Introduction
Gemcitabine (2’, 2’-difluoro 2’deoxycytidine, dFdC) is a cancer drug of the anti-metabolite class. It is a deoxycytidine analog that interferes with DNA synthesis by incorporating into elongating DNA, and indirectly interferes with DNA replication by inhibiting the nucleoside salvage pathway. Gemcitabine is a “first line treatment” in various types of solid tumors including pancreatic, non-small cell lung cancer (NSCLC), breast cancer, and some blood cancers, such as non-Hodgkin's lymphoma [1]. Gemcitabine is administered intravenously, or with injection, and it can be administered alone, or in combination with other antimetabolites such as fluorouracil, DNA damaging agents such as cisplatin, or various other chemotherapeutic agents [2]. Although the pharmacokinetic pathway of gemcitabine is similar to other deoxycytidine analogs it has certain attributes that render it a more broadly efficacious anti-metabolite [3].
Pharmacokinetics
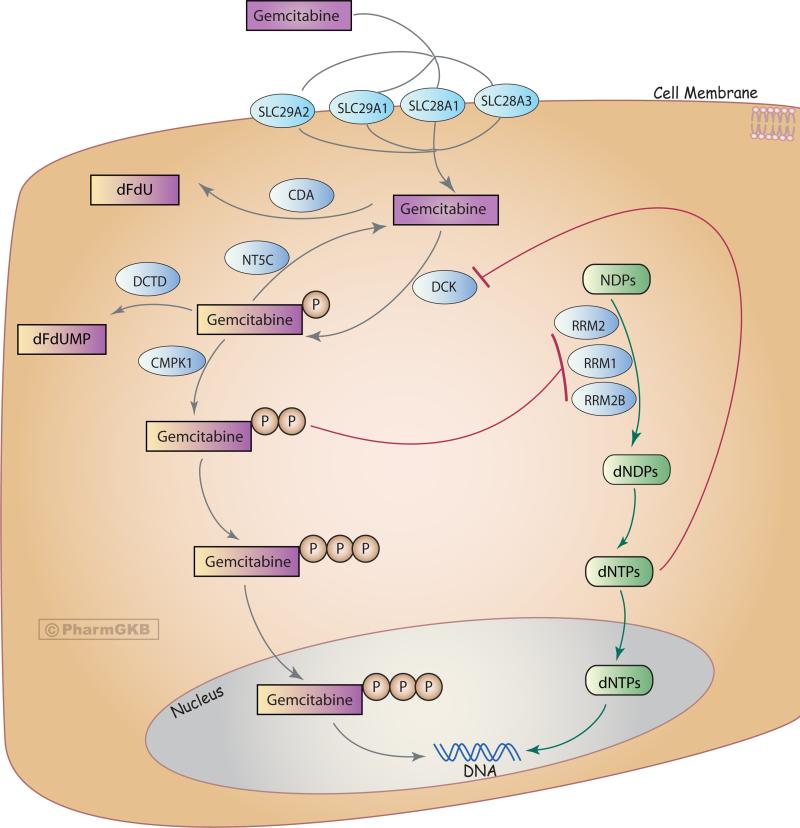
Gemcitabine is a hydrophilic molecule, and three nucleoside transporters mediate most of its uptake into cells: SLC29A1 SLC28A1, and SLC28A3 (see Figure 1). Gemcitabine is also a pro-drug that requires serial phosphorylation by multiple kinases to become pharmacologically active. Deoxycytidine kinase (DCK) catalyzes the initial, and rate-limiting monophosphorylation of gemcitabine to gemcitabine monophosphate (dFdCMP). Phosphorylation to gemcitabine di- (dFdCDP) and tri- phosphate (dFdCTP) is catalyzed by UMP/CMP kinase (CMPK1) and nucleoside-diphosphate kinase (NDPK, NME), respectively. The majority (~ 90%) of intracellular gemcitabine is inactivated by deamination by cytidine deaminase (CDA) to form 2’2’ difluorodeoxyuridine (dFdU). Additional inactivation steps include deamination of gemcitabine monophosphate by deoxycytidine deaminase (DCTD) and dephosphorylation of dFdCMP by cytosolic 5’ nucleotidases (NT5C) [3](see Figure 1).
Figure 1.
Stylized cells depicting the metabolism and mechanism of action of gemcitabine. A fully interactive version is available at PharmGKB: http://www.pharmgkb.org/pathway/PA2036.
Pharmacodynamics
Gemcitabine diphosphate (dFdCDP) depletes a cells deoxyribonucleotide (dNTP) pools via inhibition of ribonucleotide reductase 1 (RRM1). RRM1 is a sub-unit of an enzyme complex that catalyzes the formation of deoxyribonucleotides (dNTPs) from ribonucleotides (rNTPs) through the nucleoside salvage pathway [4, 5]. Gemcitabine triphosphate (dFdCTP) integrates into elongating DNA, and prevents base-excision repair by allowing for a native dNTP to be added next to it, which is termed “masked chain termination”. This irreparable error leads to inhibition of DNA synthesis, and eventually apoptosis [6, 7]. Decreasing dNTP pools increasingly favor gemcitabine uptake, and low concentrations of native deoxycytidines (dCTP) promote DCK activity, and inhibit DCTD activity. The attribute of “self-potentiation” is part of why gemcitabine is so widely used as part of a “first-line” chemotherapy treatment.
Pharmacogenomics
Given the widespread use of gemcitabine, its narrow therapeutic index, and inter-individual variability in patient response it is reasonable to expect that pharmacogenomics could be used to specifically tailor gemcitabine dosage to patients. Small study sizes, heterogeneity of the samples, including of the types and stages of cancers, and differences in chemotherapy make comparing patient responses to gemcitabine between groups very difficult.
Currently, intrinsic and acquired resistance to gemcitabine has been attributed to variation in the expression of genes involved in the transport, activation, and inactivation of gemcitabine, as well as its molecular target. Although gemcitabine is generally well-tolerated, hematological toxicities such as neutropenia are commonly reported. A certain percentage of patients also experience serious, and life-threatening complications after administration of gemcitabine. Decreased activity of the principal inactivating enzyme, CDA, is reported to be associated with toxicity, but further studies are needed to validate these findings [8]. Nucleoside diphosphate kinase (NDPKs) are less intensively studied than other genes in the gemcitabine pathway, although there are indications that alterations in their expression may affect gemcitabine resistance and prognoses in some patients [9, 10]
Variants in Transporters
Solute Carrier Family 29
The SLC29A1 transporter (also called the nitrobenzylthioinosine sensitive human equalibrative nucleoside transporter, or hENT1) is a widely expressed, plasma membrane, facilitated diffusion nucleoside transporter, with a broad selectivity for purines and pyrimidines. Kinetic studies show that the SLC29A1 transporter is the primary mediator of gemcitabine uptake into the cell and that cells that are SLC29A1 null are highly resistant to gemcitabine. SLC29A2 also has a broad affinity for purines and pyrimidines, but its gene expression is most heavily concentrated in skeletal muscle. Structurally, SLC29A1 and SLC29A2 share 46% amino acid sequence identity, and the sequences with the highest homology are their respective transmembrane helices, loops, and hydrophobic terminals [11].
Multiple studies, including a recent meta-analysis, provide strong evidence of a positive relationship between increased SLC29A1 expression and improved survival outcomes in cancer patients treated with gemcitabine [12-15]. SLC29A1 is not highly polymorphic, however, and no genetic variants in the coding region of SLC29A1 have shown significant associations with patient response to gemcitabine. Most reported associations are with genetic variants in non-coding regions. Table 1 summarizes several association studies investigating SNPs in SLC29A1, but none show consistent associations with patient outcomes. There are very few studies investigating associations between SNPs in SLC29A2 with patient response to gemcitabine, and like SLC29A1, no genetic variants in SLC29A2 are consistently associated with patient response to gemcitabine. One paper does report a trend of association between decreased protein levels of SLC29A2, and decreased incidence of complete remission in patients with acute myeloid leukemia (AML) when treated with mitoxantrone and gemcitabine [16]. Further evidence of association is needed to validate the relevance of SLC29A2 for pharmacogenomic studies of gemcitabine.
Table 1.
Single nucleotide polymorphisms in SLC29A1 and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs760370 | 201A>G | The A allele is associated with shorter overall survival compared with the G allele in people with metastatic breast cancer treated with a combination of gemcitabine and paclitaxel; this SNP is part of a haplotype with the rs747199 G allele. | 24361227 |
| rs760370 | 201A>G | The GG genotype is associated with worse tumor response compared to genotypes AA and AG in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs747199 | −706G>C | The G allele is associated with lower overall survival compared with the C allele, in people with metastatic breast cancer treated with a combination of gemcitabine and paclitaxel; this SNP is part of a haplotype with rs760370 A allele. | 24361227 |
| rs747199 | −706G>C | Both the G and C allele show a trend of association with increased expression of SLC29A1 when part of a 3 SNP haplotype with rs731780 (C) and rs70914 (G) (in vitro). | 16609362 |
| rs731780 | −1345 C>G | The C allele is associated with increased expression of SLC29A1 when part of a 3 SNP haplotype with rs70914 (G) and rs747199 (G, or C); in vitro | 16609362 |
| rs70914 | −1050G>A | The G allele is associated with increased expression of SLC29A1 when part of a 3 SNP haplotype with rs731780 (C) and rs747199 (G, or C); in vitro. | 16609362 |
| rs9394992 | 29+913C>T | The CT and TT genotypes are associated with increased risk of neutropenia, but are not associated with response compared to genotype CC in people with pancreatic cancer when treated with gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs3778504 | −52+1681G>A | The AA and AG genotypes are associated with increased time to progression compared to genotype GG in people with cancer. | 22838950 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: SNP (single nucleotide polymorphism).
Solute Carrier Family 28
SLC28A1 and SLC28A3 nucleoside transporters (also called the human concentrative nucleoside transporters, hCNTs) have a high affinity for pyrimidines, and although the SLC28A2 transporter preferentially transports purines it is also capable of transporting pyrimidines. SLC28A1 and SLC28A2 transporters are enriched in the epithelial cells of the kidney, intestine, and liver, and SLC28A3 is widely distributed in various tissues. The tissue distribution of SLC28A1 and SLC28A2 suggests that they play critical roles in the systemic absorption of nucleosides and nucleoside analogs [11].
Expression of SLC28A1 and SLC28A2 varies by tissue type, between individuals, and between cancerous and non-cancerous cells [17]. Cells derived from resected pancreatic tumors, and other cancerous cells tend to have lower SLC28A1 expression compared to non-cancerous cells [17, 18]. In addition, cells with lower levels of SLC28A1 are shown to be resistant to gemcitabine, but they can be re-sensitized with constitutive expression of SLC28A1 [18] Most reported polymorphisms in SLC28A genes are in coding regions, and not in regulatory regions [11]. Despite being more polymorphic than SLC29A genes, no SNPs in SLC28A1, or SLC28A2 are definitively associated with patient outcomes, or risk of toxicity in patients treated with gemcitabine[19-22]. A recent study reports that the C allele at rs7867504 is associated with decreased formation of gemcitabine triphosphate in patients with cancer[23]. Tables 2,3, and 4 summarize the results of pharmacogenomic studies investigating genetic variants of SLC28A genes and associations with outcomes in cancer patients treated with gemcitabine.
Table 2.
Single nucleotide polymorphisms in SLC28A1 and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs2242047 | 1528C>T (Arg51Cys) | The CC genotype is associated with slower tumor progression compared to genotype TT in people with pancreatic cancer when treated with a combination of gemcitabine and erlotinib, or fluoropyrimidines. | 22838950 |
| rs2242047 | 1528C>T (Arg51Cys) | The CC genotype is not associated with response, or risk of hematologic toxicity compared to genotypes CT and TT in people with NSCLC when treated with a combination of gemcitabine and carboplatin. | 18538445 |
| rs2242046 | +1561 G>A (Asp521Asn) | The GG genotype is associated with a decreased risk of hematologic toxicity compared to genotypes AG and AA in people with NSCLC, but is not associated with response to gemcitabine. | 18538455 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: NSCLC (non-small cell lung cancer).
Table 3.
Single nucleotide polymorphisms in SLC28A2 and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs1060896 | 225C>A (Ser75Arg) | The CC genotype is associated decreased risk of hematologic toxicity (higher neutrophil counts, but no difference in platelet counts), and decreased response (shorter median survival time, but no difference in time to progression) compared to genotypes AA and AC in people with NSCLC when treated with a combination of gemcitabine and carboplatin. This SNP was found to be in strong LD with rs11854484. | 18538445 |
| rs1060896 | 225C>A (Ser75Arg) | The AA genotype is not associated with response compared to genotypes AC and CC in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs11854484 | 65C>T (Pro22Leu) | The CC genotype is associated decreased risk of hematologic toxicity (higher neutrophil counts, but no difference in platelet counts), and decreased response (shorter median survival time, but no difference in time to progression) compared to genotypes CC and CT in people with NSCLC when treated with a combination of gemcitabine and carboplatin. This SNP was found to be in strong LD with rs1060896. | 18538445 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: SNP (single nucleotide polymorphism), NSCLC, (non-small cell lung cancer) LD (linkage disequilibrium).
Table 4.
Single nucleotide polymorphisms in SLC28A3 and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs7867504 | 267T>C (Thr89=) | The C allele is associated with decreased formation of dFdCTP compared to the T allele in people with cancer. | 24300978 |
| rs7867504 | 267T>C (Thr89=) | The CC and CT genotypes are associated with longer overall survival as compared to the TT genotype in people with metastatic breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs7867504 | 267T>C (Thr89=) | The CC genotype is not associated with tumor progression compared to genotypes CT and TT in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: dFdCTP (gemcitabine triphosphate).
Variants in Metabolizing Enzymes
Cytidine deaminase
Ninety percent of the gemcitabine that comes into a cell is deaminated by cytidine deaminase (CDA) to 2’2’ difluorodeoxyuridine (dFdU), which is much less toxic than gemcitabine. A common concern with cancer drugs is the need to balance the toxicity that is intended for cancer cells with the incidental toxicity that occurs in non-cancerous cells. Decreased expression of CDA is associated with increased efficacy of gemcitabine, but an increased risk of toxicity [24]. Three SNPs in CDA have been investigated in multiple pharmacogenomic association studies, the results of which are summarized in Table 5. The associations between these SNPs with patient response are inconsistent, or require further investigation.
Table 5.
Single nucleotide polymorphisms in CDA and related phenotypes
| rsID | Location/Effect | Phenotype | PMID |
|---|---|---|---|
| rs2072671 | 79A>C (Lys27Gln) | The C allele was not associated with CDA enzymatic activity compared to the A allele in patients with cancer when treated with gemcitabine and cisplatin. There was a toxic death case (severe hematological toxicity including leukopenia) in the single individual with the AC genotype, who also had CDA enzymatic activity less than 75% of the average of the cohort. | 17885621 |
| rs2072671 | 79A>C (Lys27Gln) | The AC and CC genotypes are associated with increased risk of severe neutropenia compared to genotype AA in people with late stage, or metastatic cancer (NSCLC, bladder, cholangiocellular carcinoma etc.). Some patients received gemcitabine monotherapy, others combination therapy with cisplatin. | 22546611 |
| rs2072671 | 79A>C (Lys27Gln) | The C allele is associated with increased frequency of nausea and vomiting compared to the A allele in people with mesothelioma that were treated with a combination of gemcitabine and platinum compounds. | 22134350 |
| rs2072671 | 79A>C (Lys27Gln) | The AA and AC genotypes are associated with increased risk of severe hematologic toxicity compared to genotype CC in people with pancreatic cancer that are treated with gemcitabine, but not in patients treated with fluorouracil. | 21625252 |
| rs2072671 | 79A>C (Lys27Gln) | The AA genotype is associated with a decreased risk of neutropenia compared to the AC and CC genotypes in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs2072671 | 79A>C (Lys27Gln) | The AA genotype is associated with increased risk of neutropenia or thrombocytopenia, and improved survival, including longer time to progression compared to genotypes AC and CC in people with stage III or IV NSCLC treated with gemcitabine and cisplatin. The A allele is also associated with decreased CDA enzymatic activity (in vitro). | 18347182 |
| rs2072671 | 79A>C (Lys27Gln) | The AA genotype is not associated with increased risk of neutropenia, or response compared to the AC and CC genotypes in people with NSCLC when treated with a combination gemcitabine and carboplatin. | 18538445 |
| rs2072671 | 79A>C (Lys27Gln) | The A allele is not associated with patient survival, or neurotoxicity compared to the C allele in people with metastatic breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs2072671 | 79A>C (Lys27Gln) | Genotype AA is associated with decreased activity of CDA in whole blood from healthy volunteers as compared to genotypes AC and CC (in vitro). | 23651026 |
| rs2072671 | 79A>C (Lys27Gln) | Genotype CC is associated with increased expression of CDA in PBMCs (in vitro). | 21521023 |
| rs2072671 | 79A>C (Lys27Gln) | The A allele is associated with decreased enzymatic activity compared to the C allele in cytoplasmic extracts from RBCs (in vitro). | 18600531 |
| rs2072671 | 79A>C (Lys27Gln) | The A allele is associated with decreased enzymatic activity compared to the C allele in in cells transfected with DNA from healthy Caucasian and African-American healthy volunteers (in vitro). | 16551864 |
| rs1048977 | 435C>T (Thr145=) | The TT genotype is associated with increased expression of CDA in peripheral mononuclear blood cells from healthy volunteers (in vitro) | 21521023 |
| rs1048977 | 435C>T (Thr145=) | The CC genotype is not associated with enzymatic activity of CDA in whole blood compared to genotypes CT and TT (in vitro). | 23651026 |
| rs1048977 | 435C>T (Thr145=) | The CC genotype is not associated with response compared to genotypes CT and TT in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs1048977 | 435C>T (Thr145=) | The C allele is not associated with response compared to the T allele in people with metastatic breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs1048977 | 435C>T (Thr145=) | The CC and CT genotypes are associated with increased elimination clearance of dFdU, and formation of dFdCTP when compared to the TT genotype in people with cancer. | 22838949 |
| rs1048977 | 435C>T (Thr145=) | The CC genotype is associated with increased response, but is not associated with hematological toxicity compared to genotypes CT and TT in people with NSCLC when treated with a combination of gemcitabine and carboplatin. | 18538445 |
| rs60369023 | 208G>A (Ala70Thr) | The AA genotype is associated with severe risk of toxicity compared to genotype GG. This is a case study of a single patient with pancreatic cancer who was treated with a combination of gemcitabine and cisplatin. | 15814642 |
| rs60369023 | 208G>A (Ala70Thr) | The A allele is associated with decreased clearance of gemcitabine, and increased risk of neutropenia compared to the G allele in people with cancer when gemcitabine is combined with platinum or fluorouracil therapy. | 17194903 |
| rs60369023 | 208G>A (Ala70Thr) | The A allele is not associated with increased risk of neurotoxicity compared to the G allele in people with metastatic breast cancer when treated with gemcitabine and paclitaxel. | 24361227 |
| rs60369023 | 208G>A (Ala70Thr) | The A allele is not associated with catalytic activity of recombinant CDA protein towards gemcitabine (in vitro). | 23230131 |
| rs4655226 | 155-3137C>T | Genotype CC is associated with increased clearance of dFdU compared to genotype CT in people with cancer. | 22838949 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: NSCLC (non-small cell lung cancer), CDA (cytidine deaminase), dFdU (2', 2' difluorodeoxyuracil), dFdCTP (gemcitabine triphosphate), PBMC (peripheral blood mononuclear cells).
Neutropenia is the most frequently reported adverse effect of gemcitabine, and risk of neutropenia is predicted to be associated with decreased CDA enzymatic activity. rs2072671 (A>C) changes the protein sequence of CDA by substituting a lysine residue for a glycine (Lys27Gln). At least five in vitro studies report that the A allele of rs2072671 is associated with decreased CDA enzymatic activity, or decreased CDA expression, compared to the C allele, although one in vitro study reports no association between the C allele and CDA enzymatic activity [25-29]. If the A allele at rs2072671 is associated with lower enzymatic activity of CDA relative to the C allele, then patients with the AA genotype should have a higher risk of neutropenia, or toxicity compared to patients with the CC genotype. Two studies support this prediction and report an increased risk of neutropenia associated with the AA genotype [25, 30]. However, three studies report an increased risk of toxicity, including neutropenia associated with the C allele compared to the A allele [22, 31, 32] and one reports no association with neutropenia [19]. In a study that reports no association between serum CDA enzymatic activity with the C allele, an AC genotype patient died after co-administration of gemcitabine and cisplatin. The patient's serum CDA enzymatic activity (measured as units of activity/mg of CDA protein) was 75% lower than the reference mean of the cohort [33], suggesting that CDA activity, perhaps more than individual SNPs, can predict risk of toxicity in patients administered gemcitabine. Associations between rs2072671 with survival outcomes, or response to gemcitabine are similarly inconclusive. Two studies report no association between rs2072671 with patient outcomes [19, 21], and one reports an association between the A allele with improved outcomes in patients with non-small cell lung cancer (NSCLC) [25].
Unlike, rs2072671, rs1048977 (C>T) is a synonymous variant (Thr145Thr). Investigations of this SNP have yielded inconclusive results regarding its effect on CDA enzymatic activity. One study reports that the TT genotype is associated with increased expression of CDA in peripheral mononuclear blood cells (PBMCs) from healthy volunteers, which would be expected to enhance CDA enzymatic activity [27]. An independent study, however, reports no association between rs1048977 and enzymatic activity of CDA when assessed in whole blood from healthy volunteers [28]. Two studies report no association between the C allele and patient outcomes [21, 22]. Another reports an association between the CC genotype with improved response to gemcitabine, including a longer median time to progression, but no association with risk of hematologic toxicity [19]. Finally, a study investigating possible associations between SNPs with changes in gemcitabine pharmacokinetics reports that there is an association between the CC and CT genotypes with increased clearance of dFdU, the product of gemcitabine metabolism by CDA [34].
The rs60369023 (G >A) variant, which has only been reported in Korean and Japanese patients [8, 21, 33, 35], changes an alanine residue to a threonine (Ala70Thr). One in vitro study reports that the A allele is associated with decreased CDA enzymatic activity, and a second in vitro study reports that the A allele is associated with decreased CDA activity towards AraC, another deoxycytidine analog, but not towards gemcitabine [26, 36]. Although this variant has not been extensively studied the evidence of an association between it and CDA enzymatic activity is compelling. In a small study of 6 Japanese cancer patients, the only person that was homozygous for the A allele suffered from severe neutropenia and abnormally high plasma levels of gemcitabine, and abnormally low levels of dFdU [37]. In another study of over 200 Japanese cancer patients, the A allele was associated with decreased dFdU levels, and elevated AUC and Cmax levelsof gemcitabine. Within the same cohort, the only patient that was homozygous for the A allele had abnormally high plasma levels of gemcitabine, and abnormally low plasma levels of dFdU. In addition, when platinum or fluorouracil were co-administered with gemcitabine, the A allele became significantly associated with neutropenia. Finally, the same study also quantified CDA enzymatic activity in a subset of patients (n=120), and reports that the A allele correlated with reduced enzymatic activity, providing further evidence of an association between the A allele with an increased risk of neutropenia, especially for patients co-administered platinum or fluorouracil [38]
Deoxycytidine Kinase
Gemcitabine is monophosphorylated by deoxycytidine kinase (DCK) to form 2′2′ difluorodeoxycytidine 5′ mono-phosphate (dFdCMP, gemcitabine monophosphate). In cancer cell lines prolonged exposure to gemcitabine correlates with decreased expression of DCK, and is associated with acquired resistance to gemcitabine [39, 40]. In vivo studies show an association between decreased DCK protein levels with shorter progression free survival in patients treated with gemcitabine [14, 41, 42]. Despite some well-established correlations between DCK protein levels with resistance to gemcitabine, no SNPs in DCK have shown a consistent association with patient response, or risk of toxicity in clinical studies [21, 22, 26, 43]. The results of those studies are summarized in Table 6.
Table 6.
Single nucleotide polymorphisms in DCK and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs4694362 | IVS6 – 1205C>T | The TT genotype was associated with an increased risk of neutropenia, but is not associated with response compared to the CT and TT genotypes in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs4694362 | IVS6 – 1205C>T | The T allele is not associated with response compared to the C allele in people with metastatic breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs4694362 | IVS6 −1205C>T | The T allele is associated with increased disease progression compared to the C allele when part of a haplotype (GGCCGTGT in rs3925058, rs7543016, rs4600090, rs9436883, rs35687416, rs1044457, rs7684954, rs4694362) in people with pancreatic cancer. | 22838950 |
| rs12648166 | 207+9846A>G | Genotype AA is not associated with response compared to genotypes AG and GG in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs12648166 | 207+9846A>G | The A allele is not associated with response compared to the G allele in people with breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs66878317 | 70A>G (Ile24Val) | The AG and GG genotypes are associated with increased clearance of gemcitabine compared to the AA genotypes in purified proteins (in vitro). | 23230131 |
| rs3775289 | IVS1 −1110T>C | Genotype CC is not associated with response, or risk of neutropenia in people with NSCLC compared to genotypes CT and TT in people with NSCLC. | 18538455 |
| rs2306744 | −201C>T | The T allele is associated with increased DCK expression both in vitro and in vivo, when part of a haplotype with the G allele of rs80143932 in people with AML. The two SNPs were found to be in strong LD. * | 15564883 |
| rs80143932 | −360C>G | The G allele is associated with increased DCK expression both in vitro and in vivo, when part of a haplotype with the T allele of rs2306744 in people with AML. The two SNPs were found to be in strong LD.* | 15564883 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: SNP (single nucleotide polymorphism), NSCLC (non-small cell lung cancer), AML (acute myeloid leukemia), DCK (deoxycytidine kinase), LD (linkage disequilibrium).
One study reports that the TT genotype at rs4694362 (C>T), an intronic SNP, is not significantly associated with patient outcomes, but that it is associated with increased risk of neutropenia in pancreatic cancer patients [22]. A second study reports that the T allele is associated with a more rapid disease progression, but only when it is part of a haplotype with seven other SNPs [20]. A third study reports that there is no association between the T allele with patient survival in metastatic breast cancer patients [21].
Despite a lack of association between variants in DCK and patient response to gemcitabine, there are well-established associations between variants in DCK and response to Ara-C, which is metabolized by DCK in a similar way to gemcitabine [44]. One study reports an association between the GT haplotype at rs80143932 and rs2306744 and a greater incidence of complete remission after the first induction of AraC with relapse free survival for six months after treatment [43]. The frequency of this haplotype is high in Asian patients and low in Caucasians (15.6% and 2%, respectively), and is also associated with increased DCK expression in vitro as well as in vivo [43, 45]. Although it has not yet been studied in patients receiving gemcitabine the associations between this haplotype with response to AraC may inform future investigations with gemcitabine.
Deoxycytidylate deaminase
Deoxycytidylate deaminase (DCTD) deaminates, and inactivates, gemcitabine monophosphate to difluorodeoxyuridine monophosphate (dFdUMP). The few studies investigating whether there are associations between genetic variants in DCTD and esponse to gemcitabine are summarized in Table 7. rs35932500 (T>C) is associated with decreased DCTD activity in vitro, but this has not been tested in clinical studies. One pharmacokinetic study reports significant associations between two SNPs in the 3’ UTR of with formation of dFdCTP [34]. A synonymous coding SNP, rs4742 (A>C), has also been evaluated in patients receiving gemcitabine, but it has not shown any significant association with either risk of neutropenia, or with patient response[19, 22].
Table 7.
Single nucleotide polymorphisms in DCTD and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs4742 | 348T>A, T>C, T>G (315 T>C) (Val105=) | Genotype TT is not associated with risk of neutropenia, or response to gemcitabine compared to genotypes CT and CC in people with NSCLC when treated with a combination gemcitabine and carboplatin. | 18538445 |
| rs4742 | 348T>A, T>C, T>G (315 T>C) (Val105=) | Genotype CC is not associated with response compared to genotypes CT and TT in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs12507552 | C>T (458+759G>A) | The TT genotype is associated with decreased survival compared to the CT and TT genotypes in people with pancreatic cancer. When the T allele is part of a haplotype (CGATAACA in rs12507552, rs3886768, rs13147196, rs13137332, rs13116598, rs6823077, rs11730323) it is associated with increased tumor progression. | 22838950 |
| rs35932500 | *T>C (172, 205A>G) (Asn69Asp, Asn58Asp) | The C allele is associated with decreased enzyme activity of DCTD compared to the T allele. The effect was seen in cells transfected with DNA from healthy Caucasian volunteers but not in cells transfected with DNA from healthy African-American volunteers (in vitro). | 16551864 |
| rs1130902 | *506G>A | The AA and AG genotypes were associated with increased formation clearance of dFdCTP compared to the GG genotype in people with cancer. | 22838949 |
| rs6834938 | *1739G>A | The CC and CT genotypes were associated with increased formation clearance of dFdCTP compared to the TT genotype in people with cancer. | 22838949 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: dFdCTP (gemcitabine triphosphate), NSCLC (non-small cell lung cancer), DCTD (deoxycytidylate deaminase).
Cytidine mono-phosphate kinase 1
Cytidine mono-phosphate kinase 1 (CMPK1) phosphorylates gemcitabine monophosphate to form 2’2’-difluorodeoxycytidine 5’ diphosphate (dFdCDP, gemcitabine diphosphate). dFdCDP inhibits the activity of ribonucleotide reductase, which replenishes cellular dNTP pools via the nucleoside salvage pathway. Table 8 summarizes the reported associations between SNPs in CMPK1 with patient response to gemcitabine. The reported associations between patient survival and SNPs in CMPK1 are inconclusive and require further investigation. The CC genotype rs1044457(C>T), a 3’UTR SNP, is associated with decreased formation of dFdCTP in patients with a variety of solid tumors [34], and a study of pancreatic cancer patients reports that the CC genotype is associated with improved overall survival, and a longer time to tumor and disease progression [20]. A third study in lung cancer patients reports no association between the CC genotype in rs1044457 and patient response [46].
Table 8.
Single nucleotide polymorphisms in CMPK1 and related phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs1044457 | *360C>T | The CC genotype was associated with improved overall survival, and a longer time to disease and tumor progression compared to the TT genotype in people with pancreatic cancer when treated with a combination of gemcitabine and erlotinib, or fluoropyrimidines. | 22838950 |
| rs1044457 | *360C>T | The CC genotype is not associated with response to gemcitabine compared to genotype TT in people with NSCLC treated with gemcitabine and cisplatin. | 21642870 |
| rs1044457 | *360C>T | The CC genotype is associated with decreased formation clearance of gemcitabine triphosphate compared to genotype TT in people with cancer. | 22838949 |
| rs4492666 | 171+1057A>C | The CC genotype is associated with shorter survival time compared to the AC and AA genotypes in people with NSCLC. | 21642870 |
| rs4492666 | 171+1057A>C | The A allele is not associated with response to gemcitabine compared to the C allele in people with breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs11211524 | 321-928A>C (172-928A>C) | The A allele is not associated with response to gemcitabine compared to allele C in people with breast cancer treated with a combination of gemcitabine and paclitaxel. | 24361227 |
| rs11211524 | 321-928A>C (172-928A>C) | The CC genotype is associated with decreased response compared to genotypes AA and AC in people with NSCLC and treated with a combination of gemcitabine and cisplatin. | 21642870 |
| rs35687416 | 240G>T (Gln80His) | The T allele is associated with improved overall survival and time to progression compared to the G allele in people with pancreatic cancer. | 22838950 |
| rs3925058 | −1995G>A | The G allele is not associated with a moderate increase in time to progression compared to the A allele in people with pancreatic cancer. | 22838950 |
| rs7543016 | 22G>C (Gly8Arg) | The G allele is associated with an increased time to progression compared to the C allele in people with pancreatic cancer. | 22838950 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: NSCLC (non-small cell lung cancer).
5’ Nucleotidases
5’ nucleotidase (NT5C) 2 and 3 catalyze the de-phosphorylation of gemcitabine mono-phosphate back to gemcitabine [1]. Increased expression of NT5C genes are associated with resistance to nucleoside analogs such as AraC and gemcitabine, and increased expression is predicted to also be associated with decreased survival in patients [47, 48]. Little is known about which specific genetic variants in NT5C or NT5C3 are responsible for variability in gene expression, much less which specific variants could influence patient outcomes. One study reports positive associations between specific genetic variants in genes that are involved in gemcitabine metabolism, including NT5C2, and NT5C3, with various pharmacokinetic endpoints including gemcitabine clearance, and dFdCTP formation clearance (the fraction of gemcitabine that forms gemcitabine triphosphate) [34]. The study reports that three SNPs in NT5C2 (rs1926029, rs3740387, rs11598702), and two SNPs in NT5C3 (rs3570117, rs6946062), are associated with inter-individual differences in gemcitabine pharmacokinetics. It is worth noting, however, that this exploratory study of 37 SNPs did not do corrections for multiple hypothesis testing, thus the reported associations merit further investigation.
Variants in drug targets
Ribonucleotide Reductase 1
Ribonucleotide reductase (RNR) catalyzes the synthesis of 2’ deoxyribonucleotides (dNTPs) from ribonucleotides. RNR is a complex that consists of a large and small subunit, and each sub-unit is composed of a ribonucleotide reductase homo-dimer. The large sub-unit is a homo-dimer of ribonucleotide reductase 1 (RRM1), and the small sub-unit is a homo-dimer of ribonulceotide reductase 2 (RRM2). RRM1 can also form an active complex with a homo-dimer of p53 inducible RRM2b, which is an isoform of RRM2 [49]. Gemcitabine diphosphate inhibits RRM1 by binding to its active site. This depletes a cell of intracellular dNTPs because RRM1 maintains cellular dNTP pools via the nucleoside salvage pathway. Less is known about the association between genetic variants in RRM2 and RRM2b gene expression, and gemcitabine resistance in vitro [24, 50]. In contrast, RRM1 has been thoroughly studied in regards to the effects of gemcitabine.
In addition to maintaining cellular dNTP pools, RRM1 also acts as a tumor suppressor. High levels of expression are associated with reduced cell migration in vitro, which has been confirmed in a syngeneic mouse model where RRM1 was also found to mediate the inhibition of cell migration, invasion, and metastasis, through PTEN, a critical tumor-suppressor [51, 52]. An in vitro study in a lung cancer cell line reports that prolonged gemcitabine exposure correlates with increased expression of RRM1, and gemcitabine resistance [51]. The association between high levels of RRM1 and gemcitabine resistance has been confirmed in multiple independent studies, which also show that gemcitabine resistant cells can be re-sensitized by siRNA knockdown of RRM1 [53-55]. Despite some conflicting results [56], current evidence supports a correlation between lower baseline RRM1 expression levels with increased time to tumor progression, and increased overall survival in patients treated with gemcitabine [49, 57, 58].
One of the most important discoveries regarding RRM1 is that higher baseline levels of RRM1 are correlated with improved outcome in cancer patients that are not treated with gemcitabine. In a study of non-small cell lung cancer (NSCLC) patients who were treated with surgery only, patients with higher baseline RRM1 protein levels in resected tumor samples had better outcomes than patients with lower baseline RRM1 protein levels [59]. Lower baseline RRM1 protein levels in tumors predicted improved response in patients treated with gemcitabine, but higher baseline RRM1 protein levels predicted improved response in patients treated with surgery only [60]. In patients treated with surgical resection, the TT genotype at rs11030918 (T>C) is associated with increased progression-free survival when it is part of a haplotype with a second RRM1 SNP, rs12806698 (C>A) [61].
The results of association studies investigating genetic variants in RRM1 with patient response to gemcitabine are summarized in Table 9. Two studies report that rs9937 (A>G) is associated with an increased risk of toxicity (hematological, and neurological toxicity), but its association with patient survival is unclear. One study reports that the G allele (when part of a haplotype with the A allele at rs1042858) is associated with a decreased risk of hematologic toxicity, but lower overall survival in breast cancer patients receiving gemcitabine monotherapy [62]. Another study reports that the AA genotype is associated with shorter progression free survival, and an increased risk of neutropenia [22]. A third study reports that the AA genotype is associated with an increased risk of neurotoxicity, but is not associated with survival outcomes in metastatic breast cancer patients [21]. All three studies provide some evidence of association between the A allele with an increased risk of adverse reactions, but the associations with survival are inconsistent. Although rs11030918 (T>C) is reported to be associated with survival in patients treated with resection, it is not associated with survival in patients administered gemcitabine. One study reports an association between the T allele at rs11030918 with an increased risk of neurotoxicity (when it is part of a haplotype with three other SNPs), and a second study reports an association between the C allele and a decreased incidence of nausea and vomiting [21, 22]. Given all the known biological functions of RNR in the development of cancer it is important to consider how the types of malignancy and the types of combination chemotherapies could result in the inconsistent associations that are reported here.
Table 9.
Single nucleotide polymorphisms in RRM1 and Related Phenotypes
| rsID | Location/Effect | Findings | PMID |
|---|---|---|---|
| rs183484 | 1082C>A (Arg284Arg) | The CC genotype is not associated with overall survival compared to the genotypes AC and AA in people with NSCLC given a combination of gemcitabine and cisplatin. | 21642870 |
| rs183484 | 1082C>A (Arg284Arg) | The AC genotype is moderately associated with a decrease in progression free survival compared to genotypes AA and CC in people with pancreatic cancer; four SNPs in this study, including rs183484, were predictive of a decrease in progression free survival when using 0 or 1 variant as a reference. The other variants included rs2072671, rs760370, rs9937. | 20665488 |
| rs1042927 | *316C>A | The C allele is associated with a decrease in overall survival compared to the A allele in people with mesothelioma. | 22134350 |
| rs1042927 | *316C>A | The A allele is associated with an increased likelihood of neurotoxicity, but not patient response compared to the C allele in people with metastatic breast cancer that are treated with a combination of gemcitabine and paclitaxel; this SNP is part of a four SNP haplotype that includes rs1042927, rs11030918, rs9937, and rs720106. | 24361227 |
| rs11030918 | −756T>C (−524T>C) | The TT genotype is not associated with risk of hematologic toxicity, or response to gemcitabine in people with NSCLC compared to genotypes CC and CT when treated with a combination of gemcitabine and cisplatin. | 18538445 |
| rs11030918 | −756T>C (−524T>C) | The C allele is associated with a decreased occurrence of nausea and vomiting compared to the T allele in people with mesothelioma. | 22134350 |
| rs11030918 | −756T>C (−524T>C) | The TT genotype is not associated with overall survival compared to the CT and CC genotypes in people with NSCLC given a combination of gemcitabine and carboplatin. | 21642870 |
| rs11030918 | −756T>C (−524T>C) | The T allele is associated with increased risk of neurotoxicity compared to the C allele in people with breast cancer; this SNP is part of a four SNP haplotype that includes rs1042927, rs11030918, rs9937, and rs720106. | 24361227 |
| rs9937 | 2455 A>G (Thr741Thr) | The G allele is associated with a decreased risk of neutropenia and leukopenia compared to the A allele when part of a haplotype with the A allele at rs1042858 in people with breast cancer. The G allele is also associated with shorter progression-free survival, and lower overall survival when part of a haplotype with the A allele at rs1042858. | 17602053 |
| rs9937 | 2455 A>G (Thr741Thr) | The AA genotype is associated with shorter progression-free survival and increased risk of neutropenia when compared to genotypes AG and GG in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs9937 | 2455 A>G (Thr741Thr) | The AA genotype is associated with increased risk of neurotoxicity compared to the AG and GG genotypes in people with metastatic breast cancer when treated with a combination of gemcitabine and paclitaxel; this SNP is part of a four SNP haplotype that includes rs1042927, rs11030918, rs9937, and rs720106. This variant was not associated with overall, or progression-free survival. | 24361227 |
| rs1042858 | 42G>A (2464G>A) (A744A) | The AA genotype is not associated with progression free survival compared to genotypes AG and GG in people with pancreatic cancer when treated with a combination of gemcitabine and radiotherapy, or platinum. | 20665488 |
| rs1042858 | 42G>A (2464G>A) (A744A) | The A allele is associated with a decreased risk of neutropenia, leukopenia, shorter progression free survival, and lower overall survival compared to the G allele in people with breast cancer when part of a haplotype with rs9937 G allele. | 17602053 |
| rs1042858 | 42G>A (2464G>A) (A744A) | The AA genotype is not associated with progression free survival compared with the genotypes AG and GG in people with NSCLC. | 20226083 |
| rs12806698 | (−37C>A) −269C>A | The CC genotype is not associated with hematological toxicity, survival, or tumor response compared to the AC or AA genotypes in people with NSCLC when treated with a combination of gemcitabine and carboplatin. | 18538445 |
| rs12806698 | (−37C>A) −269C>A | The CC genotype is not associated with overall survival compared with genotypes AC and AA in people with NSCLC. | 21642870 |
| rs12806698 | (−37C>A) −269C>A | The CC genotype is moderately associated with better overall survival and progression free survival compared to the genotypes AC and AA when part of a haplotype with genotype TT rs11030918 in people with lung cancer. | 15639717 |
| rs12806698 | (−37C>A) −269C>A | The AA genotype is associated with decreased incidence of nausea and vomiting compared to the genotypes AC and AA in people with mesothelioma. | 22134350 |
| rs12806698 | (−37C>A) −269C>A | The AC genotype is associated with improved tumor response compared with genotypes AA and CC in people with NSCLC when part of a haplotype with rs11030918 CT, although overall and progression free survival did not differ between groups. | 18483375 |
| rs12806698 | (−37C>A) −269C>A | The AC genotype is associated with longer progression free survival compared to the AA and CC genotypes in people with NSCLC. | 20226083 |
| rs720106 | 792+287T>C | The T allele is associated with increased risk of neurotoxicity compared to the C allele. This SNP is part of a four SNP haplotype that includes rs1042927, rs11030918, rs9937, and rs720106 | 24361227 |
All alleles reported in the table are on the positive strand. Variants marked by an * are the complement of what is reported in the study.
Abbreviations: SNP (single nucleotide polymorphism), NSCLC (non-small cell lung cancer).
Conclusions and Future Directions
In this review, we have summarized the results of pharmacogenomic studies investigating patient variability in response to gemcitabine. None of the SNPs covered here are consistently associated with any clinically relevant phenotypes, which could be due to one of many factors. Heterogeneity of the patients, including differences in the type of malignancy, the stage of malignancy, the type of combination chemotherapy, and differences in the ethnic background of patients, makes comparing the results between studies very difficult. In addition many studies reported here include less than one hundred patients, so if the effect size of an individual SNP is small, true associations may be missed.
As an example, the C allele at rs2072671 (A>C) in CDA changes the amino acid sequence of the protein. Because of its critical role in detoxifying gemcitabine alterations in the protein sequence might increase a patient's risk of an adverse reaction to gemcitabine (e.g. neutropenia). In the five studies that investigated associations between rs2072671 with risk of neutropenia, four report an association and one does not. Two report an association between increased risk of neutropenia with the A allele [25, 30], and two report an association with the C allele [32]. All patients were treated with gemcitabine, but some were also treated with radiotherapy, paclitaxel, erlotinib, platinum, or were sometimes treated with gemcitabine alone. In the study that reports no association between rs2072671 (A>C) with risk neutropenia, patients were a mix of Chinese, Malay, or Indian [19]. Only one of the five studies had more than 100 patients [22].
Variability of gene expression, and protein levels of SLC29A1, CDA, and RRM1 present the most compelling associations with patient response to gemcitabine. Two recent meta-analyses (12 studies, that included 875 pancreatic cancer patients, and 16 studies that included 632 pancreatic cancer patients), both concluded that a high level of SLC29A1 protein is prognostic of improved survival outcomes in patients administered gemcitabine compared to patients with low levels of SLC29A1 protein [12, 63]. Although more evidence is needed, it appears that low CDA enzymatic activity is a good predictor of whether patients are more likely to experience gemcitabine-associated toxicity [30, 33, 37, 38]. High baseline protein levels of RRM1 are also good predictors of not only whether patients are likely to develop resistance to gemcitabine, but whether they might derive more benefit from forgoing gemcitabine treatment in favor of surgical resection with curative intent [49].
In summary, gemcitabine pharmacogenomic studies investigating single nucleotide variants have yielded little in terms of clinically actionable findings. Fortunately, some relevant associations have come from association studies that investigate expression levels, or protein levels of key genes and proteins, respectively, of genes in the pharmacokinetic and pharmacodynamic pathways of gemcitabine. In addition, further research efforts extending beyond the known pharmacodynamic and pharmacokinetic pathways have uncovered additional biology associated with gemcitabine efficacy, and additional genes outside these pathways have already been identified to play a role in gemcitabine response [64, 65]. Ultimately, more research is needed to discover clinically relevant single nucleotide variants to improve upon current gemcitabine therapies for cancer patients.
Acknowledgments
The authors would like to thank Feng Liu for assistance with the graphics. PharmGKB is supported by the NIH/NIGMS R24 GM61374. Dr. Wang would also like to acknowledge support by CA138461, GM61388, and the Mayo Center for Individualized Medicine.
Abbreviations
- dFdU
2’2’ difluorodeoxyuridine
- dFdUMP
2’2’ difluorodeoxyuridine-5’- monophosphate
- NDP
ribonucleotide diphosphate
- dNDP
2’- deoxynucleotide diphosphate
- dNTP
2’- deoxynucleotide triphosphate
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
References
- 1.Wong A, Soo RA, Yong WP, Innocenti F. Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab Rev. 2009;41:77–88. doi: 10.1080/03602530902741828. [DOI] [PubMed] [Google Scholar]
- 2.Gemcitabine HCL Drug Label. [ http://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=7a753acb-5061-4d34-bc42-c2c7a9512e7a]
- 3.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Ann Oncol. Vol. 17. Suppl 5: 2006. Cellular pharmacology of gemcitabine. pp. v7–12. [DOI] [PubMed] [Google Scholar]
- 4.Heinemann V, Xu YZ, Chubb S, Sen A, Hertel LW, Grindey GB, Plunkett W. Inhibition of ribonucleotide reduction in CCRF-CEM cells by 2′,2′-difluorodeoxycytidine. Mol Pharmacol. 1990;38:567–572. [PubMed] [Google Scholar]
- 5.Heinemann V, Xu YZ, Chubb S, Sen A, Hertel LW, Grindey GB, Plunkett W. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: a mechanism of self-potentiation. Cancer Res. 1992;52:533–539. [PubMed] [Google Scholar]
- 6.Plunkett W, Huang P, Xu YZ, Heinemann V, Grunewald R, Gandhi V. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin Oncol. 1995;22:3–10. [PubMed] [Google Scholar]
- 7.Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2′,2′-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- 8.Ciccolini J, Dahan L, Andre N, Evrard A, Duluc M, Blesius A, Yang C, Giacometti S, Brunet C, Raynal C, et al. Cytidine deaminase residual activity in serum is a predictive marker of early severe toxicities in adults after gemcitabine-based chemotherapies. J Clin Oncol. 2010;28:160–165. doi: 10.1200/JCO.2009.24.4491. [DOI] [PubMed] [Google Scholar]
- 9.Li F, Hu G, Jiang Z, Guo J, Wang K, Ouyang K, Wen D, Zhu M, Liang J, Qin X, Zhang L. Identification of NME5 as a contributor to innate resistance to gemcitabine in pancreatic cancer cells. FEBS J. 2012;279:1261–1273. doi: 10.1111/j.1742-4658.2012.08521.x. [DOI] [PubMed] [Google Scholar]
- 10.Takadate T, Onogawa T, Fujii K, Motoi F, Mikami S, Fukuda T, Kihara M, Suzuki T, Takemura T, Minowa T, et al. Nm23/nucleoside diphosphate kinase-A as a potent prognostic marker in invasive pancreatic ductal carcinoma identified by proteomic analysis of laser micro-dissected formalin-fixed paraffin-embedded tissue. Clin Proteomics. 2012;9:8. doi: 10.1186/1559-0275-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Visser F, King KM, Baldwin SA, Young JD, Cass CE. The role of nucleoside transporters in cancer chemotherapy with nucleoside drugs. Cancer Metastasis Rev. 2007;26:85–110. doi: 10.1007/s10555-007-9044-4. [DOI] [PubMed] [Google Scholar]
- 12.Liu ZQ, Han YC, Zhang X, Chu L, Fang JM, Zhao HX, Chen YJ, Xu Q. Prognostic value of human equilibrative nucleoside transporter1 in pancreatic cancer receiving gemcitabin-based chemotherapy: a meta-analysis. PLoS One. 2014;9:e87103. doi: 10.1371/journal.pone.0087103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenhalf W, Ghaneh P, Neoptolemos JP, Palmer DH, Cox TF, Lamb RF, Garner E, Campbell F, Mackey JR, Costello E, et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer Inst. 2014;106:djt347. doi: 10.1093/jnci/djt347. [DOI] [PubMed] [Google Scholar]
- 14.Marechal R, Bachet JB, Mackey JR, Dalban C, Demetter P, Graham K, Couvelard A, Svrcek M, Bardier-Dupas A, Hammel P, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143:664–674. e661–666. doi: 10.1053/j.gastro.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Spratlin J, Sangha R, Glubrecht D, Dabbagh L, Young JD, Dumontet C, Cass C, Lai R, Mackey JR. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10:6956–6961. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- 16.Advani AS, Shadman M, Ali-Osman F, Barker A, Rybicki L, Kalaycio M, Sekeres MA, de Castro CM, Diehl LF, Moore JO, et al. A Phase II trial of gemcitabine and mitoxantrone for patients with acute myeloid leukemia in first relapse. Clin Lymphoma Myeloma Leuk. 2010;10:473–476. doi: 10.3816/CLML.2010.n.082. [DOI] [PubMed] [Google Scholar]
- 17.Pennycooke M, Chaudary N, Shuralyova I, Zhang Y, Coe IR. Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem Biophys Res Commun. 2001;280:951–959. doi: 10.1006/bbrc.2000.4205. [DOI] [PubMed] [Google Scholar]
- 18.Bhutia YD, Hung SW, Patel B, Lovin D, Govindarajan R. CNT1 expression influences proliferation and chemosensitivity in drug-resistant pancreatic cancer cells. Cancer Res. 2011;71:1825–1835. doi: 10.1158/0008-5472.CAN-10-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soo RA, Wang LZ, Ng SS, Chong PY, Yong WP, Lee SC, Liu JJ, Choo TB, Tham LS, Lee HS, et al. Distribution of gemcitabine pathway genotypes in ethnic Asians and their association with outcome in non-small cell lung cancer patients. Lung Cancer. 2009;63:121–127. doi: 10.1016/j.lungcan.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Woo HI, Kim KK, Choi H, Kim S, Jang KT, Yi JH, Park YS, Park JO, Lee SY. Effect of genetic polymorphisms on therapeutic response and clinical outcomes in pancreatic cancer patients treated with gemcitabine. Pharmacogenomics. 2012;13:1023–1035. doi: 10.2217/pgs.12.82. [DOI] [PubMed] [Google Scholar]
- 21.Lee SY, Im SA, Park YH, Woo SY, Kim S, Choi MK, Chang W, Ahn JS, Im YH. Genetic polymorphisms of SLC28A3, SLC29A1 and RRM1 predict clinical outcome in patients with metastatic breast cancer receiving gemcitabine plus paclitaxel chemotherapy. Eur J Cancer. 2014;50:698–705. doi: 10.1016/j.ejca.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Javle M, Dong X, Eng C, Abbruzzese JL, Li D. Gemcitabine metabolic and transporter gene polymorphisms are associated with drug toxicity and efficacy in patients with locally advanced pancreatic cancer. Cancer. 2010;116:5325–5335. doi: 10.1002/cncr.25282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatri A, Williams BW, Fisher J, Brundage RC, Gurvich VJ, Lis LG, Skubitz KM, Dudek AZ, Greeno EW, Kratzke RA, et al. SLC28A3 genotype and gemcitabine rate of infusion affect dFdCTP metabolite disposition in patients with solid tumours. Br J Cancer. 2014;110:304–312. doi: 10.1038/bjc.2013.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueno H, Kiyosawa K, Kaniwa N. Pharmacogenomics of gemcitabine: can genetic studies lead to tailor-made therapy? Br J Cancer. 2007;97:145–151. doi: 10.1038/sj.bjc.6603860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibaldi C, Giovannetti E, Vasile E, Mey V, Laan AC, Nannizzi S, Di Marsico R, Antonuzzo A, Orlandini C, Ricciardi S, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14:1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 26.Baker JA, Wickremsinhe ER, Li CH, Oluyedun OA, Dantzig AH, Hall SD, Qian YW, Ring BJ, Wrighton SA, Guo Y. Pharmacogenomics of gemcitabine metabolism: functional analysis of genetic variants in cytidine deaminase and deoxycytidine kinase. Drug Metab Dispos. 2013;41:541–545. doi: 10.1124/dmd.112.048769. [DOI] [PubMed] [Google Scholar]
- 27.Parmar S, Seeringer A, Denich D, Gartner F, Pitterle K, Syrovets T, Ohmle B, Stingl JC. Variability in transport and biotransformation of cytarabine is associated with its toxicity in peripheral blood mononuclear cells. Pharmacogenomics. 2011;12:503–514. doi: 10.2217/pgs.10.200. [DOI] [PubMed] [Google Scholar]
- 28.Carpi FM, Vincenzetti S, Ubaldi J, Pucciarelli S, Polzonetti V, Micozzi D, Mignini F, Napolioni V. CDA gene polymorphisms and enzyme activity: genotypephenotype relationship in an Italian-Caucasian population. Pharmacogenomics. 2013;14:769–781. doi: 10.2217/pgs.13.56. [DOI] [PubMed] [Google Scholar]
- 29.Giovannetti E, Laan AC, Vasile E, Tibaldi C, Nannizzi S, Ricciardi S, Falcone A, Danesi R, Peters GJ. Correlation between cytidine deaminase genotype and gemcitabine deamination in blood samples. Nucleosides Nucleotides Nucleic Acids. 2008;27:720–725. doi: 10.1080/15257770802145447. [DOI] [PubMed] [Google Scholar]
- 30.Farrell JJ, Bae K, Wong J, Guha C, Dicker AP, Elsaleh H. Cytidine deaminase single-nucleotide polymorphism is predictive of toxicity from gemcitabine in patients with pancreatic cancer: RTOG 9704. Pharmacogenomics J. 2012;12:395–403. doi: 10.1038/tpj.2011.22. [DOI] [PubMed] [Google Scholar]
- 31.Erculj N, Kovac V, Hmeljak J, Franko A, Dodic-Fikfak M, Dolzan V. The influence of gemcitabine pathway polymorphisms on treatment outcome in patients with malignant mesothelioma. Pharmacogenet Genomics. 2012;22:58–68. doi: 10.1097/FPC.0b013e32834e3572. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Zhou Y, Zhang J, Chen Y, Zhuang R, Liu T, Cai W. High incidence of severe neutropenia after gemcitabine-based chemotherapy in Chinese cancer patients with CDA 79A>C mutation. Clin Chim Acta. 2012;413:1284–1287. doi: 10.1016/j.cca.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Mercier C, Raynal C, Dahan L, Ortiz A, Evrard A, Dupuis C, Blesius A, Duluc M, Franceschini F, Giacometti S, et al. Toxic death case in a patient undergoing gemcitabine-based chemotherapy in relation with cytidine deaminase downregulation. Pharmacogenet Genomics. 2007;17:841–844. doi: 10.1097/FPC.0b013e32825ea6e3. [DOI] [PubMed] [Google Scholar]
- 34.Mitra AK, Kirstein MN, Khatri A, Skubitz KM, Dudek AZ, Greeno EW, Kratzke RA, Lamba JK. Pathway-based pharmacogenomics of gemcitabine pharmacokinetics in patients with solid tumors. Pharmacogenomics. 2012;13:1009–1021. doi: 10.2217/pgs.12.81. [DOI] [PubMed] [Google Scholar]
- 35.Iyer SN, Ankala A, Singhal RS, Hegde MR. Determination of common genetic variants in cytidine deaminase (CDA) gene in Indian ethnic population. Gene. 2013;524:35–39. doi: 10.1016/j.gene.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 36.Yue L, Saikawa Y, Ota K, Tanaka M, Nishimura R, Uehara T, Maeba H, Ito T, Sasaki T, Koizumi S. A functional single-nucleotide polymorphism in the human cytidine deaminase gene contributing to ara-C sensitivity. Pharmacogenetics. 2003;13:29–38. doi: 10.1097/00008571-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Yonemori K, Ueno H, Okusaka T, Yamamoto N, Ikeda M, Saijo N, Yoshida T, Ishii H, Furuse J, Sugiyama E, et al. Severe drug toxicity associated with a singlenucleotide polymorphism of the cytidine deaminase gene in a Japanese cancer patient treated with gemcitabine plus cisplatin. Clin Cancer Res. 2005;11:2620–2624. doi: 10.1158/1078-0432.CCR-04-1497. [DOI] [PubMed] [Google Scholar]
- 38.Sugiyama E, Kaniwa N, Kim SR, Kikura-Hanajiri R, Hasegawa R, Maekawa K, Saito Y, Ozawa S, Sawada J, Kamatani N, et al. Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J Clin Oncol. 2007;25:32–42. doi: 10.1200/JCO.2006.06.7405. [DOI] [PubMed] [Google Scholar]
- 39.Saiki Y, Yoshino Y, Fujimura H, Manabe T, Kudo Y, Shimada M, Mano N, Nakano T, Lee Y, Shimizu S, et al. DCK is frequently inactivated in acquired gemcitabine-resistant human cancer cells. Biochem Biophys Res Commun. 2012;421:98–104. doi: 10.1016/j.bbrc.2012.03.122. [DOI] [PubMed] [Google Scholar]
- 40.Achiwa H, Oguri T, Sato S, Maeda H, Niimi T, Ueda R. Determinants of sensitivity and resistance to gemcitabine: the roles of human equilibrative nucleoside transporter 1 and deoxycytidine kinase in non-small cell lung cancer. Cancer Sci. 2004;95:753–757. doi: 10.1111/j.1349-7006.2004.tb03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebastiani V, Ricci F, Rubio-Viqueira B, Kulesza P, Yeo CJ, Hidalgo M, Klein A, Laheru D, Iacobuzio-Donahue CA. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12:2492–2497. doi: 10.1158/1078-0432.CCR-05-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroep JR, Loves WJ, van der Wilt CL, Alvarez E, Talianidis I, Boven E, Braakhuis BJ, van Groeningen CJ, Pinedo HM, Peters GJ. Pretreatment deoxycytidine kinase levels predict in vivo gemcitabine sensitivity. Mol Cancer Ther. 2002;1:371–376. [PubMed] [Google Scholar]
- 43.Shi JY, Shi ZZ, Zhang SJ, Zhu YM, Gu BW, Li G, Bai XT, Gao XD, Hu J, Jin W, et al. Association between single nucleotide polymorphisms in deoxycytidine kinase and treatment response among acute myeloid leukaemia patients. Pharmacogenetics. 2004;14:759–768. doi: 10.1097/00008571-200411000-00007. [DOI] [PubMed] [Google Scholar]
- 44.Lamba JK. Genetic factors influencing cytarabine therapy. Pharmacogenomics. 2009;10:1657–1674. doi: 10.2217/pgs.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joerger M, Bosch TM, Doodeman VD, Beijnen JH, Smits PH, Schellens JH. Novel deoxycytidine kinase gene polymorphisms: a population screening study in Caucasian healthy volunteers. Eur J Clin Pharmacol. 2006;62:681–684. doi: 10.1007/s00228-006-0162-7. [DOI] [PubMed] [Google Scholar]
- 46.Ryu JS, Shin ES, Nam HS, Yi HG, Cho JH, Kim CS, Kim HJ, Lee JE. Differential effect of polymorphisms of CMPK1 and RRM1 on survival in advanced nonsmall cell lung cancer patients treated with gemcitabine or taxane/cisplatinum. J Thorac Oncol. 2011;6:1320–1329. doi: 10.1097/JTO.0b013e3182208e26. [DOI] [PubMed] [Google Scholar]
- 47.Mitra AK, Crews KR, Pounds S, Cao X, Feldberg T, Ghodke Y, Gandhi V, Plunkett W, Dolan ME, Hartford C, et al. Genetic variants in cytosolic 5′-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J Pharmacol Exp Ther. 2011;339:9–23. doi: 10.1124/jpet.111.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine). Drug Resist Updat. 2002;5:19–33. doi: 10.1016/s1368-7646(02)00002-x. [DOI] [PubMed] [Google Scholar]
- 49.Jordheim LP, Seve P, Tredan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12:693–702. doi: 10.1016/S1470-2045(10)70244-8. [DOI] [PubMed] [Google Scholar]
- 50.Sato J, Kimura T, Saito T, Anazawa T, Kenjo A, Sato Y, Tsuchiya T, Gotoh M. Gene expression analysis for predicting gemcitabine resistance in human cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2011;18:700–711. doi: 10.1007/s00534-011-0376-7. [DOI] [PubMed] [Google Scholar]
- 51.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64:3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 52.Bepler G, Sharma S, Cantor A, Gautam A, Haura E, Simon G, Sharma A, Sommers E, Robinson L. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Wonganan P, Chung WG, Zhu S, Kiguchi K, Digiovanni J, Cui Z. Silencing of ribonucleotide reductase subunit M1 potentiates the antitumor activity of gemcitabine in resistant cancer cells. Cancer Biol Ther. 2012;13:908–914. doi: 10.4161/cbt.20843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oguri T, Achiwa H, Sato S, Bessho Y, Takano Y, Miyazaki M, Muramatsu H, Maeda H, Niimi T, Ueda R. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: a role of ABCC5 in gemcitabine sensitivity. Mol Cancer Ther. 2006;5:1800–1806. doi: 10.1158/1535-7163.MCT-06-0025. [DOI] [PubMed] [Google Scholar]
- 55.Reid G, Wallant NC, Patel R, Antonic A, Saxon-Aliifaalogo F, Cao H, Webster G, Watson JD. Potent subunit-specific effects on cell growth and drug sensitivity from optimised siRNA-mediated silencing of ribonucleotide reductase. J RNAi Gene Silencing. 2009;5:321–330. [PMC free article] [PubMed] [Google Scholar]
- 56.Valsecchi ME, Holdbrook T, Leiby BE, Pequignot E, Littman SJ, Yeo CJ, Brody JR, Witkiewicz AK. Is there a role for the quantification of RRM1 and ERCC1 expression in pancreatic ductal adenocarcinoma? BMC Cancer. 2012;12:104. doi: 10.1186/1471-2407-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong W, Zhang X, Wu J, Chen L, Li L, Sun J, Lv Y, Wei X, Du Y, Jin H, Dong J. RRM1 expression and clinical outcome of gemcitabine-containing chemotherapy for advanced non-small-cell lung cancer: a meta-analysis. Lung Cancer. 2012;75:374–380. doi: 10.1016/j.lungcan.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Rosell R, Felip E, Taron M, Majo J, Mendez P, Sanchez-Ronco M, Queralt C, Sanchez JJ, Maestre J. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res. 2004;10:4215s–4219s. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- 59.Bartolucci R, Wei J, Sanchez JJ, Perez-Roca L, Chaib I, Puma F, Farabi R, Mendez P, Roila F, Okamoto T, et al. XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung Cancer. 2009;10:47–52. doi: 10.3816/CLC.2009.n.007. [DOI] [PubMed] [Google Scholar]
- 60.Akita H, Zheng Z, Takeda Y, Kim C, Kittaka N, Kobayashi S, Marubashi S, Takemasa I, Nagano H, Dono K, et al. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009;28:2903–2909. doi: 10.1038/onc.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bepler G, Zheng Z, Gautam A, Sharma S, Cantor A, Sharma A, Cress WD, Kim YC, Rosell R, McBride C, et al. Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer. 2005;47:183–192. doi: 10.1016/j.lungcan.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 62.Rha SY, Jeung HC, Choi YH, Yang WI, Yoo JH, Kim BS, Roh JK, Chung HC. An association between RRM1 haplotype and gemcitabine-induced neutropenia in breast cancer patients. Oncologist. 2007;12:622–630. doi: 10.1634/theoncologist.12-6-622. [DOI] [PubMed] [Google Scholar]
- 63.Wei CH, Gorgan TR, Elashoff DA, Hines OJ, Farrell JJ, Donahue TR. A metaanalysis of gemcitabine biomarkers in patients with pancreaticobiliary cancers. Pancreas. 2013;42:1303–1310. doi: 10.1097/MPA.0b013e3182a23ae4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou J, Wang L. FKBP5 as a selection biomarker for gemcitabine and Akt inhibitors in treatment of pancreatic cancer. PLoS One. 2012;7:e36252. doi: 10.1371/journal.pone.0036252. [DOI] [PMC free article] [PubMed] [Google Scholar]