Abstract
Template switching (TS) mediates damage-bypass via a recombination-related mechanism involving PCNA polyubiquitylation and Polymerase δ-dependent DNA synthesis. Using two-dimensional gel electrophoresis and electron microscopy, here we characterize TS intermediates arising in Saccharomyces cerevisiae at a defined chromosome locus, identifying five major families of intermediates. Single-stranded DNA gaps in the range of 150-200 nucleotides, and not DNA ends, initiate TS by strand invasion. This causes re-annealing of the parental strands and exposure of the non-damaged newly synthesized chromatid as template for replication by the other blocked nascent strand. Structures resembling double Holliday Junctions, postulated to be central double-strand break repair intermediates, but so far only visualized in meiosis, mediate late stages of TS, before being processed to hemicatenanes. Our results reveal the DNA transitions accounting for recombination-mediated DNA damage tolerance in mitotic cells and for replication under conditions of genotoxic stress.
DNA lesions compromise DNA replication, potentially leading to chromosome alterations and rearrangements 1,2. DNA damage tolerance (DDT) mechanisms uphold genome integrity by ensuring replication completion via fork recovery and gap-filling 3. Two distinct DDT modes are utilized in all organisms: an error-prone mode, largely accountable for mutagenesis and involving translesion synthesis 3, and an error-free recombination mode, known as template switching (TS), in which one newly synthesized strand serves as replication template for the other blocked nascent strand 4-7. PCNA mono- and polyubiquitylation are key regulators of DDT 8, promoting translesion synthesis 9 and mediating TS 7,10-13, respectively.
The molecular mechanism of TS is only partially understood. Studies in yeast and mammalian cells provided evidence that discontinuities are created in the newly synthesized strands during replication in the presence of damage, opposite the lesions 5,6,14. Repriming can reactivate transiently stalled forks in vitro 15, potentially providing a molecular explanation for the formation of gaps. Pulse-labeling experiments indicated that the gaps are then filled-in using the newly replicated strand as template 6. This notion was substantiated by various genetic and molecular studies that further identified the Rad5-pathway involving PCNA polyubiquitylation, as well as homologous recombination activities to be required in this process 4,5,13,16. Finally, X-shaped structures involving sister chromatid junctions and having the genetic dependencies of TS intermediates were identified using 2D gel electrophoresis during replication of damaged templates 7. Using this readout for TS, it was shown that a replication step mediated specifically by Polymerase δ is required 17, in congruence with other observations 6,18,19.
The RecQ helicase Sgs1 (BLM in mammalian cells), mutated in the cancer-prone Bloom syndrome, participates in error-free DDT by dissolving, in the context of the Sgs1-Top3-Rmi1 (STR) complex, the TS intermediates after their formation 7,19,20. Persistent TS structures can also be resolved by the Mus81-Mms4 nuclease or by overexpressing other bacterial or human resolvases 21-23. However, as all these enzymes can process a variety of substrates in vitro 24,25, the structural nature of TS intermediates has remained undefined. Two main models of TS - strand invasion via a recombination-primed mechanism, or annealing-mediated fork regression - propose as intermediates DNA structures in which the sister chromatids are paired 2,26. In vitro biochemical characterization of known TS factors provided evidence for either of these mechanisms 27,28,29, and single, double, nicked Holliday Junctions (HJs), as well as hemicatenane-like structures were proposed as potential intermediates 12,17,20,23,30.
Here we set out to identify the DNA structures and transitions involved in mediating error-free DDT by TS using a combination of 2D gel electrophoresis and electron microscopy (EM) studies. This question is biologically important for several reasons. First, TS is important for genome stability by preventing incorporation of mutations 1,3,8,31, which ultimately lead to genome instability and cancer 32. Indeed, various TS factors function as tumor suppressors 1,33. Second, DNA damaging agents, such as ultraviolet light, tobacco smoke and chemotherapeutics, which are potent inducers or occasionally target DDT pathways 8,19,34,35, cause specific mutational signatures associated with genome instability 32. Third, DNA gaps have been suggested as the predominant driver of recombination in mitotic cells, as opposed to double-strand breaks (DSBs) in meiosis 31,36-38, but the molecular mechanism through which ssDNA gaps are engaged in recombination remains elusive.
RESULTS
A linear minichromosome system to study TS-mediated DDT
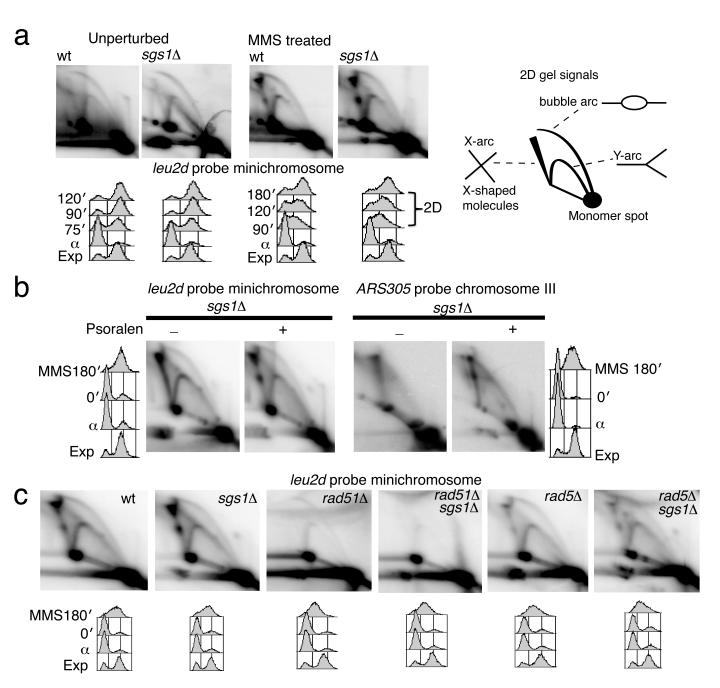
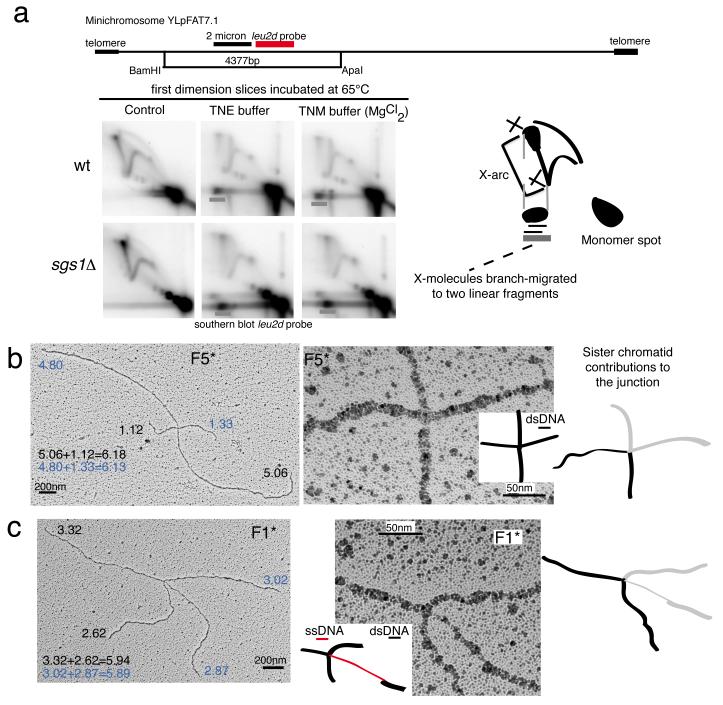
To obtain a homogenous population of TS intermediates for EM studies, we employed a 2μ–based 10.9Kb high-copy number minichromosome, YLpFAT7.1 (Supplementary Fig. 1a) 39. In Saccharomyces cerevisiae, YLpFAT7.1 minichromosomes are maintained as linear fragments, and are not entangled in concatemers (Supplementary Fig. 1b). To improve the specificity towards the intermediates of interest, we designed a restriction enzyme mix that reduces the average length of the endogenous genomic DNA to less than 5Kb (Supplementary Fig. 1c). We synchronized minichromosome-containing wt and sgs1Δ cells and then allowed them to replicate in the absence or presence of the DNA alkylating agent methyl-methanesulfonate, MMS (Fig. 1a). Similarly to endogenous chromosome replication, besides usual intermediates such as replication forks and bubbles, minichromosome replication in the presence of MMS induced the formation of X-shaped DNA structures, which further accumulated in sgs1Δ cells, proximal to the 2μ replication origin (Fig. 1a and Supplementary Fig. 1a). The migration pattern of these joint molecules (JMs) appeared as two prominent spots on the X-arc when we performed psoralen-crosslinking in vivo prior to genomic extraction, instead of the smooth X-arc signal observed in the absence of psoralen-crosslinking (Fig. 1b). TS intermediates arising on endogenous chromosomes showed the same migration pattern (Fig. 1b). These migration patterns are likely due to the fact that psoralen-crosslinking prevents migration of the junction point along the restriction fragment, stabilizing the initial X-shaped intermediates into specific spots 40. Importantly, we found that the minichromosome-derived JMs shared the same genetic requirements with classical TS intermediates 7,12, that is, their formation depended on the Rad51 and Rad5 pathways, in both wt and sgs1Δ backgrounds (Fig. 1c).
Figure 1.
TS intermediates formed on YLpFAT7.1 minichromosomes. (a) Genomic DNA samples isolated from wild-type (wt, CY11340) and sgs1Δ (CY11357) strains carrying YLpFAT7.1 minichromosomes and replicating in the presence of MMS 0.033% were analyzed by 2D gel electrophoresis using a minichromosome-specific probe, leu2d. (b) Genomic DNA samples from wt (CY11340), sgs1Δ (CY11357) cells carrying the minichromosome YLpFAT7.1, and the corresponding strains without minichromosomes, wt (CY12486) and sgs1Δ (CY13249), were analyzed by 2D gel electrophoresis using probes for the minichromosome (leu2d) or the early origin of replication on the endogenous chromosome III (ARS305). (c) The genetic dependency of minichromosome-derived X-molecules was analyzed in wt (CY11340), sgs1Δ (CY11357), rad51Δ (CY12388), rad5Δ (CY12777), sgs1Δ rad51Δ (CY12390), sgs1Δ rad5Δ (CY12775).
Isolation and assignment of TS intermediates
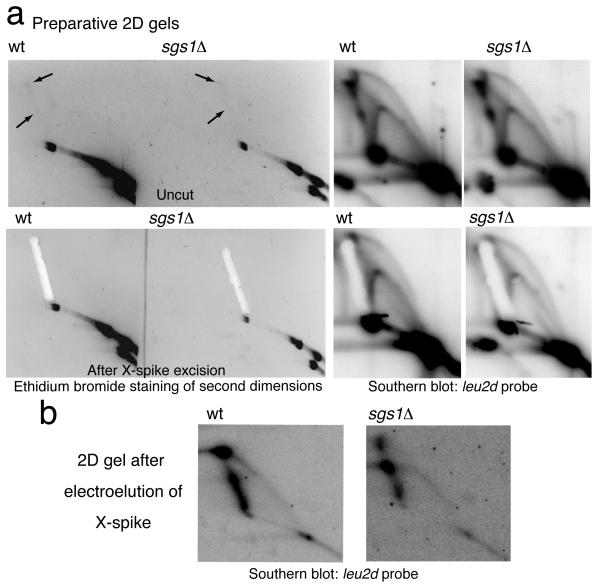
To visualize TS intermediates by EM, we psoralen-crosslinked in vivo genomic DNA of wt and sgs1Δ cells 40, enriched for minichromosome-derived DNA by cleaving the endogenous chromosomes (Supplementary Fig. 1c), and further digested with HindIII before running preparative 2D gels (Fig. 2a). We then excised the X-shaped signal from the ethidium bromide-stained second dimension gels (Fig. 2a), purified the DNA by electroelution, and confirmed via a subsequent 2D gel the migration pattern as branched molecules (Fig. 2b).
Figure 2.
X-molecule intermediate isolation and purification. (a) Typical preparative 2D gels of psoralen-crosslinked genomic DNA of wild-type (wt, CY11340) and sgs1Δ (CY11357) cells carrying the minichromosome. The black arrows indicate the X-spike spots visible after ethidium bromide staining. Agarose fragments containing the X-arcs were extracted. Uncut gels were kept and hybridized in parallel using the leu2d probe. (b) The DNA contained in the agarose fragments extracted from the 2D gels in panel (a) was isolated by electroelution and run on a subsequent 2D gel.
We proceeded with EM analysis of DNA samples obtained from two independent biological replicates. As we observed similar trends for all the crucial parameters described hereafter, in the following, we present the results obtained by pooling the number of JMs analyzed. We identified over 300 JMs for each wt and sgs1Δ cells (Supplementary Fig. 1d). To ensure strict analysis of minichromosome-derived structures, we measured the length for all the initially identified JMs (649). The assignment procedure, which allowed a 10% tolerance range in measurements, considered that minichromosome-derived TS intermediates should represent two HindIII 5.8Kb inter-connected fragments, and the branches, taken two to two, should be the size of one HindIII fragment 41. In this way, we unequivocally assigned 43% (wt) and 51% (sgs1Δ) out of the initially identified X-molecules as minichromosome-derived intermediates (Supplementary Fig. 1d). The JMs excluded by this assignment procedure are likely broken minichromosome-derived DNA or endogenous genomic DNA contaminants, caused by partial digestion. We further classified the assigned minichromosome-derived JMs into symmetric molecules—in which the junction point was stabilized by homologous pairing and two to two or all the four branches had the same length—, or asymmetric, with junction points not stabilized by homologous pairing and all four branches of different length or only two equal arms.
Structural and genetic features of symmetric JMs
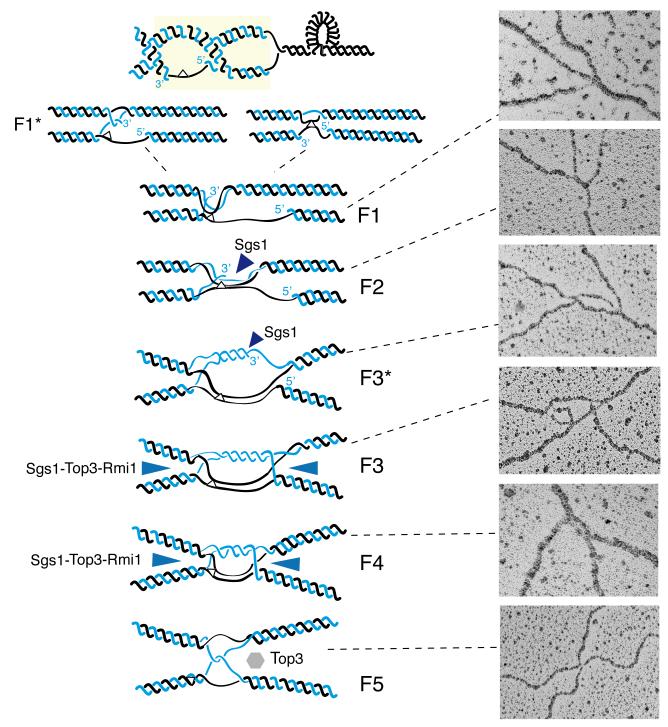
Based on the structural features of the identified symmetric JMs, which accounted for 84% of the JMs, we classified these intermediates in 5 prominent families, F1 to F5 (Table 1). Besides the F1-F5 families, a small group of single Holliday Junction (SHJ)-like structures (8/306) was observed (Table 2). We also classified the asymmetric JMs in two families, F1* and F5*. Table 2 summarizes the number and percentage of JMs belonging to each family.
Table 1.
Families of symmetric template switch intermediates.
| Family type | Schematic representation | Structural features | Described |
|---|---|---|---|
| F1 |
|
-simple cross -ssDNA gap proximal to the junction point |
Figure 3, Suppl.2 |
| F2 |
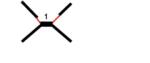
|
-double Y or double Holliday Junction (HJ)-like structure -ssDNA gap on one or two branches proximal to the junction point -thick DNA filament (1) at the junction point |
Figure 3, Suppl.3 |
| F3 |

|
-double Holliday Junction (HJ)-like structure -two dsDNA filaments (1 and 2) of the same length connecting the two HJ-like crossover points |
Figure 4, Suppl.4 |
| F4 |
|
-double Y or double Holliday Junction (HJ)-like structure -thick DNA filament (1) at the junction point |
Figure 4, Suppl.6 |
| F5 |
|
-simple cross -no ssDNA discontinuities proximal to the junction point |
Figure 4, Suppl.7 |
Table 2.
Percentage and number of joint molecule families in wt and sgs1Δ cells.
| Strains and JM families | F1* | F1 | F2 | F3* | F3 | F4 | F5 | F5* | SHJ | Total | Percentage and number of JMs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| wt |
7.3 11 |
15.9 24 |
3.3 5 |
0.7 1 |
4.6 7 |
8.6 13 |
40.4 61 |
15.2 23 |
4 6 |
100 151 |
% number |
| sgs1 Δ |
2.6 4 |
14.8 23 |
13.5 21 |
5.8 9 |
15.5 24 |
18.7 29 |
20.6 32 |
7 11 |
1.3 2 |
99.8 155 |
% number |
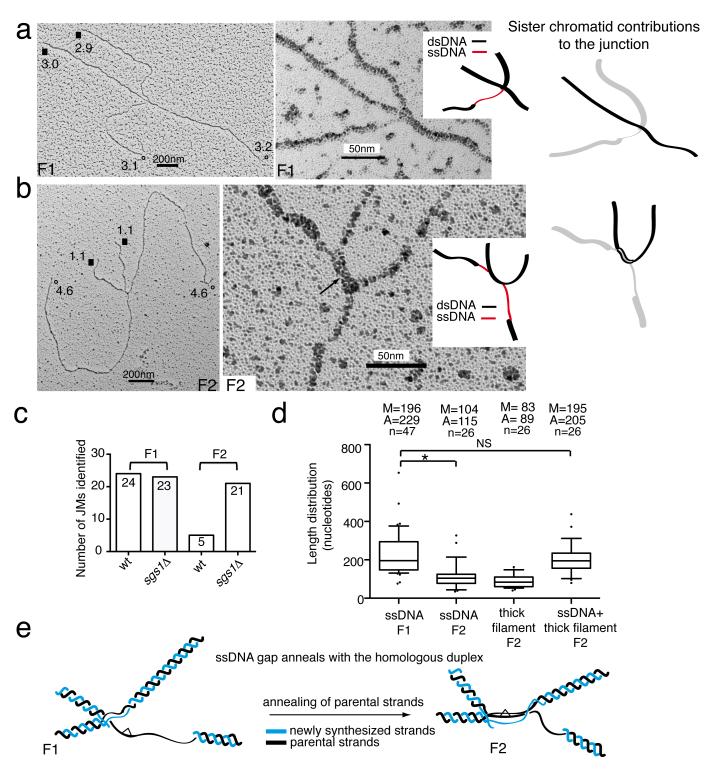
We noted that two of the identified symmetric families were characterized by the presence of a ssDNA region proximal to the junction point, on one or two of the four branches comprising the JM. Because of the wealth of genetic and molecular information associating TS with gap-filling, we reasoned that the early intermediates are likely to contain such discontinuities close to the junction point. We called these families F1 and F2 (Table 1). The junction point in F1 resembled a cross between two linear dsDNA filaments, and there was a ssDNA stretch on one branch immediately proximal to the junction point (Fig. 3a and Supplementary Fig. 2), generally of 150-200 nt in length (Supplementary Fig. 1e). This family of molecules was equally distributed in both wt and sgs1Δ cells (Table 2 and Fig. 3c). The junction point in the F2 family involved a longer homology region and contained a thick DNA filament (Table 1 and Fig. 3b). Notably, F2 molecules contained a stretch of ssDNA on one or often two branches of the JM close to the junction point (Fig. 3b and Supplementary Fig. 3) and were four times more prominent in sgs1Δ than in wt (Fig. 3c). We examined how F1 and F2 families were related by measuring the distribution of ssDNA length and other parameters (Fig. 3d). The median value of the ssDNA regions in F2 molecules (considering all molecules identified in both wt and sgs1Δ) was 104 nt, shorter than the median of the ssDNA region of F1 molecules (196 nt) (Fig. 3d). Notably, the median value of the sum of the ssDNA regions and the thick DNA filament present at the junction point in F2 molecules (195 nt) was very similar, in the 10% tolerance range, to the median value of ssDNA regions found in F1 molecules (196 nt) (Fig. 3d). When instead of the median values, we employed the average values for the parameters above the same pattern emerged (Fig. 3d). Moreover, the same was true when we performed the above calculations only for the F1 and F2 molecules found in sgs1Δ (the number of F2 molecules in wt was too low compared to the F1 molecules in the same background to make such analysis meaningful, Fig. 3c) (data not shown). These results indicate that the F2 molecules derive from F1, by partial annealing of the gap region into the homologous duplex, leading to a double Y-like DNA structure (Fig. 3e).
Figure 3.
Representative EM pictures of F1 and F2 families. The analyzed molecules derive from two independent biological experiments. (a-b) Total views of the indicated X-shaped DNA structures and enlarged views with a schematic representation of the junction point (ssDNA regions in red and dsDNA regions in black). The branch sizes until the junction point are reported in kilobases. Equal arms are labeled, respectively, with open circles (°) and black squares (■). A schematic representation of the junction point is also shown, in which the two DNA duplexes that contribute to the junction are reported, respectively, in black and light grey. The black arrow in panel b marks the thick DNA filament at the junction point that we infer to be three-stranded. Scale bars are shown. (c) Chart showing the number of F1 and F2 molecules identified in wt and sgs1Δ cells. (d) Box plot of the described parameters. Center-line represents the middle value of the data set (median), box limits represent 25th-75th percentile, whiskers indicate 10th-90th percentile, outliers are indicated by black dots. n represents the number of samples (molecules) of the data set. The median (M), average (A) and number of samples (n) values are displayed. * indicates P< 0.05 by t test, NS, not significant. (e) Schematic representation of the F1 to F2 transition. Parental strands and newly synthesized strands are shown in black and blue, respectively. A hypothetical bulky DNA lesion is represented by a white triangle.
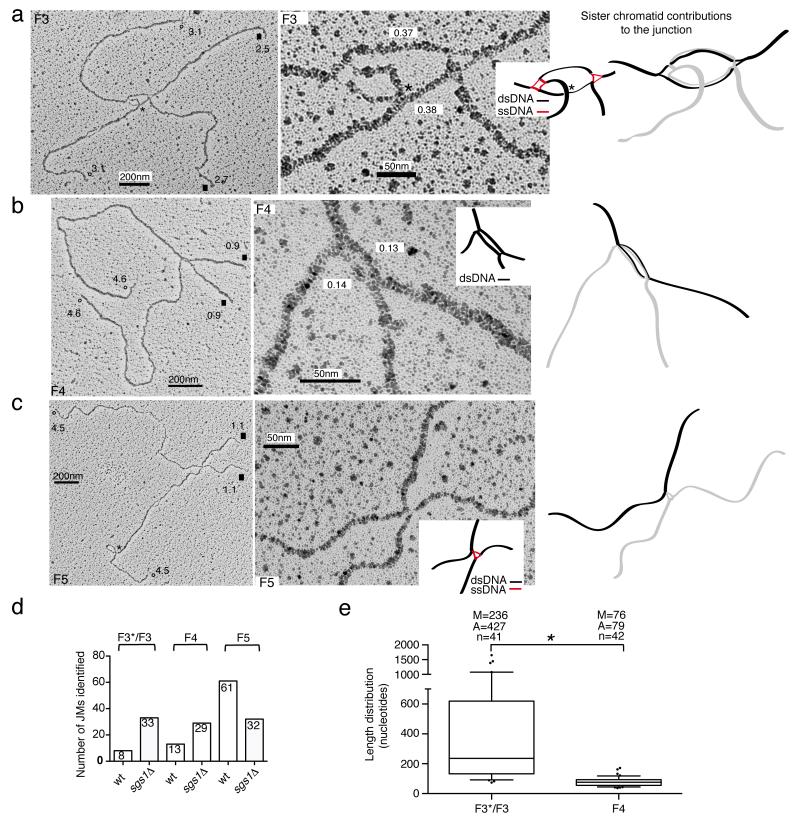
The other symmetric families we identified did not contain ssDNA discontinuities on the branches (Table 1). The F3 family structurally resembled double HJs (dHJs). The junction point was similar to a bubble and contained two dsDNA-like filaments of roughly the same length, connecting the two HJ-like crosses, occasionally seen as rhomboid structures with an open center at the crossover position (Fig. 4a and Supplementary Fig. 4). The distance between the two HJ-like centers generally ranged from 0.15 to 0.6 kb, but occasionally was longer (Fig. 4e). We also found molecules having both F2 and F3 features, called here F3*. The junction point in F3* still resembled a bubble, but we could always identify a ssDNA discontinuity on one of the two dsDNA filaments at the junction point (Supplementary Fig. 5), likely indicating ongoing DNA synthesis on one of the strands. Based on the length of the duplex filaments at the F3 junction (Fig. 4e) and the ssDNA length in F1 molecules (Fig. 3d), we inferred that the DNA synthesis step implicated in TS 6,17,19 often proceeds beyond the initial size of the gap, concomitantly leading to more extensive annealing of the parental strands (as two DNA duplexes are present between the crossover junctions in F3 and one duplex contains the newly replicated strands, it follows that the other duplex must contain the paired parental strands, a notion for which we provide further evidence below).
Figure 4.
Representative EM pictures of F3, F4 and F5 families. The analyzed molecules derive from two independent biological experiments. (a-b-c) Total views of the indicated X-shaped DNA molecules and enlarged views with a schematic representation of the junction point (ssDNA regions in red and dsDNA regions in black). Branch sizes until the junction point are reported in kilobases. Equal arms are labeled, respectively, with open circles (°) and black squares (■). The junction points of the molecules are also shown with a black or light gray code as in Fig. 3. Black asterisks mark the position of random crosses between DNA filaments. The length of the dsDNA filaments visible at the F3 and F4 junction points is reported in kilobases. Scale bars are shown. (d) Chart showing the number of F3*/F3, F4 and F5 molecules identified in wt and sgs1Δ cells. (e) Box plot showing the length distribution of the dsDNA filaments at the junction points of F3*/F3 and F4 molecules. Center-line represents the middle (median) value of the data set, box limits represent 25th-75th percentile, whiskers indicate 10th-90th percentile, outliers are indicated by dots. n represents the number of samples (molecules) of the data set. The median (M), average (A) and number of samples (n) values are displayed. * indicates P< 0.05 by t test.
F4 resembles double Y or dHJs 42 (Table 1), and the junction point contains a small, almost closed bubble (Fig. 4b and Supplementary Fig. 6). A DNA filament, generally around 80 nt, of a thickness greater than the one of a typical dsDNA filament and likely comprised by two dsDNA filaments, was present at the junction point (Figs. 4b and 4e).
The F5 molecules resemble a cross between two dsDNA filaments (Table 1, Fig. 4c and Supplementary Fig. 7). Occasionally, we detected at the junction point (and not on the branches as in F1) ssDNA filaments that appeared to connect the two DNA duplexes implicated in the junction (Supplementary Fig. 7c).
Besides the F1-F5 families, we observed a few symmetric JMs with structural features of single HJs (SHJs) (8/306, Table 2 and Supplementary Fig. 8a). We suspect that they represent the products of recombination events triggered by occasional nicks arising during replication or induced by MMS lesion processing. From a structural standpoint, it is possible that they represent reversed forks, in which the length of the reversed arm is equal to the non-replicated strand. However, besides the low likelihood for the above, we note that six out of the eight identified SHJ-like molecules were in wt, and previous EM studies of total intermediates did not identify reversed forks under these conditions 14.
As the symmetric families structurally resembled HJs, we set out to investigate their biochemical properties. Incubation at high temperature causes JMs to branch-migrate and be resolved into linear DNA when the branch points reach the ends 42-44. However, branch-migration of canonical HJs, such as synthetic HJs and meiotic recombination intermediates containing HJs, is prevented by addition of magnesium cations 45, which stabilize HJs in a stacked-X configuration 45,46. We found that the minichromosome-derived TS intermediates arising in both wt and sgs1Δ cells showed comparable branch-migration activity regardless of the presence of magnesium cations in the buffer (Fig. 5a). Based on this feature, previously reported also for TS intermediates formed on endogenous chromosomes 20,30, we conclude that the large fraction of TS intermediates are not canonical HJs.
Figure 5.
Branch migration assays and asymmetric families of JMs. (a) Branch migration assay. Genomic DNA from wt (CY11340) and sgs1Δ (CY11357) strains carrying the minichromosome YLpFAT7.1 was extracted, digested with ApaI and BamHI, and run in triplicate. The first dimension gel slices were incubated in buffer with or without magnesium at 65°C 43, before second dimension and subsequent analysis with the leu2d probe. The experiments were conducted three times with qualitatively identical results. A schematic representation of the expected position of the spot caused by branch migration of X-molecules to two linear fragments in 2D gels is shown, together with a schematic representation of the restriction fragment analyzed. (b-c) Representative EM pictures of F5* and F1* families of X-molecules. The analyzed molecules derive from two independent biological experiments. Total views of the indicated X-shaped DNA structures and enlarged views with a schematic representation of the junction point (ssDNA regions in red and dsDNA regions in black). Branch sizes until the junction point are reported in kilobases. Length values indicated with a black or blue code indicate the pairs of branches leading to the size of one minichromosome-derived HindIII fragment. The junction points of the molecules are also represented with a black and light grey code, to show the contributions of the two DNA duplexes to the junction. Scale bars are shown.
Notably, we observed the same families of DNA structures, both in what regards the symmetric and the asymmetric junctions (see below) in sgs1Δ and wt cells (Table 2). As sgs1Δ samples were specifically enriched in F2-F3-F4 families (Table 2, Figs. 3c, 4d, and Supplementary Fig. 1e), we conclude that these types of DNA structures are substrates for Sgs1 in vivo.
Structural and genetic features of asymmetric JMs
Besides the symmetric molecules, 16% of junctions were asymmetric (49/306). We could divide them in two structural categories. The JMs of one of these categories, named F5* hereafter, had four branches of different length, no ssDNA discontinuities on the branches, and the junction points resembled either crosses between two linear DNA fragments or assumed more complex structures in which three ssDNA filaments forming a triangle-like structure were visible (Fig. 5b and Supplementary Fig. 8b). As the junction point was not stabilized by homologous pairing, we could rule out F5* being SHJs, congruently with the results of the branch-migration assays (Fig. 5a). Structurally, F5* molecules resembled hemicatenanes and the symmetric F5 molecules. The relative percentage of F5 and F5* versus other JMs was increased in wt versus sgs1Δ (Table 2 and Supplementary Fig. 1e), in line with the notion that Sgs1 is not required for the last step of hemicatenane resolution 47. Moreover, the abundance of F5 and F5* structures in wt in relation to other JMs (Table 2 and Supplementary Fig. 1e) suggests that hemicatenane-processing may not be very efficient in vivo. The time window from the formation of symmetric F5 hemicatenanes to their final processing may give opportunity for slippage of the junction point, leading to the formation of asymmetric hemicatenane-like F5* molecules, wherein two dsDNA filaments are connected by non-homologous pairing (Fig. 5b and Supplementary Fig. 8b). We labeled this family F5* to illustrate their possible origin from F5 molecules. Substantiating this view, we observed the same nearly 50% decrease in number and relative percentage of F5* molecules in sgs1Δ compared to wt, as we did for F5 (Table 2 and Supplementary Fig. 1e).
We note that a subset of the F5* molecules (6/23 and 5/11 in wt and sgs1Δ, respectively, 11/306 of the total JMs) had two branches of roughly the same length, and therefore might represent reversed forks. The relative scarcity of JMs with structural properties compatible with those of reversed forks (less than 4% of the JMs) found here and their absence in previous EM studies 14, largely support the postreplicative gap-filling model of TS 6,7,17,19,35.
The other category of identified asymmetric JMs structurally resembled both hemicatenanes and F1 molecules in that they contain a ssDNA discontinuity, of about 150-200 nt, on one of the branches, proximal to the junction point (Fig. 5c and Supplementary Fig. 8c). We labeled these intermediates F1* to illustrate the possibility, inferred from their structural features, of them being F1 precursors. We envisage that replication-associated hemicatenanes 44 may become stabilized into symmetric junctions (F1) when encountering a ssDNA gap 48,49, thus facilitating strand invasion of the gap-region into the homologous sister duplex 48.
Pseudo-Double Holliday Junctions mediate TS
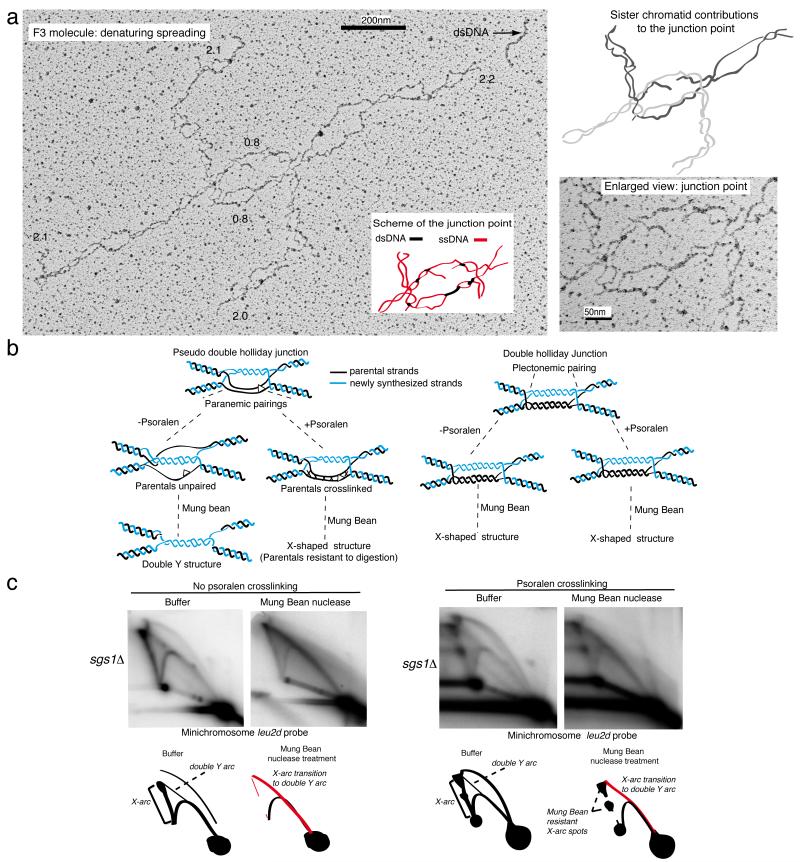
dHJ-like structures of the F3-F4 families could arise during gap-filling if besides the plectonemic pairing of the newly synthesized DNA filaments, the parental strands also anneal in vivo (paranemic pairing, Fig. 6b, left panel). To analyze if each of the dsDNA filaments present at the junction point in F3 molecules contain two ssDNA filaments paired in vivo (as opposed to them being together because of occasional nicking induced by the spreading procedure), we analyzed similarly isolated JMs under denaturing conditions. In this assay, large multi-protein complexes - mostly nucleosomal histones - tightly bound to DNA molecules protect the ssDNA filaments of DNA duplexes from being psoralen-crosslinked and, under conditions of denaturing spreading, their binding sites appear as strings of ssDNA bubbles, connected by duplex regions representing the unbound linker dsDNA that was psoralen-crosslinked 50. As a proof of principle, we identified several molecules with F3/dHJ features, whose examination revealed that the ssDNA strands composing each of the two duplexes present at the junction point in F3 molecules (see Fig. 4a) are crosslinked and thus paired in vivo (Fig. 6a).
Figure 6.
Observation of F3 molecules by denaturing spreading and biochemical identification of paranemic pairing. (a) Entire and enlarged view of the junction point of an F3 molecule following denaturing spreading. The analyzed molecules derive from two independent biological experiments. A schematic representation of the junction point is shown, with ssDNA regions in red and dsDNA filaments in black. The black arrow indicates a heavily crosslinked dsDNA filament that can be used as reference for the thickness of a dsDNA filament in this experimental condition. A schematic representation of the junction point showing the contribution of the ssDNA filaments from the two DNA duplexes (labeled, respectively, in black and light gray) to the junction point is shown. Branch sizes until the junction point and the length of the DNA filaments in the junction point are also reported (in black) in kilobases. Scale bars are shown. (b) Schematic representations of the possible outcomes of the Mung Bean nuclease treatment on pseudo double Holliday junctions or classical double Holliday junctions, with or without in vivo psoralen-crosslinking. (c) Genomic DNA from sgs1Δ (CY11357) cells carrying the minichromosome YLpFAT7.1 and replicating in the presence of MMS was extracted after psoralen-crosslinking in vivo or without psoralen-crosslinking. Mung-bean treated and non-treated samples were run on 2D gels in parallel. The experiments were conducted three times with qualitatively identical results. Schematic representations of the 2D gel signals with the transitions from X-arcs to double Y-arcs are shown.
This type of structure is compatible with F3 molecules being either canonical double HJs, in which the two dsDNA filaments connecting the two HJ crosses are stabilized by plectonemic pairing (Fig. 6b right panel, Double Holliday Junction) or with a structure in which the parental ssDNA strands are connected to each other only by paranemic pairing, for instance by the action of Rad51 (Fig. 6b, left panel, Pseudo-Double Holliday Junction). We reasoned that the two types of structures could be distinguished if the chromosomal DNA was prepared without psoralen-crosslinking in vivo: dHJs would structurally remain the same, whereas the paranemic pairing in pseudo-dHJ structures would be dissolved exposing the two parental ssDNA filaments, which upon Mung Bean nuclease treatment should be digested and yield a double Y-like structure (Fig. 6b, left panel). Consistent with this latter prediction and resembling the behavior of TS intermediates arising on endogenous chromosomes 20,30, Mung Bean nuclease treatment of minichromosome-enriched DNA prepared in the absence of in vivo psoralen-crosslinking led to a clear transition from the X-arc to a double Y structure (Fig. 6c, left panel). In contrast, the psoralen-crosslinked samples were much more resistant in this regard (Fig. 6c, right panel). Based on these biochemical and structural features, we propose that the F3 molecules accumulating in sgs1Δ and mediating TS are pseudo dHJs.
DISCUSSION
HJs are the most discussed hypothetical intermediates for recombination, modeled for DSB repair 51, but to date only visualized in meiosis 42. Here we provide evidence that non-canonical HJs (pseudo dHJs) are implicated in recombination-mediated gap-filling by TS. Analysis of the structural features of early intermediates (F1 and F2) revealed that a substantial part of the gap region becomes paired with the homologous duplex containing the newly synthesized filament (see evidence for paranemic pairing in Fig. 6), leading to a three-stranded DNA structure (F2, Figs. 3e and 7). The features of F2 molecules are consistent with a structure in which the gap region begins to be aligned with the homologous region of the sister duplex, but DNA synthesis did not begin (Figs. 3b, 3e and 7). Re-annealing of the parental strands in F2 would facilitate the displacement/exposure of the non-damaged newly synthesized filament, which would then become available as a DNA synthesis template for the stalled 3′end proximal to the gap region (Figs. 3b and 7). Subsequent extension of the 3′end would give rise to F3*-like molecules, in which DNA synthesis is ongoing only on one filament of the F3 junction point (Supplementary Fig. 5 and Fig. 7). As annotated in the features of the observed symmetric structures, measurements of the ssDNA regions and of the thick DNA filaments present at the junction point in F1-F2 molecules fully support this view (Fig. 3).
Figure 7.
Hypothetical model of TS. Replication is associated with the formation of precatenanes and hemicatenanes. Parental DNA strands are in black, newly synthesized DNA strands in blue, a hypothetical bulky DNA lesion is represented by a white triangle. Hemicatenation (in the context of precatenation) assists pairing of the gap region with the homologous duplex (F1*, F1). The damaged strand containing a ssDNA gap partly anneals with the homologous duplex (F2). The 3′ end of the newly synthesized strand proximal to the gap anneals to the non-damaged newly synthesized template and DNA synthesis is initiated (F3*). Second end capture leads to the formation of double HJ-like structures (F3) that are dissolved by the action of Sgs1-Top3-Rmi1 (represented as blue triangles) to F4 molecules (pseudo-double HJs with the crossover points close to each other) and F5 (symmetric hemicatenanes) intermediates. Representative EM pictures showing enlarged views of the junction points of the X-molecules corresponding to each major TS intermediate family are shown.
Alternative to the model and interpretation above, is that the F2 molecules already illustrate DNA synthesis-mediated gap-filling initiated by the invasion of the transiently stalled 3′ end into the homologous sister duplex and the formation of a D-loop-like structure. A caveat with this interpretation is that parental strand annealing is not necessarily expected to happen in this scenario, but it must be invoked to explain the subsequent paired parental strands observed in F3-F4 molecules (see Fig. 6a). Such pairing could happen if elongation of the D-loop—which this model envisages to happen already in F2 molecules, is coupled with annealing of the parental strands. However, such an interpretation is incongruent with the presence of ssDNA filaments on both sides of the junction point in F2 molecules (Fig. 3b). Thus, the data support the gap-mediated model of strand invasion, which is also consistent with other previous genetic deductions 17,19,37,48,52.
Regardless, however, of whether TS is initiated by the gap region or by the adjacent 3′end close to the gap, completion of TS DNA synthesis can be followed by second-end capture, with formation of dHJ-like structures, or by strand displacement that would lead to linear molecules and non-crossover products. In agreement with the inference that F3 molecules represent dHJ-like molecules that are resolved by the STR complex 20,47,53,54, we found an increased percentage of F3 molecules in sgs1Δ (15.5%) as compared to wt (4.6%) cells (Table 2). Besides F3-F4 types, F2 and F3* families also accumulated in sgs1Δ samples (Table 2), revealing that Sgs1 also potentiates D-loop displacement (Fig. 7), similar to what was proposed for the mammalian RecQ helicases RecQL5 and BLM 55,56. We thus propose that both dHJ dissolution and strand displacement account for Sgs1 roles in suppressing sister chromatid exchanges and mitotic crossover 54,57 and in facilitating TS intermediate resolution 7,22.
Dissolution of dHJ-like intermediates by STR is expected to ultimately lead to symmetric hemicatenanes or F5-like molecules (Fig. 7), later resolved by Top3. In addition to F5 structures, we detected intermediates of the F3 dissolution process (F4, Figs. 4b and 7), which still accumulated in sgs1Δ cells (Figs. 2c and 4d). This indicates that the last step of dissolution is slow, likely because of topological constrains, and that Sgs1 facilitates Top3 activity also in this context 58 (Fig. 7).
Previous work on factors that influence TS intermediate resolution failed to reveal a role for factors such as Srs2 that are important for D-loop unwinding 25,54,57. Why problems in this early step do not translate into X-molecule accumulation is not well understood. Based on these new results, it seems likely that a combination of events—redundancy of D-loop unwinding activities, very efficient pathways of second end capture and subsequent dissolution of dHJ-like intermediates, as well as sensitivity limits of previously conducted 2D gel assays, is the underlying reason. In this new light, it also appears likely that the TS-intermediate accumulation in smc5-smc6-mms21 mutants 59,60 is an indication that Smc5-Smc6-Mms21-mediated SUMOylation influences dHJ dissolution, possibly by facilitating STR activity or by enabling the formation of alternate Top3 complexes with similar in vivo activity with STR 59,60. The method used here and the advances in the structural studies of replication and recombination intermediates promise to markedly contribute at unraveling how different genome integrity pathways—which show similar phenotypic outcomes when individually ablated- are integrated to support basic chromosome functions such as DDT and replication completion.
ONLINE METHODS
Cell culture, psoralen crosslinking and genomic DNA extraction for 2D gels
The yeast strains carrying the minichromosome YLpFAT7.1 39 were grown in synthetic media without leucine, arrested in G1 using alpha factor (SIGMA, 4 μg/ml) and released in YPD media or YPD media containing 0.033% MMS. Cells were fixed in 0.1% sodium azide for 40 minutes on ice as described 7,44,49. The in vivo psoralen-crosslinking, the genomic DNA preparations (CTAB method), and the neutral-neutral 2D gels were performed as previously described 7,44. In brief, for the in vivo psoralen-crosslinking cellular pellets corresponding to 1-2 × 109 cells were washed and resuspended in 5 ml of sterile cold water. Four cycles of 10 minutes each of in vivo psoralen-crosslinking were performed by placing the cell suspensions (transferred in 6 well-plastic tissue culture plates with flat bottom) in a Stratagene Stratalinker 1800 irradiation chamber, under 365 nm (UV). Before each irradiation cycle a fresh 0.3 ml aliquot of psoralen (4, 5′, 8 trimethyl-psoralen (TMP) 0.2 mg/ml dissolved in ethanol) was added to the cell suspension, mixed with cut blue tips and incubated for 5 minutes under aluminum foil. During the entire procedure, the plates containing the cell suspensions were placed on ice. Following this procedure, the cell suspensions were transferred to 50 ml Falcon tubes, the pellets were washed twice with sterile cold water and genomic DNA was extracted as previously described 43.
Preparative 2D gels, electroelution and EM
For preparative 2D gels, 50 to 100 μg of genomic DNA was digested with a restriction enzyme mix containing PacI, XhoI, SgrAI, BssHII, SpeI, BclI, PmlI and HindIII, precipitated with potassium acetate /isopropanol and suspended in TE 1X or Tris-HCl 10mM pH 8. Digested genomic DNA was run on Owl Separation Systems, Inc Portsmouth, NH, USA, Model A2 electrophoresis chambers (gel tray 27×20cm-2.5L buffer) using TBE 1X not autoclaved and agarose gels of 500 ml. The first dimensions (0.35% agarose gels) were run at 50V for 24 hours at room temperature, and the second dimensions (0.9% agarose gels) for 12 hours at 150V (current limited at 150mA) at 4°C. For electroelution, agarose fragments were run in TBE 1X for 5 hours at 100V at room temperature in an Elutrap chamber (BioRad) with BT-1 and BT-2 membranes following the manufacturer’s instructions. Recovered DNA solutions (around 0.8 ml) were concentrated on conical Amicon Ultra centrifugal filters (0.5ml 100K-MWCO 100K) following manufacturer’s instructions. DNA samples from the electroelution were subjected to DNA spreading on carbon coated metal grids (4nm thickness) in the presence of uranyl acetate followed by platinum-based rotatory shadowing (0.4 nm without rotation and up to 8nm with rotation) and subjected to EM analysis as described 41.
Branch migration and Mung Bean nuclease digestion assays
Branch migration and Mung Bean nuclease digestion assays were conducted as previously described 43. For branch migration assays, the first dimension slices were incubated in TNE buffer (50mM NaCl, 10mM Tris-HCl pH 8, 0.1mM EDTA pH 8) or TNM buffer (50mM NaCl, 10mM Tris-HCl pH 8, 10mM MgCl2, 0.1mM EDTA pH 8), for 13 hours at 65°C. For Mung Bean nuclease assays, 12 μg of digested genomic DNA was precipitated with potassium acetate/isopropanol and resuspended in Mung Bean nuclease buffer 1X. 40 units of Mung Bean nuclease (New England Biolabs) were added to each sample followed by incubation for 75 minutes at 37°C. DNA was precipitated again, resuspended in Tris-HCl 10mM pH 8 and first and second dimension gels were run as described above.
Assignment procedure for ssDNA regions on the DNA molecules analyzed
The assignment criteria for a ssDNA region on the DNA molecules analyzed by the EM procedure used here was recently described 41. We note that in order to assign a ssDNA region on a DNA filament it is necessary to identify two points on the DNA molecule that define the borders of the ssDNA region, in which the thickness of the DNA filament decreases close to one half. The thickness of the DNA filament between these two points can slightly vary due to the positioning of the filament during the spreading procedure, but in general it is clearly lower than the average value of the thickness of a dsDNA filament, which in these experimental conditions is distributed around 20 Angstroms. While the difference between double-stranded and single-stranded DNA is promptly detected, it is generally difficult to visually distinguish thickness differences in multi-strand (>2) DNA. There are two technical explanations behind this: the first one is the reduced relative difference in thickness, a factor 2 from single- to double stranded transition, while only a factor of 1.5 from double to three stranded DNA. The second reason relates to the DNA visualization procedure: it was also previously shown that the combination of uranyl acetate staining and platinum rotatory shadowing used in this work is instrumental to appreciate the difference in thickness between dsDNA filaments and ssDNA filaments 41. This is because the Platinum rotary shadowing only stains double stranded DNA, while the Uranyl atoms positively stain both single- and double-stranded DNA. Thus, the observed difference in thickness of the stained molecules - largely determined by the amount of deposited heavy atoms - generally exceeds the difference in thickness of the biological material itself. For the same reasons, three-stranded DNA (inferred to be present at the junction point in F2 molecules) will not be covered by much more heavy atoms than double-stranded, as the latter is already abundantly recognized by both staining procedures, reducing the relative contribution to thickness of an additional DNA strand.
Length measurements
The length measurements were performed using a conversion factor expressed in graphic units per base pairs/nucleotides (automatically converted in nanometers/nucleotide according to the magnification value at which the picture was taken), obtained by the measurement in pixels of a plasmid molecule of known length used as internal standard, as detailed in 41. We note that the length of the ssDNA stretches reported here, using the same conversion factors for both dsDNA and ssDNA, might have been underestimated by a maximum experimentally determined factor of 15%, calculated for extreme conditions of long and entirely single-stranded DNA molecules that show reduced anchorage to the carbon layer and have different stretching properties.
Assignment procedure for the identified joint molecules
The assignment procedure of JMs considers that the total size of the X-molecules (the sum of the branches and, when applicable, of the extended homology region) must fall in the interval 10584bp-12953bp, which corresponds to the size of two HindIII 5.88 Kb linear fragments interconnected (see Supplementary Fig. 1a and 41). Moreover, the branches of an X-molecule taken two to two, together with the extended homology region for molecules in the category F2, F3 and F4, must have a length that falls in the interval 5292bp-6468bp, which corresponds to the size of one HindIII fragment.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by European Research Council (ERC), Associazione Italiana per la Ricerca sul Cancro (AIRC) and Fondazione Telethon grants to DB, the Swiss National Science Foundation grant PP00P3_135292 to KZ, CF and ML, AIRC and Fondazione Telethon grants to MF. We thank the Center for Microscopy and Image Analysis of the University of Zurich for technical assistance with the EM experiments, I. Psakhye for critical reading of the manuscript, W. Carotenuto at IFOM for initial assistance in drawing the model, members of our labs for helpful discussions, and Fondazione Italiana per la Ricerca sul Cancro (FIRC) for various support.
REFERENCES
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Weinert T, Kaochar S, Jones H, Paek A, Clark AJ. The replication fork’s five degrees of freedom, their failure and genome rearrangements. Curr Opin Cell Biol. 2009;21:778–784. doi: 10.1016/j.ceb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Sale JE. Competition, collaboration and coordination--determining how cells bypass DNA damage. Journal of Cell Science. 2012;125:1633–1643. doi: 10.1242/jcs.094748. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci U S A. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann AR. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- 7.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 8.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 9.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 10.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 11.Papouli E, et al. Crosstalk between SUMO and Ubiquitin on PCNA Is Mediated by Recruitment of the Helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell. 2010;38:649–661. doi: 10.1016/j.molcel.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 16.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS genetics. 2010;6:e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Ramos CA, Prakash S, Prakash L. Requirement of yeast DNA polymerase delta in post-replicational repair of UV-damaged DNA. J Biol Chem. 1997;272:25445–25448. doi: 10.1074/jbc.272.41.25445. [DOI] [PubMed] [Google Scholar]
- 19.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashton TM, Mankouri HW, Heidenblut A, McHugh PJ, Hickson ID. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol Cell Biol. 2011;31:1921–1933. doi: 10.1128/MCB.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szakal B, Branzei D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. The EMBO Journal. 2013;32:1155–1167. doi: 10.1038/emboj.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mankouri HW, Ashton TM, Hickson ID. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc Natl Acad Sci U S A. 2011;108:4944–4949. doi: 10.1073/pnas.1014240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 26.Branzei D. Ubiquitin family modifications and template switching. FEBS letters. 2011;585:2810–2817. doi: 10.1016/j.febslet.2011.04.053. [DOI] [PubMed] [Google Scholar]
- 27.Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, Livingston DM, Haracska L, Elledge SJ. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Molecular Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blastyak A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burkovics P, Sebesta M, Balogh D, Haracska L, Krejci L. Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic acids research. 2013;42:1711–1720. doi: 10.1093/nar/gkt1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glineburg MR, Chavez A, Agrawal V, Brill SJ, Johnson FB. Resolution by unassisted Top3 points to template switch recombination intermediates during DNA replication. J Biol Chem. 2013;288:33193–33204. doi: 10.1074/jbc.M113.496133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc Natl Acad Sci U S A. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hishida T, Kubota Y, Carr AM, Iwasaki H. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature. 2009;457:612–615. doi: 10.1038/nature07580. [DOI] [PubMed] [Google Scholar]
- 35.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mozlin AM, Fung CW, Symington LS. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics. 2008;178:113–126. doi: 10.1534/genetics.107.082677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci U S A. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Runge KW, Zakian VA. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol. 1989;9:1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neelsen KJ, Chaudhuri AR, Follonier C, Herrador R, Lopes M. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol Biol. 2014;1094:177–208. doi: 10.1007/978-1-62703-706-8_15. [DOI] [PubMed] [Google Scholar]
- 42.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberi G, Cotta-Ramusino C, Lopes M, Sogo J, Conti C, Bensimon A, Foiani M. Methods to study replication fork collapse in budding yeast. Methods in Enzymology. 2006;409:442–462. doi: 10.1016/S0076-6879(05)09026-9. [DOI] [PubMed] [Google Scholar]
- 44.Lopes M, Cotta-Ramusino C, Liberi G, Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- 45.Allers T, Lichten M. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic acids research. 2000;28:e6. doi: 10.1093/nar/28.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joo C, McKinney SA, Lilley DM, Ha T. Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J Mol Biol. 2004;341:739–751. doi: 10.1016/j.jmb.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 47.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nature structural & molecular biology. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Huici V, Szakal B, Urulangodi M, Psakhye I, Castellucci F, Menolfi D, Rajakumara E, Fumasoni M, Bermejo R, Jentsch S, Branzei D. DNA bending facilitates the error-free DNA damage tolerance pathway and upholds genome integrity. The EMBO Journal. 2014;33:327–340. doi: 10.1002/embj.201387425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Follonier C, Oehler J, Herrador R, Lopes M. Friedreich’s ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nature structural & molecular biology. 2013;20:486–494. doi: 10.1038/nsmb.2520. [DOI] [PubMed] [Google Scholar]
- 50.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 51.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 52.Karras GI, Fumasoni M, Sienski G, Vanoli F, Branzei D, Jentsch S. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Molecular Cell. 2013;49:536–546. doi: 10.1016/j.molcel.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 54.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes & Development. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes & development. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. The EMBO Journal. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 60.Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, Branzei D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol Biol Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.