Abstract
Objective
The goal of this study was to investigate whether there is a genotype by treatment interaction in patients experiencing stroke and treated with one of three antihypertensive drugs, i.e. chlorthalidone, amlodipine and lisinopril.
Methods
A population of 436 African Americans and 539 whites that have experienced stroke in the GenHAT study were genotyped for 768 single nucleotide polymorphisms in 280 candidate genes. To detect a genotype by treatment interaction we used the Pearson's chi-square test to assess if the genotype frequencies differed at the single SNP level for the three drug treatment groups. From these single SNP analyses we derived a summary statistic for the degree of association at the gene and gene complex levels. This was done by grouping SNPs using information on gene locations and defining gene complexes based on protein-protein interactions. To assess the statistical significance of the observed test statistic we derived an empirical p-value by simulating data under the null hypothesis.
Results
We found that, in patients that have experienced stroke, there is a significant genetic difference between hypertension drug treatment groups. In African Americans SNP rs12143842 showed significant association (p < 0.001) with drug treatment. At gene-level HNRNPA1P4 and NOS1AP in African Americans and PRICKLE1 and NINJ2 in non-Hispanic whites were significantly associated (p < 0.01) to drug treatment, while none of the gene-complexes tested showed significance.
Conclusions
Based on the genetic differences between drug treatment groups, we conclude that there may be an interaction between certain genotypes and antihypertensive treatment in stroke patients. This needs to be replicated in other studies.
Keywords: Stroke, GenHAT, genotype by treatment interaction, antihypertensive treatment
Introduction
Hypertension affects nearly one third of adults in the United States [1,2] and even more adults in Europe [3,4]. This condition increases the risk for stroke, the fourth leading cause of death in the United States [5] and second leading cause of death in Europe [6]. In clinical trials, antihypertensive therapy has been shown to reduce the risk of stroke by 35-40% [7]. Even though there are many effective pharmacologic therapies, international data suggest that blood pressure control rates are < 50% [8–12]. This low rate of hypertension control may be influenced in part by the inability to predict the antihypertensive drug likely to be most effective for an individual patient. Hypertension drug response may depend on the genetic make-up of individuals and treatment efficacy may be improved if this is considered when prescribing antihypertensive agents [13,14].
Hypertension is caused by both genetic and environmental factors. Family-based genetic studies of hypertension report a heritability of 30-60% [15]. While several common genetic variants have been associated with hypertension [16,17], they only explain a small proportion of the genetic variance of the trait. Thus, hypertension likely has a complex mode of inheritance, e.g., many low-effect genetic variants contribute to the genetic variance of the trait [15]. This is likely to also be the case with pharmacogenetic traits such as the response to antihypertensive drug treatment. Evidence collected across genome-wide association studies of complex traits suggest that although many genetic variants with small or moderate effects on the disease phenotype are detected, it appears that multiple independently associated variants are located in the same genes and that genetic variants tend to occur in genes whose products are connected in biological pathways [18– 21]. These studies support the idea that multiple mutations in the same gene (allelic heterogeneity) or multiple mutations in different members of a candidate gene complex (e.g. biological pathway) involved in the pharmacological response are more likely to influence the drug response than a random mutation.
The Genetics of Hypertension-Associated Treatment (GenHAT) study was designed to determine whether variants in hypertension susceptibility genes interact with antihypertensive medication to modify the risk of coronary heart disease and other cardiovascular endpoints, such as stroke, in hypertensives [22]. In this study we have access to a preselected set of SNPs associated with genes that are functionally related to hypertension in subjects that experienced stroke. The goal of this study was to investigate whether there is genotype by treatment interactions in patients experiencing stroke and treated with one of three antihypertensive drugs, i.e. chlorthalidone, amlodipine and lisinopril. This was assessed at the level of (1) single genetic variants (SNP level), (2) multiple genetic variants within genes calculated by grouping SNPs located within a gene (gene level), or (3) multiple genetic variants in candidate gene complexes calculated by grouping SNPs located within genes to form a complex (gene complex level). Thus, we will evaluate whether the pharmacogenetic effects of these drug treatments are determined at the single variant level, gene level or gene complex level.
Methods
Population
The subjects for this study were enrolled in GenHAT (N=39,114), an ancillary study of the Antihypertensive and Lipid Lowering Treatment to Prevent Heart Attack Trial (ALLHAT) [22,23]. The study design and methodology of GenHAT and ALLHAT have previously been described in detail [20, and 21 respectively]. Informed consent was obtained for each patient, and the protocol was approved by the institutional review board at each participating center.
ALLHAT (N=42,418) was a randomized, double blind, multicenter clinical trial of persons at least 55 years of age with hypertension and at least one other CVD risk factor recruited from 623 centers. The primary objective of ALLHAT was to examine differences by antihypertensive treatment in the incidence of fatal coronary heart disease and nonfatal myocardial infarction in persons randomized to one of four antihypertensive drug classes: a calcium channel blocker (amlodipine), an angiotensin converting enzyme (ACE) inhibitor (lisinopril), and an α-adrenergic blocker (doxazosin), each compared with a diuretic (chlorthalidone), in an assignment ratio of 1:1:1:1.7, respectively (i.e. for every 1.7 persons receiving chorthalidone, one person was randomized to each of the other drugs). The doxazosin arm was not included in our study due to the early discontinuation of the drug owing to a significant (25%) increase in CVD compared to the chlorthalidone arm [24]. Other secondary outcome measures were also evaluated, including stroke, heart failure, coronary revascularization, angina, peripheral arterial disease, end-stage renal disease and all-cause death. This analysis is focused on stroke. For this study defined as the rapid onset of persistent neurologic deficit attributable to an obstruction or rupture of the arterial system, including stroke occurring during surgery, that is not known to be secondary to brain trauma, tumor, infection, or other non-ischemic cause (ALLHAT protocol: https://ccct.sph.uth.tmc.edu/ALLHAT/Documents/Protocol.pdf).
GenHAT was designed to evaluate whether genes associated with hypertension modify the risk of primary and secondary ALLHAT outcomes in patients treated with the different anti-hypertensive drug classes. In the case-only phase of GenHAT, 11,599 ALLHAT participants who experienced adverse cardiovascular related events were successfully genotyped for 768 polymorphisms in 280 genes selected for their associations with blood pressure regulation and CVD. The focus of our study was determining whether variance in these genes could modify the risk of stroke (n=1,258 participants).
Genotyping methods
DNA isolation techniques and genotyping methods within GenHAT have been described elsewhere [22]. Briefly, DNA samples were anonymized as set forth in the Report of the Special Emphasis Panel on Opportunities and Obstacles to Genetic Research in NHLBI Clinical Studies [25].
DNA was isolated from blood clots using Gentra Puregene blood kit DNA Isolation Kits from Qiagen (Venlo, The Netherlands). For the case-only phase of GenHAT, Illumina (San Diego, CA, USA) provided custom genotyping at approximately 768 loci (see supplementary file S1 and S2) that were selected as being candidates for blood pressure regulation or CVD. Genotyping was successful for 97% of the samples (sample success rate). Replicate pairs of DNA samples were provided to Illumina to test reproducibility (number of matching allele calls), and between sample agreement was excellent (99.99%).
Statistical analysis of baseline measurements
ANOVA was used to compare the difference in baseline measurements (e.g. body mass index, gender, smoking, diabetes) between treatment groups for continuous variables, and Chi-square tests for categorical variables. Ethnicity group was determined by principal component analysis (PCA) of 64 ancestry informative markers (AIMs) [26]. For this study, only non-Hispanic whites and African Americans (excluding Hispanic African Americans) were included (these are the only groups with enough statistical power to perform ethnicity-specific analysis) and each cohort was analyzed separately.
Determining genotype-by-treatment effects
The data in this study were obtained using a case-only design. Because of the randomized design of ALLHAT, we assume there was a priori no difference in genetic profile for the three different drug treatment groups [27]. We determined whether the genetic profile of patients experiencing stroke differed between drug treatments where genetic difference between drug treatment groups is defined by 1) single genetic variants, 2) multiple genetic variants within genes, or 3) multiple genetic variants in candidate gene complexes.
For each SNP we used the chi-square test to test for independence between drug treatment and genotype frequencies. A p-value of 0.01 was considered as suggestive evidence for an association between SNP and treatment. A 10% false discovery rate (FDR) cut-off was used [28]. SNPs with a minor allele frequency of less than 0.01 were excluded.
To further improve the power of detecting genotype-by-treatment interaction effects, we determined the joint effect of the genetic variants linked to individual genes or candidate gene complexes. Candidate gene complexes among the 280 genes available were identified using STRING [29]. STRING is a database dedicated to protein-protein interactions, including both physical and functional interactions. It weighs and integrates information from numerous sources, including experimental repositories, computational prediction methods and public text collections, thus acting as a meta-database that maps all interaction evidence onto a common set of genomes and proteins. To identify interacting genes we used a cut-off of 0.90 representing a posterior probability that the interaction is a true positive. Genes with 2 or more SNPs associated were included in the gene-wide analysis. Gene complexes with 2 or more genes were included in the gene complex analysis.
Following the removal of SNPs with a minor allele frequency of less than 1%, we tested 538 SNPs in 263 genes in the African American cohort and 508 SNPs in 264 genes in the non-Hispanic white cohort. In the African American sample, 66 genes met the criteria of containing more than one SNP, and 41 complexes contained more than one gene and fitted a 0.9 cut-off for protein-protein interactions. In non-Hispanic whites, 62 genes contained more than one SNP and we identified 44 complexes that contained more than one gene and had a 0.9 cut-off for protein-protein interactions.
Determining a test statistic for individual genes and gene complexes
For each gene and gene complex we derived a summary statistic for the degree of association. Let the Chi-square (χ2) test statistic for each SNP be Ti, i = 1,... n. To determine the gene-wide test statistic, the test statistics of SNPs within the gene were averaged. Likewise the test statistics of SNPs located within genes that were part of a complex were averaged to determine the gene complex test statistic. A high value of these statistics indicated evidence for association. Under the null hypothesis of no association, Ti has a χ2 distribution (with 2 degrees of freedom (in the case of two observed genotype states: eg. AA and AB) or 4 degrees of freedom (three observed genotype states: e.g. AA, AB and BB and three treatments)). The distribution of the test statistics at the gene and gene complex level under the null hypothesis was unknown.
Determining an empirical p-value
All SNP association results are based on an empirical derived p-value. An empirical distribution of the test statistic for each SNP was derived by permuting the sample labels of the drug treatments followed by the computation of the χ2 test statistic. This was done 100,000 times. The observed test statistics for each permutation was recorded in order to obtain the empirical distribution of χ2 values under the null hypothesis. For each SNP, an empirical p-value was obtained by determining the number of empirical observations larger than the observed statistic as a fraction of the amount of permutations. Empirical p-values were derived in a similar way at the gene and gene complex levels. The empirical p-values allow us to control for the number of SNPs being tested for each gene or gene complex. To adjust for multiple testing we determined the false discovery rate [28].
All statistical analyses were performed using R (R Development Core Team 2011).
Results
Baseline characteristics
The baseline characteristics for the 1,258 participants in the GenHAT “case-only” study that all have experienced stroke are provided in Table 1. Except for a history of coronary artery bypass grafting (CABG), the three treatment arms had similar risk factor profiles. A history of CABG was most prominent in the chlorthalidone treatment arm (19.4% of the study population vs. 14.6% and 10.9% for lisinopril and amlodipine respectively).
Table 1.
Baseline characteristics for case-only African American and White participants (n=975) by treatment group
| Characteristic | Chlorthalidone | Amlodipine | Lisinopril | p-value* |
|---|---|---|---|---|
| Sample size, n (%) by treatment | 433 (44.4) | 248 (25.4) | 294 (30.2) | |
| Age (y), mean (SD) | 69.9 (7.9) | 70.0 (7.9) | 69.7 (7.9) | 0.90 |
| Women, n (col %) | 189 (43.6) | 95 (38.3) | 130 (44.2) | 0.31 |
| On antihypertensive treatment, n (col %) | 389 (89.8) | 222 (89.5) | 267 (90.8) | 0.83 |
| Race/ethnicity using SNP data (AIM): | ||||
| White, non-Hispanic, n (col %) | 256 (59.1) | 140 (56.5) | 143 (48.6) | 0.20 |
| African American, n (col %) | 177 (40.9) | 108 (43.5) | 151 (51.4) | |
| Blood pressure at baseline: | ||||
| All participants, mm Hg: SBP, mean (SD) | 148.7 (16.1) | 149.5 (15.5) | 149.1 (15.4) | 0.82 |
| DBP, mean (SD) | 83.4 (10.6) | 83.5 (11.0) | 84.1 (10.7) | 0.72 |
| Treated at baseline, mm Hg: SBP, mean (SD) | 147.5 (16.1) | 148.7 (15.5) | 148.3 (15.6) | 0.62 |
| DBP, mean (SD) | 82.7 (10.6) | 82.8 (11.1) | 83.6 (10.8) | 0.53 |
| Untreated at baseline, mm Hg: SBP, mean (SD) | 159.0 (12.2) | 155.6 (14.6) | 156.5 (12.2) | 0.52 |
| DBP, mean (SD) | 90.2 (8.0) | 89.4 (8.0) | 88.7 (9.0) | 0.76 |
| Eligibility risk factors: | ||||
| Current cigarette smoker, n (%) | 97 (22.4) | 51 (20.6) | 75 (25.5) | 0.52 |
| History of MI or stroke, n (%) | 172 (39.7) | 83 (33.5) | 116 (39.5) | 0.23 |
| History of CABG, n (%) | 84 (19.4) | 27 (10.9) | 43 (14.6) | 0.01 |
| Other atherosclerotic CVD, n (%) | 111 (25.6) | 73 (29.4) | 73 (24.8) | 0.43 |
| Major ST depression/ T-wave inversion, n (%) | 40 (9.2) | 33 (13.3) | 31 (10.5) | 0.25 |
| Type 2 diabetes, n (%) | 204 (47.1) | 115 (46.4) | 124 (42.1) | 0.40 |
| HDL-C < 35 mg/dL, n (%) | 40 (9.2) | 22 (8.9) | 23 (7.8) | 0.80 |
| LVH by electrocardiogram, n (%) | 77 (17.8) | 53 (21.4) | 48 (16.3) | 0.30 |
| Body mass index, mean (SD), kg/m2 | 29.2 (6.4) | 29.3 (5.7) | 29.3 (5.9) | 0.97 |
| Fasting glucose, mean (SD), mg/dL | 133.0 (63.8) | 123.2 (50.3) | 128.0 (59.4) | 0.20 |
| Total cholesterol, mean (SD), mg/dL | 221.6 (49.7) | 220.1 (45.3) | 217.9 (50.4) | 0.62 |
| HDL cholesterol, mean (SD), mg/dL | 45.7 (14.7) | 45.7 (13.1) | 46.3 (14.7) | 0.83 |
| LDL cholesterol, mean (SD), mg/dL | 140.5 (38.1) | 142.6 (40.9) | 138.4 (40.3) | 0.50 |
| Fasting triglycerides, mean (SD), mg/dL | 182.8 (165.2) | 160.9 (112.1) | 167.5 (121.0) | 0.19 |
| Current medication use: | ||||
| Aspirin | 184 (42.5) | 93 (37.5) | 120 (40.8) | 0.70 |
| Estrogen supplementation (women only) | 16 (3.7) | 12 (4.8) | 12 (4.1) | 0.69 |
AIM = ancestry informative marker, SBP = systolic blood pressure, DBP = diastolic blood pressure, CABG = coronary artery bypass grafting, CVD = cardiovascular disease, HDL-C = HDL cholesterol, LVH = left ventricular hypertrophy
test of differences between genotype groups: ANOVA for continuous variables, chi-square for categorical variables
Genotype by treatment effects
We measured the association of 538 and 508 SNPs in African Americans and non-Hispanic whites, respectively, with hypertension drug treatments in stroke cases. Results are shown in Table 2, 3 and 4 respectively.
Table 2.
Single SNPs demonstrating pharmacogenetic differences between treatment groups
| Race | SNP name | χ 2 | P-value1 | FDR2 | Gene ID | Gene symbol | Location |
|---|---|---|---|---|---|---|---|
| African American | rs12143842 | 21.70 | 0.0002 | 0.091 | 9722 | NOS1AP | flanking_5UTR |
| rs16847548 | 13.84 | 0.0073 | 0.854 | 9722 | NOS1AP | flanking_5UTR | |
| rs6473383 | 13.64 | 0.0091 | 0.854 | 389674 | HNRPA1P4 | flanking_5UTR | |
| non-Hispanic whites | rs4917434 | 16.35 | 0.002 | 0.546 | 22986 | SORCS3 | Intron |
| rs2607927 | 14.94 | 0.005 | 0.546 | 4815 | NINJ2 | Intron | |
| rs200148 | 14.43 | 0.006 | 0.546 | 51390 | AIG1 | flanking_5UTR | |
| rs898271 | 13.94 | 0.007 | 0.546 | 390419 | LOC390419 | flanking_3UTR |
Empirical P-values.
FDR: false discovery rate.
Table 3.
Genes associated with drug treatment assessed by grouping SNPs within a gene.
| African American | non-Hispanic white | |||||
|---|---|---|---|---|---|---|
| Gene symbol | Mean χ2 | P-value1 | FDR2 | Mean χ2 | P-value | FDR |
| NINJ2 | 4.42 | 0.2391 | 0.816 | 7.30 | <0.0001 | 0.0000 |
| HNRNPA1P4 | 12.27 | 0.0017 | 0.055 | 2.99 | 0.6562 | 0.9644 |
| PRICKLE1 | 1.67 | 0.9725 | 0.994 | 11.04 | 0.0017 | 0.0527 |
| EDN1 | 6.72 | 0.0258 | 0.517 | 5.76 | 0.0910 | 0.4701 |
| DBH | 4.53 | 0.2010 | 0.776 | 6.77 | 0.0073 | 0.1259 |
| GJB5 | 3.16 | 0.7362 | 0.994 | 6.00 | 0.0689 | 0.4272 |
| NOS1AP | 11.03 | 0.0001 | 0.009 | 6.84 | 0.0355 | 0.2448 |
| SCNN1A | 6.62 | 0.0451 | 0.572 | 3.68 | 0.3021 | 0.8918 |
| PPIAP23 | 2.70 | 0.7601 | 0.994 | 9.54 | 0.0131 | 0.1352 |
| TBC1D4 | 3.53 | 0.3936 | 0.978 | 7.35 | 0.0309 | 0.2398 |
| ATP1A2 | 3.05 | 0.7407 | 0.994 | 8.68 | 0.0102 | 0.1259 |
| ACE | 4.06 | 0.4489 | 0.978 | 6.89 | 0.0159 | 0.1411 |
| ICAM1 | 1.81 | 0.9283 | 0.994 | 9.94 | 0.0096 | 0.1259 |
| LOC440973 | 7.51 | 0.0313 | 0.517 | 2.47 | 0.8334 | 0.9644 |
Genes with empirical P-values of less than 0.05 are included.
FDR: false discovery rate.
Table 4.
Gene complexes associated with drug treatment when using each candidate gene to “fish” for known and predicted protein interactions.
| African American | non-Hispanic white | ||||||
|---|---|---|---|---|---|---|---|
| Gene symbol* | Symbols of genes in the complex | Mean χ2 | P-value1 | FDR2 | Mean χ2 | P-value | FDR |
| NPPA | NPPA, IAPP, ADM, CST3, EDN1 | 5.415 | 0.048 | 0.532 | 4.837 | 0.116 | 0.687 |
| NOS3 | NOS3, ICAM1, REN, AGTR1, PIK3CB, ACE, NOS2, EDN1 | 3.675 | 0.721 | 0.987 | 4.762 | 0.037 | 0.524 |
| ADM | ADM, IAPP, PTH, ADRB2, ADORA2A, NPPA, EDN1 | 5.163 | 0.042 | 0.532 | 4.683 | 0.093 | 0.636 |
| ATP1A2 | ATP1A2, ATP1B1 | 2.715 | 0.868 | 0.987 | 8.013 | 0.009 | 0.194 |
| ATP1B1 | ATP1B1, ATP1A2 | 2.715 | 0.868 | 0.987 | 8.013 | 0.009 | 0.194 |
Genes with empirical P-values of less than 0.05 included.
FDR: false discovery rate.
Symbol of gene that was used as “bait” in STRING database. The bait gene is included in the gene complex.
The number of significant (p<0.01) SNPs before adjusting for multiple testing was 3 and 4 for African Americans and non-Hispanic whites, while based on the number of tests performed the expected number of significant (p<0.01) SNPs is approximately 5 (0.01 × 538 and 0.01 × 508). At the gene level, we tested 66 and 62 genes (each containing more than one SNP) for African Americans and non-Hispanic whites. The number of significant (p<0.01) genes was 2 and 4 for African Americans and non-Hispanic whites (disregarding the false discovery rate). The expected number of significant (p<0.01) genes is 1. Finally, at the level of gene complexes, we tested 41 and 43 gene complexes (containing more than one gene and a protein-protein interaction cut-off of 0.9) for African American and non-Hispanic whites. The number of significant (p<0.01) gene complexes was 0 and 2 for African Americans and non-Hispanic whites. The expected number of significant (p<0.01) gene complexes is 1.
Table 2 shows the mean χ2, the gene ID and symbol of the associated gene for the significant SNPs. Supplementary files S1 and S2 show all the results for the African American and non-Hispanic white cohort respectively. The empirical distribution of the test statistics, determined by the single SNP test, is close to the distribution of the theoretical χ2 test with 4-6 degrees of freedom (see quantile-quantile plot supplementary Figure 1). SNP rs12143842 showed significance (p < 0.001) also after correcting for multiple testing (FDR < 0.1) in the African American population (Table 2). The African American and non-Hispanic white populations did not have any other significant common SNPs.
The differences in allele frequencies for each drug treatment, as well as the average across drug treatments (allele difference between cohorts) in African American and non-Hispanic white populations are shown in supplementary Table 1.
For SNP rs200148 in the African American cohort, the A allele frequency is higher in the chlorthalidone treatment group (A allele frequency = 0.58) as compared to the amlodipine and lisinopril treatment groups (0.43 and 0.49 A allele frequency respectively) (supplementary Table 1). African Americans carrying the A allele and treated with chlorthalidone rather than lisinopril or amlodipine were overrepresented among stroke patients. For the rs12143842 SNP in the African American cohort, the C allele frequency is lower in the amlodipine treatment group (C allele frequency = 0.78) as compared to chlorthalidone and lisinopril treatment groups (0.88 and 0.89 C allele frequency respectively) indicating lower stroke risk in African Americans carrying the C allele and treated with amlodipine versus chlorthalidone or lisinopril.
To improve the power of determining genotype by treatment effects, SNPs were grouped by gene and a mean test statistic determined for each gene. Using this approach we found 4 genes associated with the drug treatment after adjusting for multiple testing (Table 3): NINJ2 (non-Hispanic white cohort), HNRNPA1P4 (African American cohort), PRICKLE1 (non-Hispanic white cohort) and NOS1AP (African American cohort) (highlighted gene symbols in Table 3). Genes with a p-value < 0.05 for each cohort is included in Table 3. The corresponding genes’ p-values for the other cohorts are also included for comparison. Supplementary files S3 and S4 show all the results for the gene-wise analysis of the African American and non-Hispanic white cohort respectively. Interestingly the NOS1AP gene in the African American cohort seems to be significantly correlated with the treatments. It was also the rs12143842 SNP localized on this gene that was shown to be significant when analyzing SNPs separately.
Genes were grouped into candidate gene complexes based on known and predicted protein interactions using the STRING database (see supplementary files S5 and S6). Each candidate gene was used as “bait” to “fish” for associated genes and was also included in the complex. STRING shows proteins associated with the “bait” protein based on different types of evidence. It does not give this “complex” of associated proteins a label. Subsequently, SNPs associated with the gene complex were grouped to determine whether this would lead to a more significant association with the treatment when comparing genotype frequencies between different treatments. Although there were significant p- values (p<0.01), none of the gene complexes were below the 10% cutoff for false discovery. Complexes with a p-value < 0.05 for each cohort is included in Table 4.
Discussion
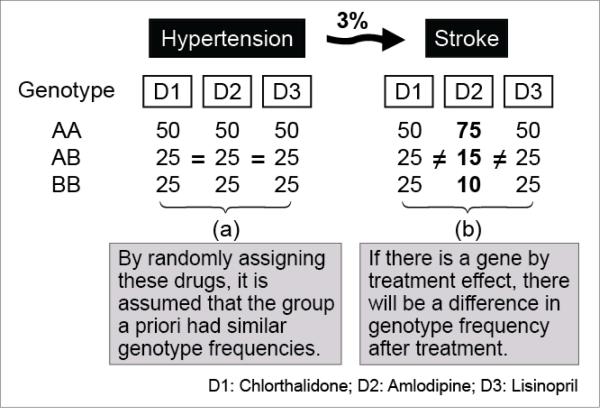
Using a case-only pharmacogenetic design, we determined whether SNPs in candidate genes for hypertension interact with antihypertensive medication to modify the risk of stroke in hypertensive patients. Our analysis was based on the assumption that patients would have similar genotype frequencies across drug treatment groups in the absence of drug by gene interaction. Given that patients were randomly assigned to their medication in this case-only study (ALLHAT was a randomized double blind, multicenter trial) [23], this is a plausible assumption. Thus, we evaluated if hypertensive patients whom all have experienced stroke and were treated with one of three drug treatments, differed in their DNA profile at the single SNP, gene, and/or gene complex level (Fig. 1).
Figure 1.
Genotype frequencies for a single SNP in an hypertesive cohort (a); and genotype frequencies after treatment in the 3% of the patients that suffered stroke (b). The illustrative SNP shows an interaction with the drug used for treatment.
Using this approach, we have shown a significant difference between drug treatment groups at the level of single SNPs as well as when grouping SNPs by genes. This significance is lost when grouping SNPs by gene complexes. Grouping SNPs by genes increased the number of SNPs that were associated with drug treatment. At the SNP level, only one (rs12143842) was significantly associated with drug treatment (African American population). In this population, the C allele frequency is lower in the amlodipine treatment group as compared to chlorthalidone and lisinopril treatment groups indicating lower stroke risk in African Americans carrying the C allele and treated with amlodipine versus chlorthalidone or lisinopril.
Grouping SNPs by genes resulted in four genes showing significance: NINJ2 (in the non-Hispanic white population), HNRNPA1P4 (African American population), PRICKLE1 (non-Hispanic white population) and NOS1AP (African American population). These results may indicate that our approach allows us to detect weaker associations exhibited by a group of SNPs located in small genomic regions such as those defined by genes. Individually these associations may not be significant, but by considering these associations jointly by defining an appropriate summary statistic we may detect the signal.
SNP rs12134842 (within 100kb of the NOS1AP gene) has earlier been identified as strongly associated to prolonging QT-interval duration [30]. A study of participants in the Rotterdam Study, a population-based, prospective cohort study of individuals of 55 years of age or older, concluded that each rs12143842(T) allele was associated with a QT-interval duration increase of 4.4-ms (p =4.4×10−28) [30]. QT-interval prolongation is an electrophysiologic phenomenon associated with sudden cardiac death. Prolongation of QT is associated with a significantly increased risk of incident stroke independent of traditional stroke risk factors [31]. Thus, it seems plausible that this SNP may interact with drug treatment to effect stroke risk.
At the gene level, NOS1AP (encoding for Nitric oxide synthase 1 adaptor protein) also showed significance (p=0.0001, African American population) implicating the other SNPs at this gene as relevant to stroke pharmacogenetics. SNP rs10494366 has previously been shown to be associated with QT-interval in several studies [32–37]. NINJ2 appears to be a strong candidate for harboring pharmacogenetic variants since this gene has been previously associated with ischemic stroke [38]. The NINJ2 gene encodes an adhesion molecule expressed in glia and shows increased expression after nerve injury [39]. According to Ikram et al. [38] both rs11833579 and rs12425791 were shown to be significantly associated with ischemic stroke and, in particular, the atherothrombotic stroke subtype. The combined effect of these SNPs was associated with drug treatment effects in our study (Table 4). The two SNPs (rs6473383 and rs11997468) in the HNRNPA1P4 (heterogeneous nuclear ribonucleoprotein A1 pseudogene 4) gene associated with drug treatment has previously been shown to be associated to heart failure (p = 3.1×10−6 and p = 3.4×10−6 respectively), although they did not reach genome wide significance (set at 5.0×10−7) [40].
Even though they did not reach genome wide significance, SNPs rs1520832 and rs1033264 within the PRICKLE1 gene (located at 12q12) have been associated with incident heart failure (p-values 1.2×10−6 and 1.4×10−6 respectively, measured in the European ancestry population) [40]. Several studies suggest that PRICKLE genes play an important role in the central nervous system. PRICKLE1 is a component of the planar cell polarity (PCP) pathway that regulates cell migration and polarity in various contexts [41]. Mutations in components of the PCP pathway lead to a spectrum of neurological phenotypes and disorders. For example, a missense mutation in PRICKLE1 is associated with progressive myoclonus epilepsy in humans, and its reduced gene dosage increases sensitivity to induced seizure in mice [42,43]. Additionally, overexpression of PRICKLE1 promotes neurite outgrowth in neuroblastoma cells [44]. These studies suggest that Prickle genes have broader roles in the CNS.
Although stroke risk reduction on antihypertensive treatment is mainly due to blood pressure control level [45] a component of stroke protection may be exerted by other drug specific mechanisms of action such as improvements in endothelial function and anti-platelet activity [46]. Evaluating main effects of variables of interest as well as controlling for main effects of covariates (such as blood pressure control level during follow-up) cannot be tested in the context of the case-only design [47]. Therefore, testing whether the gene drug interaction effects on stroke identified in this study is mediated by differential blood pressure control level by treatment class or other drug related mechanisms was beyond the scope of this study. Further investigations are required to understand the exact mechanisms underlying these findings. Additionally, we are aware of the importance of replication to verify our results, however we were not able to find an appropriate replication study considering our population, the treatments used, the polymorphisms assayed and the end points tested.
The modeling approach described here goes beyond the search for single variants by evaluating the significance of the participation of entire genes and pathways, by adding evidence of association of polymorphisms with the phenotype. Our results suggests that grouping variants at the gene level will contribute in finding novel associations by joining the contribution from multiple near-significant variants in the same gene. The significance gained by grouping SNPs by genes was lost when grouping SNPs by gene complexes in this dataset. Even though the entire gene complex consists of genes that are functionally related to hypertension, many of the variants located in these genes may not interact with the drug treatment. Thus more “noise” could be added to the near-significant variants and thereby diluting their contribution. To avoid this dilution effect a more flexible statistical modeling approach is required to allow the variants to contribute unequally to the summary statistic. Our gene complex approach relies on the availability of known and predicted protein-protein interaction data. We used only high quality protein-protein interactions, thus limiting the number of gene complexes that could be formed. It may have led to an overrepresentation of well-studied genes in the complexes studied. We do not expect this to influence our false discoveries because our testing procedure is based on p-values obtained from a large number of permutations.
The variants used in this study are located in genes that have previously been reported to be associated with blood pressure regulation thus making them all plausible candidates for interacting with blood pressure medication. Some of the variants we highlight are located in genes that have previously been associated with stroke in external studies helping to lend support to the validity of our results. In summary, from this work we can conclude that, in patients that have experienced stroke, there is a significant genetic difference between hypertension drug treatment groups at the level of single SNPs. The two populations did not show the same SNPs to be significantly associated to treatment. This implies that replicate studies need to be carefully designed with respect to the genetic background of the study population. Our results indicate that taking advantage of prior biological knowledge when interpreting genotype association studies may lead to the discovery of new genes/genotypes and thereby contributing to our understanding of the genetic components that play an important role in pharmacogenetics.
Supplementary Material
Acknowledgments
Sources of Funding
This work was supported in part by R01 HL63082 (GenHAT) and N01-HC-35130 (ALLHAT) from the National Heart, Lung and Blood Institute, National Institutes of Health, US Department of Health and Human Services, Bethesda, MD.
Footnotes
This material has not been published or accepted for publication elsewhere nor is it under consideration by another publication.
Conflicts of Interest
No conflicts of interest has been declared.
REFERENCES
- 1.Yoon SS, Otschega Y, Louis T. Recent trends in the prevalence of high blood pressure and its treatment and control, 1999-2008. US Department of Health and Human Services, CDC, National Center for Health Statistics; Hyattsville, MD: 2010. [PubMed] [Google Scholar]
- 2.Morbidity and mortality: 2009 chart book on cardiovascular, lung and blood diseases. Vol. 54. US Department of Health and Human Services, National Institutes of Health; Rockville, MD: 2009. National Heart, Lung, and Blood Institute Chart 3-67. Chart 3-67. p. 114. [Google Scholar]
- 3.Wolf-Maier K, Cooper RS, Banegas JR, Giampaoli S, Hense H-W, Joffres M, et al. Hypertension prevalence and blood pressure levels in 6 European countries, Canada, and the United States. JAMA. 2003;289:2363–2369. doi: 10.1001/jama.289.18.2363. [DOI] [PubMed] [Google Scholar]
- 4.Joffres M, Falaschetti E, Gillespie C, Robitaille C, Loustalot F, Poulter N, et al. Hypertension prevalence, awareness, treatment and control in national surveys from England, the USA and Canada, and correlation with stroke and ischaemic heart disease mortality: a cross-sectional study. BMJ Open. 2013;3:e003423. doi: 10.1136/bmjopen-2013-003423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics--2014 update: a report from the american heart association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nichols M, Townsend N, Luengo-Fernandez R, Leal J, Gray A, Scarborough P, et al. Table 1.1. European Cardiovascular Disease Statistics 2012. European Heart Network; European Society of Cardiology; Brussels: Sophia Antipolis: 2012. [Google Scholar]
- 7.Neal B, MacMahon S, Chapman N. Effects of ACE inhibitors, calcium antagonists, and other blood-pressure-lowering drugs: results of prospectively designed overviews of randomised trials. The Lancet. 2000;356:1955–1964. doi: 10.1016/s0140-6736(00)03307-9. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie C, Kuklina EV, Briss PA, Blair NA, Hong Y. Division for Heart Disease and Stroke Prevention, National Center for Chronic Disease Prevention and Health Promotion, CDC. Vital Signs: Prevalence, Treatment, and Control of Hypertension - United States, 1999-2002 and 2005-2008. Circulation. 2011;123:933–944. [Google Scholar]
- 9.Ma J, Stafford RS. Screening, Treatment, and Control of Hypertension in US Private Physician Offices, 2003–2004. Hypertension. 2008;51:1275–1281. doi: 10.1161/HYPERTENSIONAHA.107.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori H, Ukai H, Yamamoto H, Saitou S, Hirao K, Yamauchi M, et al. Current status of antihypertensive prescription and associated blood pressure control in Japan. Hypertens Res. 2006;29:143–151. doi: 10.1291/hypres.29.143. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez-Roca GC, Pallarés-Carratalá V, Alonso-Moreno FJ, Escobar-Cervantes C, Barrios V, Llisterri JL, et al. Working group of arterial hypertension of the Spanish Society of Primary Care physicians (Group HTA/SEMERGEN); PRESCAP 2006 investigators Blood pressure control and physicians’ therapeutic behavior in a very elderly Spanish hypertensive population. Hypertens Res. 2009;32:753–758. doi: 10.1038/hr.2009.102. [DOI] [PubMed] [Google Scholar]
- 12.Thoenes M, Neuberger H-R, Volpe M, Khan BV, Kirch W, Böhm M. Antihypertensive drug therapy and blood pressure control in men and women: an international perspective. J Hum Hypertens. 2010;24:336–344. doi: 10.1038/jhh.2009.76. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari P, Bianchi G. Genetic mapping and tailored antihypertensive therapy. Cardiovasc Drugs Ther. 2000;14:387–395. doi: 10.1023/a:1007860115489. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JA. Advancing management of hypertension through pharmacogenomics. Ann Med. 2012;44(Suppl 1):S17–22. doi: 10.3109/07853890.2011.653399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navis G, Bakker SJL, van der Harst P. Dissecting the genetics of complex traits: lessons from hypertension. Nephrol Dial Transplant. 2010;25:1382–1385. doi: 10.1093/ndt/gfq091. [DOI] [PubMed] [Google Scholar]
- 16.Hottenga J-J, Whitfield JB, de Geus EJC, Boomsma DI, Martin NG. Heritability and stability of resting blood pressure in Australian twins. Twin Res Hum Genet. 2006;9:205–209. doi: 10.1375/183242706776382455. [DOI] [PubMed] [Google Scholar]
- 17.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lage K, Greenway SC, Rosenfeld JA, Wakimoto H, Gorham JM, Segrè AV, et al. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci U. S. A. 2012;109:14035–14040. doi: 10.1073/pnas.1210730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnett DK, Boerwinkle E, Davis BR, Eckfeldt J, Ford CE, Black H. Pharmacogenetic approaches to hypertension therapy: design and rationale for the Genetics of Hypertension Associated Treatment (GenHAT) study. Pharmacogenomics J. 2002;2:309–317. doi: 10.1038/sj.tpj.6500113. [DOI] [PubMed] [Google Scholar]
- 23.Davis BR, Cutler JA, Gordon DJ, Furberg CD, Wright JT, Jr, Cushman WC, et al. Rationale and design for the Antihypertensive and Lipid Lowering Treatment to Prevent Heart Attack Trial (ALLHAT). ALLHAT Research Group. Am J Hypertens. 1996;9:342–360. doi: 10.1016/0895-7061(96)00037-4. [DOI] [PubMed] [Google Scholar]
- 24.The ALLHAT Officers and Coordinators for the ALLHAT Collaborative Research Group Major cardiovascular events in hypertensive patients randomized to doxazosin vs chlorthalidone: The antihypertensive and lipid-lowering treatment to prevent heart attack trial (allhat). JAMA. 2000;283:1967–1975. [PubMed] [Google Scholar]
- 25.National Heart, Lung, and Blood Institute, NHLBI Report of the Special Emphasis Panel on Opportunities and Obstacles to Genetic Research in NHLBI Clinical Studies. National Heart, Lung, and Blood Institute, NIH; Bethesda, Md: 1997. [Google Scholar]
- 26.Lynch AI, Irvin MR, Boerwinkle E, Davis BR, Vaughan LK, Ford CE, et al. RYR3 gene polymorphisms and cardiovascular disease outcomes in the context of antihypertensive treatment. Pharmacogenomics J. 2013;13:330–334. doi: 10.1038/tpj.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Little J, Sharp L, Khoury MJ, Bradley L, Gwinn M. The epidemiologic approach to pharmacogenomics. Am J Pharmacogenomics. 2005;5:1–20. doi: 10.2165/00129785-200505010-00001. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 29.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eijgelsheim M, Aarnoudse ALHJ, Rivadeneira F, Kors JA, Witteman JCM, Hofman A, et al. Identification of a common variant at the NOS1AP locus strongly associated to QT-interval duration. Hum Mol Genet. 2009;18:347–357. doi: 10.1093/hmg/ddn341. [DOI] [PubMed] [Google Scholar]
- 31.Soliman EZ, Howard G, Cushman M, Kissela B, Kleindorfer D, Le A, et al. Prolongation of QTc and Risk of Stroke: The REGARDS (REasons for Geographic and Racial Differences in Stroke) Study. J Am Coll Cardiol. 2012;59:1460–1467. doi: 10.1016/j.jacc.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aarnoudse A-JLHJ, Newton-Cheh C, de Bakker PIW, Straus SMJM, Kors JA, Hofman A, et al. Common NOS1AP Variants Are Associated With a Prolonged QTc Interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 33.Post W, Shen H, Damcott C, Arking DE, Kao WHL, Sack PA, et al. Associations between genetic variants in the NOS1AP (CAPON) gene and cardiac repolarization in the old order Amish. Hum Hered. 2007;64:214–219. doi: 10.1159/000103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao WHL, Arking DE, Post W, Rea TD, Sotoodehnia N, Prineas RJ, et al. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119:940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehtinen AB, Newton-Cheh C, Ziegler JT, Langefeld CD, Freedman BI, Daniel KR, et al. Association of NOS1AP genetic variants with QT interval duration in families from the Diabetes Heart Study. Diabetes. 2008;57:1108–1114. doi: 10.2337/db07-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raitakari OT, Blom-Nyholm J, Koskinen TA, Kähönen M, Viikari JSA, Lehtimäki T. Common variation in NOS1AP and KCNH2 genes and QT interval duration in young adults. The Cardiovascular Risk in Young Finns Study. Ann Med. 2009;41:144–151. doi: 10.1080/07853890802392529. [DOI] [PubMed] [Google Scholar]
- 37.Arking DE, Pfeufer A, Post W, Kao WHL, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 38.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide Association Studies of Stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araki T, Milbrandt J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J Neurosci. 2000;20:187–195. doi: 10.1523/JNEUROSCI.20-01-00187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, et al. Association of Genome-Wide Variation With the Risk of Incident Heart Failure in Adults of European and African Ancestry A Prospective Meta-Analysis From the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. Circ Cardiovasc Genet. 2010;3:256–266. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seifert JRK, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- 42.Bassuk AG, Wallace RH, Buhr A, Buller AR, Afawi Z, Shimojo M, et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy ataxia syndrome. Am J Hum Genet. 2008;83:572–581. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao H, Manak JR, Sowers L, Mei X, Kiyonari H, Abe T, et al. Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am J Hum Genet. 2011;88:138–149. doi: 10.1016/j.ajhg.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimura L, Watanabe-Takano H, Sato Y, Tokuhisa T, Hatano M. Prickle promotes neurite outgrowth via the Dishevelled dependent pathway in C1300 cells. Neurosci Lett. 2009;467:6–10. doi: 10.1016/j.neulet.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 45.Lawes CMM, Bennett DA, Feigin VL, Rodgers A. Blood pressure and stroke: an overview of published reviews. Stroke. 2004;35:1024. [PubMed] [Google Scholar]
- 46.Roush GC, Buddharaju V, Ernst ME, Holford TR. Chlorthalidone: mechanisms of action and effect on cardiovascular events. Curr Hypertens Rep. 2013;15:514–521. doi: 10.1007/s11906-013-0372-1. [DOI] [PubMed] [Google Scholar]
- 47.Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: case-control studies with no controls! Am J Epidemiol. 1996;144:207–213. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.