Abstract
Locus mapping has uncovered diverse etiologies for familial atrial fibrillation (AF), dilated cardiomyopathy (DCM), and mixed cardiac phenotype syndromes, yet the molecular basis for these disorders remains idiopathic in most cases. Whole-exome sequencing (WES) provides a powerful new tool for familial disease gene discovery. Here, synergistic application of these genomic strategies identified the pathogenic mutation in a familial syndrome of atrial tachyarrhythmia, conduction system disease (CSD), and DCM vulnerability. Seven members of a three-generation family exhibited the variably expressed phenotype, three of whom manifested CSD and clinically significant arrhythmia in childhood. Genome-wide linkage analysis mapped two equally plausible loci to chromosomes 1p3 and 13q12. Variants from WES of two affected cousins were filtered for rare, predicted-deleterious, positional variants, revealing an unreported heterozygous missense mutation disrupting the highly conserved kinase domain in TNNI3K. The G526D substitution in troponin I interacting kinase, with the most deleterious SIFT and Polyphen2 scores possible, resulted in abnormal peptide aggregation in vitro and in silico docking models predicted altered yet energetically favorable wild-type mutant dimerization. Ventricular tissue from a mutation carrier displayed histopathological hallmarks of DCM and reduced TNNI3K protein staining with unique amorphous nuclear and sarcoplasmic inclusions. In conclusion, mutation of TNNI3K, encoding a heart-specific kinase previously shown to modulate cardiac conduction and myocardial function in mice, underlies a familial syndrome of electrical and myopathic heart disease. The identified substitution causes a TNNI3K aggregation defect and protein deficiency, implicating a dominant-negative loss of function disease mechanism.
INTRODUCTION
Atrial fibrillation (AF) is the most common sustained arrhythmia in clinical practice (1). Typically attributable to the cumulative effects of acquired cardiovascular risk factors in the elderly, AF is an idiopathic, heritable disorder in a subset of younger individuals (2). In certain families, AF segregates as a Mendelian trait enabling genomic strategies for disease gene discovery. Unlike candidate gene approaches that require a priori knowledge, functionally diverse and unsuspected underpinnings of AF may be revealed. For example, locus mapping has led to identification of ion channel (KCNQ1, MIM 607542) (3) and circulating hormone (NPPA, MIM 108780) (4) defects in autosomal dominant AF, and nuclear protein defects (5,6) in autosomal recessive (NUP155, MIM 606694) and X-linked recessive (EMD, MIM 300384) AF. Further, ion channel (SCN5A, MIM 600163) (7) or nuclear protein (LMNA, MIM 150330) (8) defects have been linked to autosomal dominant mixed-phenotype syndromes characterized by variably expressed AF, conduction system disease (CSD), and dilated cardiomyopathy (DCM). Notwithstanding, the molecular bases for arrhythmogenesis and heart failure in AF and DCM, respectively, remain idiopathic in a majority of cases. To this end, the recent advent of whole-exome sequencing (WES) provides a powerful complementary tool for familial cardiovascular disease gene discovery (9).
Here, we studied a multi-generational family with a unique cardiac phenotype characterized by variably expressed atrial tachyarrhythmia, CSD, and vulnerability to DCM. Linkage analysis and WES were used as synergistic genomic strategies to identify a novel mutation in TNNI3K, encoding troponin I interacting kinase (TNNI3K, MIM 613932). TNNI3K has been proposed as a therapeutic target for heart disease based on its cardiac specificity (10) and established role as a modulator of myopathic (11–13) and electrical (14,15) heart disease in mice, underscoring its merit as a candidate gene. In our study, in vitro and in silico investigations implicated a propensity for abnormal aggregation of mutant TNNI3K, confirmed by marked reduction of TNNI3K protein staining in situ and the presence of unique amorphous inclusions in ventricular tissue. These findings establish a direct link between TNNI3K perturbation and human cardiac disease.
RESULTS
Familial cardiac phenotype
Seven living family members were diagnosed with a syndrome of variably expressed electrical and myopathic heart disease (Fig. 1; Supplementary Material, Table S1), with CSD and atrial arrhythmia as the most consistent, earliest onset traits. The proband (III.4) presented with supraventricular tachycardia at Age 20 and was treated with beta-receptor blocker therapy. He had a normal echocardiogram but unexplained left anterior fascicular block. His family history was remarkable for arrhythmia in several relatives, chronic DCM and congestive heart failure in his mother and maternal grandmother, and childhood sudden unexpected death in a maternal aunt. At Age 31, surveillance Holter monitoring documented paroxysmal multifocal atrial tachycardia and frequent premature ventricular contractions. His echocardiogram remained unremarkable. At Age 39, he was hospitalized for dyspnea and diagnosed with left ventricular enlargement and reduced ejection fraction of 45%. Angiography ruled out coronary artery disease but endomyocardial biopsy demonstrated mild focal myocyte hypertrophy and interstitial fibrosis, typical yet non-specific, histopathologic features of DCM. No significant arrhythmia and a normal heart rate range of 68–88 beats per minute were documented during 48 h of telemetric monitoring, excluding tachycardia-induced cardiomyopathy. To effect reverse ventricular remodeling, treatment with an angiotensin-converting enzyme inhibitor was initiated, and nadolol was switched to metoprolol. Echocardiographic features of DCM resolved over the ensuing 18 months. At Age 50, he was physically active and asymptomatic with an ejection fraction of 55% but CSD had progressed to bifascicular block. Six other living relatives had infra-Hisian CSD with variable expression of atrial tachyarrhythmia, including AF/flutter (n = 4) or ectopic/multifocal atrial tachycardia (n = 2). In three children, the arrhythmia was successfully suppressed with combination medical therapy and catheter-based ablation of right atrial foci. The proband's mother had chronic DCM and ultimately died in congestive heart failure, but DCM resolved with medical therapy (n = 2) or was confined to isolated left ventricular enlargement (n = 1) in other relatives. Only one of seven affected family members had DCM associated with atrial tachycardia and inadequate ventricular rate control.
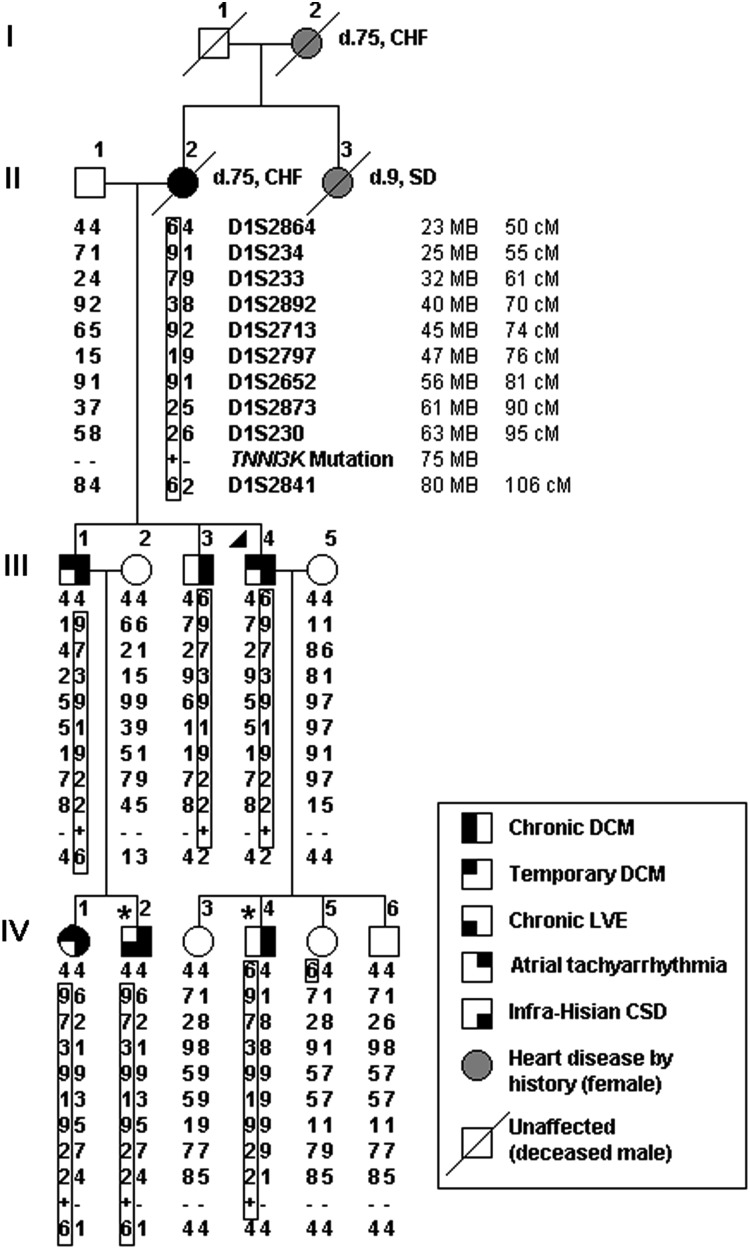
Figure 1.
Pedigree structure and haplotypes at 1p31-p36.1 locus for family with autosomal dominant electrical and myopathic heart disease. Phenotypic traits are indicated by shaded quadrants within the pedigree symbol. A triangle (▸) designates the proband whereas an asterisk (*) indicates individuals who underwent WES. Short tandem repeat DNA markers are listed from p telomere to centromere, with map locations according to the National Center for Biotechnology Information Web site (hg19 human reference genome) and given in megabases (MB) and centimorgans (cM). Haplotypes for marker genotypes are shown in columns below pedigree symbols with identical disease-associated haplotypes (boxed) inherited by all seven affected family members. A recombination event in III.1 defines D1S2864 as the upper-flanking marker whereas the lower-flanking marker, D1S2841, is defined by recombination events in III.3 and III.4. TNNI3K maps within this 57 MB locus. Presence of the identified TNNI3K-G526D mutant allele is indicated by a plus symbol (+) and its absence by a minus (−) symbol. CHF, congestive heart failure; CSD, conduction system disease; d., age at death; DCM, dilated cardiomyopathy; LVE, left ventricular enlargement; SD, sudden unexpected death.
Locus mapping identifies potential chromosomal loci
Genome-wide genotyping and linkage analysis, specifying a dominant mode of inheritance, mapped two equally plausible disease gene loci on chromosomes 13q12 and 1p31-36.1. Each locus had a peak multipoint logarithm of the odds (LOD) score of 2.4 (1:250 odds of false positive linkage), the maximum achievable for this pedigree. Construction of haplotypes revealed linkage to a 10 MB region at 13q12 (data not shown) and a 57 MB region at 1p31-36.1 (Fig. 1). Together, these loci were comprised of over 800 positional candidate genes, posing a daunting task for selection of genes for mutation scanning by Sanger sequencing. Accordingly, WES was employed as a synergistic strategy for disease gene discovery.
Exome sequencing reveals pathogenic TNNI3K variant
Genomic DNA samples from two affected cousins (IV.2 and IV.4) underwent WES following targeted exome capture. Each sample yielded >70 million 101 base paired-end reads and passed quality control standards. Over 99% of the reads mapped to the genome and over 65% to the targeted regions. Exome coverage was similar in both individuals, with at least 80% coverage of the targeted regions demonstrating a minimal read depth of 20 reads. To verify that known arrhythmia genes were excluded as candidates, including the prototypic CSD/DCM genes LMNA and SCN5A, targeted analyses were performed for 33 genes present on commercially available gene panels from Ambrygen and Gene Dx. None of these genes were located within the 1p3 or 13q12 loci and locus-free analysis of WES data did not reveal rare, shared variants within these genes.
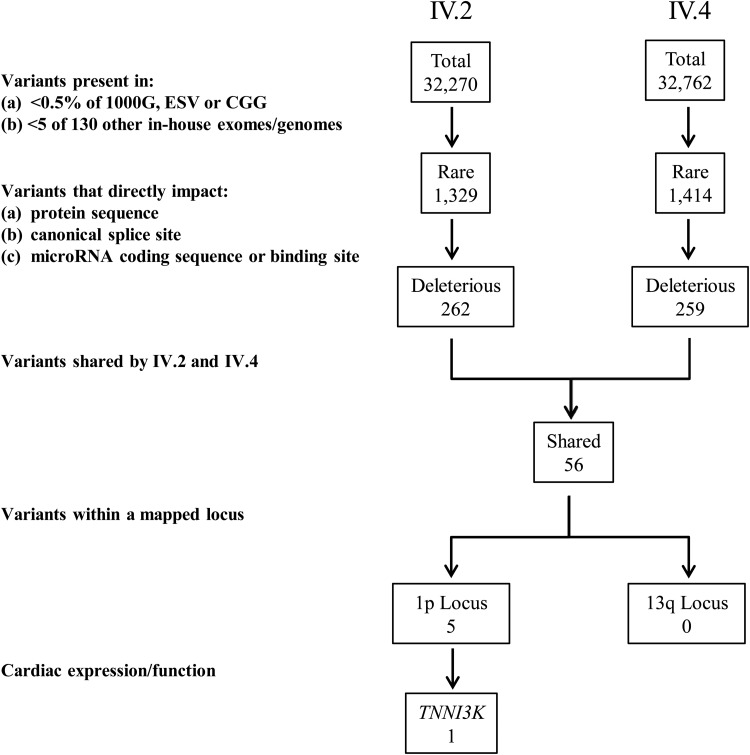
Variant call format (VCF) files with single nucleotide variant (SNV) and insertion/deletion (INDEL) calls from each individual were loaded into the Ingenuity® Variant Analysis™ server, enabling an iterative filtering process of annotated variants to identify the pathogenic mutation (Fig. 2). False positive subtractive filtering was done by eliminating any variants that were present in >5 WES or WGS datasets collected from 130 individuals not affected with DCM or AF. Rarity of variants was then considered with elimination of all variants present in >0.5% of 7663 publicly available exome and genome databases. Next, variants were restricted to those that directly impacted a protein sequence, canonical splice site, or microRNA coding sequence/binding sites. The list of remaining variants was subsequently narrowed to those shared by the affected cousins and localized to either the 13q12 or 1p31-36.1 locus. This filtering process culminated in a short list of five heterozygous variants at the 1p31-36.1 locus. Four of the variants were ruled out based on the corresponding gene function, expression, and/or known role in non-cardiac diseases. Two of the four were somatic missense variants identified in cancer tissue: COSM177577 [NM_020379.2: c.592 G > A (p.E198K)] was identified in MAN1C1 and rs121908688 [NM_003579.3: c.188 C > A (p.P63H)] in RAD54L (MIM 603615). A third previously reported variant (rs201699710) was identified in RPS6KA1 (MIM 601684) which encodes for a protein that has a ubiquitous expression pattern. This missense variant, p.R21G (NM_001006665.1:c.61 A > G) was reported with a minor allele frequency of 0.05% and predicted to be benign by both SIFT (16) and Polyphen2 (17). The fourth missense variant, p.A145T (NM_000329.2:c.433 G > A) in RPE65 (MIM 180069), was a poor candidate for heart disease due to tissue specific expression in the retinal pigment epithelium and an established genetic link to retinitis pigmentosa. A novel, predicted-deleterious missense variant in TNNI3K (MIM 613932) remained the sole candidate for disease causality in this family. Evidence supporting TNNI3K as a candidate gene were compelling, including high, cardiac-specific expression and an established role in cardiac physiology (10–15,18,19).
Figure 2.
Variant filtering scheme for WES data. An iterative filtering approach was applied to SNVs and insertion/deletions identified in individuals IV.2 and IV.4 by WES. The population frequency of individual variants and their impact on protein structure or expression were assessed as inclusion criteria. While there were no variants at the 13q locus, 5 variants resided within the 1p locus. One of these occurred in TNNI3K, a gene that is highly and specifically expressed in the heart and has an established role in cardiac physiology.
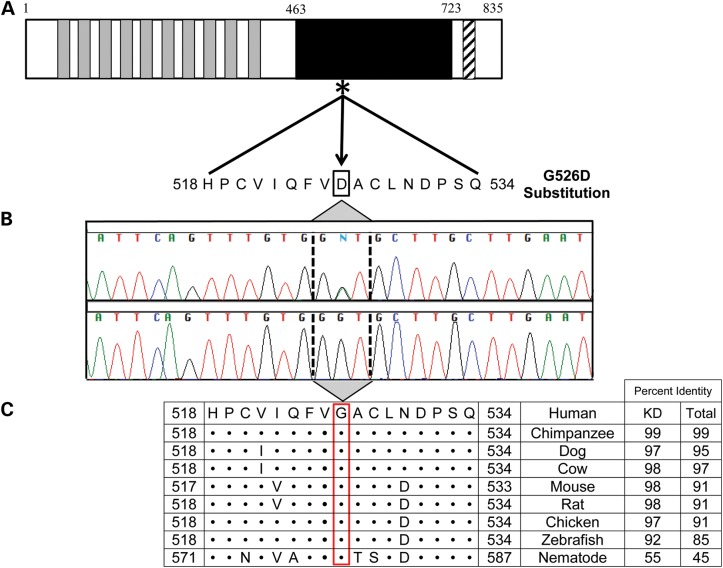
The identified missense mutation in TNNI3K, encoding TNNI3 interacting kinase, was a c.1577 G > A (NM_015978.2) nucleotide transition in exon 16 (Fig. 3). This resulted in a p.G526D substitution within the kinase domain that replaced a small, hydrophobic glycine with a bulky, negatively charged aspartic acid. G526D had the most deleterious score possible in SIFT (16) (score = 0) and Polyphen2 (17) (score = 1) and is unreported in >7600 exomes/genomes comprising publicly available databases. TNNI3K also has a Residual Variation Intolerance Score of 27%, indicating its tolerance of variation is <73% of other genes (20). The mutation demonstrated complete cosegregation with disease—all seven affected individuals were heterozygous carriers and the three unaffected family members were not. To determine if heterozygous mutations in TNNI3K underlie non-syndromic heart disease, the 25 exons were screened in 125 unrelated probands with either familial DCM (n = 64) or familial AF (n = 61). Additionally, the 9 exons encoding the kinase domain were screened in 40 individuals with sporadic DCM or AF with CSD manifest by left QRS axis deviation. Three missense variants previously reported in public databases were identified, but excluded as pathogenic based upon lack of segregation with disease in other family members.
Figure 3.
Location and conservation of TNNI3K-G526D mutation. (A) The protein topology is shown for TNNI3K, including 10 functional ankyrin repeat domains (gray), a serine-rich domain (diagonal hatch lines) and a single functional kinase domain (black) where the heterozygous c.1577 G > A resides (asterisk). (B) Sanger sequencing verified the mutation, which resulted in a G526D substitution. (C) Conservation of this residue is illustrated whereby • indicates identical residues. Overall, the amino acid sequence of TNNI3K is highly conserved with the amino acid identity to the human TNNI3K ortholog indicated for both the kinase domain (KD) and the entire protein.
In vitro expression of TNNI3K-G526D demonstrates abnormal aggregation
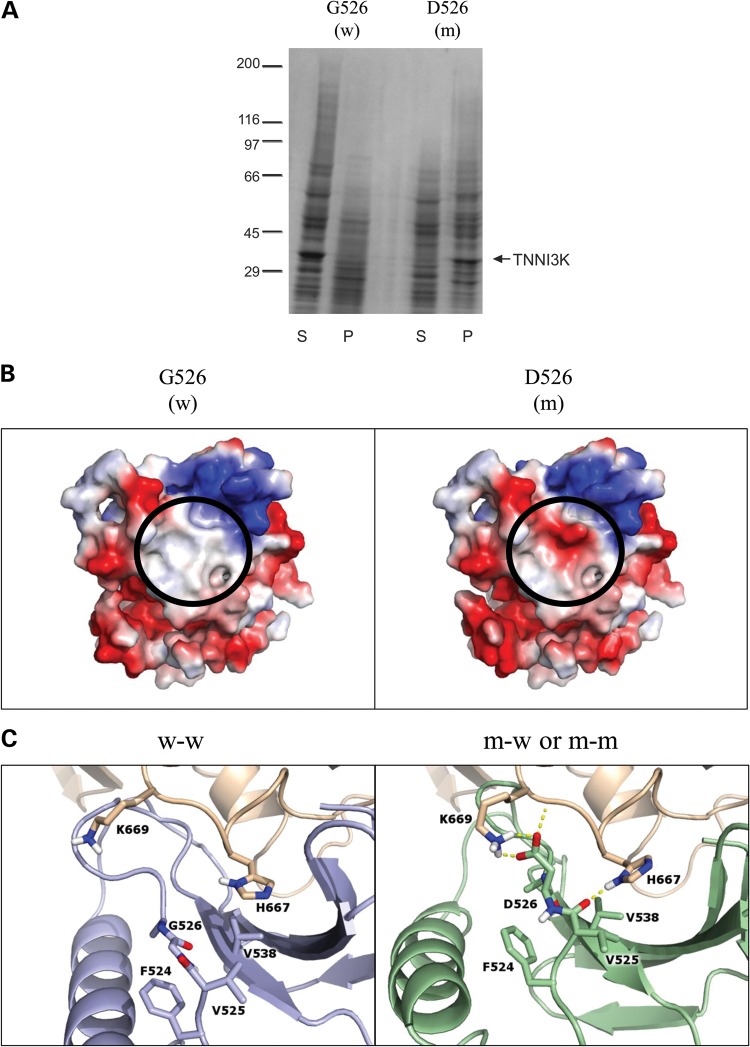
We hypothesized that TNNI3K-G526D would disrupt kinase activity of the protein and generated constructs of wild-type (G526) and mutant (D526) kinase domains for an in vitro kinase assay. Mutant peptide was insoluble, even in the presence of Triton X (Fig. 4A), precluding assessment of enzymatic activity. By contrast, wild-type peptide was completely soluble.
Figure 4.
In vitro expression and in silico modeling of mutant peptide. (A) Kinase domains of wild-type (w; G526) and mutant (m; D526) TNNI3K were expressed and the resulting pellet (P) and supernatant (S) fractions were analyzed by SDS-Page and Coomassie Blue, demonstrating poor solubility of the mutant peptide. The arrow indicates the 32 kilodalton kinase domain of TNNI3K protein and the molecular weight standards (×10−3) are shown on the left. (B) Electrostatic protein modeling (21) highlights a large hydrophobic patch on the surface of the kinase domain (white), which becomes negatively charged (red) in the presence of the G526D mutation. (C) The left panel depicts docking of wild-type (blue) to another wild-type TNNI3K (tan) (w–w) whereas the right panel shows the docking pose for mutant TNNI3K (green) to either wild-type or mutant TNNI3K (both represented in tan) (m-w and m–m). Residues within 3.5 Å of residue 526 in either pose are shown in atomic detail. Hydrogen bonds involving these indicated residues are drawn as dashed yellow lines. When mutated, the loop containing residue 526 shifts toward the binding partner to make more contacts including an electrostatic lysine-aspartic acid interaction (depicted as h-bonds).
In silico structural modeling of TNNI3K-G526D reveals alterations in binding interfaces
Because the in vitro assay supported a global effect on protein assembly conferred by the G526D substitution, rather than a focal effect on kinase activity, protein models were created to predict the effect of the mutation on protein structure and dimerization. Although it was not predicted to directly impact functional sites within the ATP-binding catalytic domain, substitution of the neutral glycine for a negatively charged aspartic acid at residue 526 markedly altered the hydrophobic surface of the protein in this region (Fig. 4B). Tertiary structure models revealed that the inherent stability of the kinase domain was unchanged in the presence of the mutation. Further support from in silico models imposing substitution with each of the other 19 amino acids at residue 526 confirmed that aspartic acid resulted in the smallest change in stability (ΔΔG) of any substituted residue.
Subsequent models focused on the mutation's predicted impact on dimerization. Oligomerization of TNNI3K has been previously demonstrated (22), but details regarding the binding interfaces have not been defined. Here, in silico modeling of kinase domain (AA 463–723) docking identified four classes of binding interfaces that were consolidated into two principal scenarios; an interface involving the region where the mutation resides (head) and an interface on the opposite side (body) (Supplementary Material, Fig. S1A). While the same basic set of docking scenarios was observed in the presence of the mutation, D526 altered the predicted probability of each interaction as indicated by the number of poses in each cluster (Supplementary Material, Fig. S1B). In the presence of the mutation, the head-to-head pose was abolished due to added steric constraints, resulting in an amplification of the other poses (Supplementary Material, Fig. S1C). Furthermore, within the body-to-head pose, there were an increased number of contacts utilizing backbone and side chain atoms in the context of mutant D526 interacting with the body of either the wild-type (G526) or mutant (D526) kinase domain, enhanced interactions that are not present in the wild-type–wild-type interactions (Fig. 4C). To evaluate the differential interaction strength in the presence of these additional contacts, FoldX (23) was utilized and demonstrated that interaction strength was slightly increased (0.3 kcal/mol after including entropic and solvation terms) for the mutant. Introduction of the aspartic acid at reside 526 led to an increase in the affinity of the protein–protein interaction due to hydrogen bonding (−0.5 kcal/mol), Van der Waals contacts (−3 kcal/mol), chain entropy (−1 kcal/mol), and electrostatics (−0.4 kcal/mol), all of which are partially compensated by less favorable solvation energy (+4.5 kcal/mol).
In summary, protein modeling suggested that the G526D substitution would have minimal effect on monomeric TNNI3K structure, but alter the relative affinity of docking configurations while maintaining mutant–wild-type protein interaction. These findings supported the observed in vitro aggregation of mutant TNNI3K and predicted a dominant-negative effect of the heterozygous mutation.
Immunohistochemistry of cardiac tissue with mutant TNNI3K reveals marked reduction in TNNI3K protein staining
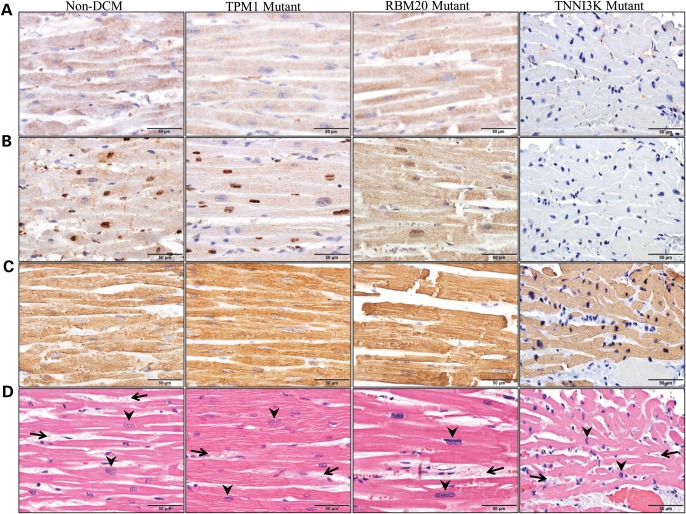
TNNI3K is highly and specifically expressed in both adult and fetal myocardium (10,13). To determine the mutation's effect on protein expression and localization, immunohistochemical staining for TNNI3K protein was performed on histologic sections of ventricular tissue. Representative photomicrographs utilizing anti-TNNI3K antibodies raised against the N-terminus (Fig. 5A) or the C-terminus (Fig. 5B), showed uniform distribution of TNNI3K protein throughout the sarcoplasm of cardiomyocytes from a patient without DCM (non-DCM). The antibody recognizing the C-terminus (Fig. 5B) also produced a strong yet patchy nuclear staining pattern, with only a subset of nuclei showing this feature. Two individuals with genetically defined familial DCM due to a missense mutation in TPM1, encoding tropomyosin 1 (alpha) (TPM Mutant) or RBM20, encoding RNA binding motif protein 20 (RBM20 Mutant) each showed a distribution of TNNI3K expression similar to non-DCM. In contrast, heterozygosity for TNNI3K-G526D was associated with markedly reduced TNNI3K protein staining in the myocardium, a feature that was observed with both antibodies (Fig. 5A and B). Actin expression (Fig. 5C) in the sarcoplasm was retained in all four individuals, and complete absence of staining was observed with omission of the primary antibody in a negative control study (data not shown). Standard hematoxylin and eosin (H&E) staining (Fig. 5D) demonstrated non-specific histopathologic changes in all subjects, including cardiomyocyte hypertrophy (as evidenced by nuclear enlargement) and mild patchy interstitial fibrosis surrounding individual cardiomyocytes, although the degree of cardiomyocyte hypertrophy was only mild in the TNNI3K mutant, in contrast with the moderate-to-severe hypertrophy present in the other three patients.
Figure 5.
Detection of TNNI3K protein in ventricular tissue by immunohistochemistry. Immunohistochemistry was performed using three antibodies on ventricular tissue from four individuals. Column 1 illustrates cardiac tissue from an individual without DCM (non-DCM), Columns 2 and 3 illustrate cardiac tissue from individuals with genetically defined familial DCM due to mutations in TPM1 (TPM1 Mutant) or RBM20 (RBM20 Mutant), respectively. The fourth column illustrates cardiac tissue from individual III.4 who carries the G526D mutation in TNNI3K (TNNI3K Mutant). Antibodies that recognize (A) the N-terminus of TNNI3K or (B) the C-terminus of TNNI3K reveal markedly reduced TNNI3K protein expression in the sarcoplasm (A and B) and the nuclei (B) of cardiomyocytes in the individual with the G526D mutation as compared with control tissue. (C) In contrast, actin expression was retained in all individuals. (D) H&E staining demonstrated non-specific histopathologic changes, including cardiomyocyte hypertrophy (evidenced by enlargement of cardiomyocyte nuclei, arrowheads) and mild interstitial fibrosis (increased fibrous connective tissue between individual cardiomyocytes, arrows), although the degree of cardiomyocyte hypertrophy was only mild in the TNNI3K mutant, in contrast with the moderate-to-severe hypertrophy in the other three patients.
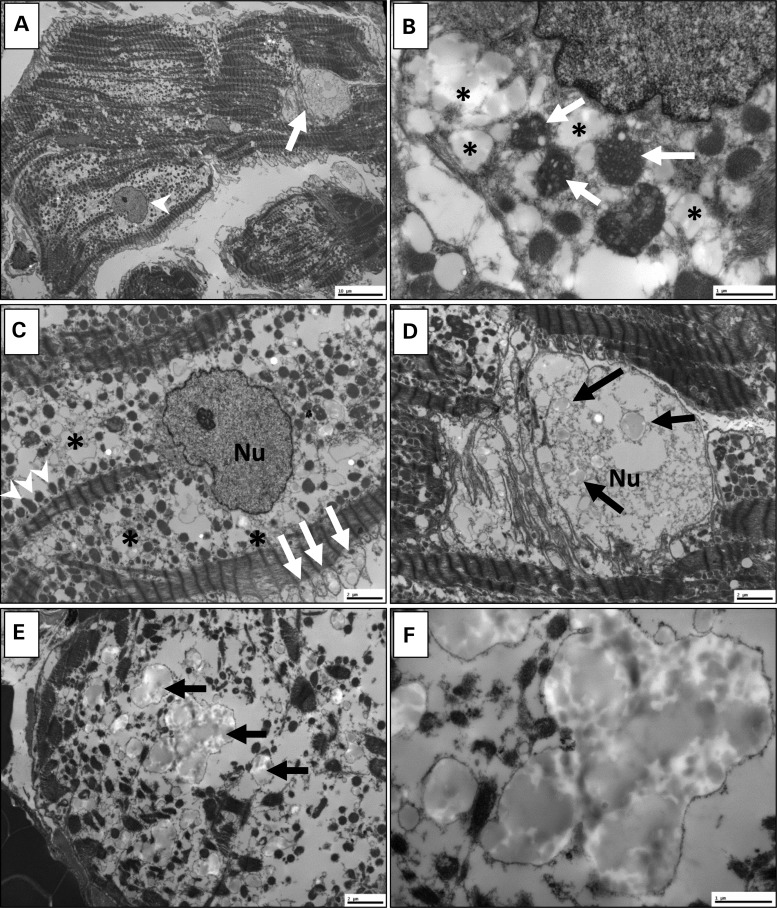
Transmission electron microscopy of cardiac tissue reveals amorphous intranuclear and intrasarcoplasmic inclusions
Ultrastructural analysis of a right ventricular endomyocardial biopsy from III.4 demonstrated several ultrastructural abnormalities, including non-specific findings and abnormalities that appeared specific to the presence of the TNNI3K mutation. Non-specific features of myocardial disease included variable loss of myofilaments and mitochondriosis (Fig. 6A) with some mitochondria exhibiting architectural disorganization of cristae with vacuolization and degenerative changes (Fig. 6B). While some nuclei appeared morphologically normal (Fig. 6C), others were distinctly abnormal with numerous amorphous intranuclear inclusions (Fig. 6D), a finding that has not been reported in other myocardial diseases. In addition to many severely swollen mitochondria (Fig. 6C), the sarcoplasm also contained inclusions that were morphologically similar to the inclusions observed in the nuclei. Notably, inclusions were particularly associated with areas of marked myofilament loss (Fig. 6E), suggesting that they may represent a cardinal ultrastructural feature of TNNI3K cardiomyopathy. At higher magnification, the globular, electron-opaque nature of the inclusions indicated that they were likely proteinaceous in nature. Inclusions appeared to be membrane-bound, possibly representing distended mitochondria or lysosomes (Fig. 6F).
Figure 6.
Transmission electron microscopy of ventricular tissue. Ultrastructure of ventricular myocardium was determined for individual III.4. (A) At scanning magnification, non-specific mitochondriosis (increased number of mitochondria) is apparent with variable loss of myofilaments. Some nuclei are normal (arrowhead); others are abnormal (arrow). (B) Some mitochondria (arrows) show architectural disorganization of cristae with vacuolization and degenerative changes, and are surrounded by sarcoplasmic inclusions (*). (C) The sarcoplasm contains many severely swollen mitochondria (arrows), as well as scattered preserved mitochondria (arrowheads) within extensive zones (*) of complete mitochondrial disruption. In this cardiomyocyte, the nucleus (Nu) is intact. (D) Other nuclei are abnormal, containing numerous amorphous intranuclear inclusions (arrows). (E) Similar inclusions (arrows) are present in the sarcoplasm, particularly in areas of marked myofilament loss. (F) At higher magnification, inclusions are amorphous, globular and electron-opaque. Some appear to be membrane-bound, possibly representing distended mitochondria or lysozomes. Scale bar: 10 µm (A); 2 µm (C–E); 1 µm (B and F).
DISCUSSION
The cardiac-specific expression of TNNI3K and its established role in murine cardiac physiology has prompted great interest in modulation of TNNI3K expression as a potential therapeutic strategy for heart disease (13). Here, we demonstrate for the first time the essential role of TNNI3K in human hearts by characterizing the clinical and functional consequences of TNNI3K perturbation in a three-generation family. Synergistic genomic strategies of locus mapping and WES enabled discovery of a pathogenic mutation in TNNI3K, a kinase that is highly conserved across species.
The observed mixed cardiac phenotype in our family appears to be specific to genetic perturbation of TNNI3K, as additional mutations were not found in non-syndromic cases of familial DCM and AF. TNNI3K expression has been noted in all four chambers of the heart and, while transcript levels are more pronounced in ventricles (10,13), protein levels are greatest in the atria (14). Notably, atrial tachyarrhythmia was a manifestation of disease among all TNNI3K mutation carriers. Three children had arrhythmogenic foci in the right atrium, which were successfully ablated with subsequent reduction in arrhythmia burden despite persistence of the underlying TNNI3K defect throughout the atrial myocardium. The phenomenon of heritable forms of atrial arrhythmia being amenable to ablation of focal triggers has been previously reported (24). TNNI3K expression in the cardiac conduction system has not been reported, yet it is strongly implicated by the infra-Hisian CSD uniformly observed among affected family members. In contrast to susceptibility to high grade SA and AV node disease reported in mixed cardiac phenotypes caused by SCN5A (7) and LMNA (8) mutations, none of the TNNI3K mutation carriers had sinus node dysfunction or >1° AV block. Notwithstanding, a role of Tnni3k in murine cardiac electrical signaling has been implicated by its localization to a quantitative trait locus for PR interval duration, an established intermediate phenotype of atrial fibrillation (14). Furthermore, introduction of missense mutations in the C-terminus of Tnni3k have been shown to precipitate arrhythmic outbreaks in cultured murine cardiomyocytes and whole hearts upon direct intramyocardial injection (15).
Clinical and echocardiographic manifestations of overt, chronic DCM occurred at relatively advanced ages in TNNI3K mutation carriers, yet several family members demonstrated a vulnerability to myocardial dysfunction that was responsive to medical therapy. Moreover, ventricular myocardium of the proband had a marked reduction in TNNI3K staining and examination by light and electron microscopy demonstrated a degenerative myopathy. While the precise substrate(s) and signaling pathway of this kinase are still under investigation, TNNI3K was recently shown to phosphorylate two serine residues of cardiac troponin I, suggesting a direct role in the regulation of cardiac contraction (19) This role is further supported by its direct interaction with troponin I, cardiac α-actin and myosin binding protein C (10), three sarcomeric proteins with an established genetic link to DCM (25). Several investigations support a modulatory role for TNNI3K in mechanical heart function. TNNI3K is overexpressed in human heart failure (13), but it is unknown if this is an adaptive or pathological response. Murine studies support both beneficial and deleterious effects of TNNI3K, depending on context (11–15,18,19). Overexpression of TNNI3K has been shown to enhance cardiomyocyte differentiation and cardiac function in vitro (18) and promote physiological cardiac hypertrophy in vivo (19) suggesting a beneficial role in cardiac physiology in non-disease states. Conversely, TNNI3K exacerbates ischemia/reperfusion injury in mice, an effect which was markedly reduced with the administration of a TNNI3K inhibitor (13). Consistent with this observation, high levels of TNNI3K in mice that are predisposed to heart failure due to overexpression of calsequestrin (11) or transverse aortic constriction (13) lead to a more rapid disease progression and decline in cardiac function.
The potential consequences of point mutations on protein expression, localization and degradation are varied, and depend on the nature of the altered amino acid sequence and its impact on the tertiary and quaternary structure of the protein. Herein, in vitro, in silico and in situ data collectively implicate a dominant-negative effect of TNNI3K-G526D, resulting in protein aggregation and reduction in protein concentration or bioavailability in the cardiomyocyte. Immunohistochemical staining was performed with two TNNI3K antibodies, one raised against the N-terminus and the other raised against the C-terminus. While each antibody showed uniform cytoplasmic staining in myocardial tissue from controls, the C-terminal antibody also demonstrated a patchy, nuclear staining. Similar differences in staining patterns of TNNI3K have been described in other studies (10,12,13), potentially attributable to antibody specificity, recognition of unique isoforms or age-dependent localization patterns. Notwithstanding, we observed reduced staining with either antibody in tissue from the TNNI3K mutant. Ultrastructural analysis of myocardial tissue from the proband revealed the presence of amorphous, intranuclear and cytoplasmic inclusions, which may represent aggregates of TNNI3K protein. If confirmed, this would suggest that the observed reduction in TNNI3K protein staining may be caused either by enhanced degradation, decreased antigenicity, or a combination of both as a consequence of aggregation.
Protein aggregation has previously been shown to be a pathological substrate for electrical heart disease (26). Heat shock proteins (HSPs) maintain the proper folding of other proteins to prevent toxic aggregation and the protective role of HSPs has led to an interest in therapeutic agents that induce the heat shock response in the setting of AF (26). Moreover, a direct link to protein aggregation has also been established in DCM with mutations in a HSP protein (CRYAB, MIM 123590) causing protein aggregation and maladaptive reductive stress (27). The present study provides additional evidence that pathologic aggregation of proteins essential for cardiac conduction and myocyte function may underlie development of heritable forms of heart disease.
In summary, we demonstrate that mutation of TNNI3K, encoding a heart-specific kinase known to modulate murine cardiac conduction and myocardial function, underlies a familial syndrome of electrical and myopathic heart disease in humans. Identification of additional TNNI3K mutations would be required to further establish this genotype–phenotype relationship. Consistent with a dominant-negative loss of function effect, the identified substitution causes a TNNI3K aggregation defect with secondary protein deficiency. The specific mechanisms remain to be elucidated whereby mutant TNNI3K leads to CSD, electrical instability, myofilament loss, and ultimately DCM.
MATERIALS AND METHODS
Study subjects
The multi-generational family described in this study is White and of German ancestry. Subjects provided written informed consent under a research protocol approved by the Mayo Clinic Institutional Review Board. Phenotypic assessment and diagnoses in symptomatic and at-risk family members were based on clinically indicated testing and abstracted from medical records. Rhythm and conduction abnormalities were detected by standard 12-lead and ambulatory electrocardiography. Diagnostic criteria for DCM were: lack of coronary artery disease (in adults), left ventricular diastolic and/or systolic dimensions >95th percentile indexed for body surface area by echocardiography (28), and left ventricular ejection fraction <50%. Spouses of affected subjects were assumed to have normal cardiac phenotypes. Genomic deoxyribonucleic acid (DNA) was isolated from peripheral-blood white cells or saliva. For TNNI3K mutation scanning, genomic DNA samples from 64 unrelated probands with non-syndromic familial DCM and 61 unrelated probands with non-syndromic familial AF were utilized. In addition, the kinase domain of TNNI3K was analyzed in 40 individuals with sporadic DCM or AF and CSD manifest by left QRS axis deviation. WES or whole-genome sequences (WGS) from 130 individuals not affected with DCM or AF were used for false positive subtraction filtering of exome variant calls.
Locus mapping
Thirteen family members underwent genome-wide genotyping with the ABI PRISM Linkage Mapping Set HD5, Version 2.5 (Applied Biosystems, Foster City, CA) comprised of primer pairs that flank 811 polymorphic short tandem repeat DNA markers. Markers were amplified from DNA samples by the polymerase chain reaction (PCR), resolved on an ABI PRISM 3130xl, and scored by GeneMapper Software (Applied Biosystems) for subsequent linkage analysis. Two-point and multipoint linkage analyses were performed with Simwalk2 Version 2.91 (29), modeling dominant inheritance and specifying the following variables: phenocopy rate 0.001, equal marker allele frequencies and dichotomous liability classes (‘affected’ and ‘unaffected’). LOD scores were determined using a 100% penetrance model.
Exome sequencing and bioinformatics analysis
WES and variant annotation were performed on DNA samples from two clinically affected individuals, utilizing the Medical Genome Facility and Bioinformatics Core at the Mayo Clinic. Preparation and 38 MB exome capture of each DNA sample was performed utilizing TruSeq PE cBot V1 cluster reagents (Illumina, San Diego, CA) and the SureSelect Target Enrichment System (Version 1.0; Agilent, Santa Clara, CA). Each sample was run in a single lane and 101 bp paired-end sequencing was carried out on Illumina's HiSeq2000 platform with the TruSeq SBS Kit V1. The reads were aligned to the hg19 reference genome using Novoalign (http://novocraft.com) followed by the sorting and marking of duplicate reads using Picard (http://picard.sourceforge.net). Local realignment of INDELs and base quality score recalibration were then performed using the Genome Analysis Toolkit (GATK) (30). SNVs and INDELs were called across both samples simultaneously using GATK's Unified Genotyper with variant quality score recalibration (31). The resultant variant call format (VCF) files were analyzed using Ingenuity® Variant Analysis™ software (www.ingenuity.com/variants) from Ingenuity Systems, where variants were functionally annotated and filtered by an iterative process. To determine the rarity of variants, minor allele frequencies from three publicly available databases were utilized, collectively comprised of 7663 whole-genome or whole-exome datasets: 1000 Genomes (WGS data from 1092 individuals) (32), the Exome Variant Server (WES data from 6503 individuals) (33), and Complete Genomics Genome (WGS data from 69 individuals) (34).
Mutation scanning and sanger sequencing
PCR primer pairs were designed to encompass the 25 translated exons and flanking splice junctions of TNNI3K and are available in Supplementary Material, Table S2. PCR-amplified products of 23 of the 25 exons were evaluated for heterozygous sequence variation in familial DCM and AF cohorts using denaturing high-performance liquid chromatography (DHPLC) heteroduplex analysis (WAVE DHPLC System, Transgenomic, Omaha, NE). Chromatographic elution profiles of amplified fragments were compared against the wild-type homoduplex pattern; samples yielding anomalous traces were Sanger sequenced to determine the precise genetic variant. Because PCR reagents for optimal amplification of exons 10 and 18 were not compatible with DHPLC, Sanger sequencing of these exons was performed on all samples. For identified variants, genotyping by Sanger sequencing was performed on all affected family members to determine segregation with disease.
In vitro analysis: expression of wild-type and mutant TNNI3K
DNA fragment encoding kinase domain of TNNI3K was PCR amplified from a clone containing full-length human cardiac troponin I interacting kinase (Open Biosystems/Thermo Scientific). For site-directed mutagenesis, QuikChange kit (Stratagene, La Jolla, CA) was used to change the endogenous G526 residue to a D. The sequence verified PCR products were cloned into BamHI and HindIII sites of pFastBac1 transfer plasmid (Invitrogen/Life Technologies). The resulting vectors encoding wild-type and mutated TNNI3K kinase domain were used for the site-specific transposition of an expression cassette into bacmids according to manufacturer's instructions. Working baculovirus stocks were produced by infecting 2.2 × 107 Sf9 cells on a 15 cm plate with 0.3 ml of post-transfectional supernatant and virus collection at 72 h post-infection. For protein expression, 0.3 ml working stock was used to infect Sf9 cell monolayers (2.2 × 107 cells per 15 cm plate). Cells were collected 70–86 h post-infection. To assess the solubility of the expressed proteins, harvested insect cells were solubilized in 10 mm Tris–HCl, pH 8.0, 0.15 M NaCl, 1% Triton, and centrifuged at 15 000g. The equal volumes of resulting supernatants (S) and pellets (P) were subjected to 3–20% gradient SDS-PAGE and stained with Coomassie Blue.
In silico analysis: protein structural modeling
Homology modeling was performed to generate a 3D model of the kinase domain of TNNI3K using the SWISS-MODEL server (35). The target sequence was comprised of residues 463–723 from SwissProt entry Q59H18 with the 35% sequence identity template, 3PPZ. Modeling of the wild-type (G526) and mutant (D526) kinase domain was refined using FoldX (23) prior to more detailed analysis of the structural stability, either with FoldX or Molecular Dynamics energy minimization. For molecular docking, the ClusPro server (36) was employed to rank poses and discriminate real poses from false positives (37). ClusPro generated 109 poses (candidate positioning of protein–protein interactions), each beginning from a different set of initial conditions, and refines, clusters and ranks the top 1000.
In situ analysis: immunohistochemistry of cardiac tissue
Right ventricular endomyocardial biopsy tissue from the proband was formalin fixed and paraffin embedded prior to sectioning at 5 μm for immunohistochemical staining. Three additional paraffin embedded specimens served as controls. Heart tissue was obtained at autopsy from the left ventricle of a 61-year-old male who died from non-cardiac causes and had no autopsy evidence of DCM. In addition, left ventricular tissue was available from two individuals with genetically defined familial DCM—a 10-year-old female with a mutation in TPM1, encoding tropomyosin 1 (alpha) (38), and a 29-year-old female with a mutation in RBM20, encoding RNA binding motif protein 20 (39). Tissue sections were deparaffinized prior to antibody optimization, which was carried out by an automated immunohistochemistry-staining machine (DAKO Autostainer Plus, DAKO, Carpinteria, CA). TNNI3K protein expression was evaluated with two rabbit polyclonal anti-human TNNI3K antibodies, previously shown to be specific for TNNI3K by Western blot analysis: ab86564 recognizes amino acids 756–805 and ab111140 recognizes amino acids 200–250 which includes ankyrin repeat domains 5 and 6 (Abcam, Cambridge, MA). A mouse monoclonal anti-human antibody against α and γ isoforms of muscle actin (HHF35, DAKO) was used as a positive control. For all antibodies, the chromogen used for detection was diaminobenzidine (DAKO). A negative control underwent the same procedure, but without the addition of primary antibody. All slides were counterstained with hematoxylin prior to microscopic examination. In addition, all tissue sections were stained with standard H&E for histopathological analysis.
Electron microscopy
The fixed tissue was rinsed three times in 0.1 M sodium phosphate buffer, post-fixed, and stained in 1% osmium tetroxide. The tissue was then rinsed three times in distilled water, en bloc stained in 2% aqueous uranyl acetate, dehydrated in a graded series of ethanol then absolute acetone. The tissues were subsequently infiltrated and embedded in epoxy resin. Semi-thin (0.6 µ) sections for light microscopy were cut with an ultramicrotome and stained with toluidine blue. Thin sections, ∼100 nm in thickness, were cut with an ultramicrotome, mounted on a copper grid and post-stained with 0.3% aqueous lead citrate prior to examination in a FEI Tecnai G2 12 transmission electron microscope operated at 80 kV. Digital images were captured with a Gatan Model 785 ES1000W Erlangshen side mount CCD camera and Digital Micrograph software.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (R01 HL071225 to T.O.), the Mayo Clinic Center for Individulized Medicine, and a generous gift from the Marriott Family.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the family and patients who participated in this study and the physicians who referred them. We thank Rhianna Sundsbak for her role in performing immunohistochemical experimentation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lloyd-Jones D.M., Wang T.J., Leip E.P., Larson M.G., Levy D., Vasan R.S., D'Agostino R.B., Massaro J.M., Beiser A., Wolf P.A., et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110:1042–1046. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 2.Darbar D., Herron K.J., Ballew J.D., Jahangir A., Gersh B.J., Shen W.-K., Hammill S.C., Packer D.L., Olson T.M. Familial atrial fibrillation is a genetically heterogeneous disorder. J. Am. Coll. Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y.H., Xu S.J., Bendahhou S., Wang X.L., Wang Y., Xu W.Y., Jin H.W., Sun H., Su X.Y., Zhuang Q.N., et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 4.Hodgson-Zingman D.M., Karst M.L., Zingman L.V., Heublein D.M., Darbar D., Herron K.J., Ballew J.B., de Andrade M., Burnett J.C., Olson T.M. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. New Engl. J. Med. 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Chen S., Yoo S., Chakrabarti S., Zhang T., Ke T., Oberti C., Yong S.L., Fang F., Li L., et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Karst M.L., Herron K.J., Olson T.M. X-linked nonsyndromic sinus node dysfunction and atrial fibrillation caused by Emerin mutation. J. Cardiovasc. Electrophysiol. 2005;19:510–515. doi: 10.1111/j.1540-8167.2007.01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson T.M., Michels V.V., Ballew J.D., Reyna S.P., Karst M.L., Herron K.J., Horton S.C., Rodeheffer R.J., Anderson J.L. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Spudich S., De Girolami U., et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 9.Theis J.L., Sharpe K.M., Matsumoto M.E., Chai H.S., Nair A.A., Theis J.D., de Andrade M., Wieben E.D., Michels V.V., Olson T.M. Homozygosity mapping and exome sequencing reveal GATAD1 mutation in autosomal recessive dilated cardiomyopathy. Circ. Cardiovasc. Genet. 2011;4:585–594. doi: 10.1161/CIRCGENETICS.111.961052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Meng X.M., Wei Y.J., Zhao X.W., Liu D.Q., Cao H.Q., Liew C.C., Ding J.F. Cloning and characterization of a novel cardiac-specific kinase that interacts specifically with cardiac troponin I. J. Mol. Med. 2003;81:297–304. doi: 10.1007/s00109-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 11.Wheeler F.C., Tang H., Marks O.A., Hadnott T.N., Chu P.L., Mao L., Rockman H.A., Marchuk D.A. TNNI3K modifies disease progression in murine models of cardiomyopathy. PLoS Genet. 2009;5:1–11. doi: 10.1371/journal.pgen.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H., Xiao K., Mao L., Rockman H.A., Marchuk D.A. Overexpression of TNNI3K, a cardiac-specific MAPKKK, promotes cardiac dysfunction. J. Mol. Cell. Cardiol. 2013;54:101–111. doi: 10.1016/j.yjmcc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vagnozzi R.J., Gatto G.J., Jr, Kallander L.S., Hoffman N.E., Mallilankaraman K., Ballard V.L.T., Lawhorn B.G., Stoy P., Philp J., Graves A.P., et al. Inhibition of the cardiomyocyte-specific kinase TNNI3K limits oxidative stress, injury, and adverse remodeling in the ischemic heart. Sci. Transl. Med. 2013;5:1–12. doi: 10.1126/scitranslmed.3006479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodder E.M., Scicluna B.P., Milano A., Sun A.Y., Tang H., Remme C.A., Moerland P.D., Tanck M.W.T., Pitt G.S., Marchuk D.A., et al. Dissection of a quantitative trait locus for PR interval duration identifies TNNI3K as a novel modulator of cardiac conduction. PLoS Genet. 2012;8:1–8. doi: 10.1371/journal.pgen.1003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai Z.F., Chen Y.Z., Chen J., Tsuda H., Kitamoto Y., Kim-Mitsuyama S. Mutation of TNNI3K gene increased incidence of arrhythmias in culture cardiomyocytes and in vivo mouse hearts. Circulation. 2012;126:A31. Resuscitation Science Symposium. Abstract. [Google Scholar]
- 16.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai Z.F., Chen Y.Z., Feng L.P., Meng X.M., Ding J.F., Wang L.Y., Ye J., Li P., Cheng X.S., Kitamoto Y., et al. Overexpression of TNNI3K, a cardiac-specific MAP kinase, promotes P19CL6-derived cardiac myogenesis and prevents myocardial infarction-induced injury. Am. J. Physiol. Heart Circ. Physiol. 2008;295:H708–H716. doi: 10.1152/ajpheart.00252.2008. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Wang J., Su M., Wang C., Chen J., Wang H., Song L., Zou Y., Zhang L., Zhang Y., et al. TNNI3K, a cardiac-specific kinase, promotes physiological cardiac hypertrophy in transgenic mice. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0058570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:1–13. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y., Cao H.Q., Liu Z., Ding J.F., Meng X.M. Identification of the dual specificity and the functional domains of the cardiac-specific protein kinase TNNI3K. Gen. Physiol. Biophys. 2007;26:104–109. [PubMed] [Google Scholar]
- 23.Van Durme J., Delgado J., Stricher F., Serrano L., Schymkowitz J., Rousseau F. A graphical interface for the FoldX forcefield. Bioinformatics. 2011;27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- 24.Olson T.M., Alekseev A.E., Moreau C., Liu X.K., Zingman L.V., Miki T., Seino S., Asirvatham S.J., Jahangir A., Terzic A. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat. Clin. Pract. Cardiovasc. Med. 2007;4:110–116. doi: 10.1038/ncpcardio0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally E.M., Golbus J.R., Puckelwartz M.J. Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Invest. 2013;123:19–26. doi: 10.1172/JCI62862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijering R.A.M., Zhang D., Hoogstra-Berends F., Henning R.H., Brundel B.J.J.M. Loss of proteostatic control as a substrate for atrial fibrillation: a novel target for upstream therapy by heat shock proteins. Front. Physiol. 2012;3:1–11. doi: 10.3389/fphys.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajasekaran N.S., Connell P., Christians E.S., Yan L.J., Taylor R.P., Orosz A., Zhang X.Q., Stevenson T.J., Peshock R.M., Leopold J.A., et al. Human αβ-crystallin mutation causes oxido-reductive stress and protein aggregation in cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry W.L., Gardin J.M., Ware J.H. Echocardiographic measurements in normal subjects from infancy to old age. Circulation. 1980;62:1056–1061. doi: 10.1161/01.cir.62.5.1054. [DOI] [PubMed] [Google Scholar]
- 29.Sobel E., Lange K. Descent graphs in pedigree analysis: application to haplotyping, location scores, and marker sharing statistics. Am. J. Hum. Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePristo M.A., Banks E., Poplin R.E., Garimella K.V., Maquire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The 1000 Genomes Consortium. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://eversusgs.washington.edu/EVS/ ) [October 2013]
- 34.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 35.Kiefer F., Arnold K., Kunzli M., Bordoli L., Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozakov D., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Vajda S. How good is automated protein docking? Proteins. 2013;81:2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olson T.M., Kishimoto N.Y., Whitby F.G., Michels V.V. Mutations that alter the surface of alpha-tropomyosin are associated with dilated cardiomyopathy. J. Mol. Cell Cardiol. 2001;33:723–732. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 39.Brauch K.M., Karst M.L., Herron K.J., de Andrade M., Pellikka P.A., Rodeheffer R.J., Michels V.V., Olson T.M. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.