rDNA, possibly through competitive interactions with Ubf1. To test whether this mechanism is supported by findings in BBDS, we tested for changes in the nucleolar localization of RUNX2 in BBDS growth plate cells. Immunofluorescence detected RUNX2 in the nuclear matrix and nucleoli of control growth plate cells; however, in BBDS growth plate cells, RUNX2 was absent from the nucleoli and strictly localized to the nuclear matrix (Fig. 7C and D). Thus, increased nucleolar activity of FGFR2 in BBDS leads to nucleolar exclusion of RUNX2, elevated pre-rRNA expression, increased osteoprogenitor cell proliferation and decreased osteoblast differentiation.
DISCUSSION
By investigating the pathophysiology of BBDS, we have revealed a nucleolar role for FGFR2 in osteoprogenitor cells. We show that the mutations in FGFR2 that cause BBDS augment a normal function for FGFR2 in the nucleolus where it interacts at the rDNA promoter with FGF2 and UBF1 to promote transcription by limiting transcriptional repression by RUNX2. The resultant increase in rRNA promotes proliferation and reduces osteoblast differentiation in osteoprogenitor cells. While previous gain- and loss-of-function studies have shown that Fgfr2 promotes both proliferation and differentiation of osteoprogenitor cells, our work demonstrates that these activities are distinct and are controlled at the level of the subcellular localization of FGFR2. Two FGFR2 signaling routes, one at the plasma membrane and one in the nucleolus, can partition activities of RUNX2 to maintain a proper balance for osteoprogenitor cell proliferation and differentiation (Fig. 7E). Canonical FGFR2 signaling initiated at the plasma membrane promotes osteoblast differentiation by increasing the RUNX2 expression, stabilization and transcriptional activity at bone-specific genes (12,36,37). On the other hand, we show that nucleolar FGFR2 signaling promotes osteoprogenitor cell proliferation by opposing RUNX2, which functions as a repressor of rDNA transcription. Thus, we propose that BBDS is a consequence of an imbalance in canonical and nucleolar FGFR2 signaling.
The disease-causing FGFR2 mutations in BBDS are the first genetic changes in a FGFR that demonstrate a role for nuclear FGF signaling during vertebrate development. Evidence for nuclear FGF signaling emerged nearly two decades ago. However, no genetic approach to date has fully delineated the role of FGFRs in the nucleus. While nuclear Fgfr2 has been observed during differentiation in vertebrate development (39), this phenomenon has remained largely unstudied because the mouse models used to study the function of Fgfr2, which include a variety of knock-outs and knock-ins, contain genetic alterations that do not distinguish between the nuclear and membrane functions of the receptor.
Our finding that nucleolar FGFR2 promotes rDNA transcription in BBDS has implications for other congenital disorders. FGFR2 mutations are responsible for at least 10 distinct birth defects, and it will be important to examine if altered nucleolar activity of FGFR2 contributes to the skeletal abnormalities in these disorders. We expect that increased osteoprogenitor cell proliferation and differentiation caused by FGFR2 gain-of-function will be explained by increased FGFR2 signaling at the plasma membrane and in the nucleolus. Conversely, we anticipate that reduced osteoprogenitor cell proliferation and differentiation resulting from FGFR2 loss-of-function will be explained by decreased FGFR2 signaling at both the plasma membrane and in the nucleolus. Our findings will also shed light on a class of birth defects with ribosome dysfunction known as ribosomopathies. Ribosomes, as the protein-producing machines of the cell, perform a ubiquitous and essential function in mRNA translation. Yet, unexpectedly, human disorders with ribosomal dysfunction, including Treacher Collins syndrome and Diamond Blackfan Anemia, exhibit specific skeletal defects similar to BBDS (40). Our discovery that FGFR2 regulates rDNA transcription in preosteoblasts identifies new specificity in the upstream regulation of ribosome biogenesis during skeletal development and can explain this phenotypic overlap.
Defining a role for FGFR2 in rDNA transcription will also lend new insight into the pathogenesis and treatment of cancer. Many of the FGFR2 germline mutations found in skeletal birth defects are identical to somatic FGFR2 mutations seen in cancer. Cancers of the endometrium, breast, lung, gastrointestinal tract and kidney carry FGFR2-activating mutations found in craniosynostosis disorders, including the p.Met391Arg in BBDS (41). Such merging of germline and somatic genetics strongly suggests that these diseases share a common etiology. FGFR2′s ability to regulate rDNA transcription could be this molecular connection, as increased rDNA transcription induces neoplastic transformation (42). Currently, inhibitors that nonspecifically target FGFRs are in clinical use to treat cancer, yet because FGFR2 acts as both an oncogene and a tumor suppressor, this approach may prove to be problematic (43). Our discovery that FGFR2 has two distinct signaling routes could aid development of safer cancer therapies that specifically target the pro-proliferative role of FGFR2 in the nucleolus while avoiding potentially dangerous side effects associated with the non-selective FGFR inhibitors.
By showing the biological significance of nucleolar FGFR2 signaling in human development, our findings in BBDS help to expand the paradigm for FGF signaling. Most signaling pathways regulate development by directing gene expression profiles through pathway-specific transcription factors: the TGFβ pathways rely on Smads, Wnt signaling uses TCF/LEF and the Hedgehog pathway employs Gli. While a dedicated transcription factor has not been identified for the FGF pathway, our study shows that the pathway can instead control gene expression by utilizing the nuclear FGFR2–FGF2 complex. Together, FGFR2 and FGF2 exhibit the traits of a transcriptional regulator: FGF2 has sequence-specific DNA-binding activity (44,45) and FGFR2 modifies interacting proteins through its tyrosine kinase activity. A fundamental shift in our view of the FGF pathway to include a nuclear arm that directly regulates gene expression can reveal the mechanisms underlying different cellular responses to FGF signaling in tissues both within and outside the skeleton and also lend new insight into the diseases that result from FGF signaling dysfunction, such as in birth defects and cancer.
MATERIALS AND METHODS
Cell culture and transient transfections
HEK 293T cells and MC3T3-E1 cells were transiently transfected using jetPrime (PolyPlus) or FuGene (Promega) according to the manufacturer's instructions. Human primary growth plate cells and the surrounding periosteum were previously isolated from the distal femurs of a BBDS patient (ISDR ID# R07-401) and a normal stage-matched control (18). Mouse primary calvarial preosteoblasts were isolated at postnatal Day 4 from Fgfr2flx/flx mice using an established procedure (46) and transduced with either Ad5-CMV-Cre or Ad5-CMV-eGFP control virus (Baylor College of Medicine) as previously described (47). After 72 h, transduced cells were harvested and assayed for recombination by genomic PCR using previously described primer sets and protocol (6).
Immunofluorescence
Cells were seeded on chamber slides (Lab-Tek), fixed in 4% paraformaldehyde, permeabilized, blocked and incubated with anti-Bek (FGFR2) (Santa Cruz, 1:200), anti-FGF2 (Santa Cruz, 1:500), anti-B23 (Santa Cruz, 1:200), anti-UBF1 (Santa Cruz, 1:400), anti-FLAG-M2 (Sigma, 1:2000) or anti-RUNX2 (Santa Cruz, 1:200) antibody overnight at 4°C. Primary antibodies were detected using Alexa Fluor 568 conjugated goat anti-rabbit and/or Alexa Fluor 488 conjugated goat anti-mouse (Invitrogen) diluted 1:400 in 1% goat serum/PBS at room temperature for 1 h. Coverslips were mounted on slides using Vectashield with DAPI (Vector Laboratories), and confocal images were taken on a Leica TCS-SP2 AOBS inverted confocal microscope.
Cellular fractionation
For cytoplasmic and nuclear fractions, cells were processed using the Thermo Scientific NE-PER Nuclear and Cytoplasmic Extraction Kit according to the manufacturer's instructions. For nucleolar extracts, HEK 293T cells were transfected with FGFR2WT-V5-pcDNA3.1, FGFR2M391R-V5-pcDNA3.1 or FGFR2Y381D-V5-pcDNA3.1 and fractionated using sucrose gradient centrifugation (48). Isolated nucleoli were resuspended in RIPA lysis buffer and analyzed by western blot.
Western blot analysis
RIPA protein extracts were normalized by microBCA (Pierce), resolved on a 10% SDS–PAGE gel, transferred to a nitrocellulose membrane and probed with anti-Bek (FGFR2/Fgfr2) (Santa Cruz, 1:200), anti-FGF2 (Santa Cruz, 1:750), anti-β-actin (Cell Signaling, 1:1000), anti-UBF1 (Santa Cruz, 1:500), anti-RUNX2 (Santa Cruz, 1:1000), anti-α-tubulin (Cell Signaling, 1:10 000), anti-FLAG-M2 (Sigma, 1:5000) or anti-V5-E10 (Invitrogen, 1:5000) overnight at 4°C. Immunoreactivity was detected using a Phototope-HRP western blot detection system according to the manufacture's instructions (Cell Signaling). The fold difference was calculated using values for band density calculated by ImageJ.
Quantitative PCR
RNA was isolated from tissue and cells using Trizol (Invitrogen) or RNeasy Mini Kit (Qiagen), respectively. cDNA was synthesized by reverse transcription (Qiagen), and qPCR was performed using PerfeCTa SYBR Green FastMix (Quanta Biosciences) and the primer sets as listed in Supplementary Material, Table S1. The following TaqMan primers (Applied Biosystems) were also used: mouse Runx2 (Assay ID: mm00501584_m1*), mouse osteocalcin (Assay ID: mm03413826_mH) and mouse actin (Assay ID: mm00455685_m1*).
Immunoprecipitation
Cells were transfected with either V5-tagged or FLAG-tagged versions of the mutant and wild-type receptor, lysed in RIPA buffer, and processed using the Dynabeads Protein G Immunoprecipitation Kit according to the manufacture's instructions (Invitrogen) with the following antibodies: anti-V5-E10 (Santa Cruz), anti-B23 (Santa Cruz), anti-importin-β (Abcam) or IgG control. Eluted protein G bead complexes were analyzed by western blot.
Signaling experiments
Cells were transiently transfected with GFP-pcDNA3.1, FGFR2WT-pcDNA3.1, FGFR2M391R-pcDNA3.1, FGFR2Y381D-pcDNA3.1, pCMV-FGFR2WT-FLAG, pCMV-NoLS-FGFR2WT-FLAG or pCMV-FGFR2M391R-FLAG. After 24 h, cells were serum-starved, stimulated with 100 ng/ml FGF2 with 10 μg/ml Heparin for 10 min and lysed in RIPA buffer. Lysates were analyzed by western blot probed with the following antibodies (Cell Signaling): anti-p-ERK1,2 (1:1000), anti-ERK (1:1000), anti-p-p38 (1:1000), anti-p38 (1:1000), anti-p-AKT (1:1000), and anti-AKT (1:1000), anti-p-PLCγ1 (1:1000), anti-PLCγ1 (1:1000), anti-p-STAT1 (1:1000), anti-STAT1 (1:1000), p-FRS2a (1:000), FRS2a (1:500) and anti-α-tubulin (1:10 000). The fold difference was calculated using values for band density calculated by ImageJ.
Reporter assays
For 45S pre-rRNA reporter assays, MC3T3-E1 cells were co-transfected with pMr1930-BH reporter plasmid (kind gift from I. Grummt) and GFP-pcDNA3.1, FGFR2WT-pcDNA3.1, FGFR2M391R-pcDNA3.1, FGFR2Y381D-pcDNA3.1 and/or pcDNA3.1-HA-Runx2 (kind gift from B. Frenkel). After 24 h, reporter activation was detected by qPCR for pUC9 (29,30) and mouse β-actin (Supplementary Material, Table S1). For luciferase reporter assays, MC3T3-E1 cells were transiently transfected with 6XOSE2-luciferase reporter (kind gift from G. Karsenty) and GFP-pcDNA3.1, FGFR2WT-pcDNA3.1 or FGFR2M391R-pcDNA3.1. After 24 h, lysates were mixed with luciferase assay reagent (Promega) and read with a luminometer.
Cell proliferation analysis
MC3T3-E1 cells were seeded at 4 × 104 cells/well in 12-well plates. After 24 h, cells were transiently transfected with FGFR2WT-pcDNA3.1, FGFR2M391R-pcDNA3.1 or FGFR2Y381D-pcDNA3.1. Cells were grown for 24 h, either left untreated or treated for an additional 24 h with 300 nm CX-5461 (49), and counted in duplicate using the TC10 Automated Cell Counter (Bio-Rad).
Osteoblast differentiation assays
Doxycycline-inducible lentiviral vectors expressing V5-tagged FGFR2WT, FGFR2M391R or FGFR2Y381D (UCLA vector core) were used to generate stably transduced MC3T3-E1 preosteoblasts. Preosteoblasts were cultured with 250 ng/ml doxycycline and induced to differentiate with 50 μg/ml ascorbic acid and 10 mm β-glycerophosphate. After 6 days of growth, preosteoblasts were fixed and incubated with NBT/BCIP substrate solution to detect alkaline phosphatase or collected for qPCR analysis as described. After 21 days of culture, cells were fixed and stained with 2% alizarin red S pH 4.2 to detect mineralized bone matrix.
Chromatin immunoprecipitation
ChIP was performed as previously described with slight modifications (Hitchler and Rice 2011). Briefly, cells were cross-linked, nuclei isolated and sonicated to generate DNA fragments of ∼500 bp. Chromatin was diluted and 10% of the supernatant was kept for input. The rest of the supernatant was pre-cleared and incubated overnight with 5 μg anti-UBF1 (Santa Cruz), anti-Bek (FGFR2/Fgfr2) (Santa Cruz), anti-FGF-2 (Santa Cruz) or anti-V5-E10 (Santa Cruz) followed by a 4-h incubation with Protein G Dynabeads. Protein G bead complexes were washed, protein-associated chromatin was eluted and cross-linking reversed. DNA was then purified, precipitated and quantified by qPCR using primers previously published (Supplementary Material, Table S1). ChIP enrichment was determined as percentage of input and/or normalized to a negative control sequence in β-actin.
Statistical analysis
The Student's t-test was used to test for significance in each set of values, assuming equal variance. Mean values plus or minus standard error are plotted. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the March of Dimes Basil O’ Connor Starter Scholar (grant number 5-FY12-166 to A.E.M.); National Institute of Health (grant numbers 5 P30 DE020750-02 to A.E.M., T90DE021982 to C.L.N., 5T32HD060549-02 to J.E.S. and 5T32CA009320 to C.T.T.); James H. Zumberge USC Faculty Research and Innovation Fund to A.E.M.; and the American Cancer Society (RSG117619 to J.C.R.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ingrid Grummt for the pMr1930-BH rDNA minigene reporter, Pascal Roussel for the NoLS-pEGFP-C2 plasmid, Baruch Frenkel for the MC3T3-E1 preosteoblasts and HA-Runx2 plasmid, Gerard Karsenty for the 6XOSE2-luciferase plasmid, Nan Hatch for the FGFR2-pcDNA3.1 and FGFR2-V5-pcDNA3.1 plasmids and the UCLA Vector Core for lentivirus production. We also thank Ryan Roberts, Sean Brugger and Robert Maxson for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stoll C., Dott B., Roth M.P., Alembik Y. Birth prevalence rates of skeletal dysplasias. Clin. Genet. 1989;35:88–92. doi: 10.1111/j.1399-0004.1989.tb02912.x. [DOI] [PubMed] [Google Scholar]
- 2.Wilkie A.O. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Lew E.D., Bae J.H., Rohmann E., Wollnik B., Schlessinger J. Structural basis for reduced FGFR2 activity in LADD syndrome: Implications for FGFR autoinhibition and activation. Proc. Natl. Acad. Sci. USA. 2007;104:19802–19807. doi: 10.1073/pnas.0709905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eswarakumar V.P., Horowitz M.C., Locklin R., Morriss-Kay G.M., Lonai P. A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:12555–12560. doi: 10.1073/pnas.0405031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Xiao R., Yang F., Karim B.O., Iacovelli A.J., Cai J., Lerner C.P., Richtsmeier J.T., Leszl J.M., Hill C.A., et al. Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development. 2005;132:3537–3548. doi: 10.1242/dev.01914. [DOI] [PubMed] [Google Scholar]
- 6.Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E.N., Towler D.A., Ornitz D.M. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 7.Eswarakumar V.P., Monsonego-Ornan E., Pines M., Antonopoulou I., Morriss-Kay G.M., Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129:3783–3793. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- 8.Moore R., Ferretti P., Copp A., Thorogood P. Blocking endogenous FGF-2 activity prevents cranial osteogenesis. Dev. Biol. 2002;243:99–114. doi: 10.1006/dbio.2001.0533. [DOI] [PubMed] [Google Scholar]
- 9.Ohbayashi N., Shibayama M., Kurotaki Y., Imanishi M., Fujimori T., Itoh N., Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim B.G., Kim H.J., Park H.J., Kim Y.J., Yoon W.J., Lee S.J., Ryoo H.M., Cho J.Y. Runx2 phosphorylation induced by fibroblast growth factor-2/protein kinase C pathways. Proteomics. 2006;6:1166–1174. doi: 10.1002/pmic.200500289. [DOI] [PubMed] [Google Scholar]
- 11.Miraoui H., Oudina K., Petite H., Tanimoto Y., Moriyama K., Marie P.J. Fibroblast growth factor receptor 2 promotes osteogenic differentiation in mesenchymal cells via ERK1/2 and protein kinase C signaling. J. Biol. Chem. 2009;284:4897–4904. doi: 10.1074/jbc.M805432200. [DOI] [PubMed] [Google Scholar]
- 12.Xiao G., Jiang D., Gopalakrishnan R., Franceschi R.T. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J. Biol. Chem. 2002;277:36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- 13.Dailey L., Ambrosetti D., Mansukhani A., Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Sorensen V., Nilsen T., Wiedlocha A. Functional diversity of FGF-2 isoforms by intracellular sorting. Bioessays. 2006;28:504–514. doi: 10.1002/bies.20405. [DOI] [PubMed] [Google Scholar]
- 15.Arese M., Chen Y., Florkiewicz R.Z., Gualandris A., Shen B., Rifkin D.B. Nuclear activities of basic fibroblast growth factor: potentiation of low-serum growth mediated by natural or chimeric nuclear localization signals. Mol. Biol. Cell. 1999;10:1429–1444. doi: 10.1091/mbc.10.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reilly J.F., Maher P.A. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J. Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stachowiak M.K., Maher P.A., Stachowiak E.K. Integrative nuclear signaling in cell development – a role for FGF receptor-1. DNA Cell Biol. 2007;26:811–826. doi: 10.1089/dna.2007.0664. [DOI] [PubMed] [Google Scholar]
- 18.Merrill A.E., Sarukhanov A., Krejci P., Idoni B., Camacho N., Estrada K.D., Lyons K.M., Deixler H., Robinson H., Chitayat D., et al. Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am. J. Hum. Genet. 2012;90:550–557. doi: 10.1016/j.ajhg.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouche G., Gas N., Prats H., Baldin V., Tauber J.P., Teissie J., Amalric F. Basic fibroblast growth factor enters the nucleolus and stimulates the transcription of ribosomal genes in ABAE cells undergoing G0 – G1 transition. Proc. Natl. Acad. Sci. USA. 1987;84:6770–6774. doi: 10.1073/pnas.84.19.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng Z., Liang Y., Lin C.Y., Comai L., Chirico W.J. Direct regulation of rRNA transcription by fibroblast growth factor 2. Mol. Cell. Biol. 2005;25:9419–9426. doi: 10.1128/MCB.25.21.9419-9426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hisaoka M., Ueshima S., Murano K., Nagata K., Okuwaki M. Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol. Cell. Biol. 2010;30:4952–4964. doi: 10.1128/MCB.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedlocha A., Falnes P.O., Madshus I.H., Sandvig K., Olsnes S. Dual mode of signal transduction by externally added acidic fibroblast growth factor. Cell. 1994;76:1039–1051. doi: 10.1016/0092-8674(94)90381-6. [DOI] [PubMed] [Google Scholar]
- 23.Gaubert F., Escaffit F., Bertrand C., Korc M., Pradayrol L., Clemente F., Estival A. Expression of the high molecular weight fibroblast growth factor-2 isoform of 210 amino acids is associated with modulation of protein kinases C delta and epsilon and ERK activation. J. Biol. Chem. 2001;276:1545–1554. doi: 10.1074/jbc.M001184200. [DOI] [PubMed] [Google Scholar]
- 24.Lechertier T., Sirri V., Hernandez-Verdun D., Roussel P. A B23-interacting sequence as a tool to visualize protein interactions in a cellular context. J. Cell Sci. 2007;120:265–275. doi: 10.1242/jcs.03345. [DOI] [PubMed] [Google Scholar]
- 25.Moss T., Stefanovsky V., Langlois F., Gagnon-Kugler T. A new paradigm for the regulation of the mammalian ribosomal RNA genes. Biochem. Soc. Trans. 2006;34:1079–1081. doi: 10.1042/BST0341079. [DOI] [PubMed] [Google Scholar]
- 26.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 27.Laferte A., Favry E., Sentenac A., Riva M., Carles C., Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montanaro L., Trere D., Derenzini M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budde A., Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 30.Young D.W., Hassan M.Q., Pratap J., Galindo M., Zaidi S.K., Lee S.H., Yang X., Xie R., Javed A., Underwood J.M., et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y.H., Tang Z., Kundu R.K., Wu L., Luo W., Zhu D., Sangiorgi F., Snead M.L., Maxson R.E. Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: a possible mechanism for MSX2-mediated craniosynostosis in humans. Dev. Biol. 1999;205:260–274. doi: 10.1006/dbio.1998.9114. [DOI] [PubMed] [Google Scholar]
- 32.Satokata I., Ma L., Ohshima H., Bei M., Woo I., Nishizawa K., Maeda T., Takano Y., Uchiyama M., Heaney S., et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 33.Rice R., Rice D.P., Olsen B.R., Thesleff I. Progression of calvarial bone development requires Foxc1 regulation of Msx2 and Alx4. Dev. Biol. 2003;262:75–87. doi: 10.1016/s0012-1606(03)00355-5. [DOI] [PubMed] [Google Scholar]
- 34.Lee B., Thirunavukkarasu K., Zhou L., Pastore L., Baldini A., Hecht J., Geoffroy V., Ducy P., Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 35.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 36.Kim H.J., Kim J.H., Bae S.C., Choi J.Y., Kim H.J., Ryoo H.M. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J. Biol. Chem. 2003;278:319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- 37.Park O.J., Kim H.J., Woo K.M., Baek J.H., Ryoo H.M. FGF2-activated ERK mitogen-activated protein kinase enhances Runx2 acetylation and stabilization. J. Biol. Chem. 2010;285:3568–3574. doi: 10.1074/jbc.M109.055053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ducy P., Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmahl J., Kim Y., Colvin J.S., Ornitz D.M., Capel B. Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development. 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- 40.Hannan K.M., Sanij E., Rothblum L.I., Hannan R.D., Pearson R.B. Dysregulation of RNA polymerase I transcription during disease. Biochim. Biophys. Acta. 2013;1829:342–360. doi: 10.1016/j.bbagrm.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollock P.M., Gartside M.G., Dejeza L.C., Powell M.A., Mallon M.A., Davies H., Mohammadi M., Futreal P.A., Stratton M.R., Trent J.M., et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 43.Turner N., Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 44.Amalric F., Bouche G., Bonnet H., Brethenou P., Roman A.M., Truchet I., Quarto N. Fibroblast growth factor-2 (FGF-2) in the nucleus: translocation process and targets. Biochem. Pharmacol. 1994;47:111–115. doi: 10.1016/0006-2952(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 45.Sosnowski B.A., Gonzalez A.M., Chandler L.A., Buechler Y.J., Pierce G.F., Baird A. Targeting DNA to cells with basic fibroblast growth factor (FGF2) J. Biol. Chem. 1996;271:33647–33653. doi: 10.1074/jbc.271.52.33647. [DOI] [PubMed] [Google Scholar]
- 46.Bakker A.D., Klein-Nulend J. Osteoblast isolation from murine calvaria and long bones. Methods Mol. Biol. 2012;816:19–29. doi: 10.1007/978-1-61779-415-5_2. [DOI] [PubMed] [Google Scholar]
- 47.Fouletier-Dilling C.M., Bosch P., Davis A.R., Shafer J.A., Stice S.L., Gugala Z., Gannon F.H., Olmsted-Davis E.A. Novel compound enables high-level adenovirus transduction in the absence of an adenovirus-specific receptor. Hum. Gene Ther. 2005;16:1287–1297. doi: 10.1089/hum.2005.16.1287. [DOI] [PubMed] [Google Scholar]
- 48.Busch H., Muramatsu M., Adams H., Steele W.J., Liau M.C., Smetana K. Isolation of nucleoli. Exp. Cell Res. 1963;24(Suppl. 9):150–163. [PubMed] [Google Scholar]
- 49.Drygin D., Lin A., Bliesath J., Ho C.B., O'Brien S.E., Proffitt C., Omori M., Haddach M., Schwaebe M.K., Siddiqui-Jain A., et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.