Abstract
The t(1; 11) translocation appears to be the causal genetic lesion with 70% penetrance for schizophrenia, major depression and other psychiatric disorders in a Scottish family. Molecular studies identified the disruption of the disrupted-in-schizophrenia 1 (DISC1) gene by chromosome translocation at chromosome 1q42. Our previous studies, however, revealed that the translocation also disrupted another gene, Boymaw (also termed DISC1FP1), on chromosome 11. After translocation, two fusion genes [the DISC1-Boymaw (DB7) and the Boymaw-DISC1 (BD13)] are generated between the DISC1 and Boymaw genes. In the present study, we report that expression of the DB7 fusion gene inhibits both intracellular NADH oxidoreductase activities and protein translation. We generated humanized DISC1-Boymaw mice with gene targeting to examine the in vivo functions of the fusion genes. Consistent with the in vitro studies on the DB7 fusion gene, protein translation activity is decreased in the hippocampus and in cultured primary neurons from the brains of the humanized mice. Expression of Gad67, Nmdar1 and Psd95 proteins are also reduced. The humanized mice display prolonged and increased responses to the NMDA receptor antagonist, ketamine, on various mouse genetic backgrounds. Abnormal information processing of acoustic startle and depressive-like behaviors are also observed. In addition, the humanized mice display abnormal erythropoiesis, which was reported to associate with depression in humans. Expression of the DB7 fusion gene may reduce protein translation to impair brain functions and thereby contribute to the pathogenesis of major psychiatric disorders.
INTRODUCTION
Over 20 years ago, St Clair et al. (1) reported a large Scottish family with major psychiatric disorders. In this family, a balanced chromosome translocation t(1;11) co-segregates with schizophrenia, major depression and other psychiatric disorders, suggesting the translocation as a causal genetic mutation (2). The disrupted-in-schizophrenia 1 (DISC1) gene was subsequently identified from the breakpoint of the translocation on chromosome 1 (3). Since then, the DISC1 gene has been extensively studied and proposed to play important roles in neuronal cell proliferation, differentiation, migration, maturation and synaptic functions (4–8). In a comprehensive summary of DISC1 studies, Brandon and Sawa (9) provided a recent review on the potential roles of DISC1 in the pathogenesis of psychiatric disorders.
Our previous studies, however, found that the translocation also disrupted another gene, Boymaw (also termed DISC1 fusion partner 1, DISC1FP1), on chromosome 11 in the Scottish family (10). After translocation, the DISC1-Boymaw (DB7) and the Boymaw-DISC1 (BD13) fusion genes are generated. The DB7 fusion protein is insoluble (11). Insoluble DISC1 proteins were reported in the postmortem brains of sporadically collected patients with schizophrenia, major depression and bipolar disorder (12). These findings prompted us to study the functions of insoluble DB7 fusion protein in hopes of elucidating molecular mechanisms underlying the major psychiatric disorders.
In this study, we investigated the effects of full-length DISC1 (DISC1-FL), truncated DISC1, DISC1-Boymaw (DB7) and Boymaw-DISC1 (BD13) protein expression in different cell lines. We found that expression of the DB7 fusion gene inhibits oxidoreductase activities, rRNA synthesis and protein translation. Humanized DISC1-Boymaw mice were generated via gene targeting to examine the in vivo functions of the fusion genes. We confirmed inhibition of protein translation by the DISC1-Boymaw fusion gene in the humanized mice. Molecular and behavioral characterizations of the humanized mice were also conducted.
RESULTS
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) reduction and DB7 fusion gene expression
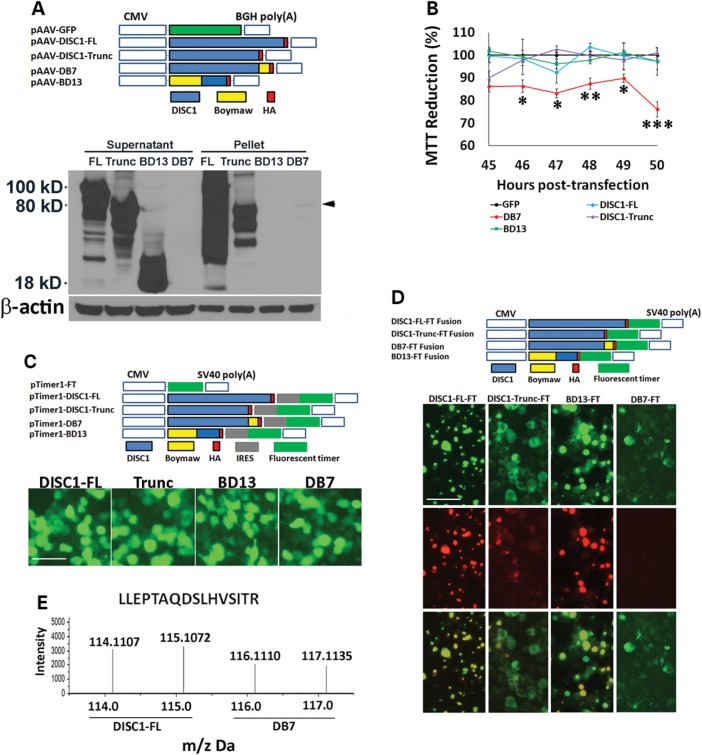
The DISC1 gene is mostly expressed in neuronal cells (13,14). To examine potential functions of the DB7 fusion gene, we conducted our studies in HEK293T cells that share 90% of expressed genes with human brain (15, 16). The cDNA genes encoding DISC1-FL, truncated DISC1 (1–597 amino acids), DISC1-Boymaw (DB7) and Boymaw-DISC1 (BD13) are tagged with an HA epitope at their C-terminal (Fig. 1A). Full-length DISC1-FL, truncated DISC1 and BD13 fusion proteins were abundantly expressed in contrast to low levels of DB7 fusion proteins exclusively found in the pellet fraction (Fig. 1A, overexposure in Supplementary Material, Fig. S1A). Similar expression patterns were also observed in transfected COS-7 cells (Supplementary Material, Fig. S1B and C). Since we do not know what exact functions the DB7 protein may serve, we first chose an assay to measure the broad cellular activity of the cells transfected with these genes. MTT reduction assays were therefore conducted (17–20). Surprisingly, only the DB7 fusion gene expression significantly decreased MTT reduction while MTT reduction did not differ between cells expressing DISC1-FL, truncated DISC1, BD13 and green fluorescent protein (GFP) control genes (Fig. 1B). Decreased MTT reduction by the DB7 fusion gene expression was also observed in both COS-7 and neuroblastoma SH-SY5Y cells (Supplementary Material, Fig. S1D and E). We were perplexed by the low expression of the DB7 fusion proteins because all expression constructs contain the same promoter and transcription terminator. One possible explanation could be that the DB7 mRNA is less stable than the mRNA transcripts of the other three genes. To examine such a possibility, bi-cistronic gene constructs were generated (Fig. 1C). If the DB7 mRNA were unstable, we would expect little expression of the fluorescence marker gene downstream of the DB7 open reading frame (ORF). Surprisingly, expression of the fluorescence marker gene was not different between cells expressing any of the four bi-cistronic genes (Fig. 1C), indicating no difference in their mRNA abundance. To examine whether low level expression of the DB7 fusion proteins may be caused by rapid protein degradation, we constructed fusion genes in which each gene was fused with a fluorescence timer (FT) E5 (Fig. 1D) (21). The FT E5 proteins change color from green to red, 9 h after translation. All fusion genes, except the DB7-FT, produced strong co-localized green and red fluorescence (Fig. 1D). However, we could only detect weak green fluorescence in the absence of any red fluorescence in cells expressing the DB7-FT proteins, suggesting possible rapid protein degradation. In further support of this conclusion, addition of proteasome inhibitor N-acetyl-l-leucyl-l-leucyl-L-norleucinal (LLnL) to HEK293T cells transfected with the DB7 fusion gene remarkably increased the level of DB7 protein expression (Supplementary Material, Fig. S1F). To provide direct evidence for rapid protein degradation, we performed a proteomic analysis of cells expressing the DB7 fusion gene using the isobaric tags for relative and absolute quantification (iTRAQ) (Supplementary Material, Fig. S2A, B and C). Total proteins from the DISC1-FL and the DB7 over-expression cells were labeled with 114, 115, 116 and 117 Da mass tags, respectively. Hundreds of unique DISC1 peptides were identified in cells expressing the DB7 fusion gene, and their average abundance was ∼70% of that in cells expressing the DISC1-FL gene. The abundance of a representative DISC1 peptide is shown between cells expressing full-length DISC1 and the DB7 fusion genes (Fig. 1E). We conclude that these abundant DISC1 peptides from cells expressing the DB7 fusion gene are likely peptide fragments generated by rapid protein degradation within the cell.
Figure 1.
MTT reduction and rapid degradation of DB7 fusion protein. (A) pAAV plasmid constructs. Each expression construct was tagged with HA epitope. Western blot analyses revealed abundant expression of full-length DISC1 (FL), truncated DISC1 (Trunc) and BD13 (Boymaw-DISC1). The expression of the DB7 (DISC1-Boymaw) was very low (black arrowhead), and restricted to the pellet fraction. Equal amounts of proteins were loaded for the western blot (see β-actin control). (B) MTT reduction assays at multiple time points. There were 4–6 replica wells per construct per time point. The mean value of the MTT reduction in cells transfected with GFP was used as the control (100%) to calculate relative MTT reduction for all other constructs at each time point. ANOVA analysis revealed a significant gene effect [F(4,126) = 28.09, P < 0.0001]. Post hoc analyses (Tukey studentized range test) revealed that cells transfected with the DISC1-Boymaw (DB7) construct displayed a significant decrease in MTT reduction at all time points except 45 h post-transfection. Error bar: SEM. (*P < 0.05, **P < 0.01, ***P < 0.001). (C) Bi-cistronic constructs in pTimer1 plasmid vector. Each gene was linked with a fluorescence marker (timer) gene via an IRES site. Green fluorescence was examined 48 h after transfection of HEK293T cells. There was no difference between any of the four gene constructs. Scale bar: 25 µm. (D) The FT E5 was in-frame fused with DISC1-FL, truncated DISC1, DB7 and BD13 in pTimer plasmid vector. Expression of the fusion proteins was examined 48 h after transfection of HEK293T cells. Both green and red fluorescence were readily detected and co-localized in DISC1-FL-FT, DISC1-trunc-FT and BD13-FT. Weak green, and no red, fluorescence was detected in comparable number of cells expressing DB7-FT only after overexposure. Scale bar: 25 µm. (E) In iTRAQ experiments, proteins from cells expressing the full-length DISC1 gene were labeled with 114 and 115 Da mass tags; proteins from cells expressing the DB7 gene were labeled with 116 and 117 Da mass tags. Abundant human DISC1 peptides were also detected in cells expressing the DB7 gene.
Characterization of the Boymaw gene
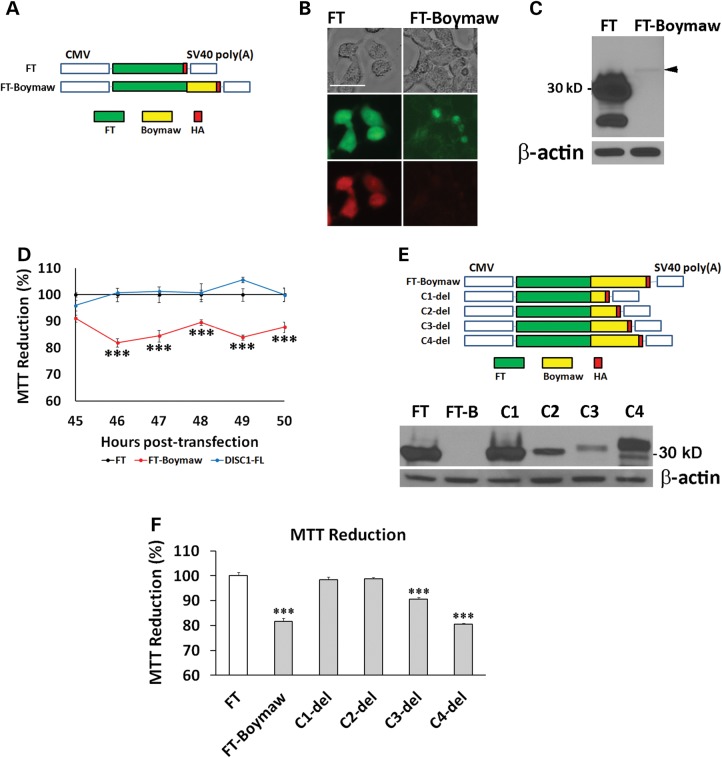
Since the truncated DISC1 proteins did not decrease MTT reduction, we wondered whether the Boymaw, rather than the DISC1, was responsible for both decreased MTT reduction and rapid protein turnover of the DB7 fusion proteins. Boymaw was therefore in-frame fused at the C-terminal of a randomly selected gene (the FT E5 proteins, FT-Boymaw) to follow the same fusion pattern of the DISC1-Boymaw (DB7) gene (Fig. 2A). In contrast to strong green and red fluorescence in cells expressing the control construct FT, weak green fluorescence was detected in the absence of red fluorescence in cells expressing the FT-Boymaw fusion gene (Fig. 2B). Consistent with the fluorescence images, western blot analyses detected abundant control FT proteins with little FT-Boymaw fusion proteins (Fig. 2C). A significant decrease of MTT reduction was also observed in cells expressing the FT-Boymaw fusion gene in comparison with the controls (Fig. 2D). These data indicate that the Boymaw cause decreased MTT reduction as well as rapid protein turnover of both the FT-Boymaw and the DISC1-Boymaw (DB7) fusion proteins. To investigate whether the inhibition of MTT reduction causes rapid protein turnover or vice versa, a series of C-terminal deletions of the FT-Boymaw gene were generated (Fig. 2E). The C4-del was as effective as the FT-Boymaw in decreasing MTT reduction (Fig. 2F). However, deletion of a short C-terminal sequence in the C4-del dramatically improved protein stability in contrast to barely detectable FT-Boymaw (FT-B) fusion proteins (Fig. 2E). The C2-del had a comparable MTT reduction to the controls (FT) (Fig. 2F), but was less abundant (Fig. 2E). These data indicate that different, although overlapping, regions of the Boymaw gene contribute to the regulation of rapid protein turnover and inhibition of MTT reduction.
Figure 2.
Characterization of the Boymaw gene. (A) Boymaw was fused with a FT tagged with HA in pTimer plasmid vector. The pTimer-HA plasmids (FT) were used as controls. (B) Expression of the FT-Boymaw fusion proteins was examined 48 h after transfection of HEK293T cells. While green and red fluorescence of the control FT proteins were very strong, little green and no red fluorescence of FT-Boymaw was detected. Scale bar: 15 µm. (C) Western blot analysis barely detected expression of the FT-Boymaw fusion proteins (black arrowhead) in comparison with abundant control FT proteins. (D) MTT assays revealed decreased MTT reduction by expression of the FT-Boymaw fusion gene in comparison with the controls and DISC1-FL [F(2,66) = 145.12, P < 0.0001]. Post hoc analyses (Tukey studentized range test) revealed that cells transfected with the FT-Boymaw construct displayed a significant decrease in MTT reduction at all time points except 45 h post-transfection. There were 4–6 replica wells per construct per time point. The mean value of the MTT reduction in cells transfected with FT was used as the control (100%) to calculate relative MTT reduction for the other two constructs at each time point. (E) A series of C-terminal deletion of FT-Boymaw fusion gene. Western blot analysis revealed expression of each deletion construct. (F) MTT assays were conducted 48 h after transfection of each deletion construct. There was a significant group effect on MTT reduction [F(8, 27) = 92.14, P < 0.000001]. Post hoc analyses (Tukey studentized range test) revealed significant MTT reduction in cells transfected with FT-Boymaw, C3-del and C4-del. There were four replica wells per construct. The mean value of the MTT reduction in cells transfected with FT was used as the control (100%) to calculate relative MTT reduction for all other constructs. Error bar: SEM. (*P < 0.05, **P < 0.01, ***P < 0.001).
Reduced NADH oxidoreductase activities
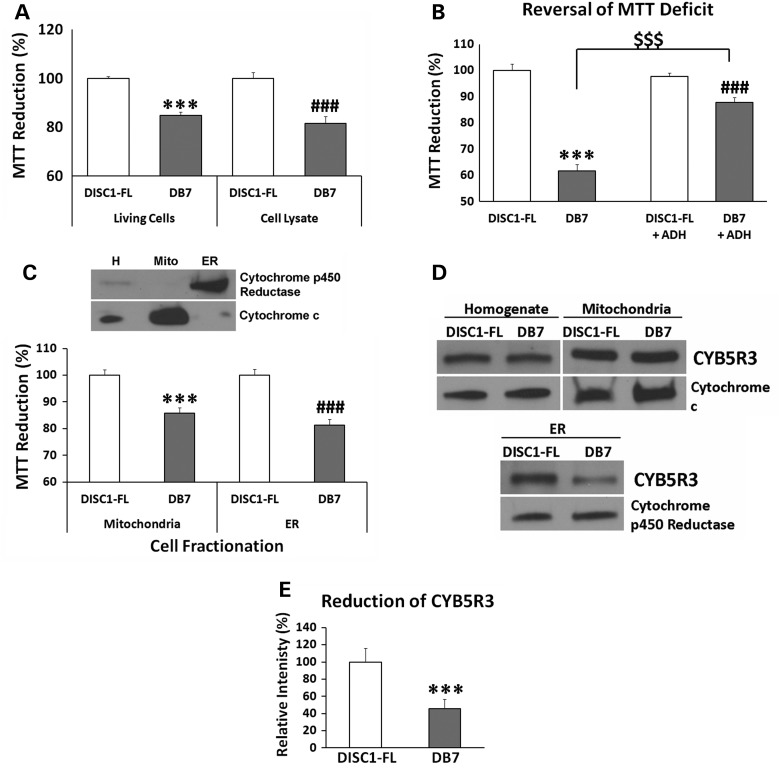
Our studies found that decreased MTT reduction is not caused by reduced proliferation of the cells expressing the DISC1-Boymaw (DB7) fusion gene (Supplementary Material, Fig. S3A). To rule out potential differences in MTT uptake in living cells, the transfected cells were partially lysed for the MTT assay. DB7 cell lysate display a similar magnitude of MTT reduction deficit to the living cells (Fig. 3A). These data therefore indicate that intracellular MTT reduction activities are decreased in DB7 expressing cells. NADH oxidoreductases are major intracellular electron donors in the reduction of MTT (20). We therefore first examined whether NADH concentration was decreased in cells expressing the DB7 gene. No significant difference in intracellular NADH was found between cells expressing DISC1-FL and DB7 genes (Supplementary Material, Fig. S3B). Cytosolic NADH concentration was also examined with NADH fluorescence sensor Peredox (22), and no difference was found either (Supplementary Material, Fig. S3C). To further examine whether NADH oxidoreductase activities were reduced in DB7 expressing cells, recombinant Escherichia coli NADH alcohol dehydrogenase (ADH), which catalyzes MTT reduction in the presence of NADH, was added to the DISC1-FL and DB7 cell lysates, respectively. Addition of the ADH dramatically increased MTT reduction in the DB7 cell lysate, but not in the DISC1-FL cell lysate (Fig. 3B). These data indicate that the DB7 cell lysate have decreased activities of NADH oxidoreductases. To localize the NADH oxidoreductases, we conducted density-gradient ultracentrifugation to isolate both mitochondria and microsomes (fragmented endoplasmic reticulum). Western blot analyses demonstrated enriched expression of cytochrome c in the mitochondrial fraction and cytochrome p450 reductase in the microsomal fraction (Fig. 3C). A significant decrease of MTT reduction was observed in both mitochondrial and microsomal fractions from the cells expressing the DB7 gene. Further analysis identified a significant reduction of cytochrome b5 reductase 3 (CYB5R3) proteins in the microsomal fraction of the cells expressing DB7 (Fig. 3D and E). No differential CYB5R3 expression was detected between DISC1-FL and DB7 in either cell homogenate or mitochondria.
Figure 3.
Reduced NADH oxidoreductase activities. (A) HEK293T cells were transfected with DISC1-FL and DB7, respectively. Two days after transfection, MTT reduction was performed in both living cells and cell lysate, respectively. Student's t-tests (unpaired, two-tailed) were used to compare MTT reduction between DISC1-FL and DB7 in living cells and cell lysate, respectively. The mean value of the MTT reduction in cells transfected with DISC1-FL was used as the control (100%) to calculate relative MTT reduction for DB7 in each group. Significantly decreased MTT reduction in DB7 transfection was observed in living cells (n = 3) [t(4) = 10.74, P < 0.001].and cell lysate (n = 6) [t(10) = 4.92, ###P < 0.001]. (B) Reversal of MTT reduction deficit in cell lysate. There were 12 replica wells for each gene and treatment. Unpaired student's t-test (two-tailed) was used for statistical analysis. Significantly decreased MTT reduction was observed in HEK293T cell lysate from DB7 transfection in comparison with DISC1-FL [t(22) = 11.50, P < 0.001]. E. coli ADH can catalyze MTT reduction in the presence of NADH. Addition of the ADH did not completely reverse the deficit of MTT reduction in DB7 lysate in comparison with DISC1-FL lysate [t(22) = 4.26, ###P < 0.001]. However, addition of recombinant E. coli ADH significantly improved MTT reduction in DB7 cell lysate [t(22) = 8.66, $$$P < 0.001 (comparison between DB7 lysate with and without ADH)]. No difference was found in MTT reduction of the DISC1-FL cell lysate with or without ADH. The mean value of the MTT reduction of DISC1-FL samples without ADH was used as the control (100%) to calculate relative MTT reduction for DB7, DISC1-FL+ADH and DB7+ADH. (C) Purification of mitochondria and endoplasmic reticulum (ER) from transfected HEK293T cells using density-gradient ultracentrifugation. Large amount of cells was transfected for each purification experiment. Cytochrome c and cytochrome p450 reductase were used as marker proteins for mitochondria and ER respectively. MTT reduction assays were conducted in purified mitochondria and ER from HEK293T cells transfected with DISC1-FL and DB7 constructs. The value of MTT reduction was further normalized with total amount of proteins in mitochondria and ER, respectively. Significantly reduced MTT reduction was observed in both mitochondria (n = 6)[t(10) = 5.53, two-tailed, P < 0.001) and ER (n = 6)[t(10) = 6.59, two-tailed, ###P < 0.001] isolated from cells transfected with DB7 expression construct. (D) The level of CYB5R3 protein expression was examined with western blot analysis, and normalized with the expression of ER marker cytochrome p450 reductase (CYPOR). CYB5R3 expression was reduced in the ER of the cells expressing the DB7 fusion gene. No significant difference in CYB5R3 expression was observed between DISC1-FL and DB7 in either cell homogenate or mitochondria after normalization with mitochondrial marker cytochrome c. (E) CYB5R3 expression was quantified in ER preparations of each construct (n = 4). Unpaired student's t-test (two-tailed) analysis revealed a significant reduction of CYB5R3 in DB7 [t(6) = 18.84, P < 0.001] Error bar: SEM. (*P < 0.05, **P < 0.01, ***P < 0.001).
To fully characterize the effects of the DB7 fusion gene expression, we conducted proteomic analysis of cells expressing the DB7 fusion gene. We did not find additional candidate NADH oxidoreductases whose expression was significantly decreased (Supplementary Material, Fig. S2A, S2B, S2C, and Supplementary Material, Table S1). Several reasons could account for the results. First, proteomic analysis can only survey a portion of total cellular proteins. Second, we found that expression of several NADH oxidoreductases was decreased, but did not reach statistical significance because of a limited number of identified peptides. Third, NADH oxidoreductase activities are modulated not only by protein expression but also by protein modifications. The molecular nature of the reduced activities of NADH oxidoreductases in cells expressing the DB7 fusion gene remains to be fully investigated. Among the top 18 differentially expressed proteins ranked according to their p values, however, seven were directly involved in the regulation of protein translation (Supplementary Material, Fig. S2C).
Inhibition of rRNA expression and protein translation in vitro
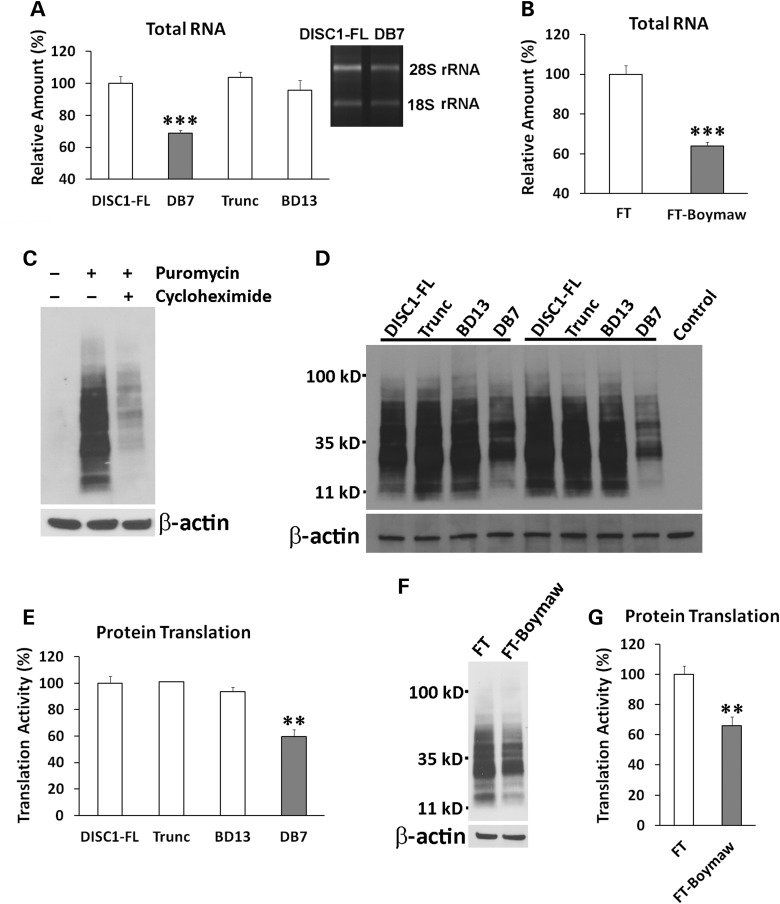
To investigate whether expression of the DISC1-Boymaw (DB7) fusion gene alters protein translation, we first examined rRNA expression in cells expressing the DISC1-FL, truncated DISC1, DB7 and BD13 genes, respectively. Interestingly, we found a significant reduction of total RNA in cells expressing the DB7 fusion gene (Fig. 4A). Reduction of the total RNA was not caused by differential cell proliferation (Supplementary Material, Fig. S3D). Since rRNA makes up more than 80% of total RNA, we quantified the expression of both 28S and 18S rRNAs. Reduction of both 28S and 18S rRNAs was found only in cells expressing the DB7, not the other three genes. Consistent with the effects of the DB7 fusion gene expression, expression of the FT-Boymaw fusion gene also decreased total RNA and rRNA expression (Fig. 4B). To provide direct evidence for inhibition of protein translation, we conducted SUnSET (surface sensing of translation) experiments to measure protein translation in the transfected cells (23). In the presence of cycloheximide, an inhibitor of protein translation, we detected little puromycin incorporation in newly synthesized proteins in western blot analysis (Fig. 4C). In pulse-labeling experiments, we found significant reduction of puromycin labeling from cells expressing the DB7 fusion gene, suggesting decreased protein translation activities (Fig. 4D and E). There was no significant difference in puromycin labeling between cells expressing the other three genes. We confirmed that expression of the FT-Boymaw fusion gene also inhibits protein translation similar to the DB7 fusion gene (Fig. 4F and G). All together, it appears that it is the Boymaw gene, rather than the DISC1 gene, that contributes to the inhibition of oxidoreductase activities, rRNA expression and protein translation in both the DISC1-Boymaw and FT-Boymaw fusion genes. We did not observe any changes of these cellular activities in cells expressing the full-length DISC1, truncated DISC1 or the Boymaw-DISC1 fusion genes.
Figure 4.
Inhibition of rRNA expression and protein translation. (A) Reduction of total RNA in cells expressing the DB7 gene [unpaired, two-tailed student's t-test, t(10) = 9.84, P < 0.001]. There were three to six replica wells for each gene construct. Significant reduction of rRNA expression was observed between DB7 and DISC1-FL. The same amount of rRNA reduction was further confirmed in later transfection experiments. Expression of total RNA and rRNA was normalized against the number of cells. The level of total RNA and rRNA expression of the DISC1-FL was used as the reference (100%) to calculate relative expression of the other three groups. (B) Reduction of total RNA and rRNA in cells expressing FT-Boymaw fusion gene compared with the control FT gene [unpaired, two-tailed student's t-test, t(8) = 7.6, P < 0.001). There were five replica wells for each construct. Significant reduction of rRNA was observed in cells expressing the FT-Boymaw fusion gene. (C) Pulse-labeling of new protein synthesis via incorporation of puromycin. Anti-puromycin antibody has no cross-reactions with HEK293T cellular proteins. A 10 min pulse-labeling generated massive incorporation of puromycin in western blot analysis. However, the presence of cycloheximide, an inhibitor of protein translation, dramatically reduced the puromycin labeling. (D) Inhibition of protein translation. Protein translation activities were measured with SUnSET in cells at 48 h post-transfection. Remarkable reduction of puromycin labeling was observed in cells expressing the DB7 construct in two representative sets of transfection experiments. (E) Quantification of reduction of puromycin incorporation. There were four replica dishes for each construct. There was a significant reduction of protein translation in cells expressing the DB7 construct in comparison with the DISC1-FL construct [unpaired, two-tailed student's t-test, t(6) = 5.03, P < 0.01]. The mean value of the incorporation of puromycin in DISC1-FL samples was used as the control (100%) to calculate relative intensity of puromycin incorporation in the other three genes. No difference was found in protein translation activity between the other three constructs. (F) There was a reduction of protein translation in cells expressing the FT-Boymaw fusion gene compared with the control FT gene in SUnSET. (G) There were six replica dishes for each construct. Significant reduction of puromycin incorporation was observed in cells expressing the FT-Boymaw fusion gene [unpaired, two-tailed student's t-test, t(10) = 4.48, P < 0.01]. Error bar: SEM. (*P < 0.05, **P < 0.01, ***P < 0.001).
Generation of humanized DISC1-Boymaw mice
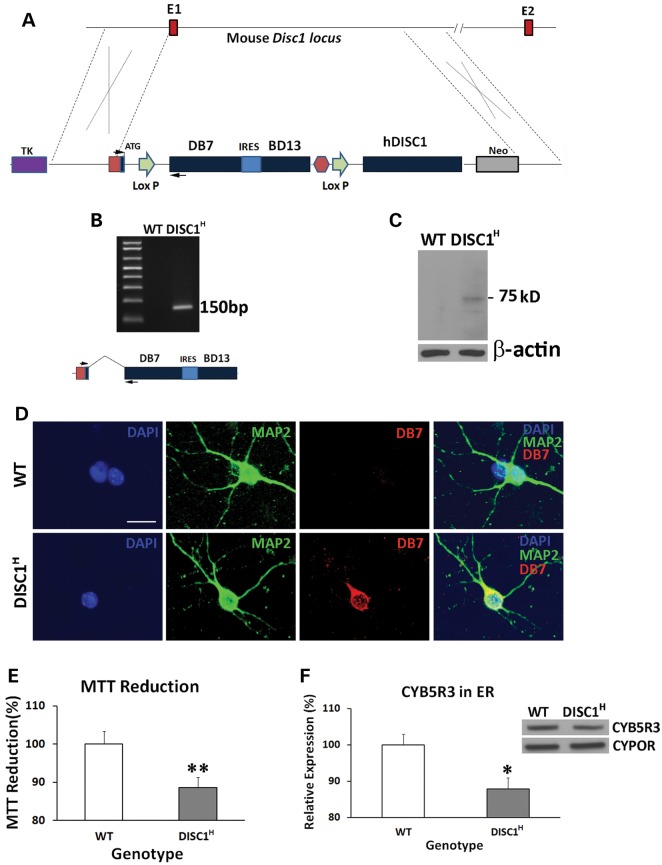
To investigate the in vivo functions of the DB7 fusion gene, we generated humanized DISC1-Boymaw mice. Human DISC1-Boymaw fusion gene (DB7) and Boymaw-DISC1 fusion gene (BD13) were knocked-in to replace mouse endogenous disc1 gene (Fig. 5A). A bi-cistronic gene, consisting of the two fusion genes connected with an internal ribosome entry site (IRES), was designed to splice with exon 1 of human DISC1 gene. The bi-cistronic cassette was also flanked with two loxP sites upstream of a human DISC1 gene for future genetic rescue experiments. Germ-line transmission of the bi-cistronic gene was obtained. To examine whether the bi-cistronic gene is expressed and correctly spliced with exon 1, we conducted reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of total RNA extracted from the brains of humanized DISC1-Boymaw mice (DISC1H) (Fig. 5B). A 150 bp DNA fragment was specifically amplified. Further sequencing confirmed the correct splicing (data not shown). Before examination of the expression of the DB7 proteins of the bi-cistronic gene in the humanized mice, we first evaluated eight different anti-DISC1 antibodies using our DISC1 over-expressing cell lysates (Supplementary Material, Fig. S4A). Five antibodies (ab55808, ab62069, ab59017, 3D4 and 14F2) with the highest affinities to human DISC1 proteins were selected for western blot detection of the DB7 fusion protein. Unfortunately, there were extensive non-specific cross-reactions of the antibodies with proteins from mouse brain homogenate in western blot analysis (data not shown), preventing the detection of the fusion proteins. However, the antibodies have less background in western blot analyses of mouse primary neuron culture (H. Song, personal communication). We therefore performed our analyses in cultured primary neurons from neonatal mice. The antibody against the N-terminal residues of human DISC1 protein recognized a 75 kDa protein band in western blot analysis of the primary neurons from the humanized DISC1-Boymaw mice, but not wild-type mice (Fig. 5C). Two reasons support this band as the DB7 fusion protein. First, the protein band is about the calculated size of the DB7 fusion protein (73 kDa). Second, the absence of this band in wild-type mice rules it out as a non-specific cross-reaction of the antibody. To detect the expression of the BD13 proteins of the bi-cistronic gene, we further screened the DISC1 antibodies in our BD13 over-expressing cell lysate. Two antibodies recognizing the C-terminal residues of human DISC1 proteins displayed some, but not high (compared with anti-HA antibody), affinity to the BD13 proteins (Supplementary Material, Fig. S4B). Due to the low affinity of the two antibodies, we could not detect the expression of the BD13 proteins expected to be ∼17 kDa (Supplementary Material, Fig. S4C, S4D). To further confirm the expression of the DB7 fusion proteins, immunocytochemical analyses were conducted. Co-localization of the DB7 proteins with neuronal cell marker MAP2 was observed in the primary neurons from the heterozygous DISC1-Boymaw mice (Fig. 5D). Expression of the DB7 protein did not cause gross anatomical abnormalities in the brain of the heterozygous DISC1-Boymaw mice (Supplementary Material, Fig. S5). To examine whether expression of the DB7 fusion protein decreases MTT reduction in vivo, we performed MTT reduction assays in the brain homogenate of neonatal heterozygous DISC1-Boymaw and wild-type control mice. A significantly decreased MTT reduction was observed in the heterozygous DISC1-Boymaw mice (Fig. 5E). Western blot analysis also confirmed a significant decrease of Cyb5r3 proteins in ER isolated from the brains of the heterozygous DISC1-Boymaw mice (Fig. 5F).
Figure 5.
Generation of humanized DISC1-Boymaw Mice. (A) Both the DISC1-Boymaw and the Boymaw-DISC1 fusion genes were knocked-in to replace mouse endogenous disc1 gene. E1 and E2 are exon 1 and 2 of mouse disc1 gene. (B) RT-PCR analysis of the expression of the DB7 fusion genes in humanized DISC1-Boymaw mice (DISC1H). The two PCR primers were localized in exon 1 and DB7, respectively (arrows). The expression of the bi-cistronic gene generated a 150 bp cDNA fragment after RT-PCR. Correct splicing was confirmed after sequencing. (C) Western blot analysis of the DB7 fusion protein in cultured primary neurons. Expression of the DISC1-Boymaw (DB7) fusion proteins was detected from the primary neurons isolated from the humanized DISC1-Boymaw mice. The 75 kDa protein band is very close to predicted molecular size (∼73 kDa). (D) Immunocytochemical staining of the DB7 fusion proteins in the primary neurons. Primary neurons were isolated from postnatal Day 1 mice, and cultured 4 days in vitro before immunostaining. Co-localization between MAP2 and DB7 proteins was observed. Scale bar: 20 µm. (E) Decreased MTT reduction in brain homogenate of neonatal heterozygous mice (DISC1H) (n = 14) in comparison with wild-type control littermates (n = 12) after normalization with total amount of proteins (t(24) = 2.984, P < 0.01, unpaired, two-tailed student's t-test). (F) Reduced expression of Cyb5r3 in the ER isolated from the brains of neonatal DISC1-Boymaw mice (n = 16) and wild-type littermates (n = 18) after normalization with ER marker Cypor (unpaired, two-tailed student's t-test, t(32) = 2.58, P < 0.05). Error bar: SEM. (*P < 0.05, **P < 0.01).
Inhibition of rRNA expression and protein translation in vivo
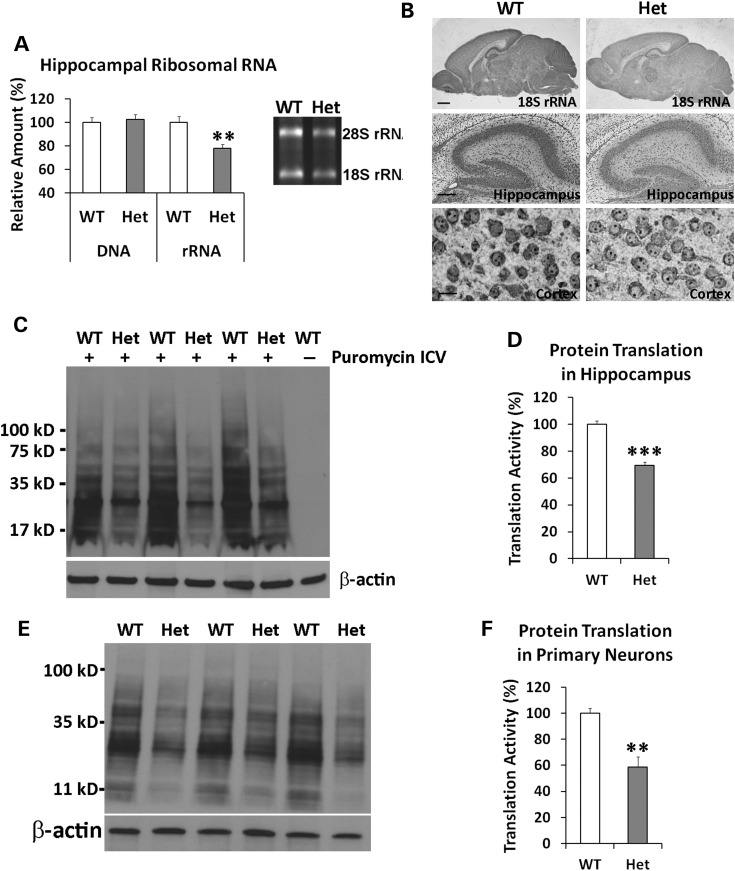
We were particularly interested in the inhibition of both rRNA expression and protein translation in HEK293T cells expressing the DB7 fusion gene. To investigate whether expression of the DB7 fusion gene also decreases rRNA expression in vivo, we extracted total RNA from mouse hippocampus and cortex. Heterozygous DISC1-Boymaw mice had significantly less hippocampal rRNA than their wild-type siblings (Fig. 6A). No difference in hippocampal DNA was found between the two genotypes. Reduction of rRNA expression was also observed in the cortex of the heterozygous DISC1-Boymaw mice (Supplementary Material, Fig. S6A). To confirm whether rRNA expression is reduced in neuronal cells, we conducted non-radioactive chromogenic RNA in situ hybridization to visualize 18S rRNA expression (Fig. 6B) (Supplementary Material, Fig. S6B). Reduced expression of 18S rRNA was observed across mouse brain including both hippocampal and cortical neuronal cells in the heterozygous DISC1-Boymaw mice. Consistent with reduction of rRNA expression, we found significant reduction of protein translation in the hippocampus of the heterozygous DISC1-Boymaw mice after intracerebroventricular (ICV) injection of puromycin in in vivo SUnSET experiments (Fig. 6C and D) (24). To investigate whether the inhibition of protein translation is independent of potential differential brain activities, we examined protein translation activities in cultured primary neurons isolated from postnatal Day 1 mice. Protein translation activity was significantly lower in the primary neurons isolated from the heterozygous DISC1-Boymaw mice than their wild-type siblings (Fig. 6Eand F).
Figure 6.
Decreased rRNA expression and protein translation. (A) Hippocampal DNA and RNA were isolated from 3-week old heterozygous DISC1-Boymaw (n = 8) and wild-type control mice (n = 12) respectively. No difference in total amount of DNA was observed between the two genotypes. However, significant reduction of total RNA was observed [unpaired, two-tailed student's t-test, t(18) = 3.33, P < 0.01]. After gel electrophoresis, 28S and 18S rRNA displayed significant reduction in the hippocampus of the heterozygous DISC1-Boymaw mice. (B) Chromogenic RNA in situ hybridization of 18S rRNA was conducted in the mouse brains at postnatal Day 7 (scale bar: 400 µm). Reduction of 18S rRNA was observed in both hippocampal neuronal cells (scale bar: 100 µm) as well as cortical neuronal cells (scale bar: 10 µm). Nuclear rRNA was readily detected in the nucleolus of the cortical neurons. (C) Protein translation activities in hippocampus. Intracerebroventricular injection of puromycin generated massive labeling of new protein synthesis in mouse hippocampus. Decreased puromycin labeling can be readily observed in the heterozygous mice. (D) Significant reduction of protein synthesis activities was found in the hippocampus of the heterozygous DISC1-Boymaw mice in comparison with wild-type sibling mice [unpaired, two-tailed student's t-test, t(4) = 9.28, P < 0.001]. (E) Primary neurons were isolated from postnatal Day 1 mice and cultured at 4 days in vitro before SUnSET experiments. After pulse-labeling, western blot revealed less puromycin labeling in the primary neurons from the heterozygous DISC1-Boymaw mice than their wild-type siblings. (F) Significant reduction of protein synthesis was detected [unpaired, two-tailed student's t-test, t(4) = 4.74, P < 0.01]. Error bar: SEM. (*P < 0.05, **P < 0.01, ***P < 0.001).
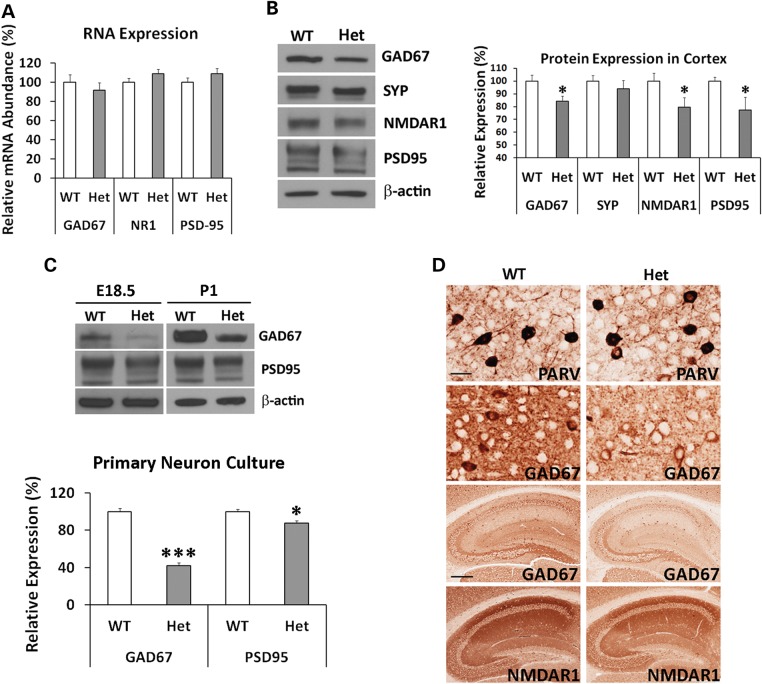
Decreased Gad67, Nmdar1 and Psd95 expression
We next examined whether general inhibition of protein translation in the humanized mice reduces protein expression of genes critical for pathophysiology of schizophrenia and other psychiatric disorders. Genes encoding Gad67, Nmdar1 and Psd95 proteins were examined because of their central roles in GABAergic and glutamatergic neurotransmission as well as synaptogenesis. There are also many reports on their down-regulation in psychiatric disorders (25–32), and degradation of PSD95 is proposed to play a key role in autism (33). We first conducted real-time RT-PCR quantification of their RNA expression, and did not find significant reduction in the humanized mice (Fig. 7A). To examine their protein expression, we conducted western blot analyses. Significant reduction of Gad67, Nmdar1 and Psd95 was observed in the heterozygous DISC1-Boymaw mice (Fig. 7B). No significant difference was found in synaptophysin. It has been reported that neural circuitry activity can regulate Gad67 gene expression at the system level (34). To rule out any potential effects from differential neural circuitry activity of individual mice, we examined Gad67 expression in primary neuron culture. Reduction of Gad67 proteins was remarkable in the primary neurons isolated from both E18.5 embryos and postnatal Day 1 (P1) DISC1-Boymaw mice (Fig. 7C). Moderate reduction of Psd95 was also observed. Expression of Nmdar1 was very low and inconclusive. Immunohistochemical analyses were also conducted to examine expression pattern of these proteins in adult mouse brains (Fig. 7D). Gad67 and Nmdar1 proteins displayed similar expression patterns between the heterozygous DISC1-Boymaw mice and wild-type siblings. In the heterozygous DISC1-Boymaw mouse brains, there is no obvious loss of Gad67-positive cortical GABAergic interneurons (data not shown). Anti-Psd95 antibody did not work on mouse paraffin sections. We also included anti-parvalbumin antibody in the analysis to examine whether this major subgroup of cortical GABAergic interneurons may be abnormal in the heterozygous DISC1-Boymaw mice. We did not find differences in either the number of positive neurons or intensity of parvalbumin expression between the two genotypes.
Figure 7.
Reduction of Gad67, Nmdar1 and Psd95 Proteins. (A) Q-PCR analysis of mRNA transcripts of Gad67, Nmdar1 and Psd95 genes in adult mouse brain. There was no significant difference between wild-type and the heterozygous DISC1-Boymaw mice in Gad67 [unpaired, two-tailed student's t-test, t(19) = 0.73, n.s.], Nmdar1 [unpaired, two-tailed student's t-test, t(19) = −1.44, n.s.] and Psd95 [unpaired, two-tailed student's t-test, t(19) = −1.17, n.s.] expression. (B) Western blot analysis and quantification of Gad67, synaptophysin (Syp), Nmdar1 and Psd95 in the brains of the adult heterozygous DISC1-Boymaw (n = 9) and wild-type (n = 9) mice. Unpaired two-tailed student's t-test was used for statistical analysis. No significant reduction of synaptophysin was observed between the two genotypes. Significant reduction of Gad67 [t(16) = 2.37, P < 0.05], Nmdar1 [t(16) = 2.55, P < 0.05] and Psd95 [t(16) = 2.58, P < 0.05] proteins was observed after normalization with either total amount of loading protein or β-actin expression. (C) In primary neurons isolated from cortex and striatum of postnatal Day 1 mice, significant reduction of both Gad67 (t(4) = 11.92, P < 0.001) and Psd95 [t(4) = 3.30, P < 0.05) proteins was observed in cultured primary neurons at 4 days in vitro from the DISC1-Boymaw mice (n = 3) in comparison with wild-type controls (n = 3) (unpaired, two-tailed student's t-test). Reduction of both proteins was further confirmed in the primary neuron culture (isolated from cortex and striatum) at 4 days in vitro from E18.5 embryos. (D) Immunohistochemical analysis of parvalbumin, Gad67, and Nmdar1 proteins in the heterozygous DISC1-Boymaw mice (anti-Psd95 antibodies did not work on paraffin sections). Reduction of Gad67 and Nmdar1 was observed in both hippocampus (scale bar: 100 µm) and cortex (scale bar: 20 µm). No difference was detected in parvalbumin expression. White ‘spots’ were nuclei not stained by antibodies. Error bar: SEM. (*P < 0.05, **P < 0.01, ***P < 0.001).
Behavioral analyses of the humanized DISC1-Boymaw mice
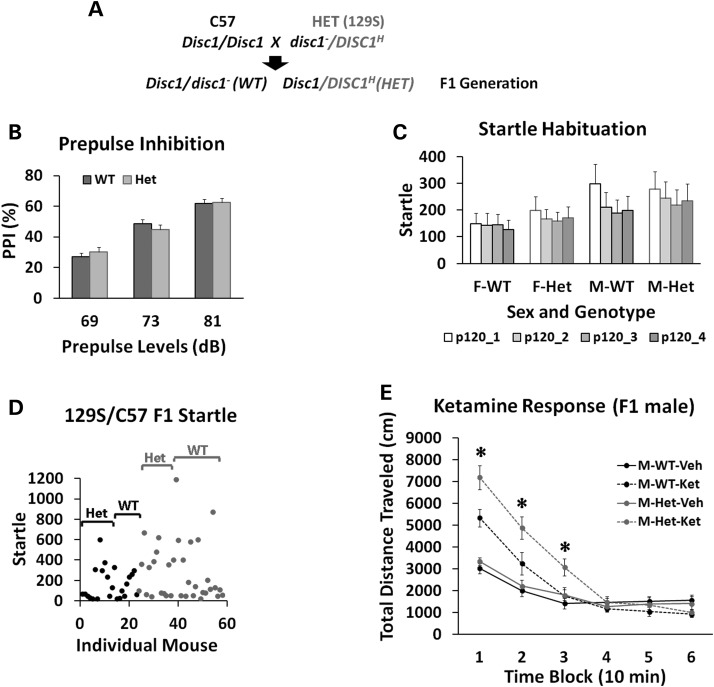
Since the DB7 fusion gene is a gain-of-function mutation in inhibiting protein translation, the function of the fusion gene was therefore studied in the heterozygous DISC1-Boymaw mice. Mice on the C57 background carry wild-type Disc1 genes, but 129S mice are natural null mutants of the full length disc1 gene (6, 35) (Supplementary Material, Fig. S6C and D). By following the recommendation of using F1 genetic background by Banbury Conference on Genetic Background in Mice (36), we generated heterozygous DISC1-Boymaw mice on F1 129S/C57 genetic background for behavioral studies (Fig. 8A). In the F1 generation, both wild-type and the DISC1-Boymaw heterozygous mice carry one allele of mouse wild-type Disc1 gene. The DISC1-Boymaw heterozygous mice were healthy and indistinguishable from their wild-type sibling mice. Prepulse inhibition (PPI) and habituation of acoustic startle were examined because they are both deficient in patients with schizophrenia and/or other major psychiatric disorders (37, 38). No differences in PPI (100 ms interstimulus interval; ISI), startle, or startle habituation were found between the genotypes (Fig. 8B and C). However, we noticed that there was a large startle variation between individuals with a significant number of mice with low startle (<50) (Fig. 8D), which could complicate the measurement of PPI. The source of such a large startle variation remains unknown since there is no genetic segregation in F1 generation. Schizophrenia patients are reported to have high sensitivity and prolonged responses to ketamine, an NMDA receptor antagonist (39–41). Cortical GABAergic inhibitory interneurons have been proposed to be the primary targets of NMDAR antagonists (42, 43). In light of our findings of reduced Nmdar1 and Gad67 proteins, we examined responses of the heterozygous DISC1-Boymaw male mice to ketamine in the mouse behavioral pattern monitor (BPM) (44). A significant gene by ketamine interaction was observed [F(1,27) = 4.30, P < 0.05]. Post hoc analyses revealed that the heterozygous DISC1-Boymaw male mice display a larger increase in total distance traveled (Fig. 8E), counts (Supplementary Material, Fig. S7A) and transitions (Supplementary Material, Fig. S7B) than sibling wild-type mice in locomotion after ketamine injection. It is unlikely that less locomotion of the wild-type mice results from ketamine overdose, since we did not observe initial suppression and later increase of locomotion by ketamine overdosing in the wild-type mice. However, multiple dosages of ketamine are needed to fully address the ketamine sensitivity of the heterozygous DISC1-Boymaw mice in the future.
Figure 8.
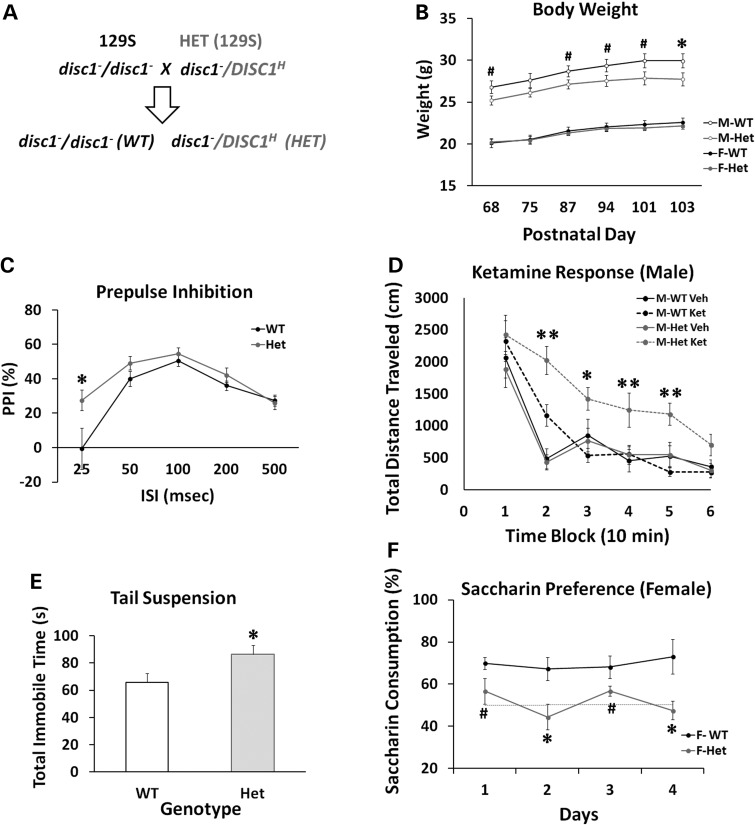
Behavioral analyses of the humanized DISC1-Boymaw mice on F1 129S/C57 genetic background. (A) The breeding scheme of mice on F1 129S/C57 background. In the F1 generation, there is no genetic segregation between individual mice. Both wild-type and the heterozygous DISC1-Boymaw mice carry one wild-type copy of mouse Disc1 gene. (B) The cohort of F1 mice consists of 28 wild-type and 26 heterozygous DISC1-Boymaw mice. There was no sex effect or gene effect on PPI [F(1,52) = 0.00, n.s.]. (C) There was no gene effect on startle habituation [F(1,50) = 0.21, n.s.]. (D) A large startle variation was observed between individual mice despite the fact that they are genetically identical. Dots represent individual mice (black = females, pale = males). (E) Males were tested for their responses to ketamine in the BPM. There were 15 wild-type and 14 heterozygous age-matched males. A significant gene by ketamine interaction was observed [F(1,27) = 4.30, P < 0.05]. Post hoc analyses (Tukey studentized range test) revealed that the heterozygous mice displayed significantly increased responses to ketamine in comparison with wild-type male controls in the first three blocks. Error bar: SEM. #P < 0.10; *P < 0.05; **P < 0.01.
We next examined the behavioral phenotypes of the humanized mice in the absence of full-length wild-type Disc1 gene on 129S genetic background (Fig. 9A). Because wild-type 129S mice carry mutant disc1 genes, we define that ‘wild-type mice’ in our studies refer to mice that do not carry the knocked-in DISC1-Boymaw fusion gene, rather than refer to the status of mouse endogenous disc1 gene. Both wild-type and heterozygous mice were healthy; however, male heterozygous mice were slightly, but significantly, smaller than their wild-type littermates (Fig. 9B). No significant body weight difference was observed in females between the two genotypes. In contrast to mice on mixed 129S/C57 genetic background, mice on 129S pure genetic background have higher startle with less variation between individuals (Supplementary Material, Fig. S8A), which makes the 129S genetic background more suitable for PPI studies. To extend the PPI studies with ISI at 100 ms in the F1 mice, we performed PPI tests with various ISIs in this cohort. When PPI was conducted with a short ISI at 25 ms, however, enhanced PPI, or impaired prepulse facilitation, was observed in the heterozygous DISC1-Boymaw mice, suggesting abnormal information processing (Fig. 9C). After ketamine administration in the BPM, significant gene [F(1,52) = 11.42, P < 0.01], ketamine [F(1,52) = 24.06, P < 0.0001] and gene X ketamine X sex interaction [F(1,52) = 5.40, P < 0.05] effects were observed in total distance traveled. In male mice, a significant gene X ketamine interaction [F(1,26) = 8.61, P < 0.01] was observed in the distance traveled (Fig. 9D) and other measures of locomotion activity (Supplementary Material, Fig. S8B, S8C, S8D). Post hoc analyses revealed significantly increased locomotion in the heterozygous DISC1-Boymaw mice injected with ketamine in all blocks except the first and last, suggesting increased and prolonged responses to ketamine. In female mice, a significant gene effect [F(1,26) = 5.63, P < 0.05] was detected without gene X ketamine interaction (Supplementary Material, Fig. S8E). To examine whether the heterozygous mice display behavioral phenotypes related to depression, a tail suspension test was conducted to assess mouse behavioral despair (45, 46). The heterozygous DISC1-Boymaw mice displayed significantly more immobility time than wild-type sibling controls without sex effects (P < 0.05) (Fig. 9E). A saccharin preference test was also conducted to examine anhedonia-like behavior, as seen in depression in humans, in the heterozygous mice. Wild-type female mice displayed significantly higher preference for saccharin than the heterozygous female mice (Fig. 9F). Because wild-type male mice had no significant preference for saccharin, the heterozygous DISC1-Boymaw male mice cannot be assessed for depression-like behavior using this behavioral paradigm (Supplementary Material, Fig. S8F). It has been reported that some rodent strains display a sex difference in saccharin preference in which females have strong preference while males have little preference or even some avoidance (47). In general, 129S mice are reported to have only slight saccharin preference (http://www.criver.com/SiteCollectionDocuments/129.pdf). These findings may explain why wild-type male mice on the 129S background did not display saccharin preference. Taken together, the humanized DISC1-Boymaw male mice consistently displayed a prolonged response to ketamine on different genetic backgrounds, mimicking phenotypes of schizophrenia and potentially depression. Replication of this phenotype in different cohorts of mice helped rule out type I error by multiple comparisons. Abnormal information processing of startle and depressive-related behaviors has also been suggested, and further confirmation is needed in future.
Figure 9.
Behavioral characterization of the humanized DISC1-Boymaw mice on 129S background. (A) Breeding of wild-type and the heterozygous DISC1-Boymaw mice on 129S pure genetic background. 129S mice carry a frame-shift mutation in exon 6 of the disc1 gene. (B) The cohort consisted of 14 wild-type females, 14 heterozygous females, 13 wild-type males and 15 heterozygous males. The age range of the birth date between individual mice was less than a week. Body weight was compared between the wild-type and the heterozygous mice. Heterozygous males displayed slightly, but significantly, reduced body weight at postnatal Day 103 [unpaired, two-tailed student's t-test, t(26) = 2.132, P < 0.05]. No difference was found in body weight between female wild-type and heterozygous mice. (C) There was a significant gene effect [F(1,54) = 4.41, P < 0.05] in PPI at 69 dB prepulse. A significant gene X ISI interaction was also observed [F(4,216) = 2.90, P < 0.05]. Post hoc analysis revealed that the heterozygous DISC1-Boymaw mice displayed increased PPI (or impaired prepulse facilitation) with ISI at 25 ms. No sex difference was observed. (D) The male heterozygous DISC1-Boymaw mice display a significant gene effect [F(1,26) = 6.26, P < 0.05] as well as a gene X ketamine interaction [F(1,26) = 8.61, P < 0.01]. Post hoc analyses (Tukey studentized range test) revealed that the male heterozygous mice displayed significantly increased and prolonged responses to ketamine in comparison with wild-type male controls in all time blocks except the first and last one. (E) Total immobility time (seconds) was recorded during a 6 min suspension test. Five wild-type male mice were lost because of fighting in cages after previous tests. The heterozygous DISC1-Boymaw mice (n = 29) displayed significantly more immobility time than the wild-type control mice (n = 22) [unpaired, two-tailed student's t-test, t(49) = 2.22, P < 0.05). (F) Two or three mice were grouped in a single cage according to genotype. Saccharin preference test was conducted for four consecutive days. Consumption of saccharin was measured each day, and compared between the wild-type and the heterozygous mice in each sex. Significant gene effect was detected [F(1,11) = 17.85, P < 0.01] between female wild-type (n = 7) and the DISC1-Boymaw heterozygous mouse cages (n = 6). Post hoc analysis revealed that the female heterozygous mice displayed significantly less saccharin consumption than female wild-type controls mice at Day 2 and Day 4. Error bar: SEM. #P < 0.10; *P < 0.05; **P < 0.01.
Abnormal erythropoiesis in humanized DISC1-Boymaw mice
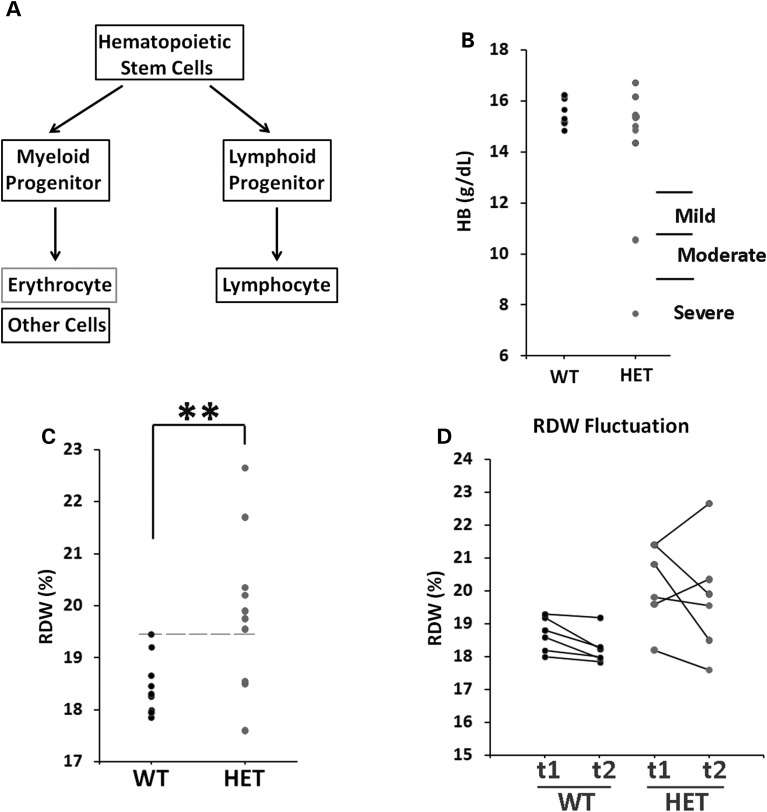
Mutations of ribosomal protein genes, which impair protein translation, have been demonstrated to cause Diamond–Blackfan anemia (48). Since the human DISC1 gene is also expressed in lymphoblastoid cells, we conducted hematological analysis of the heterozygous DISC1-Boymaw mice to investigate whether inhibition of protein translation by the DISC1-Boymaw fusion gene may cause anemia (Fig. 10A). Interestingly, 2 out of 10 heterozygous DISC1-Boymaw male mice displayed lower concentrations of hemoglobin (Fig. 10B) (Supplementary Material, Table S2), indicating different degrees of anemia (49). A significant difference between the two genotypes is an increase of red blood cell distribution width (RDW), which measures the variation of red blood cell volume, in the heterozygous DISC1-Boymaw mice (Fig. 10C). We re-examined RDW in some of these mice 1 week later to investigate RDW stability. RDW displayed little change in wild-type mice, but higher RDW fluctuation was observed in their sibling heterozygous DISC1-Boymaw mice (Fig. 10D). These data indicated that the DISC1-Boymaw fusion gene impairs the stability of erythropoiesis in the heterozygous DISC1-Boymaw mice.
Figure 10.
Increased RDW in the humanized DISC1-Boymaw mice. (A) Schematic pathway of erythropoiesis. (B) Concentration of hemoglobin (HB) was measured in the blood of individual male mice. Severe, moderate and mild anemia was diagnosed according to hemoglobin concentrations (49). Two heterozygous DISC1-Boymaw mice suffered from severe and moderate anemia. All wild-type mice were normal. Each dot represents an individual mouse. (C) Homogeneity of variances of RDW data was first assessed with Levene's test (P = 0.057). Significantly higher RDW was found in the heterozygous DISC1-Boymaw mice [F(1,19) = 8.451, **P < 0.01]. Most of these mice have RDW above the upper limit of the normal RDW range of their wild-type sibling mice (dashed line). (D) Six wild-type and six heterozygous DISC1-Boymaw male mice were re-examined 1 week later (t2) for their RDW stability, and compared with their initial (t1) RDW measurement. There was little change in RDW of the wild-type mice during this period. However, high magnitude of RDW fluctuation was observed in the heterozygous DISC1-Boymaw mice, indicating unstable erythropoiesis.
DISCUSSION
The t(1;11) chromosome translocation in the Scottish family co-segregates with schizophrenia and major depression with 70% penetrance (1–3). Despite extensive studies on the DISC1 gene, the role of the disruption of the DISC1 gene alone remains unclear in the pathogenesis of major psychiatric disorders (35,50,51). Our previous studies revealed that the DISC1 gene disruption is accompanied by disruption of the Boymaw gene and generation of the DISC1-Boymaw and Boymaw-DISC1 fusion genes (10,11). In the present study, we found that expression of the DB7 fusion gene decreases NADH oxidoreductase activities, rRNA synthesis and protein translation in both in vitro cell studies and in vivo humanized DISC1-Boymaw mice. Despite inhibition of protein translation by the DB7 fusion gene, however, we did not find gross anatomical abnormalities in the brain of the heterozygous DISC1-Boymaw mice. Inhibition of protein translation does not always cause obvious developmental defects. For example, reduced protein translation by silencing translational initiation factor eIF4E isoform (IFE-2) has no effects on Caenorhabditis elegans development and body size at 20°C (52). Even though we did not find obvious developmental abnormalities, reduced expression of Gad67, Nmdar1 and Psd95 proteins was observed in the brain of the heterozygous DISC1-Boymaw mice. Behavioral studies showed that the humanized mice display phenotypes related to both schizophrenia and depression. Our studies therefore suggest that the DB7 fusion gene could be one of the causal gene mutation(s) in the pathogenesis of major psychiatric disorders in the Scottish family. It should be kept in mind, however, that the humanized mice also harbor the BD13 fusion gene in addition to the DB7 fusion gene. Reduction of protein translation in the brains of the humanized mice is likely caused by the expression of the DB7 fusion gene, according to our in vitro cell transfection studies. However, we cannot rule out a potential contribution from the BD13 fusion gene to the behavioral phenotypes, although we did not find any cellular phenotypes in HEK293T cells expressing the BD13 fusion gene.
Our in vitro studies revealed that the DB7 fusion protein is unstable and its expression decreases intracellular oxidoreductase activities, rRNA synthesis and protein translation. These phenotypes are unlikely caused by the insolubility of the DB7 proteins per se, since the much more abundant insoluble DISC1 and truncated DISC1 proteins did not generate any of these cellular phenotypes in the over-expressing HEK293T cells. However, the insoluble DISC1 proteins in HEK293T cells may be structurally different from the insoluble proteins found in postmortem human brains (12). Surprisingly, fusion of the Boymaw gene to a randomly selected gene (fluorescence marker) generated the same cellular phenotypes as the DB7 fusion gene. It will be interesting to know whether the human Boymaw gene encodes any protein or peptide. Regardless of the Boymaw gene being a coding or non-coding RNA the DB7 fusion gene appears to be a gain-of-function mutation. The role of the Boymaw gene merits further studies. Unfortunately, there is no human Boymaw gene orthologue in mouse genome.
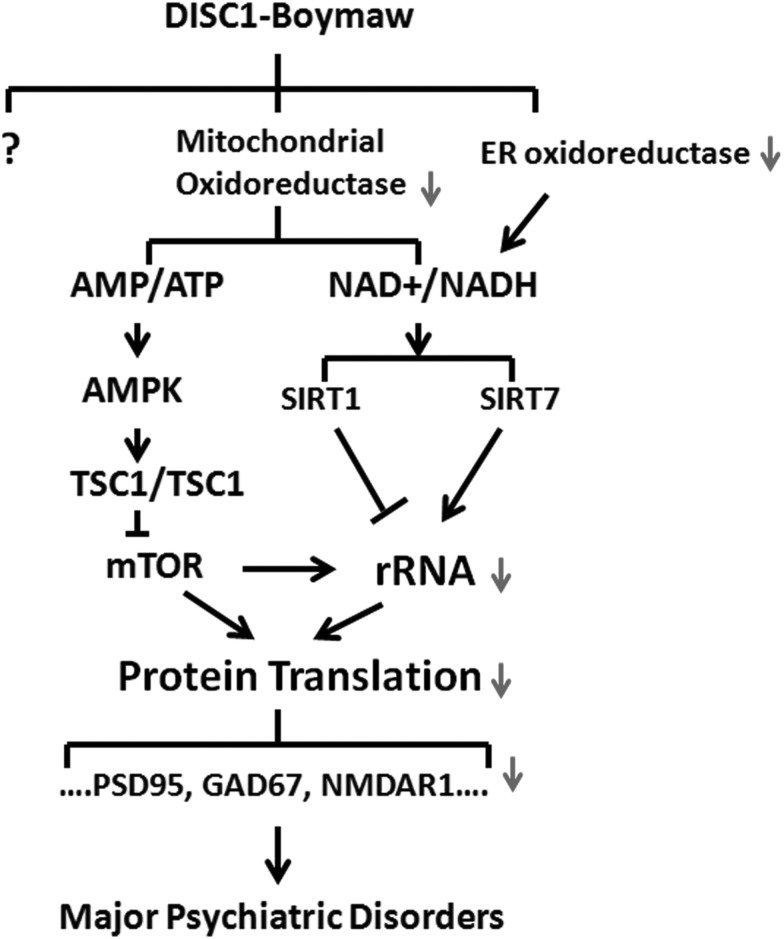
Eykelenboom et al (53) identified three different fusion transcripts from the patient lymphoblastoid cells from the Scottish family. The DB7 fusion gene used in our studies encodes exactly the same fusion protein as their CP60 transcript. The second CP69 fusion transcript is generated by alternative splicing 12 nucleotides upstream of the stop codon of the CP60 transcript. Our deletion analysis demonstrated that the last few amino acid residues of Boymaw are not required for inhibition of MTT reduction. Therefore, it is likely that the CP69 fusion protein is also capable of inhibiting MTT reduction. The third CP1 fusion transcript encodes a protein with just one more amino acid addition to the C-terminus of the truncated DISC1 protein that has been used in the generation of several mouse DISC1 models (54,55). The CP1 protein does not display any detectable toxicity to mitochondria, which is consistent with our observation that over-expression of truncated DISC1 proteins has no effect on MTT reduction, rRNA expression and protein translation. Our immunocytochemical analysis confirmed localization of the DB7 fusion protein (but not DISC1-FL, truncated DISC1 or BD13) in mitochondria (Supplementary Material, Fig. S9). Interestingly, MTT reduction was also reduced in mitochondria of cells expressing the DB7 fusion gene (Fig. 3C). Decreased activities of mitochondrial NADH oxidoreductases may relate to loss of mitochondrial membrane potential in cells expressing the DB7 fusion protein (53). We therefore hypothesize that reduction of oxidoreductase activity may decrease cell metabolism to down-regulate rRNA expression and protein translation. One potential pathway could be that inhibition of the mitochondrial oxidoreductase activities reduces adenosine triphosphate (ATP) production (Fig. 11), which is supported by the finding of loss of mitochondrial membrane potential in cells expressing the DB7 fusion protein (53). Reduction of ATP can trigger activation of the AMP-activated protein kinase (AMPK), a sensor of cell energy status (56). The AMPK pathway is a well-established pathway to regulate protein translation. In this pathway, the activated AMPK phosphorylates TSC1/TSC2 to inhibit mammalian target of rapamycin (mTOR). mTOR kinase plays a central role to stimulate protein translation either directly via activation of translational initiation factors (57) or indirectly via phosphorylating transcription initiation factor TIF-1A to promote rRNA transcription (58). Mutation of TSC1/TSC2 leads to autism (59), which shares many common psychopathological features with major psychiatric disorders. Another potential pathway linking cellular metabolism to rRNA transcription could be initiated through the Sirtuin family of proteins, which are NAD+-dependent deacetylase to sense cellular NAD+ changes. SIRT1 (Sirtuin 1) has been demonstrated to epigenetically silence rDNA chromatin (60, 61). However, it is also reported that nucleolar SIRT7 (Sirtuin 7) functions as an activator for rRNA transcription (62). It is clear that the potential underlying mechanisms are complex and much more work is needed.
Figure 11.
Hypothetical pathways of the DB7 fusion gene. Cell metabolism is tightly coupled with protein synthesis. Reduction of NADH oxidoreductase activity may alter production of ATP and NAD+ which can trigger at least two known pathways (AMPK and Sirtuin) to down-regulate rRNA transcription and protein translation. AMPK, AMP-activated protein kinase; TSC1 and TSC2, tuberous sclerosis complex 1 and 2; mTOR, mammalian target of rapamycin.
One of the key questions is how expression of the DB7 fusion protein reduces oxidoreductase activity. Identification of these NADH oxidoreductases in mitochondrion and cytoplasm is critical to unravel the signaling pathways from cellular metabolic activity to protein translation. In contrast to inhibition of protein translation by the DB7 fusion gene, exaggerated protein translation has been suggested as the core pathophysiology of Fragile X Syndrome (63) and other autism genes (24). It is possible that an optimal protein translation is necessary for neural plasticity (64), and excessive or less protein translation are harmful to brain functions. Translational control of protein synthesis may play important roles in many other psychiatric disorders. Indeed, it was reported that hypermethylation of ribosomal RNA gene promoter was associated with suicide (65), mild cognitive impairment and Alzheimer's disease (66), as well as Huntington's diseases (67). Considering that rRNA synthesis is a rate-limiting step of ribosome biogenesis (68), reduction of rRNA will likely result in a decrease of protein translation. Our SUnSET experiments clearly demonstrated inhibition of protein translation in both the cells transiently expressing the DB7 fusion gene and the heterozygous DISC1-Boymaw mice. Since we did not find significant reduction of mRNA transcripts of Gad67, Nmdar1 and Psd95 genes, the reduction of general protein translation therefore could be responsible for the decrease of Gad67, Nmdar1 and Psd95 protein expression in the heterozygous DISC1-Boymaw mice. Indeed, it was reported that cellular abundance of proteins is predominantly controlled at the level of protein translation (69,70). However, it is also possible that protein turnover may be altered and contributes to the reduction of these proteins in the humanized mice. The effects of the expression of the DB7 gene may not be restricted to the inhibition of protein translation. For example, we observed reduced expression of Cyb5r3 reductase in endoplasmic reticulum (ER), where the enzyme plays an important role in lipid biosynthesis and has been demonstrated to be essential for brain development (71–73).
Impaired NMDAR-mediated neurotransmission has been well established in the pathophysiology of schizophrenia. Prolonged ketamine effects and exacerbation of symptoms are observed in schizophrenia patients (39–41) and our mouse genetic model (74). Consistent with the findings in human patients, we found reduction of both Nmdar1 and Gad67 proteins in heterozygous DISC1-Boymaw mice which display prolonged responses to the non-competitive NMDAR antagonist, ketamine. This phenotype has been replicated in different cohorts of mice on various genetic backgrounds regardless of the presence or absence of wild-type Disc1 gene. It is unknown why the female humanized mice do not show prolonged responses to ketamine. The female humanized mice are not symptom-free, since they display abnormal phenotypes in other behavioral paradigms. We found increased PPI in the heterozygous DISC1-Boymaw mice at a short ISI (25 ms) in both sexes. In mice, PPI diminishes with decreasing ISI starting from 100 ms, and eventually transforms into prepulse facilitation. Abnormally enhanced PPI at short ISI is indicative of altered information processing in the heterozygous DISC1-Boymaw mice. Although this alteration in PPI differs from that seen in patients with schizophrenia (75), enhanced PPI at short ISI has been reported in rats treated with dopamine receptor agonists (76, 77).
Inhibition of protein translation by the fusion gene impacts not only the brain but also erythropoiesis of the heterozygous DISC1-Boymaw mice. We found an incomplete penetrance of the DISC1-Boymaw fusion gene to anemia, consistent with the role of impaired protein translation demonstrated by ribosomal protein gene mutations in Diamond–Blackfan anemia (48). Interestingly, the heterozygous DISC1-Boymaw mice also display increased RDW. In human, over 2 million red blood cells are generated every second with small variation in cell volume (human RDW: 11.5–14.5%). It is possible that such an extremely active and precise biological process in mouse is highly susceptible to the DISC1-Boymaw mutation which confers even mild inhibition of protein translation. Recently, an association between increased RDW and depression was reported from a multi-year large scale study in humans (78). The study found a gradient positive correlation between increased RDW and the risk of developing depression in thousands of patients. It will be interesting to know whether patients with major depression in the Scottish family may display increased RDW.
It should be kept in mind that our studies are focused on the investigation of the biological functions of the DISC1-Boymaw fusion genes. To that end, the 129S genetic background, which contains a frame-shift mutation in endogenous disc1 gene, provides a unique advantage for our studies to avoid Disc1 gene dosage difference between wild-type and the heterozygous DISC1-Boymaw mice. However, the chromosome translocation in the Scottish family generates not only the fusion genes but also loss of one allele of the DISC1 gene. Therefore, our humanized DISC1-Boymaw mice on 129S background should be viewed as a genetic model for the DISC1-Boymaw fusion genes rather than the chromosome translocation in the Scottish family. In the future, we could generate mice carrying full-length human DISC1 genes on mouse Disc1 locus using the Cre recombinase. After breeding these DISC1 mice with the heterozygous DISC1-Boymaw mice, we could generate wild-type mice carrying two human DISC1 (DISC1/DISC1) genes and heterozygous mice carrying one human DISC1 and one DISC1-Boymaw fusion gene (DISC1/DISC1H). Comparisons between wild-type (DISC1/DISC1) and the heterozygous (DISC1/DISC1H) will provide a more complete model for the chromosome translocation in the Scottish family.
MATERIALS AND METHODS
DNA constructs
Full length DISC1, truncated DISC1, DB7 and BD13 fusion genes were generated through PCR amplification and ligation. All expression constructs were tagged with HA epitopes as described (11). The adeno-associated virus (AAV) shuttle and pTimer plasmid vectors were used to generate expression constructs, bi-cistronic constructs connected with the IRES of encephalomyocarditis virus (EMCV) virus (79), and FT fusion protein constructs, respectively. Boymaw was also fused with FT in pTimer plasmid vector. All constructs were confirmed with complete sequencing. A series of FT-Boymaw C-terminal deletion were generated by using PCR amplification and subsequent In-Fusion ligation (Clontech). The following pairs of PCR primers were used: C1-del: Boymaw-C1-del-f: 5′CCTTGAGTAC CCATACGACG TTCCAGACT3′; Boymaw-C1-del-r: 5′TATGGGTACT CAAGGCTTCT ATCTGTGGT3′; C2-del: Boymaw-C2-del-f: 5′TCTACCATAC CCATACGACG TTCCAGACT3′; Boymaw-C2-del-r: 5′TATGGGTATG GTAGAAGCCT GCAGACAAAG3′; Boymaw-C3-del-f: 5′GAAGTCATAC CCATACGACG TTCCAGACT3′; Boymaw-C3-del-r: 5′TATGGGTATG ACTTCAGGTA ACTGAAACC3′; Boymaw-C4-del-f: 5′GAGACATTAC CCATACGACG TTCCAGACT3′; Boymaw-C4-del-r: 5′TATGGGTAAT GTCTCTTATT CCAGTCTGT3′.
In vitro cell culture
HEK293T, COS-7 and SH-SY5Y cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or DMEM/F12 medium supplemented with 10% fetal bovine serum, penicillin and streptomycin (Life Technologies, CA) at 37°C in a humidified atmosphere containing 5% CO2. HEK293T, COS-7 cell transfections were performed using Mirus TransIT® reagent (Mirus, WI), according to manufacturer's instructions. SH-SY5Y cells were transduced with recombinant AAV virus. Primary culture of mouse neuronal cells was established from both wild-type and heterozygous DISC1-Boymaw embryos at E18.5 and P1 as described in our previous studies (80). The mouse cerebrum were dissected and cut into small pieces in Neurobasal A medium on ice. The tissue was triturated 10 times with a fire polished 9-in. Pasteur pipette and allowed to settle on ice for 1 min. The supernatant was then transferred to a new tube and centrifuged gently at 600–700 rpm for 5 min to pellet the cells. The cells were resuspended in B27/Neurobasal medium (B27/Neurobasal with 0.5 mM glutamine, no glutamate, 5 ng/ml FGF2).
MTT reduction assays
HEK293T cells and COS-7 cells were seeded at cell density of 1 × 105 cells per well in 24-well plates, 24 h prior to transfection. Cell proliferation was measured by counting cell numbers with 0.04% Trypan blue 48 h after transfection. MTT reduction assay was modified from Mosmann T (81). MTT (Sigma, MO) was dissolved in 1× PBS at the concentration of 5 mg/ml, and filtered through 0.2 µm membrane. The stock solution was stored at −20°C. For MTT reduction in living cells, MTT stock solution was added at the final concentration of 0.5 mg/ml, and the cells were incubated at 37°C for 1.25 h. For MTT assay in cell lysate, transfected cells were first lysed in lysis buffer (18) (100 mM NaCl, 10 mM Tris, pH 7.5, 1 mM EDTA, 0.01%Triton X-100, protease inhibitors cocktail) at 4°C for 30 min with gentle shaking. MTT was added to the lysate at the final concentration of 0.5 mg/ml, and the reaction was incubated at 37°C for 1 h. For MTT assay in purified ER and mitochondria, NADH (Sigma, MO) was added in the assay at the final concentration of 0.5 mM, and the reactions were incubated at 37°C for 10 min for mitochondria, and 30 min for ER, respectively. For the reversal of deficient MTT reduction, 2.5 Unit of E. coli ADH (Cat# 49641, Sigma, MO) was added into the DISC1-FL and DB7 cell lysate, respectively, and further incubated at 37°C for 2 h. MTT crystal were collected and dissolved in DMSO. Absorbance at 540 nm was measured with SpectraMax M5 Multi-Mode Microplate Readers (Molecular Devices, PA). Neonatal mouse brains were dissected on postnatal Day 1. After washing three times with pre-chilled PBS, brains were cut into small pieces in 1.5 ml isotonic extraction buffer (IEB) (10 mM HEPES, pH 7.8 with 0.25 M sucrose, 1 mM EGTA, and 25 mM KCl) containing protease inhibitor cocktail (Sigma, MO), and further homogenized with Dounce homogenizer on ice. Equal amounts of protein homogenate were used for MTT reduction assays.
Western blot analysis
Supernatant soluble proteins were extracted from HEK293T cells and COS-7 cells in Passive Lysis Buffer (PLB) (Promega, WI) containing 0.2% Sarkosyl and 1× protease inhibitor cocktail (Sigma, MO) at room temperature for 15 min on a shaker. Total proteins from cell pellets, endoplasmic reticulum, mitochondria and brain homogenate of neonatal mice were solubilized with sonication in PLB containing 1× protease inhibitor cocktail. Protein concentration was measured with Bradford (Abs. 595 nm) method using Coomassie Plus Protein Assay (Thermo Scientific, IL). After electrophoresis, proteins were transferred onto PVDF membranes which were subsequently blocked with 5% non-fat dry milk in TBST buffer (pH 7.5, 10 mM Tris–HCl, 150 mM NaCl, and 0.1% Tween 20) at room temperature for 1 h. The membranes were incubated with the following primary antibodies at 4°C overnight: mouse monoclonal anti-cytochrome C (1:5000), ab110325 (Abcam, MA); rabbit polyclonal anti-Cyb5r3 (1:600), sc-67284; mouse monoclonal anti-Cypor (1:400); sc-25270; mouse monoclonal anti-β-actin (1:5000); sc-47778 (Santa Cruz Biotechnology, CA). After washing three times, the membranes were further incubated with horseradish peroxidase-conjugated anti-mouse IgG (1:5000, Cell signaling, MA), anti-rabbit IgG (1:5000, Santa cruz, CA) for 1.5 h at room temperature. Rat monoclonal anti-HA antibody (1:500) conjugated with peroxidase (3F10) (Roche, CA) was used to directly detect HA epitope. Quantification of protein expression was conducted with Image J.
Proteomic analysis
HEK293T cells were transfected with DISC1-FL and DB7 genes, respectively. Two days later, cells were harvested and sent to UCSD Mass Spectrometry Facility Core for iTRAQ analysis. The samples were labeled with 114, 115, 116 and 117 Da mass tags, respectively. Protein identification and quantitation were performed using the Paragon™ Algorithm in ProteinPilot™ Software, searching against the NCBInr database.
NADH concentration
Intracellular NADH extraction was conducted as described in Cyclex NAD+/NADH colorimetric Assay Kit (CycLex, Japan). In brief, 2 × 105 cells were seeded in each well of a12-well plate one day before transfection. Cells were collected 48 h post-transfection. NADH extraction buffer [50 mM NaOH and 1 mM ethylenediaminetetraacetic acid (EDTA)] was added to cell pellets and gently vortexed 2–3 times. To reduce viscosity, samples were incubated at 60°C for 30 min. Equal volume of neutralization buffer (0.3 M potassium phosphate buffer, pH 7.4) was added and incubated at least 5 min on ice. NADH was extracted from the supernatant solution after spinning down the neutralized samples at 15 000 rpm for 5 min at 4°C. The cell pellets were solubilized to measure protein concentration for the normalization of NADH concentration. E. coli ADH (Sigma) was used to measure NADH concentration in MTT reduction assays.
Density-gradient ultracentrifugation
Mitochondria and ER were purified from HEK293T cells and neonatal mouse brains via density-gradient ultracentrifugation as described in Endoplasmic Reticulum Isolation Kit (Sigma, MO). In brief, HEK293 T cells (∼5 × 107) were incubated in hypotonic buffer (10 mM HEPES pH7.8, 1 mM EGTA and 25 mM KCl) for 20 min at 4°C. The cells were pelleted with 600 × g for 5 min, and resuspended with appropriate volume of the IEB (10 mM HEPES, pH 7.8, with 0.25 M sucrose, 1 mM EGTA and 25 mM KCl). For mouse brains, dissected brains were cut into small pieces in the IEB. The cells and the tissue were further homogenized with Dounce homogenizer on ice with 2 ml of IEB. Large cellular debris and nuclei were pelleted after centrifuging at 1000 × g for 10 min at 4°C. The supernatant was transferred into a new centrifuge tube and further centrifuged at 12 000 × g for 15 min at 4°C. Crude mitochondria were pelleted, and the supernatant fraction containing ER was transferred to a new ultracentrifuge tube and centrifuged at 150 000 × g in a Beckman Optima™ LE-80K (Beckman Coulter, CA) at 4°C for 1 h to purify ER. ER pellets were resuspended in IEB containing protease inhibitors. Crude mitochondrion pellet was further purified as described (82). In brief, mitochondrion pellet was resuspended in 1 ml of mitochondrion isolation buffer (MIB) (70 mM sucrose, 210 mM D-mannitol, 1 mM EDTA-K2, 0.23 mM PMSF, 5 mM HEPES–Tris, pH 7.4, and protease inhibitors cocktail) and layered onto 1.5 ml of 7.5% (w/v) Ficoll-sucrose medium (7.5% Ficoll-MIB) on top of 1 ml of 10% (w/v) Ficoll-sucrose medium (10% Ficoll-MIB) and centrifuged in a Beckman Optima™ LE-80K (Beckman Coulter, CA) for 1.5 h at 100 000 × g at 4°C. Mitochondria were pelleted at the bottom of the ultracentrifuge tube. The pellets were suspended in 1 ml of MIB and then centrifuged for 5 min at 15 000 × g at 4°C. The mitochondrion pellets were washed once in MIB and finally resuspended in appropriate volume of MIB. The purified mitochondria were either used for MTT reduction or stored at −80°C for subsequent analyses.
SUnSET
HEK293 cells seeded at cell density of 1 × 105 cells per well in 24-well plates, 24 h prior to transfection. After transfection, cells were pulse-labeled with puromycin at the concentration of 10 µg/ml for 10 min in the presence or absence of cycloheximide at final concentration of 25 µM. Total proteins were extracted and load on polyacrylamide electrophoresis gel electrophoresis (PAGE) gel (10 µg per lane) for western blot analysis. Anti-puromycin antibody (1:10 000) (12D10, Millipore) was used to detect incorporation of puromycin in newly synthesized proteins. Blots were stripped later and re-probed with anti-β-actin antibody. Image J was used to quantify the incorporation of puromycin.
Generation of humanized DISC1-Boymaw mice
Both DB7 and BD13 fusion genes were connected with IRES of EMCV virus (11, 79). The bi-cistronic gene was terminated with SV40 poly (A). A splicing acceptor was added to generate in-frame splicing of the bi-cistron with the first exon of human DISC1 gene. The bi-cistron gene cassette was flanked with two loxP sites. Downstream of the bi-cistron, human DISC1 cDNA was capped with a splicing acceptor. Such a design aimed for conditional restoration of DISC1 gene expression in genetic rescue experiments in the future. Two homology arms with respective sizes of 4 and 3 kb were amplified from the mouse disc1 gene of 129S ES cells (129S2/SvPasCrl) to assemble the final gene targeting construct. The final gene targeting construct was electroporated into mouse embryonic stem (ES) cells. After positive and negative selections with G418 and ganciclovir, ∼1500 ES cell colonies were picked for further screening. Two homologous recombinant ES cell colonies were identified. After blastocyst injection, chimeric mice were obtained and bred with 129S females for germline transmission of the targeted DISC1 allele.
Mouse breeding
Heterozygous DISC1-Boymaw mice were bred with wild-type mice to generate wild-type and heterozygous mice on 129S genetic background (129S2/SvPasCrl, Charles River) for all molecular studies. Different cohorts of mice were generated on the F1 hybrid (129S/C57) and 129S genetic backgrounds for behavioral studies. PCR was used for DISC1-Boymaw mouse genotyping (DISC1-Common-R: 5′TAACAACAGCCAGTGTGCAAG 3′; DISC1-Neo-F: 5′GGTGGGCTCTATGGCTTCTGA3′; DISC1-F: 5′TGCATTCACATGTGTTGCCTT TG 3′). Mice were housed in groups (two to four mice per cage) in a climate-controlled animal colony with a reversed day/night cycle. Behavioral testing occurred during the dark cycle. Food (Harlan Teklab, Madison, WI) and water were available ad libitum, except during behavioral testing. All behavioral testing procedures were approved by the UCSD Institutional Animal Care and Use Committee (permit number: A3033–01) prior to the onset of the experiments. Mice were maintained in American Association for Accreditation of Laboratory Animal Care (AALAC) approved animal facilities. For behavioral studies, all mice were 2- to 6-month old siblings, and the age range of the birth date between all mice was less than a week.
Hematological analysis
A cohort of wild-type and heterozygous DISC1-Boymaw mice was generated on 129S background. Male mice were anesthetized with isofluorane before blood collection. Submandibular venous lancets (Goldenrod 5 mm) were used to collect ∼100 µL into EDTA anticoagulated 0.5 ml Microtainer tubes (Becton-Dickinson). Hematological analysis was conducted using the Hemavet 950 FS at the UC San Diego Hematology Core Laboratory. Six adult wild-type male mice and six heterozygous male DISC1-Boymaw siblings were first used for hematological analysis. To replicate the findings, another independent group of wild-type mice and their heterozygous DISC1-Boymaw siblings (N = 4, for each genotype) was used.
RNA, DNA extraction and RT-PCR analysis
DNA and RNA were extracted from cells and mouse brains using Trizol method. RNA expression of the DB7 fusion gene was examined with RT-PCR using the primers: human DISC1 e1-f: 5′GCGGCGTGAGCCACCGCGCAG3′ human DISC1 e2-r: 5′GAACCGGAACAGTGTGCCCAC3′. Mouse endogenous disc1 RNA expression was examined for alternative splicing of exon 6 using primers:
mouse Disc1 exon2f: 5′CCATGCTGCAGGACTACCTA3′
mouse Disc1 exon3f: 5′GAAAAAGTGGTCGAGGATGG3′
mouse Disc1 exon4f: 5′GCAGCTTCTTGGGTTACCTG3′
mouse Disc1 exon5f: 5′ACCAGGAGGGACTGGCTTAT3′
mouse Disc1 exon6f: 5′GCTGTGACCTGATGGCACT3′
mouse Disc1 exon10r: 5′ACCTTCCAACACTTCCATGC3′
For the real-time RT-PCR quantification of mRNA transcripts of Gad67, Nmdar1 and Psd95 genes, cDNA was first synthesized with random hexamers using the Superscript III First-Strand Synthesis Kit (Invitrogen). A comparative Ct method was used for quantification with SYBR Green, according to the manufacturer's protocol (Bio-Rad CFX384). Beta-actin was used as a reference control for normalization. The following primers were used in Q-PCR:
Mouse Gad67 f: 5′TCACCCTCGATTTTTCAACC3′
Mouse Gad67 r: 5′TTGACCATCCAACGATCTCTC3′
Mouse Nmdar1 f: 5′GGCAGTAAACCAGGCCAATA3′
Mouse Nmdar1 r: 5′GTGGGAGTGAAGTGGTCGTT3′
Mouse Psd95 f: 5′AAGTCAGAGCCCCCTACTCG3′
Mouse Psd95 r: 5′AGGGTGTCCGTGTTGACAAT3′
Mouse β-Actin f: 5′TTCTACAATGAGCTGCGTGTG3′
Mouse β-Actin r: 5′GGGGTGTTGAAGGTCTCAAA3′
Immunohistochemical and immunocytochemical staining and western blot
Immunohistochemical staining of parvalbumin, Gad67, Nmdar1 were conducted in mouse brain paraffin sections as described in our previous publication (83). For immunocytochemical staining, primary neurons were fixed in 3.7% formalin/PBS at room temperature for 10 min. After 1 h of blocking with 5% goat serum containing 0.2% Triton X-100 at room temperature, the slides were incubated with mouse monoclonal anti-DISC1 (3D4) (1:1000 dilution, gift from Dr Carsten Korth) and rabbit polyclonal anti-MAP2 (1:2,000 dilution, PRB-547C, Covance) antibodies at 4°C overnight. After rinsing three times with PBST, the slides were incubated with Alexa Fluor® 568 goat anti-mouse IgG (1:500 dilution, Invitrogen) and Alexa Fluor® 488 goat anti-rabbit IgG (1:500 dilution, Invitrogen) for 1 h at room temperature. The slides were washed three times with PBST each for 5 min, and mounted with VECTASHIELD® Hard Set™ Mounting Medium with DAPI (Vector, CA). The DB7 fusion proteins were detected in western blot analysis of primary neuron culture using rabbit polyclonal anti-DISC1 antibody (1:500 dilution, ab55808, Abcam) recognizing N-terminal residues of human DISC1proteins. Expression of Gad67, Nmdar1, Psd95, synaptophysin and β-actin in mouse brain and primary culture of neuronal cells was examined using the antibodies: mouse monoclonal anti-Gad67 (1:10 000 dilution, G5419, sigma); mouse monoclonal anti-Nmdar1 (1:5000 dilution, 556308, BD Biosciences); mouse monoclonal anti-Psd95 (1:3000 dilution, Ab99009, Abcam); rabbit polyclonal anti-synaptophysin (1:3000 dilution, 5461S, Cell signaling); mouse monoclonal anti-β-actin (1:5000 dilution, sc-47778, Santa Cruz Biotechnology).
SUnSET in mice
The in vivo SUnSET experiments were conducted as described (24). Sex-, and age-matched 2-month old sibling wild-type and heterozygous mice were anesthetized. Unilateral ICV injection of puromycin (25 µg/2.5 µl) was performed with coordinates: −0.22 mm anterioposterior, −1.0 mm mediolateral, and −2.4 mm dorsoventral. Mice were sacrificed by cervical dislocation 1 h after the injection. The hippocampus was dissected for total protein extraction. Equal amounts of protein (70 μg) were loaded on PAGE gel for western blot analysis. Anti-puromycin antibody (1:1000)(12D10, Millipore) was used to detect incorporation of puromycin in newly synthesized proteins. Blots were then stripped and re-probed with ant-β-actin antibody. Image J was used to quantify the incorporation of puromycin.
RNA in situ hybridization
We developed a modified protocol for chromogenic RNA in situ hybridization according to branched DNA amplification assays (84, 85). Oligonucleotide adaptors were added to the 5′ end of three 18S rRNA probes (5′TCACTGTACC GGCCGTGCGT3′; 5′CCTAGCTGCGGTATCCAGGC3′; 5′GCGATCGGCCCGAGGTTATC3′). In brief, paraffin sections were dewaxed, rehydrated and partially digested with pepsin (4 mg/ml) at 37°C for 10 min. After PBS washing, sections were pre-hybridized with Quick-hyb Solution (Stratagene, 201220) with 0.1 M DTT, and 50 µg/ml denatured ssDNA. Pre-hybridization was conducted at 40°C for 15 min in pre-heated wet plastic chamber. After pre-hybridization, probes were added to the final concentration of 200 ng/ml. Hybridization was conducted at 40°C overnight in pre-heated wet pyrex chamber. After hybridization, sections were washed, and hybridized with oligonucleotide amplifier labeled with biotin. Horseradish peroxidase-conjugated avidin (Thermo Scientific, 21123) was used to bind biotin at final concentration of 5.5 µg/ml. Chromogenic reaction was performed using DAB Peroxidase Substrate Kit (Catalog# SK-4100, Vector Labs).
Acoustic startle
Startle reactivity and PPI were measured with startle chambers (SR-LAB, San Diego Instruments, San Diego, CA) using 65 dB background noise, three types of prepulse-pulse trials (69, 73 and 81 dB) and pulse-alone (120 dB) (86). The session started with five pulse-alone trials at 120 dB intensity to stabilize startle. The prepulse testing block contained three prepulse trial types in which the 69 dB prepulse onset preceded the 120 dB pulse by 25, 50, 100, 200 and 500 ms. Prepulse trials and pulse alone trials were presented in a pseudorandom order. The session ended with five pulse-alone trials. There was an average of 15 s (range: 12–30 s) between trials. Mice were assigned in a pseudo-random order, and placed in the startle chambers, where a 65-dB background noise level was presented for a 10-min acclimation period and continued throughout the test session.
Tail suspension
The tail suspension test was conducted as described previously with some modifications (45, 46). In brief, mice were suspended by the tail with adhesive tape placed 1–2 cm from the end of the tail. The mice were visually isolated at the center of a white box. Mice were suspended for 6 min, and the duration of immobility—during which the mice did not produce any apparent voluntary movements—was recorded by an observer blind to genotype.
Saccharin preference
The saccharin preference test was conducted as described with some modifications (87). In brief, mice were separated and housed, two or three per cage, by genotype. Saccharin sodium salt hydrate (Sigma) was dissolved in tap water to 1.0%. Each cage was supplied with one bottle of saccharin solution and one bottle of normal drinking water. The bottles were weighed before placement in each cage and 24, 48, 72 and 96 h thereafter. Preference for the saccharin solution was calculated as the percentage of saccharin solution consumed out of the total liquid consumption each day. Food was available ad libitum during the course of the experiment.
Behavioral pattern monitor
Spontaneous behavioral data were recorded using the mouse BPM (San Diego Instruments, San Diego, CA), as described previously (44, 88). In brief, a single chamber consists of a 30.5 × 61 × 38-cm area, with a Plexiglas hole board floor that was equipped with floor holes in the front, middle, and rear parts of the floor and eight wall holes (three along each side of the long walls and two holes in the front and back walls). Mice were tested during the dark phase of their light cycle. During testing, a white noise generator produced background noise at 65 dB. The measurement of transitions, center time and spatial coefficient of variation were based on the nine divided regions of the chambers. The status of the photobeams was sampled every 0.1 s. The session lasted 60 min. Raw data were transformed into the location of the animal (in X–Y coordinates), whether holepoking or rearing occurred (events), and the duration of each event (time). The chambers were cleaned thoroughly between testing sessions.
Ketamine treatment
Ketamine was dissolved in saline, and administered i.p. at a volume of 5 ml/kg with a dosage of 60 mg/kg immediately prior to the start of the BPM session. A within-subjects design was used for the ketamine studies. After administration of ketamine, mouse locomotor activities were recorded for 60 min in BPM chambers.
Statistical analysis
All data were first tested for normal distribution using the Kolmogorov–Smirnov test before calculation of differences. For statistical analyses, repeated-measures analysis of variance (ANOVA) with genotypes as a between-subjects factor and drug treatment, block and prepulse intensity as within-subjects factors were performed on the %PPI data and locomotor activity (e.g., total distance). Post hoc analyses were carried out using Newman–Keuls or Tukey's test. Alpha level was set to 0.05. All statistical analyses were carried out using the BMDP statistical software (Statistical Solutions Inc., Saugus, MA).
AUTHORS’ CONTRIBUTIONS
X.Z. conceived, performed, analyzed and wrote the studies. B.J., K.H., M.K., L.Z. and J.Y. performed and analyzed the research. M.G. provided critical discussion and comments on the studies.
SUPPLEMENTARY MATERIAL
FUNDING
These studies were supported by grants from the National Institute of Mental Health MH073991 (X.Z.), MH086075 (X.Z.), NARSAD Young Investigator Award (X.Z.), and the Veterans Affairs VISN 22 Mental Illness Research, Education, and Clinical Center (M.G.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Carsten Korth and Nick Brandon for generously providing anti-DISC1 antibodies.
Conflict of Interest statement. None declared.
REFERENCES
- 1.St Clair D., Blackwood D., Muir W., Carothers A., Walker M., Spowart G., Gosden C., Evans H.J. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood D.H., Fordyce A., Walker M.T., St Clair D.M., Porteous D.J., Muir W.J. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millar J.K., Wilson-Annan J.C., Anderson S., Christie S., Taylor M.S., Semple C.A., Devon R.S., Clair D.M., Muir W.J., Blackwood D.H., et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 4.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 5.Duan X., Chang J.H., Ge S., Faulkner R.L., Kim J.Y., Kitabatake Y., Liu X.B., Yang C.H., Jordan J.D., Ma D.K., et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kvajo M., McKellar H., Arguello P.A., Drew L.J., Moore H., MacDermott A.B., Karayiorgou M., Gogos J.A. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao Y., Ge X., Frank C.L., Madison J.M., Koehler A.N., Doud M.K., Tassa C., Berry E.M., Soda T., Singh K.K., et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q., Charych E.I., Pulito V.L., Lee J.B., Graziane N.M., Crozier R.A., Revilla-Sanchez R., Kelly M.P., Dunlop A.J., Murdoch H., et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol. Psychiatry. 2011;16:1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandon N.J., Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X., Geyer M.A., Kelsoe J.R. Does disrupted-in-schizophrenia (DISC1) generate fusion transcripts? Mol. Psychiatry. 2008;13:361–363. doi: 10.1038/sj.mp.4002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X., Chen Q., Schaukowitch K., Kelsoe J.R., Geyer M.A. Insoluble DISC1-Boymaw fusion proteins generated by DISC1 translocation. Mol. Psychiatry. 2010;15:669–672. doi: 10.1038/mp.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leliveld S.R., Bader V., Hendriks P., Prikulis I., Sajnani G., Requena J.R., Korth C. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 2008;28:3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L., Liu Y., Ky B., Shughrue P.J., Austin C.P., Morris J.A. Cloning and characterization of Disc1, the mouse ortholog of DISC1 (disrupted-in-Schizophrenia 1) Genomics. 2002;80:662–672. doi: 10.1006/geno.2002.7012. [DOI] [PubMed] [Google Scholar]
- 14.Lipska B.K., Peters T., Hyde T.M., Halim N., Horowitz C., Mitkus S., Weickert C.S., Matsumoto M., Sawa A., Straub R.E., et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 15.Shaw G., Morse S., Ararat M., Graham F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 16.Ascano M., Jr., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M., et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Peterson D.A., Kimura H., Schubert D. Mechanism of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J. Neurochem. 1997;69:581–593. doi: 10.1046/j.1471-4159.1997.69020581.x. [DOI] [PubMed] [Google Scholar]
- 18.Trevisi L., Pighin I., Bazzan S., Luciani S. Inhibition of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) endocytosis by ouabain in human endothelial cells. FEBS Lett. 2006;580:2769–2773. doi: 10.1016/j.febslet.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 19.Berridge M.V., Tan A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993;303:474–482. doi: 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 20.Stockert J.C., Blazquez-Castro A., Canete M., Horobin R.W., Villanueva A. MTT assay for cell viability: Intracellular localization of the formazan product is in lipid droplets. Acta Histochem. 2012;114:785–796. doi: 10.1016/j.acthis.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Terskikh A., Fradkov A., Ermakova G., Zaraisky A., Tan P., Kajava A.V., Zhao X., Lukyanov S., Matz M., Kim S., et al. ‘Fluorescent timer’: protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 22.Hung Y.P., Albeck J.G., Tantama M., Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 24.Santini E., Huynh T.N., MacAskill A.F., Carter A.G., Pierre P., Ruggero D., Kaphzan H., Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krystal J.H., Karper L.P., Seibyl J.P., Freeman G.K., Delaney R., Bremner J.D., Heninger G.R., Bowers M.B.z, Jr., Charney D.S. Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch. Gen. Psychiatry. 1994;51:199–214. doi: 10.1001/archpsyc.1994.03950030035004. [DOI] [PubMed] [Google Scholar]
- 26.Dalmau J., Gleichman A.J., Hughes E.G., Rossi J.E., Peng X., Lai M., Dessain S.K., Rosenfeld M.R., Balice-Gordon R., Lynch D.R. Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. Lancet Neurol. 2008;7:1091–1098. doi: 10.1016/S1474-4422(08)70224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iizuka T., Sakai F., Ide T., Monzen T., Yoshii S., Iigaya M., Suzuki K., Lynch D.R., Suzuki N., Hata T., et al. Anti-NMDA receptor encephalitis in Japan: long-term outcome without tumor removal. Neurology. 2008;70:504–511. doi: 10.1212/01.wnl.0000278388.90370.c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X.M., Sakai K., Roberts R.C., Conley R.R., Dean B., Tamminga C.A. Ionotropic glutamate receptors and expression of N-methyl-D-aspartate receptor subunits in subregions of human hippocampus: effects of schizophrenia. Am. J. Psychiatry. 2000;157:1141–1149. doi: 10.1176/appi.ajp.157.7.1141. [DOI] [PubMed] [Google Scholar]
- 29.Law A.J., Deakin J.F. Asymmetrical reductions of hippocampal NMDAR1 glutamate receptor mRNA in the psychoses. Neuroreport. 2001;12:2971–2974. doi: 10.1097/00001756-200109170-00043. [DOI] [PubMed] [Google Scholar]
- 30.Volk D.W., Austin M.C., Pierri J.N., Sampson A.R., Lewis D.A. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch. Gen. Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto T., Volk D.W., Eggan S.M., Mirnics K., Pierri J.N., Sun Z., Sampson A.R., Lewis D.A. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J. Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curley A.A., Arion D., Volk D.W., Asafu-Adjei J.K., Sampson A.R., Fish K.N., Lewis D.A. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am. J. Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai N.P., Wilkerson J.R., Guo W., Maksimova M.A., DeMartino G.N., Cowan C.W., Huber K.M. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Z.J. Activity-dependent development of inhibitory synapses and innervation pattern: role of GABA signalling and beyond. J. Physiol. 2009;587:1881–1888. doi: 10.1113/jphysiol.2008.168211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuroda K., Yamada S., Tanaka M., Iizuka M., Yano H., Mori D., Tsuboi D., Nishioka T., Namba T., Iizuka Y., et al. Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum. Mol. Genet. 2011;20:4666–4683. doi: 10.1093/hmg/ddr400. [DOI] [PubMed] [Google Scholar]
- 36.Banbury. Mutant mice and neuroscience: recommendations concerning genetic background. Banbury Conference on genetic background in mice. Neuron. 1997;19:755–759. doi: 10.1016/s0896-6273(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 37.Braff D.L., Geyer M.A., Swerdlow N.R. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- 38.Geyer M.A., Braff D.L. Habituation of the Blink reflex in normals and schizophrenic patients. Psychophysiology. 1982;19:1–6. doi: 10.1111/j.1469-8986.1982.tb02589.x. [DOI] [PubMed] [Google Scholar]
- 39.Lahti A.C., Koffel B., LaPorte D., Tamminga C.A. Subanesthetic doses of ketamine stimulate psychosis in schizophrenia. Neuropsychopharmacology. 1995;13:9–19. doi: 10.1016/0893-133X(94)00131-I. [DOI] [PubMed] [Google Scholar]
- 40.Holcomb H.H., Lahti A.C., Medoff D.R., Cullen T., Tamminga C.A. Effects of noncompetitive NMDA receptor blockade on anterior cingulate cerebral blood flow in volunteers with schizophrenia. Neuropsychopharmacology. 2005;30:2275–2282. doi: 10.1038/sj.npp.1300824. [DOI] [PubMed] [Google Scholar]
- 41.Malhotra A.K., Pinals D.A., Adler C.M., Elman I., Clifton A., Pickar D., Breier A. Ketamine-induced exacerbation of psychotic symptoms and cognitive impairment in neuroleptic-free schizophrenics. Neuropsychopharmacology. 1997;17:141–150. doi: 10.1016/S0893-133X(97)00036-5. [DOI] [PubMed] [Google Scholar]
- 42.Homayoun H., Moghaddam B. NMDA receptor hypofunction produces opposite effects on prefrontal cortex interneurons and pyramidal neurons. J. Neurosci. 2007;27:11496–11500. doi: 10.1523/JNEUROSCI.2213-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moghaddam B., Adams B., Verma A., Daly D. Activation of glutamatergic neurotransmission by ketamine: a novel step in the pathway from NMDA receptor blockade to dopaminergic and cognitive disruptions associated with the prefrontal cortex. J. Neurosci. 1997;17:2921–2927. doi: 10.1523/JNEUROSCI.17-08-02921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risbrough V.B., Masten V.L., Caldwell S., Paulus M.P., Low M.J., Geyer M.A. Differential contributions of dopamine D1, D2, and D3 receptors to MDMA-induced effects on locomotor behavior patterns in mice. Neuropsychopharmacology. 2006;31:2349–2358. doi: 10.1038/sj.npp.1301161. [DOI] [PubMed] [Google Scholar]
- 45.Steru L., Chermat R., Thierry B., Simon P. The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology (Berl) 1985;85:367–370. doi: 10.1007/BF00428203. [DOI] [PubMed] [Google Scholar]
- 46.Cryan J.F., Mombereau C., Vassout A. The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neurosci. Biobehav. Rev. 2005;29:571–625. doi: 10.1016/j.neubiorev.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Valenstein E.S., Kakolewski J.W., Cox V.C. Sex differences in taste preference for glucose and saccharin solutions. Science. 1967;156:942–943. doi: 10.1126/science.156.3777.942. [DOI] [PubMed] [Google Scholar]
- 48.Horos R., von Lindern M. Molecular mechanisms of pathology and treatment in Diamond Blackfan Anaemia. Br. J. Haematol. 2012;159:514–527. doi: 10.1111/bjh.12058. [DOI] [PubMed] [Google Scholar]
- 49.Raabe B.M., Artwohl J.E., Purcell J.E., Lovaglio J., Fortman J.D. Effects of weekly blood collection in C57BL/6 mice. J. Am. Assoc. Lab. Anim. Sci. 2011;50:680–685. [PMC free article] [PubMed] [Google Scholar]
- 50.Duff B.J., Macritchie K.A., Moorhead T.W., Lawrie S.M., Blackwood D.H. Human brain imaging studies of DISC1 in schizophrenia, bipolar disorder and depression: a systematic review. Schizophr. Res. 2013;147:1–13. doi: 10.1016/j.schres.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 51.Thomson P.A., Parla J.S., McRae A.F., Kramer M., Ramakrishnan K., Yao J., Soares D.C., McCarthy S., Morris S.W., Cardone L., et al. 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: analysis for association with psychiatric disorder and cognitive traits. Mol. Psychiatry. 2013;19:668–675. doi: 10.1038/mp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Syntichaki P., Troulinaki K., Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 53.Eykelenboom J.E., Briggs G.J., Bradshaw N.J., Soares D.C., Ogawa F., Christie S., Malavasi E.L., Makedonopoulou P., Mackie S., Malloy M.P., et al. A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum. Mol. Genet. 2012;21:3374–3386. doi: 10.1093/hmg/dds169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hikida T., Jaaro-Peled H., Seshadri S., Oishi K., Hookway C., Kong S., Wu D., Xue R., Andrade M., Tankou S., et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc. Natl. Acad. Sci. USA. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pletnikov M.V., Ayhan Y., Nikolskaia O., Xu Y., Ovanesov M.V., Huang H., Mori S., Moran T.H., Ross C.A. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol .Psychiatry. 2008;13:173–186. doi: 10.1038/sj.mp.4002079. , 115. [DOI] [PubMed] [Google Scholar]
- 56.Hardie D.G. The AMP-activated protein kinase pathway--new players upstream and downstream. J. Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 57.Topisirovic I., Sonenberg N. mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb. Symp. Quant. Biol. 2011;76:355–367. doi: 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- 58.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 59.Ehninger D., de Vries P.J., Silva A.J. From mTOR to cognition: molecular and cellular mechanisms of cognitive impairments in tuberous sclerosis. J. Intellect. Disabil. Res. 2009;53:838–851. doi: 10.1111/j.1365-2788.2009.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K., et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 61.Grummt I., Ladurner A.G. A metabolic throttle regulates the epigenetic state of rDNA. Cell. 2008;133:577–580. doi: 10.1016/j.cell.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 62.Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santoro M.R., Bray S.M., Warren S.T. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 64.Bhakar A.L., Dolen G., Bear M.F. The pathophysiology of fragile X (and what it teaches us about synapses) Annu. Rev. Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McGowan P.O., Sasaki A., Huang T.C., Unterberger A., Suderman M., Ernst C., Meaney M.J., Turecki G., Szyf M. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS ONE. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pietrzak M., Rempala G., Nelson P.T., Zheng J.J., Hetman M. Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease. PLoS ONE. 2011;6:e22585. doi: 10.1371/journal.pone.0022585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J., Hwang Y.J., Boo J.H., Han D., Kwon O.K., Todorova K., Kowall N.W., Kim Y., Ryu H. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington's disease. Cell Death Differ. 2011;18:1726–1735. doi: 10.1038/cdd.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grewal S.S., Li L., Orian A., Eisenman R.N., Edgar B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 69.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 70.Vogel C. Translation's coming of age. Mol. Syst. Biol. 2011;7:498. doi: 10.1038/msb.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keyes S.R., Cinti D.L. Biochemical properties of cytochrome b5-dependent microsomal fatty acid elongation and identification of products. J. Biol. Chem. 1980;255:11357–11364. [PubMed] [Google Scholar]
- 72.Uauy R., Hoffman D.R., Peirano P., Birch D.G., Birch E.E. Essential fatty acids in visual and brain development. Lipids. 2001;36:885–895. doi: 10.1007/s11745-001-0798-1. [DOI] [PubMed] [Google Scholar]
- 73.Ewenczyk C., Leroux A., Roubergue A., Laugel V., Afenjar A., Saudubray J.M., Beauvais P., Billette de Villemeur T., Vidailhet M., Roze E. Recessive hereditary methaemoglobinaemia, type II: delineation of the clinical spectrum. Brain. 2008;131:760–761. doi: 10.1093/brain/awm337. [DOI] [PubMed] [Google Scholar]
- 74.Ji B., Wang X., Pinto-Duarte A., Kim M., Caldwell S., Young J.W., Behrens M.M., Sejnowski T.J., Geyer M.A., Zhou X. Prolonged Ketamine Effects in Hypomorphic Mice: Mimicking Phenotypes of Schizophrenia. PLoS ONE. 2013;8:e66327. doi: 10.1371/journal.pone.0066327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Braff D.L., Grillon C., Geyer M.A. Gating and habituation of the startle reflex in schizophrenic patients. Arch. Gen. Psychiatry. 1992;49:206–215. doi: 10.1001/archpsyc.1992.01820030038005. [DOI] [PubMed] [Google Scholar]
- 76.Swerdlow N.R., Shoemaker J.M., Auerbach P.P., Pitcher L., Goins J., Platten A. Heritable differences in the dopaminergic regulation of sensorimotor gating. II. Temporal, pharmacologic and generational analyses of apomorphine effects on prepulse inhibition. Psychopharmacology (Berl) 2004;174:452–462. doi: 10.1007/s00213-003-1480-4. [DOI] [PubMed] [Google Scholar]
- 77.Swerdlow N.R., Lelham S.A., Sutherland Owens A.N., Chang W.L., Sassen S.D., Talledo J.A. Pramipexole effects on startle gating in rats and normal men. Psychopharmacology (Berl) 2009;205:689–698. doi: 10.1007/s00213-009-1577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.May H.T., Anderson J.L., Muhlestein J.B., Bair T.L., Knight S., Horne B.D. Red Cell Distribution Width and Depression Among Patients Undergoing Angiography. Circulation. 2013;128:S314. [Google Scholar]
- 79.Bochkov Y.A., Palmenberg A.C. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. BioTechniques. 2006;41:283–284. doi: 10.2144/000112243. , 286, 288 passim. [DOI] [PubMed] [Google Scholar]
- 80.Zhou X., Barrett T.B., Kelsoe J.R. Promoter variant in the GRK3 gene associated with bipolar disorder alters gene expression. Biol. Psychiatry. 2008;64:104–110. doi: 10.1016/j.biopsych.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J .Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 82.Ji B., La Y., Gao L., Zhu H., Tian N., Zhang M., Yang Y., Zhao X., Tang R., Ma G., et al. A comparative proteomics analysis of rat mitochondria from the cerebral cortex and hippocampus in response to antipsychotic medications. J. Proteome Res. 2009;8:3633–3641. doi: 10.1021/pr800876z. [DOI] [PubMed] [Google Scholar]
- 83.Kim M., Soontornniyomkij V., Ji B., Zhou X. System-wide immunohistochemical analysis of protein co-localization. PLoS ONE. 2012;7:e32043. doi: 10.1371/journal.pone.0032043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collins M.L., Irvine B., Tyner D., Fine E., Zayati C., Chang C., Horn T., Ahle D., Detmer J., Shen L.P., et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Player A.N., Shen L.P., Kenny D., Antao V.P., Kolberg J.A. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J. Histochem. Cytochem. 2001;49:603–612. doi: 10.1177/002215540104900507. [DOI] [PubMed] [Google Scholar]
- 86.Acheson D.T., Stein M.B., Paulus M.P., Geyer M.A., Risbrough V.B. The effect of pregabalin on sensorimotor gating in ‘low’ gating humans and mice. Neuropharmacology. 2012;63:480–485. doi: 10.1016/j.neuropharm.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flaisher-Grinberg S., Overgaard S., Einat H. Attenuation of high sweet solution preference by mood stabilizers: a possible mouse model for the increased reward-seeking domain of mania. J. Neurosci. Methods. 2009;177:44–50. doi: 10.1016/j.jneumeth.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 88.Tanaka S., Young J.W., Halberstadt A.L., Masten V.L., Geyer M.A. Four factors underlying mouse behavior in an open field. Behav. Brain Res. 2012;233:55–61. doi: 10.1016/j.bbr.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.