Abstract
The function of neural circuits depends on the precise connectivity between populations of neurons. Increasing evidence indicates that disruptions in excitatory or inhibitory synapse formation or function lead to excitation/inhibition (E/I) imbalances and contribute to neurodevelopmental and psychiatric disorders. Leucine-rich repeat (LRR)-containing surface proteins have emerged as key organizers of excitatory and inhibitory synapses. Distinct LRR proteins are expressed in different cell types and interact with key pre- and postsynaptic proteins. These protein interaction networks allow LRR proteins to coordinate pre- and postsynaptic elements during synapse formation and differentiation, pathway-specific synapse development, and synaptic plasticity. LRR proteins thus play a critical role in organizing synaptic connections into functional neural circuits, and their dysfunction may contribute to neuropsychiatric disorders.
Keywords: Synaptogenesis, Synaptic Adhesion, Excitation/Inhibition Balance, Connectivity, Synaptic Transmission, Glutamate Receptor
LRR proteins and the organization of functional neural circuits
The function of neural circuits depends on the precise connectivity between populations of neurons. In the central nervous system (CNS) this is mediated by glutamatergic and GABAergic synapses, and there is emerging evidence that disruptions in the formation or function of excitatory or inhibitory synapses lead to excitation/inhibition (E/I) imbalances, which characterize several psychiatric and neurodevelopmental disorders [1–8]. These considerations underscore the importance of understanding the molecular control of excitatory and inhibitory synapse formation, and the signals that allow cell type-specific control of E/I balance.
Here, we review recent evidence indicating that cell surface proteins containing an extracellular leucine-rich repeat (LRR) domain [9] are key organizers of excitatory and inhibitory synapses in the CNS. Distinct LRR proteins are expressed in different cell types, and are generally localized to the postsynaptic membrane, from where they exert a strong influence on the development of synaptic connections. LRR proteins interact with key components of the postsynaptic machinery and trans-synaptically couple to essential presynaptic receptors, which places them in an ideal position to coordinate pre- and postsynaptic differentiation during circuit formation. New insights indicate that the function of LRR proteins extends beyond the initial formation of synaptic contacts and point to a role in regulating functional properties and activity-dependent plasticity of synapses. These findings indicate that LRR proteins play a critical role in the organization and function of neural circuits.
LRR proteins as regulators of synapse development
Recent studies have identified several closely related LRR protein families as regulators of synapse development. A simple in vitro assay, which tests the ability of neurons to form synapses onto cocultured heterologous cells expressing candidate genes [10, 11], has been instrumental in identifying synapse-organizing or synaptogenic LRR proteins. The elucidation of their trans-synaptic interactions has highlighted a common theme of diverse postsynaptic ligands coupling to a limited repertoire of presynaptic receptors. Below, we provide an inventory of these LRR proteins and discuss their roles in synapse development and function, focusing on the vertebrate system.
LRRTMs control excitatory synapse development via distinct presynaptic partners
Since their original identification as synaptogenic proteins in a coculture assay-based expression screen [12], Leucine-Rich Repeat Transmembrane Neuronal proteins (LRRTM1-4) have rapidly become the most intensively studied family of LRR-containing synaptic organizers. LRRTMs are type I transmembrane proteins with an extracellular LRR domain and a C-terminal PDZ interaction site (Fig. 1A), and distinct, partially overlapping expression patterns in the brain [13] (Fig. 4). LRRTM2, the best characterized family member, localizes to the postsynaptic density of excitatory synapses and regulates postsynaptic differentiation by recruiting key elements of the synaptic machinery, including the scaffolding protein PSD-95 and glutamate receptor subunits [12, 14]. LRRTM2 expressed on the surface of non-neuronal cells induces presynaptic differentiation in contacting axons of co-cultured neurons, a property shared with LRRTM1, LRRTM4, and to a lesser degree, LRRTM3 [12, 14, 15]. LRRTM1 and LRRTM2 induce presynaptic differentiation by trans-synaptically binding to neurexins [14–16], a family of alternatively spliced receptors that organize presynaptic development and function [17, 18]. Neurexins interact with a multitude of postsynaptic ligands, including the neuroligins and the cerebellin-glutamate receptor δ complex [19–21]. LRRTM binding to neurexin is regulated by alternative splicing: LRRTM1 and -2 only bind to neurexins lacking a small insert at splice site 4 (S4) [15, 16]. Surprisingly, LRRTM4 uses a different mechanism to induce presynaptic differentiation. LRRTM4 binds to heparan sulfate proteoglycans (HSPGs), most prominently glypicans, and requires HS on the presynaptic cell surface to induce synapse formation [22, 23] (Fig. 1A). Unlike neurexins, GPI-anchored glypicans lack a cytoplasmic domain, raising the possibility of a transmembrane co-receptor to trigger presynaptic differentiation. The receptor protein tyrosine phosphatase LAR (leukocyte common antigen-related) binds glypican in Drosophila [24], and is a candidate for such a co-receptor.
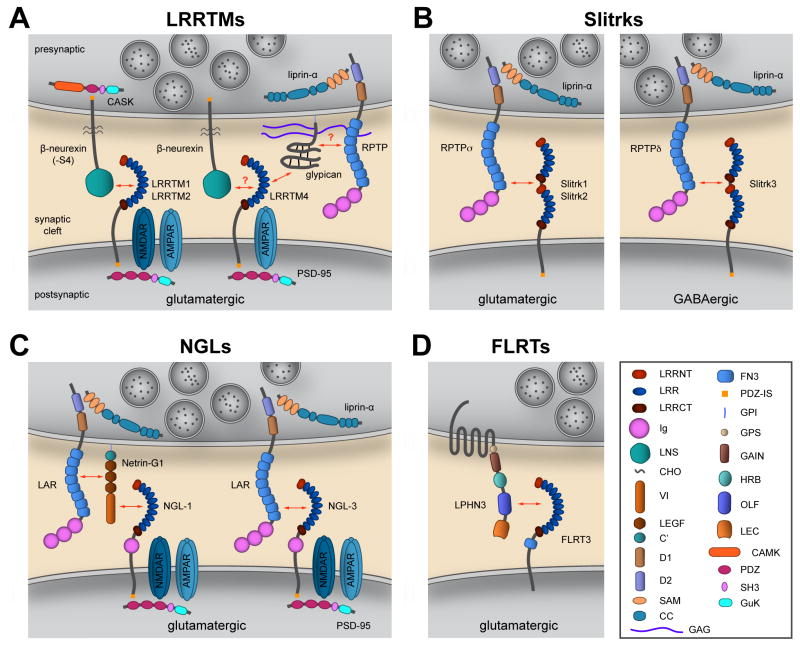
Figure 1. Synaptic LRR proteins and their interactions.
Postsynaptic LRR proteins and their presynaptic partners form a trans-synaptic complex that bridges the synaptic cleft and recruits essential scaffolding molecules and neurotransmitter receptors to the synapse. A, Regulation of excitatory synapse development by LRRTMs. LRRTM1 and LRRTM2 bind to presynaptic α- (long) and β- (short) neurexins that lack a small insert at splice site 4 (S4) in the LNS domain. Only β-neurexin is shown. LRRTM4 binds to presynaptic HSPGs, including glypican. Glypican may act via a co-receptor such as LAR family RPTPs to induce presynaptic differentiation. LRRTM4 can also bind neurexin under certain conditions, but the functional significance of this interaction is not clear. The cytoplasmic tails of LRRTMs, neurexins and RPTPs couple to the scaffolding proteins PSD-95, CASK and liprin-α, respectively. B, Slitrk1 and Slitrk2 bind to presynaptic RPTPσ and regulate excitatory synapse formation in cultured neurons. Slitrk3 binds to presynaptic RPTPδ and regulates inhibitory synapse formation. C, NGL-1 binds to presynaptic Netrin-G1, and this interaction induces recruitment of LAR to the complex. NGL-2 binds to presynaptic Netrin-G2 (not shown). NGL-3 binds to the first two FN3 repeats in LAR. D, FLRT3 binds to the adhesion GPCR latrophilin.
Domain abbreviations: LRRNT, LRRCT, LRR N- and C-terminal flanking domains; Ig, immunoglobulin-like; LNS, laminin-α/neurexin/sex-hormone-binding globulin (also known as Laminin-G domain); CHO, carbohydrate attachment; VI, laminin N-terminal; LEGF, laminin EGF-like motifs 1–3; C′, C-terminal domain; D1, D2, membrane-proximal (catalytically active) and -distal (inactive) tyrosine phophatase domains; SAM, sterile alpha motif; CC, coiled coil; GAG, glycosaminoglycan (heparan sulfate); FN3, fibronectin type III; PDZ-IS, post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (DlgA), and zonula occludens-1 protein (zo-1) (PDZ) interaction site; GPI, glycosylphosphatidylinositol; GPS, GPCR proteolytic site; GAIN, GPCR autoproteolysis-inducing domain; HRB, hormone-binding domain; OLF, olfactomedin; LEC, lectin domain; CaMK, Ca2+/calmodulin-dependent kinase; SH3, Src homology 3; GuK, guanylate kinase domain.
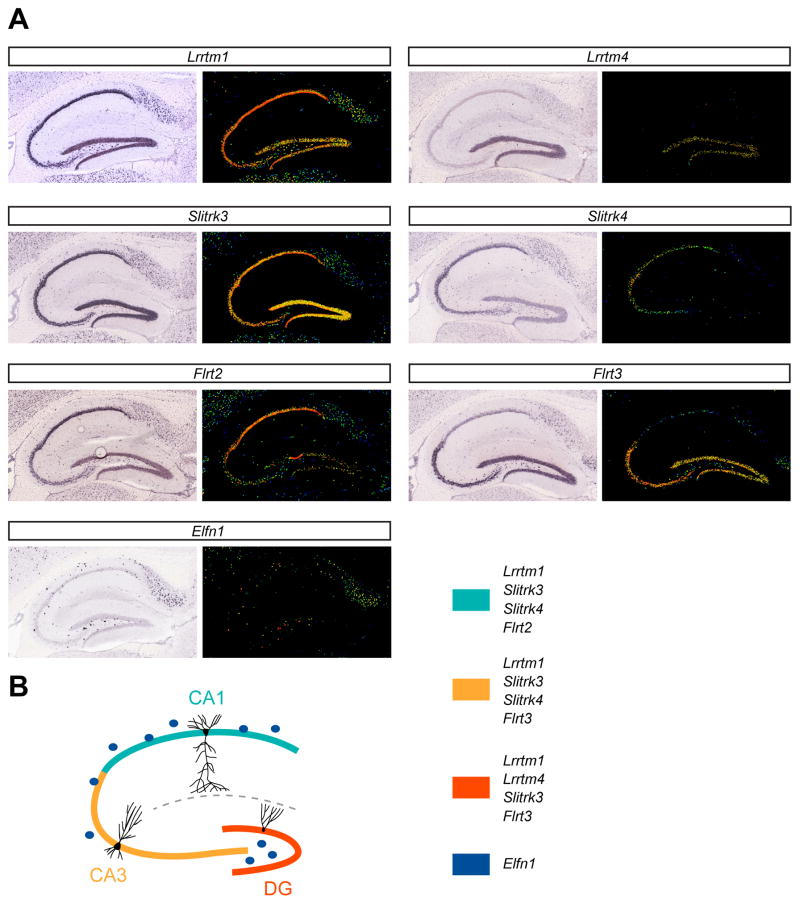
Figure 4. Cell type-specific expression patterns of synaptic LRR genes.
A, In situ hybridizations showing gene expression patterns in P56 sagittal mouse hippocampal sections for a limited set of synaptic LRR genes discussed in this review. Lrrtm1 (http://mouse.brain-map.org/experiment/show/68162201) is broadly expressed in the hippocampus, whereas Lrrtm4 (http://mouse.brain-map.org/experiment/show/70437762) expression is restricted to DG. Slitrk3 (http://mouse.brain-map.org/experiment/show/69874150) is broadly expressed in the hippocampus, whereas Slitrk4 (http://mouse.brain-map.org/experiment/show/69874156) is expressed in the CA region, but is absent from DG. Flrt2 (http://mouse.brain-map.org/experiment/show/70565569) and Flrt3 (http://mouse.brain-map.org/experiment/show/71656690) show complementary expression patterns in the hippocampus: Flrt2 is expressed in CA1, whereas Flrt3 is expressed in CA3 and DG. Elfn1 (http://mouse.brain-map.org/experiment/show/77791978) is expressed in somatostatin-positive interneurons in the hippocampus. All images in (A) are obtained from the Allen Mouse Brain Atlas (http://mouse.brain-map.org) [124]. For each gene, the left panel shows the original in situ hybridization signal, and the right panel shows the expression mask image using a heat map color scale to indicate intensity of expression (with red signal indicating high expression and blue signal low expression). B, Schematic summary representation of gene expression patterns in the hippocampus based on (A). Different hippocampal cell types express different (combinations of) synaptic LRR genes.
Overexpression of LRRTM2 or LRRTM4 in cultured hippocampal neurons increases the density of excitatory, but not inhibitory, synapses [14, 15, 22, 23]. LRRTMs most likely promote excitatory synapse development by interacting with their respective presynaptic binding partners, since overexpression of an LRRTM2 mutant lacking the LRR domain fails to increase the density of excitatory synapses [14], and enzymatic removal of HS abolishes the increase in presynaptic input density following LRRTM4 overexpression in hippocampal neurons [23]. In agreement with a role in regulating excitatory synapses, LRRTM2 knockdown [14, but see 25] and LRRTM4 knockout or knockdown [22, 23] selectively decrease excitatory synapse density in cultured neurons.
In vivo analyses of LRRTM function have revealed subtle pre- and postsynaptic anatomical defects. LRRTM1 knockout mice display increased VGluT1 immunofluorescence and an altered, more dispersed, distribution of synaptic vesicles [12, 26]. Postsynaptically, Golgi stained hippocampal CA1 pyramidal neurons in LRRTM1 knockout mice have longer, possibly immature, dendritic spines, but no overall change in spine density, although EM analysis indicates a small decrease in asymmetric synapse density [26]. Knockout or knockdown of LRRTM4 decreases spine density in dentate gyrus and cortex, respectively [22, 23]. Functionally, LRRTM2 knockdown decreases evoked AMPA receptor (AMPAR)- and NMDAR-mediated glutamatergic synaptic transmission in dentate granule cells [14], and combined LRRTM1/LRRTM2 knockdown in early postnatal CA1 neurons selectively reduces evoked AMPAR-mediated synaptic transmission [27]. Despite the altered organization of synaptic vesicles in LRRTM1 knockout mice, presynaptic functional defects have not been detected following LRRTM knockout or knockdown [14, 23, 27]. Together, these anatomical and functional changes are consistent with a role for LRRTMs in regulating synapse formation, maturation or maintenance, or possibly all of these, in vivo.
Slitrks regulate excitatory and inhibitory synapse development via RPTPs
Based on their structural similarity to LRRTMs (Fig. 1B), Slit- and Trk-like proteins (Slitrk1-6) were predicted to have synaptogenic properties in a bioinformatics search [12]. When tested in the coculture assay, Slitrk2 expressed on the surface of fibroblasts indeed induced presynaptic differentiation in contacting axons [12]. Subsequent studies showed that all Slitrk family members have similar activity [28]. Slitrks are broadly expressed in the hippocampus (Fig. 4), and are postsynaptically localized [28–30]. Whereas Slitrk1, -2, -4, -5 and -6 induce both excitatory and inhibitory presynaptic differentiation in cocultures [28], overexpression of Slitrk1, -2, -4, or -5 in cultured hippocampal neurons increases the density of excitatory, but not inhibitory, synapses [29]. Conversely, knockdown of Slitrk1, -2, -4, or -5 selectively decreases excitatory synapse density [29], indicating that these Slitrk family members regulate excitatory synapse development in cultured neurons. In vivo, the role of Slitrks in excitatory synapse development has been best characterized for Slitrk5. Loss of Slitrk5 decreases dendritic arbor complexity of striatal neurons, reduces AMPAR and NMDAR levels, and decreases synaptic transmission in corticostriatal circuits without affecting presynaptic properties, indicating a role for Slitrk5 in the development of corticostriatal excitatory synapses [30]. Slitrk6 mutants display a reduction in the synaptic marker Ribeye in the retina during postnatal development, but apparently normal levels in adult, suggesting a developmental delay in synaptogenesis in the absence of Slitrk6 [31].
Slitrk3 is unique in that it is the only LRR protein identified thus far to function exclusively at inhibitory synapses. Slitrk3 selectively induces inhibitory presynaptic differentiation in cocultures as well as in cultured neurons, and Slitrk3 knockdown decreases inhibitory, but not excitatory, synapse density [28, 29]. Consistent with a role in regulating inhibitory synapse development, loss of Slitrk3 in vivo results in a decreased numbers of functional inhibitory, but not excitatory hippocampal synapses [28]. Anatomically, there is a select reduction in inhibitory synapse density in specific hippocampal laminae or even subregions of laminae in Slitrk3 knockout mice [28], suggesting the loss of subsets of inhibitory inputs, possibly belonging to distinct interneuron types.
The effects of Slitrks on excitatory or inhibitory synapse development depend on their interactions with presynaptic receptors. Candidate-based approaches and affinity chromatography of brain extract with recombinant Slitrk1 identified the LAR family receptor protein tyrosine phophatase δ (RPTPδ) as a Slitrk interactor, and subsequent experiments confirmed that Slitrk1, -2 and -3 interact with RPTPδ and RPTPσ, but not with LAR itself [28, 29] (Fig. 1B). RPTPs interact with a host of postsynaptic receptors, including the LRR proteins NGL-3 and TrkC (see below), interleukin-1-receptor accessory protein-like 1 (IL1RAPL1) and interleukin-1 receptor accessory protein (IL1RAcP) [32–36]. Thus, analogous to LRRTMs and their presynaptic partners neurexins, Slitrks bind to presynaptic receptors that interact with multiple postsynaptic ligands [reviewed in 37].
Although Slitrks are capable of interacting with both RPTPδ and RPTPσ, their functional effects on excitatory and inhibitory synapse development are mediated by more selective interactions. Slitrk2 and -3 require RPTPδ but not RPTPσ, to induce inhibitory presynaptic differentiation, and conversely, excitatory presynaptic differentiation induced by Slitrk1 and -2 requires RPTPσ, but not RPTPδ [28, 29]. Endogenous RPTPσ appears to be selectively expressed on glutamatergic axons [32], which would explain the functional requirement for RPTPσ in Slitrk-mediated excitatory synapse development. Less clear is why Slitrk1 and -2 interact with both RPTPδ and RPTPσ receptors and induce both excitatory and inhibitory synapse formation in fibroblast-neuron cocultures, but selectively promote excitatory synapse development in neurons. Possibly, RPTP-Slitrk1 or -2 interactions activate postsynaptic signaling pathways that only stabilize excitatory synapses, but little is known about Slitrk signaling [38]. Like Slitrk1 and -2, Slitrk3 interacts with both RPTPδ and RPTPσ, yet Slitrk3 only induces inhibitory presynaptic differentiation in cocultures and in neurons. Slitrk3 may require additional receptors to induce inhibitory synaptic differentiation, or glutamatergic axons may express cues that specifically inhibit Slitrk3. To complicate things further, the RPTP ligand IL1RAPL1 induces excitatory synapse formation via RPTPδ [34, 36], indicating that binding of postsynaptic partners to presynaptic RPTPδ does not always trigger inhibitory synapse formation. Binding of IL1RAPL1 to RPTPs is modulated by alternative splicing [34], which could be a mechanism to differentially regulate ligand-receptor interactions in glutamatergic and GABAergic axons. Clearly, determining how the interactions of Slitrks with their RPTP receptors shape excitatory and inhibitory synapses constitutes a major challenge.
NGLs regulate input-specific synapse development
Netrin-G Ligands (NGL1-3) are structurally related to LRRTMs and Slitrks, but contain an additional Ig domain in their extracellular region (Fig. 1C). Their identification as PSD-95 interactors in yeast two hybrid screens suggested a role at synapses, and biochemical fractionation and immuno-EM confirmed that NGL-2 and -3 localize to the postsynaptic density [33, 39]. All NGLs induce presynaptic differentiation in coculture assays, with NGL-3 having markedly stronger effects than NGL-1 or -2 [33, 39]. NGL-3 binds to all LAR family RPTPs [33, 40]. NGL-1 and -2 interact with the GPI-anchored proteins Netrin-G1 and Netrin G2, respectively [39, 41, 42] (Fig. 1C). The lack of a cytoplasmic domain in Netrin-Gs suggests the existence of a transmembrane co-receptor to mediate NGL-1- and NGL-2-induced presynaptic differentiation. Indeed, NGL-1 binding to presynaptic Netrin-G1 induces a cis interaction of Netrin-G1 with LAR to promote presynaptic differentiation [43]. Netrin-G1 binding to LAR is modulated by alternative splicing of Netrin-G1 [43], which can generate at least 10 isoforms [44–46]. Alternative splicing of Netrin-G2 does not affect binding to NGL-2 [47], suggesting that Netrin-G splicing serves to regulate cis interactions. Thus, like LRRTMs, different NGL family members interact with transmembrane or GPI-anchored presynaptic partners, and as suggested for the LRRTM4-glypican interaction, the recruitment of additional cis co-receptors is required for Netrin-G-mediated presynaptic differentiation. It will be of interest to determine whether such trans-induced formation of cis protein complexes constitutes a general theme in synapse development.
What might be the advantage of the indirect interaction of NGL-1 with LAR via Netrin-G1, as opposed to NGL-3’s direct interaction with LAR RPTPs? One possibility could be that through alternative splicing, Netrin-Gs may couple trans-synaptic interaction with NGLs to a network of multiple cis receptors, providing additional layers of control over synapse development. Remarkably, NGL-2 binding to Netrin-G2 does not recruit LAR to the complex [43], suggesting the existence of additional cis interactors. Whether these are other RPTPs or unrelated synaptic receptors remains to be determined.
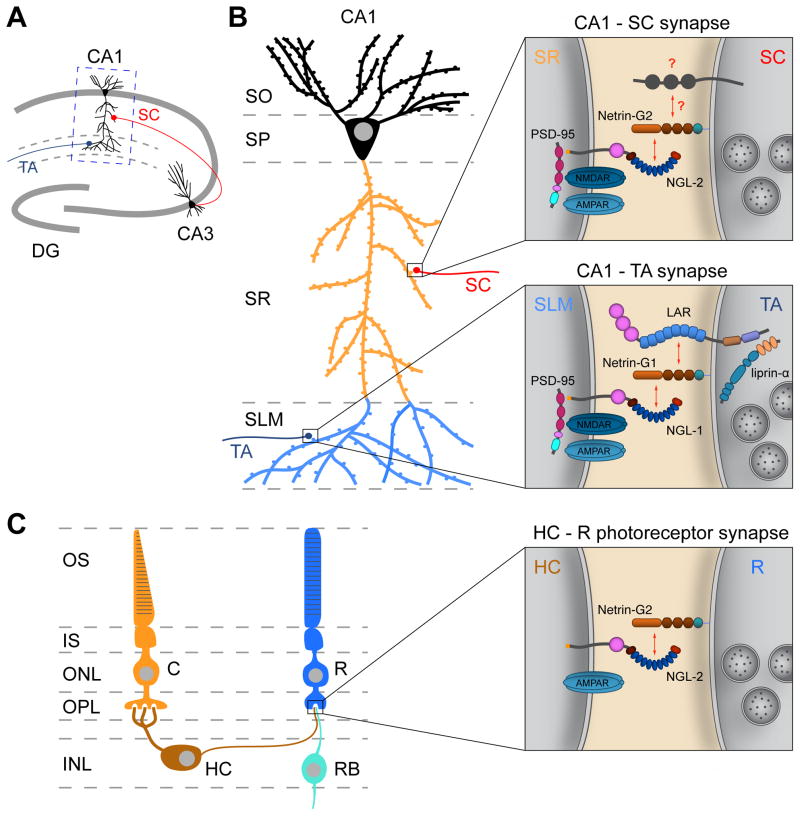
An additional advantage of the Netrin-G/NGL interaction is increased specificity. Netrin-Gs are expressed on the surface of distinct axonal populations [41, 44, 45, 48], and determine dendritic clustering of their respective postsynaptic NGLs in a pathway-specific manner [41]. In the hippocampal CA1 region, Netrin-G1 is expressed on entorhinal cortex-derived temporoammonic fibers in the stratum lacunosum moleculare (SLM), and NGL-1 distribution is restricted to the corresponding distal dendritic segment of the CA1 neuron (Fig. 2A, B). Loss of axonal Netrin-G1 in Ntng1 knockout mice results in a diffuse distribution of NGL-1 along the entire CA1 dendrite. Similarly, loss of Ntng2 in Schaffer collateral axons in the stratum radiatum (SR) results in dispersal of NGL-2, which is normally restricted to the proximal CA1 dendritic segment [41] (Fig. 2A, B). In agreement with the pathway-specific distribution of NGL-2, knockout or knockdown of NGL-2 in CA1 neurons decreases the density of dendritic spines and the strength of glutamatergic transmission in SR, but not in SLM [49]. The decrease in SR spine density following NGL-2 knockdown can be rescued by restoring the interaction with presynaptic Netrin-G2, indicating that the Netrin-G2/NGL-2 complex regulates synapse development in an input-specific manner. Furthermore, loss of NGL-2 impairs the ability of CA1 neurons to functionally integrate coincident subthreshold SR and SLM stimuli, resulting in reduced CA1 spiking probability and thus affecting CA1 output in the hippocampal circuit [49]. A similar pathway-specific role for NGL-2 was discovered in the horizontal cell (HC) axon – rod photoreceptor connection in the retina [50]. NGL-2 localizes to horizontal cell axon tips, which form postsynaptic specializations onto Ntng2-expressing rod photoreceptor cells (Fig. 2C). In NGL-2 knockout mice, HC axons overshoot into neighboring laminae and form fewer synapses with rods. Presynaptic release sites in rod photoreceptors display immature characteristics, suggesting that NGL-2/Netrin-G2 signaling regulates presynaptic maturation. Loss of NGL-2 does not affect HC dendrite – cone photoreceptor connectivity, and impairs visual function in a pathway-specific manner [50]. Together, these studies suggest that the Netrin-G/NGL trans-synaptic interaction may broadly regulate pathway-specific synapse development.
Figure 2. Regulation of pathway-specific synapse development by NGLs.
A, Schematic representation of CA1 hippocampal circuits. CA1 pyramidal neurons receive input on their proximal dendrites from hippocampal CA3 axons (the Schaffer collaterals, SC), and input on their distal dendrites from temporoammonic (TA) axons originating from entorhinal cortex. B, Schematic representation of a CA1 neuron receiving SC and TA inputs. SC axons are confined to the stratum radiatum (SR) and TA axons to the stratum lacunosum moleculare (SLM). Netrin-G2 and NGL-2 localize to the pre- and postsynaptic side, respectively, of the SC inputs in SR, and NGL-2 is functionally required for the development of these inputs. Netrin-G1 and NGL-1 localize to the pre- and postsynaptic side, respectively, of the TA inputs in SLM. Analogous to the NGL-1/Netrin-G1/LAR complex, NGL-2 binding to Netrin-G2 may also induce recruitment of a presynaptic co-receptor (indicated with ‘?’). Cartoon CA1 neuron based on [123]. DG, dentate gyrus; CA, cornu ammonis; SO, stratum oriens; SP, stratum pyramidale. C, Schematic representation of B-type horizontal cell (HC) connectivity in the retina. Photoreceptors form triad synapses with HCs and bipolar cells (RB) in the outer plexiform layer (OPL). HC axon terminals synapse with rod (R) photoreceptors, and HC dendrites with cone (C) photoreceptors. NGL-2 selectively localizes to the tips of HC axons, which form a postsynaptic specialization at the HC-R photoreceptor synapse. Loss of NGL-2 results in fewer HC-R synapses, but does not affect HC-C connectivity. OS, photoreceptor outer segment; IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; RB, rod bipolar cell.
The mechanisms by which NGLs regulate postsynaptic differentiation are not well understood. Cyclin-dependent kinase-like 5 (Cdkl5) interacts with NGL-1 and phosphorylates NGL-1 on a serine residue close to the PDZ interaction site. This phosphorylation promotes NGL-1 binding to PSD-95 and formation of dendritic spines in cultured hippocampal neurons [51]. The PDZ interaction site in NGL-2 was previously shown to recruit PSD-95 to dendritic protrusions in cultured neurons [39] and is also required for NGL-2-mediated regulation of dendritic spines in vivo [49]. Thus, interactions of NGL with synaptic scaffolding proteins regulate spine morphogenesis in vitro and in vivo.
FLRTs interact with latrophilin, a presynaptic adhesion GPCR
Similar to LRRTMs, Slitrks, and NGLs, Fibronectin Leucine-Rich Repeat Transmembrane proteins (FLRT1-3) bind to a presynaptic receptor that interacts with multiple postsynaptic ligands. In the hippocampus, FLRT2 and FLRT3 show complementary expression patterns (Fig. 4), whereas FLRT1 is only weakly expressed. The role of FLRTs in synaptic development is best characterized for FLRT3, an adhesion molecule present at excitatory synapses and in postsynaptic density fractions [52]. Affinity chromatography on brain extracts using the recombinant FLRT3 extracellular domain identified the G protein-coupled receptor (GPCR) latrophilin 3 (LPHN3) [52] (Fig. 1D). Latrophilins (LPHN1-3) are adhesion-GPCRs [53] containing large extracellular domains with multiple interaction motifs [54]. They were originally identified, together with neurexins, as receptors for the neurotransmitter release-inducing black widow spider venom α-latrotoxin [55–59]. Affinity chromatography with recombinant LPHN1 or LPHN3 ectodomains also identified FLRTs and an additional family of surface receptors, the Teneurins, as the main latrophilin interactors [52, 60, 61]. Teneurins (Ten1-4) are cell adhesion molecules that regulate connectivity in invertebrate and vertebrate olfactory, neuromuscular and visual systems [62–64]. Immuno-EM on purified synaptosomes indicates that endogenous LPHN1 is mostly presynaptically localized, and that LPHN1 binding activity is largely postsynaptic [60]. Thus, like the neurexins and RPTPs, latrophilins interact with multiple postsynaptic ligands. Furthermore, these interactions can be modulated by alternative splicing, as the presence of a small insert between the LPHN1 lectin and olfactomedin domains reduces the affinity for Teneurins without affecting binding to FLRT3 [61].
FLRTs differ from LRRTMs, Slitrks, and NGLs in that they do not induce presynaptic differentiation in coculture assays [52]. Remarkably, Teneurins do induce presynaptic differentiation in cocultures [60], suggesting that FLRT3 or Teneurin binding to LPHN mediates differential effects. The two LPHN ligands may bind to different domains in LPHN, activate different intracellular signaling pathways, or induce recruitment of additional receptors in cis, all of which could account for their differential effects. FLRT3 binds to the LPHN3 olfactomedin domain [65], whereas Teneurins bind to both olfactomedin and lectin domains and minimally require the lectin domain [61, 65], suggesting that the two ligands bind to different regions of the LPHN ligand-binding domain. Little is known about downstream effectors of LPHN, although the addition of a soluble extracellular fragment of Ten-2 increases Ca2+ signaling in cultured neurons, which could indicate activation of G protein signaling [60]. LPHN1 also binds to S4-lacking neurexins [66], raising the possibility that Teneurins could induce a cis complex of LPHN and neurexin to trigger presynaptic differentiation.
Although FLRTs do not induce presynaptic differentiation in cocultures, manipulating FLRT3 and LPHN3 levels does affect glutamatergic synapse development. Knockdown of FLRT3 in cultured hippocampal granule cells reduces excitatory synapse density, and competition experiments with excess soluble LPHN3 ectodomains or culture-wide knockdown of LPHN3 similarly reduces the density of excitatory synapses [52]. In vivo, knockdown of FLRT3 reduces dendritic spine density and evoked perforant path synaptic transmission onto hippocampal granule cells [52]. Knockdown of LPHN3 in cortical layer 2/3 (L2/3) neurons reduces the density of synapses formed by L2/3 axons in L5 [65]. This defect can be rescued by co-expressing shRNA-resistant mutant LPHN3 lacking the lectin domain, but not the olfactomedin domain [65], suggesting that the interaction with FLRTs, but not Teneurins, is required for synapse formation by L2/3 axons. Thus, FLRT3 regulates glutamatergic synapse development in cultured neurons and in vivo, likely via a trans-synaptic interaction with LPHN3.
The molecular mechanisms by which FLRT3 regulates postsynaptic development are unclear. Unlike LRRTMs, Slitrks, and NGLs, FLRT proteins lack a PDZ interaction site, suggesting that they do not recruit synaptic scaffolding proteins. Work in other systems indicates that FLRT3 acts as a modulator of cell surface receptors regulating various developmental processes including growth factor signaling, adhesion and axon pathfinding. In Xenopus, FLRT3 is required for FGF signaling, and interacts in cis with cotransfected FGF receptors [67–69]. FLRT3 further interacts with cadherin and protocadherin in coexpression experiments, and interferes with cadherin-mediated cell adhesion via the small GTPase RndI, possibly by modulating cadherin surface levels [70–72]. In mouse rostral thalamic neurons, FLRT3 acts as a co-receptor of the Robo1 receptor and, in the presence of the Robo1 ligand Slit1, determines attraction to the axon guidance cue Netrin-1 via its DCC receptor [73]. Intermediate thalamic neurons expressing Robo1 and DCC, but are not attracted by Netrin-1 because they lack FLRT3. Thus, FLRT3 can engage in cis interactions with other surface receptors and thereby modulate cellular responsiveness, depending on context. Whether similar interactions occur at the synapse, and whether they might be regulated by trans interactions of FLRT3 with LPHN, or with Unc5, receptors that also interact with FLRT3 in trans [74], remains to be determined.
Other LRR proteins regulating excitatory and inhibitory synapse development
A surprising discovery from a coculture-based expression screen is TrkC [32], the tyrosine kinase receptor for the neurotrophin NT-3, which broadly regulates neural development [75]. TrkC is the only Trk receptor that induces presynaptic differentiation in cocultures, and this requires the LRR and first Ig domain, which are not involved in NT-3 binding. TrkC trans-synaptically interacts with RPTPσ, but not with RPTPδ or LAR, and this interaction is required for excitatory synapse development, independent of TrkC kinase activity [32].
Leucine-Rich Repeat and Fibronectin Type III Domain Containing proteins (also known as Synaptic Cell Adhesion-Like proteins; LRFN/SALM1-5) were identified as PSD-family-interacting proteins [76–78]. LRFN4/SALM3 and LRFN5/SALM5 are the only LRFN/SALM family members with synaptogenic activity in coculture assays and promote both excitatory and inhibitory synapse formation [79]. LRFN1/SALM2 regulates excitatory synapse maturation [76]. The LRFN/SALM trans-synaptic binding partners have not been identified, although LRFN3/SALM4 and LRFN5/SALM5 members can interact homophilically in trans [80].
LRR proteins as regulators of synaptic function and plasticity
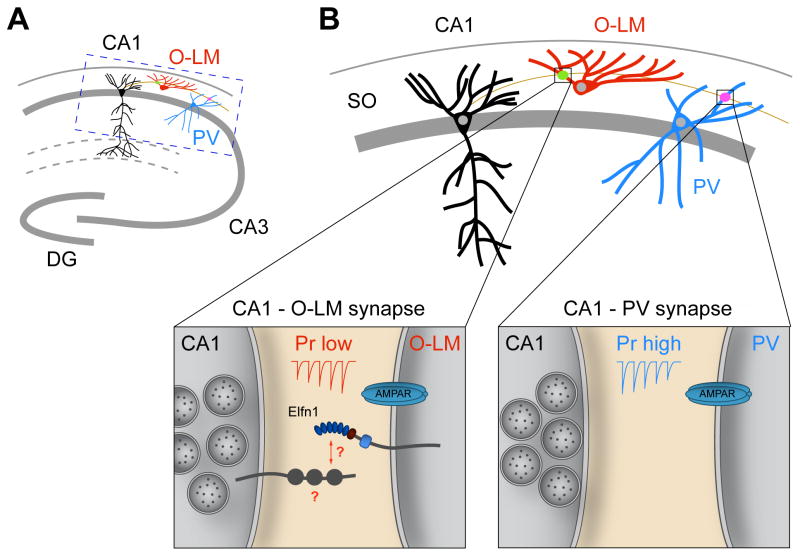
The LRR proteins discussed thus far all have synaptogenic effects in cultured neurons. An exception is Elfn1, an LRR protein with a domain organization similar to FLRTs, but containing a longer cytoplasmic tail. Elfn1 does not induce presynaptic differentiation in coculture assays or affect synapse number when overexpressed in cultured neurons [81]. Rather, Elfn1 trans-synaptically instructs presynaptic neurotransmitter release properties. Elfn1 is expressed in somatostatin-positive hippocampal interneurons called oriens-lacunosum moleculare (O-LM) cells [9, 81] (Fig. 4). O-LM cells receive excitatory input from CA1 axons, and Elfn1 localizes to the postsynaptic density of these inputs [81] (Fig. 3A, B). CA1 axons target a second population of interneurons, which are parvalbumin (PV)-positive but do not express Elfn1. The two classes of synapses made by the same axon, the CA1 - O-LM synapse and the CA1 - PV synapse, have strikingly different functional characteristics: CA1 - O-LM synapses are strongly facilitating with a low initial probability of release, whereas CA1 - PV synapses are depressing, with a high probability of release (Fig. 3A, B). Knockdown of Elfn1 in O-LM cells reduces facilitation and increases release probability, suggesting that postsynaptic Elfn1 controls presynaptic release probability at CA1 – O-LM synapses. Indeed, expressing Elfn1 at CA1 - PV synapses turns these normally depressing synapses into mildly facilitating ones [81]. Thus, Elfn1 trans-synaptically regulates target-specific release properties and contributes to the functional diversity of synapses. Elfn1-mediated facilitation requires presynaptic GluK6 kainate receptors [81], but whether Elfn1 interacts with a presynaptic receptor like other LRR proteins, requires additional receptors in cis, or activates a retrograde signal is unknown.
Figure 3. Elfn1 regulates release properties at a specific synapse.
A, CA1 axons synapse on two interneuron populations in the stratum oriens (SO): somatostatin-positive O-LM (oriens-lacunosum moleculare) interneurons and parvalbumin (PV)-positive interneurons. B, CA1 – O-LM synapses have a low probability of release (Pr) and are facilitating; CA1 – PV synapses have a high Pr and are depressing. Elfn1 specifically localizes to the postsynaptic side of the CA1 – O-LM synapse and controls release probability in CA1 axons in a target cell-specific manner. Elfn1 may interact with a presynaptic receptor (indicated with ‘?’), require additional receptors in cis, or activate a retrograde signal to regulate presynaptic release properties.
The LRRTMs are another class of LRR proteins with a role in shaping synaptic properties. Combined knockdown of LRRTM1/LRRTM2 at both developing and mature CA1 synapses blocks LTP at Schaffer collateral synapses [82]. The block in LTP can be rescued by re-expressing the extracellular domain of LRRTM2, but not the extracellular domain in which two key residues that mediate the interaction with presynaptic neurexin [16] have been mutated. LRRTM1/LRRTM2 knockdown decreases GluA1 AMPAR surface expression under basal conditions and prevents the increase in synaptic GluA1 surface levels following chemical induction of LTP (cLTP) in cultured neurons. Intriguingly, the initial delivery of surface GluA1 upon cLTP is not impaired by LRRTM1/LRRTM2 knockdown, suggesting that LRRTM1 and -2 are mainly required for maintaining AMPARs at synapses during LTP [82]. LRRTMs can directly interact with AMPARs [14, 83], which could be a mechanism for stabilizing AMPARs at synaptic sites. LRRTM4 has similar, though not identical, effects to LRRTM1 and -2. Knockout of LRRTM4 also prevents the cLTP-induced increase in GluA1 synaptic surface levels in dentate granule cells, but does not affect basal GluA1 synaptic surface levels [23]. Together, these results suggest that LRRTMs, and possibly other LRR proteins, are important regulators of plasticity-induced changes in synaptic properties.
Concluding Remarks
There has been tremendous progress in the past few years in the identification of novel LRR-containing synaptic adhesion molecules, the elucidation of their trans-synaptic interactions, and the understanding of their role in the development and plasticity of synapses. Several families of postsynaptic LRR proteins, with similar domain organization, but with distinct cell type-specific expression, have now been identified. The significance of such diversity of LRR proteins, especially at glutamatergic synapses, is not well understood, but an attractive possibility is that they may provide the underlying basis of structural and functional diversity of CNS synapses. Since individual neurons likely express several LRR proteins, it will be important to determine how LRR proteins, possibly in combination, control the development and plasticity of functional neural circuits. Many LRR proteins have multiple binding partners, both across the synaptic cleft as well as in cis, and it will be important to fully characterize these protein interaction networks and determine how they are modulated by alternative splicing and activity. Furthermore, very little is known about the downstream signaling pathways to which LRR proteins and their binding partners couple. The role of LRR proteins in the plasticity and functional properties of synapses is only beginning to be understood. Addressing these questions should enhance our understanding of this important family of synaptic proteins (Box 1).
Box 1. Outstanding questions.
Why are there so many LRR proteins involved in regulating excitatory synapses? Do they act at specific synapses, regulate different aspects of glutamatergic synapse development, or act in combination to define synaptic identity?
What is the cellular and subcellular localization of LRR proteins in vivo? Do individual synapses contain more than one LRR protein?
Are there other LRR proteins that selectively function at GABAergic synapses?
How are LRR protein interaction networks modulated by activity and alternative splicing?
How can LRR proteins interact with multiple receptors yet selectively regulate one class of synapses?
How do mutations in LRR proteins affect neural circuit development and function?
Finally, it is worth noting that the central role of LRR proteins in regulating excitatory and inhibitory connections may provide insight into processes that are disrupted in neurodevelopmental and psychiatric disorders. Some LRR genes have been directly linked to human brain disorders, including autism and schizophrenia (Box 2 and Table 1) [84], and we expect that deep sequencing approaches will likely reveal additional associations that are not detected by common variant (SNP) analysis. More generally, the fact that LRR proteins can regulate excitatory and inhibitory synapses in a cell type-specific manner should enhance our understanding of how the synaptic substructure contributes to circuit function and behavior, and that understanding could provide a framework for developing modulators of LRR interactions to correct synaptic dysfunction associated with mental illness.
Box 2. LRR genes in neurodevelopmental and psychiatric disorders.
Recent genetic studies of disorders such as autism, schizophrenia, and attention deficit hyperactivity disorder (ADHD) suggest that LRR proteins and their binding partners are important for the organization and function of neural circuits in humans as well. Table 1 gives an overview of mutations associated with neurodevelopmental and psychiatric disorders in genes encoding LRR proteins and binding partners discussed in this review. A recent, comprehensive overview of the extensive literature on mutations in NRXN1 associated with neurodevelopmental disorders is given in [85]. A recent, detailed overview of mutations in PTPR, SLITRK and NTRK3 (TrkC) genes associated with neuropsychiatric disorders is provided in [37].
The mutations in genes encoding LRR proteins and binding partners summarized in Table 1 are rare, often affecting only a few individuals in that study’s patient cohort, and are not always reproduced in other studies. Several of the LRR genes discussed in this review have been linked to multiple disorders; mutations in LRRTM3 for instance have been associated with disorders as diverse as autism and late onset Alzheimer’s disease. Conversely, a disorder often involves mutations in multiple LRR genes and their binding partners (as well as other, unrelated genes). Given their critical role in the organization and function of neural circuits in vertebrate model systems, it seems likely that human mutations in LRR genes or their binding partners could compromise synapse development or function, perturb E/I balance, and lead to a range of neurodevelopmental and psychiatric disorders. However, the effects of these mutations may be subtle. In many cases, patients still carry a functional copy of the gene, and for many of the mutations described in Table 1, such as single nucleotide polymorphisms (SNPs) in introns, it is not yet clear how they affect gene expression. In the case of the LRRTM genes (with the exception of LRRTM4), which are nested in the introns of α-catenin genes [13], mutations in LRRTMs might not only affect expression of LRRTM genes, but could conceivably also affect CTNNA gene expression.
In a few cases, the effects of disease-associated mutations on gene expression levels have been determined. A rare disruption in NTNG1 in a single patient with Rett syndrome was reported to decrease the abundance of a specific NTNG1 splice variant [86]. Decreased expression of specific NTNG1 splice variants has also been observed in patients with schizophrenia and bipolar disorder [87, 88]. This could conceivably affect binding of Netrin-G1 to LAR, which is modulated by alternative splicing of Netrin-G1 [43], and thus perturb synapse development. Rare missense mutations in NTNG1 and LRRTM1 (Table 1) do not seem to affect critical regions of the proteins. A rare missense mutation (A247S) has been identified in the LPHN3 olfactomedin domain [89], the domain required for FLRT3 binding [65], but whether this mutation is ADHD-specific and affects the interaction with FLRT3 is not clear. Thus, more work is needed to obtain a better understanding of the impact of disease-associated mutations on gene expression, gene function, and the function of neural circuits.
Table 1.
Postsynaptic LRR proteins and their presynaptic binding partners implicated in neurodevelopmental and psychiatric disorders.
| Gene/Protein | Mutation | Disorder | References |
|---|---|---|---|
| LRRTM1/LRRTM1 | Rare missense mutation N330S; SNP (upstream region) | Schizophrenia | [90–92] |
| LRRTM2/LRRTM2 | CNV (duplication) | Bipolar disorder | [93] |
| CNV (deletion) | Developmental delay | [94] | |
| LRRTM3/LRRTM3 | SNP (5′UTR, promoter and intronic) | Alzheimer’s disease, late onset (LOAD) | [95–103] |
| SNP (intronic) | Autism | [104, 105] | |
| CNV (deletion and duplication) | Autism | [106, 107] | |
| LRRTM4/LRRTM4 | CNV (deletion) | Autism | [107–109] |
| SNP (intronic) | Autism | [105] | |
| GPC1/GPC1 | CNV (deletion) | Autism | [107, 110] |
| GPC4/GPC4 | CNV (deletion) | Autism | [107] |
| GPC5/GPC5 | CNV (deletion and duplication) | Autism | [107, 109] |
| CNV | Bipolar disorder | [111] | |
| GPC6/GPC6 | CNV (deletion and duplication) | Autism | [108, 109] |
| NTNG1/Netrin-G1 | Rare missense mutations (Y23C, T135I) | Autism | [112, 113] |
| Rare gene disruption affecting alternative splicing | Rett syndrome | [86] | |
| SNP (intronic) | Schizophrenia | [87, 114, 115] | |
| NTNG2/Netrin-G2 | SNP (intronic) | Schizophrenia | [87] |
| GWAS | Autism | [108] | |
| FLRT1/FLRT1 | CNV (duplication) | Autism | [116] |
| FLRT2/FLRT2 | CNV (duplication) | Autism | [106, 108] |
| LPHN1/LPHN1 | Silent mutation | Autism | [117] |
| LPHN3/LPHN3 | SNP (intronic); Rare missense mutations | Attention-deficit/hyperactivity disorder (ADHD) | [89, 118–120] |
| CNV (deletion) | Autism | [107, 108] | |
| LRFN2/LRFN2 (SALM1) | CNV (duplication) | Autism | [107] |
| LRFN5/LRFN5 (SALM5) | SNP (intronic) | Autism | [121] |
| CNV (deletion) | Autism; developmental delay | [107, 108, 122] |
Sources: PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and the following disease-specific gene databases: Autism: SFARI, Simons Foundation Autism Research Initiative (http://www.sfari.org), AutismKB (http://www.autismkb.cbi.pku.edu.cn); Schizophrenia: SZGene (http://www.szgene.org); Bipolar Disorder: BDGene (http://www.bdgene.psych.ac.cn); ADHD: ADHDgene (http://www.adhd.psych.ac.cn); Alzheimer’s disease: Alzgene (http://www.alzgene.org). SNP: single nucleotide polymorphism; CNV: copy number variation; GWAS: genome-wide association study.
Highlights.
LRR proteins regulate the development, function and plasticity of excitatory and inhibitory synapses
Postsynaptic LRR proteins interact with key presynaptic receptors to promote pre- and postsynaptic differentiation
Distinct LRR proteins are expressed in different cell types in the CNS
LRR protein dysfunction may disrupt the excitation/inhibition balance and contribute to neuropsychiatric disorders
Acknowledgments
Work in the authors’ labs on LRR proteins is supported by an ERC Starting Grant (#311083) and FWO Odysseus Grant (JdW), and NIH grant R01NS067216 (AG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tabuchi K, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etherton M, et al. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci U S A. 2011;108:13764–13769. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudouin SJ, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- 4.Etherton MR, et al. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmeisser MJ, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 6.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penagarikano O, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolan J, et al. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics. 2007;8:320. doi: 10.1186/1471-2164-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biederer T, Scheiffele P. Mixed-culture assays for analyzing neuronal synapse formation. Nat Protoc. 2007;2:670–676. doi: 10.1038/nprot.2007.92. [DOI] [PubMed] [Google Scholar]
- 11.Scheiffele P, et al. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 12.Linhoff MW, et al. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauren J, et al. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 14.de Wit J, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko J, et al. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui TJ, et al. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci. 2010;30:7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudouin S, Scheiffele P. SnapShot: Neuroligin-neurexin complexes. Cell. 2010;141:908, 908 e901. doi: 10.1016/j.cell.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Ichtchenko K, et al. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 20.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 21.Krueger DD, et al. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr Opin Neurobiol. 2012;22:412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 22.de Wit J, et al. Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron. 2013;79:696–711. doi: 10.1016/j.neuron.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqui TJ, et al. An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron. 2013;79:680–695. doi: 10.1016/j.neuron.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 24.Johnson KG, et al. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006;49:517–531. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 25.Ko J, et al. Neuroligins/LRRTMs prevent activity- and Ca2+/calmodulin-dependent synapse elimination in cultured neurons. J Cell Biol. 2011;194:323–334. doi: 10.1083/jcb.201101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takashima N, et al. Impaired cognitive function and altered hippocampal synapse morphology in mice lacking Lrrtm1, a gene associated with schizophrenia. PLoS One. 2011;6:e22716. doi: 10.1371/journal.pone.0022716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soler-Llavina GJ, et al. The neurexin ligands, neuroligins and leucine-rich repeat transmembrane proteins, perform convergent and divergent synaptic functions in vivo. Proc Natl Acad Sci U S A. 2011;108:16502–16509. doi: 10.1073/pnas.1114028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H, et al. Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat Neurosci. 2012;15:389–398. S381–382. doi: 10.1038/nn.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yim YS, et al. Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 2013;110:4057–4062. doi: 10.1073/pnas.1209881110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shmelkov SV, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. 591p. doi: 10.1038/nm.2125. following 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tekin M, et al. SLITRK6 mutations cause myopia and deafness in humans and mice. J Clin Invest. 2013;123:2094–2102. doi: 10.1172/JCI65853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi H, et al. Postsynaptic TrkC and presynaptic PTPsigma function as a bidirectional excitatory synaptic organizing complex. Neuron. 2011;69:287–303. doi: 10.1016/j.neuron.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo J, et al. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida T, et al. IL-1 receptor accessory protein-like 1 associated with mental retardation and autism mediates synapse formation by trans-synaptic interaction with protein tyrosine phosphatase delta. J Neurosci. 2011;31:13485–13499. doi: 10.1523/JNEUROSCI.2136-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida T, et al. Interleukin-1 receptor accessory protein organizes neuronal synaptogenesis as a cell adhesion molecule. J Neurosci. 2012;32:2588–2600. doi: 10.1523/JNEUROSCI.4637-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valnegri P, et al. The X-linked intellectual disability protein IL1RAPL1 regulates excitatory synapse formation by binding PTPdelta and RhoGAP2. Hum Mol Genet. 2011;20:4797–4809. doi: 10.1093/hmg/ddr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi H, Craig AM. Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: presynaptic hubs for synapse organization. Trends Neurosci. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kajiwara Y, et al. SLITRK1 binds 14-3-3 and regulates neurite outgrowth in a phosphorylation-dependent manner. Biol Psychiatry. 2009;66:918–925. doi: 10.1016/j.biopsych.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, et al. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 40.Kwon SK, et al. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J Biol Chem. 2010;285:13966–13978. doi: 10.1074/jbc.M109.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura-Akiyoshi S, et al. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci U S A. 2007;104:14801–14806. doi: 10.1073/pnas.0706919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin JC, et al. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- 43.Song YS, et al. Trans-induced cis interaction in the tripartite NGL-1, netrin-G1 and LAR adhesion complex promotes development of excitatory synapses. J Cell Sci. 2013;126:4926–4938. doi: 10.1242/jcs.129718. [DOI] [PubMed] [Google Scholar]
- 44.Yin Y, et al. Laminets: laminin- and netrin-related genes expressed in distinct neuronal subsets. Mol Cell Neurosci. 2002;19:344–358. doi: 10.1006/mcne.2001.1089. [DOI] [PubMed] [Google Scholar]
- 45.Nakashiba T, et al. Netrin-G1: a novel glycosyl phosphatidylinositol-linked mammalian netrin that is functionally divergent from classical netrins. J Neurosci. 2000;20:6540–6550. doi: 10.1523/JNEUROSCI.20-17-06540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meerabux JM, et al. Human netrin-G1 isoforms show evidence of differential expression. Genomics. 2005;86:112–116. doi: 10.1016/j.ygeno.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Brasch J, et al. Crystal structure of the ligand binding domain of netrin G2. J Mol Biol. 2011;414:723–734. doi: 10.1016/j.jmb.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakashiba T, et al. Complementary expression and neurite outgrowth activity of netrin-G subfamily members. Mech Dev. 2002;111:47–60. doi: 10.1016/s0925-4773(01)00600-1. [DOI] [PubMed] [Google Scholar]
- 49.DeNardo LA, et al. NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron. 2012;76:762–775. doi: 10.1016/j.neuron.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soto F, et al. NGL-2 regulates pathway-specific neurite growth and lamination, synapse formation, and signal transmission in the retina. J Neurosci. 2013;33:11949–11959. doi: 10.1523/JNEUROSCI.1521-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ricciardi S, et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol. 2012;14:911–923. doi: 10.1038/ncb2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Sullivan ML, et al. FLRT Proteins Are Endogenous Latrophilin Ligands and Regulate Excitatory Synapse Development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langenhan T, et al. Sticky signaling--adhesion class G protein-coupled receptors take the stage. Sci Signal. 2013;6:re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- 54.Silva JP, Ushkaryov YA. The latrophilins, “split-personality” receptors. Adv Exp Med Biol. 2010;706:59–75. doi: 10.1007/978-1-4419-7913-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ushkaryov YA, et al. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 56.Krasnoperov VG, et al. alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 57.Davletov BA, et al. Isolation and biochemical characterization of a Ca2+-independent alpha-latrotoxin-binding protein. J Biol Chem. 1996;271:23239–23245. doi: 10.1074/jbc.271.38.23239. [DOI] [PubMed] [Google Scholar]
- 58.Sugita S, et al. alpha-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. J Biol Chem. 1998;273:32715–32724. doi: 10.1074/jbc.273.49.32715. [DOI] [PubMed] [Google Scholar]
- 59.Krasnoperov VG, et al. The calcium-independent receptor of alpha-latrotoxin is not a neurexin. Biochem Biophys Res Commun. 1996;227:868–875. doi: 10.1006/bbrc.1996.1598. [DOI] [PubMed] [Google Scholar]
- 60.Silva JP, et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci U S A. 2011;108:12113–12118. doi: 10.1073/pnas.1019434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boucard AA, et al. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J Biol Chem. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mosca TJ, et al. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong W, et al. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antinucci P, et al. Teneurin-3 specifies morphological and functional connectivity of retinal ganglion cells in the vertebrate visual system. Cell Rep. 2013;5:582–592. doi: 10.1016/j.celrep.2013.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Sullivan ML, et al. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Dev. 2014;9:7. doi: 10.1186/1749-8104-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boucard AA, et al. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem. 2012;287:9399–9413. doi: 10.1074/jbc.M111.318659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bottcher RT, et al. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- 68.Haines BP, et al. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol. 2006;297:14–25. doi: 10.1016/j.ydbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 69.Wheldon LM, et al. Critical role of FLRT1 phosphorylation in the interdependent regulation of FLRT1 function and FGF receptor signalling. PLoS One. 2010;5:e10264. doi: 10.1371/journal.pone.0010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogata S, et al. TGF-beta signaling-mediated morphogenesis: modulation of cell adhesion via cadherin endocytosis. Genes Dev. 2007;21:1817–1831. doi: 10.1101/gad.1541807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, et al. A protocadherin-cadherin-FLRT3 complex controls cell adhesion and morphogenesis. PLoS One. 2009;4:e8411. doi: 10.1371/journal.pone.0008411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karaulanov E, et al. Unc5B interacts with FLRT3 and Rnd1 to modulate cell adhesion in Xenopus embryos. PLoS One. 2009;4:e5742. doi: 10.1371/journal.pone.0005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leyva-Diaz E, et al. FLRT3 Is a Robo1-Interacting Protein that Determines Netrin-1 Attraction in Developing Axons. Curr Biol. 2014;24:494–508. doi: 10.1016/j.cub.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 74.Yamagishi S, et al. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. Embo J. 2011;30:2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ko J, et al. SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 77.Wang CY, et al. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morimura N, et al. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 79.Mah W, et al. Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J Neurosci. 2010;30:5559–5568. doi: 10.1523/JNEUROSCI.4839-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seabold GK, et al. The SALM family of adhesion-like molecules forms heteromeric and homomeric complexes. J Biol Chem. 2008;283:8395–8405. doi: 10.1074/jbc.M709456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sylwestrak EL, Ghosh A. Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science. 2012;338:536–540. doi: 10.1126/science.1222482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soler-Llavina GJ, et al. Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation. Neuron. 2013;79:439–446. doi: 10.1016/j.neuron.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 84.de Wit J, et al. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu Rev Cell Dev Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 85.Bena F, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- 86.Borg I, et al. Disruption of Netrin G1 by a balanced chromosome translocation in a girl with Rett syndrome. European journal of human genetics : EJHG. 2005;13:921–927. doi: 10.1038/sj.ejhg.5201429. [DOI] [PubMed] [Google Scholar]
- 87.Aoki-Suzuki M, et al. A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol Psychiatry. 2005;57:382–393. doi: 10.1016/j.biopsych.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 88.Eastwood SL, Harrison PJ. Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008;33:933–945. doi: 10.1038/sj.npp.1301457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Domene S, et al. Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:11–18. doi: 10.1002/ajmg.b.31141. [DOI] [PubMed] [Google Scholar]
- 90.Francks C, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry. 2007;12:1129–1139. 1057. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ludwig KU, et al. Supporting evidence for LRRTM1 imprinting effects in schizophrenia. Mol Psychiatry. 2009;14:743–745. doi: 10.1038/mp.2009.28. [DOI] [PubMed] [Google Scholar]
- 92.Leach EL, et al. The imprinted gene LRRTM1 mediates schizotypy and handedness in a nonclinical population. Journal of human genetics. 2014 doi: 10.1038/jhg.2014.30. [DOI] [PubMed] [Google Scholar]
- 93.Malhotra D, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kleffmann W, et al. 5q31 Microdeletions: Definition of a Critical Region and Analysis of LRRTM2, a Candidate Gene for Intellectual Disability. Molecular syndromology. 2012;3:68–75. doi: 10.1159/000341252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Majercak J, et al. LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103:17967–17972. doi: 10.1073/pnas.0605461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liang X, et al. Effect of heterogeneity on the chromosome 10 risk in late-onset Alzheimer disease. Hum Mutat. 2007;28:1065–1073. doi: 10.1002/humu.20567. [DOI] [PubMed] [Google Scholar]
- 97.Martin ER, et al. Interaction between the alpha-T catenin gene (VR22) and APOE in Alzheimer’s disease. Journal of medical genetics. 2005;42:787–792. doi: 10.1136/jmg.2004.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thornton-Wells TA, et al. Confronting complexity in late-onset Alzheimer disease: application of two-stage analysis approach addressing heterogeneity and epistasis. Genetic epidemiology. 2008;32:187–203. doi: 10.1002/gepi.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 100.Edwards TL, et al. An association analysis of Alzheimer disease candidate genes detects an ancestral risk haplotype clade in ACE and putative multilocus association between ACE, A2M, and LRRTM3. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:721–735. doi: 10.1002/ajmg.b.30899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reitz C, et al. Effect of genetic variation in LRRTM3 on risk of Alzheimer disease. Arch Neurol. 2012;69:894–900. doi: 10.1001/archneurol.2011.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lincoln S, et al. LRRTM3 interacts with APP and BACE1 and has variants associating with late-onset Alzheimer’s disease (LOAD) PLoS One. 2013;8:e64164. doi: 10.1371/journal.pone.0064164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J, et al. Association of LRRTM3 polymorphisms with late-onset Alzheimer’s disease in Han Chinese. Exp Gerontol. 2014;52:18–22. doi: 10.1016/j.exger.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 104.Sousa I, et al. Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism. 2010;1:7. doi: 10.1186/2040-2392-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Autism Genome Project, C., et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hussman JP, et al. A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol Autism. 2011;2:1. doi: 10.1186/2040-2392-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McQuillin A, et al. Analysis of genetic deletions and duplications in the University College London bipolar disorder case control sample. European journal of human genetics : EJHG. 2011;19:588–592. doi: 10.1038/ejhg.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fukasawa M, et al. Case-control association study of human netrin G1 gene in Japanese schizophrenia. Journal of medical and dental sciences. 2004;51:121–128. [PubMed] [Google Scholar]
- 115.Ohtsuki T, et al. Association of polymorphisms in the haplotype block spanning the alternatively spliced exons of the NTNG1 gene at 1p13.3 with schizophrenia in Japanese populations. Neurosci Lett. 2008;435:194–197. doi: 10.1016/j.neulet.2008.02.053. [DOI] [PubMed] [Google Scholar]
- 116.Bucan M, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arcos-Burgos M, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- 119.Jain M, et al. A cooperative interaction between LPHN3 and 11q doubles the risk for ADHD. Mol Psychiatry. 2012;17:741–747. doi: 10.1038/mp.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ribases M, et al. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav. 2011;10:149–157. doi: 10.1111/j.1601-183X.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 121.Wang K, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mikhail FM, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- 123.Megias M, et al. Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience. 2001;102:527–540. doi: 10.1016/s0306-4522(00)00496-6. [DOI] [PubMed] [Google Scholar]
- 124.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]