Abstract
RNA-binding proteins (RBPs) regulate numerous aspects of gene expression, thus identification of endogenous targets of RBPs is important for understanding their functions in cells. Here we identified transcriptome-wide targets of Rbfox3 in neuronally differentiated P19 cells and mouse brain using Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP). Although Rbfox3 is known to regulate pre-mRNA splicing through binding to the UGCAUG motif, PAR-CLIP analysis revealed diverse Rbfox3 targets including primary-microRNAs (pri-miRNAs) which lack the UGCAUG motif. Induced expression and depletion of Rbfox3 led to changes in the expression levels of a subset of PAR-CLIP-detected miRNAs. In vitro analyses revealed that Rbfox3 functions as a positive and a negative regulator at the stage of pri-miRNA processing to precursor-miRNA. Rbfox3 binds directly to pri-miRNAs and regulates the recruitment of the microprocessor complex to pri-miRNAs. Our study proposes a novel function for Rbfox3 in miRNA biogenesis.
INTRODUCTION
RNA-binding proteins (RBPs) play important roles in many aspects of gene expression regulation including splicing and other processing, translation, and stability of RNA transcripts. An RBP often interacts with multiple target RNAs at its own specific yet divergent RNA element and an RNA transcript is bound by many different RBPs in a dynamic fashion during its lifetime. Cell type and tissue-specific RBPs often regulate tissue-dependent expression and diversity of target genes, and help to establish specific cellular functions. There are hundreds of RBPs in the human genome and many of them have not been well characterized with respect to function.
The family of RNA binding protein fox-1 (C. elegans) homolog (Rbfox) contains a single conserved RNA recognition motif (RRM) in the middle of the protein molecule. Systematic Evolution of Ligands by Exponential Enrichment (SELEX) using zebrafish rbfox1l and human RBFOX1 yielded a common (U)GCAUG sequence as a high affinity binding motif1,2. Since the UGCAUG element had been known as a splicing regulatory element in alternative splicing of a number of genes3–6, studies on Rbfox proteins have focused on their roles in alternative splicing. The importance of the interaction between Rbfox and UGCAUG on pre-mRNAs in the alternative splicing regulation has been well-documented for individual genes and at a genome-wide level1,2,7–14. The Rbfox family in mammals consists of three members, Rbfox1, Rbfox2, and Rbfox3. Rbfox1 is expressed in heart, skeletal muscle, and neuronal tissues, while Rbfox2 is ubiquitously expressed in many tissues from the embryonic stem cell stage through adulthood1,8,13,14. Rbfox3 is expressed specifically in postmitotic neurons15. Each Rbfox gene undergoes extensive alternative splicing, generating many isoforms with a common RRM. The C-terminal splice variants of Rbfox1 and Rbfox2 are differentially expressed in tissues and show differences in intracellular localization and splicing activity7,16. Although the Rbfox proteins and their splice isoforms can regulate alternative splicing of the same exons to some extent when exogenously expressed, their in vivo targets may differ due to the differences in expression profile and interaction with other proteins. Recent studies following depletion of Rbfox in animals and cultured cells have shown that Rbfox plays important roles in a number of biological processes17–22. However the exact function of Rbfox in these biological processes is largely unknown.
To understand the biological function of RBPs, it is necessary to determine their binding targets in a specific cellular context. Crosslinking and immunoprecipitation of RNA–RBP complexes followed by high-throughput sequencing (CLIP-seq, HITS-CLIP) has been widely used to obtain a snapshot of where an RBP binds in intact cells23–26. A modified version, Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP), uses photoreactive ribonucleoside analogs such as 4-thiouridine (4SU) to obtain high-resolution data. Irradiation of cells by low-energy 365 nm UV-light to crosslink RBPs with photoreactive 4SU incorporated into nascent RNAs leads to more efficient and specific crosslinking27. In addition to cultured cells, in vivo PAR-CLIP has also been successfully used in C. elegans28. Comprehensive analyses of RBP targets by CLIP-seq have provided transcriptome-wide targets related to a known function of the RBP and have yielded mechanistic insights into RBP function23–25. They also have yielded unexpected targets implicating previously unrecognized functions of the RBP23,26. Although to date most of the studies on Rbfox have focused on splicing regulation, the roles of Rbfox may not be limited to splicing. Rbfox3 studies to date were based on selected individual genes, thus a genome-wide analysis is needed to better understand the Rbfox3 function.
MiRNAs are short single-stranded RNA molecules that posttranscriptionally regulate gene expression by inhibiting translation and triggering degradation of target mRNAs and are essential for normal development and cellular homeostasis29,30. Dysregulation of miRNA expression is linked to numerous human diseases including cancer and heart disease31,32. Thus, studying miRNA processing is important for understanding the regulatory architecture and the biological function of miRNAs. The long primary miRNAs (pri-miRNAs) are transcribed by RNA polymerase II from the genome, and then cleaved into hairpin-structured precursor miRNAs (pre-miRNAs) by the microprocessor complex composed of the RNase III enzyme Drosha and the double-stranded RNA-binding protein Dgcr8 in the nucleus. Pre-miRNAs are then exported by Xpo5 to the cytoplasm, where they are processed by another RNase III Dicer-containing complex to produce mature miRNAs29,30. Interestingly, expression levels of pri-miRNAs are not always correlated with those of their pre-miRNAs, suggesting that each pri-miRNA is differentially processed. Although recent studies have identified several proteins including RBPs that regulate pri-miRNA processing by interacting with the microprocessors or by binding to pri-miRNAs33–42, detailed mechanisms how the associated RBPs regulate pri-miRNA processing are largely unknown.
In this study, we set out to identify the endogenous targets of Rbfox3 in neuronal cells at the genome-scale using PAR-CLIP. Unexpectedly, we found that Rbfox3 interacts with a wide-range of RNA sequences other than UGCAUG, and that pri-miRNAs are over-represented in Rbfox3 targets. Rbfox3 binds specifically to a subset of pri-miRNAs and functions as either a positive or negative regulator in pri-miRNA processing.
RESULTS
PAR-CLIP identifies RNA-binding sites of endogenous Rbfox3
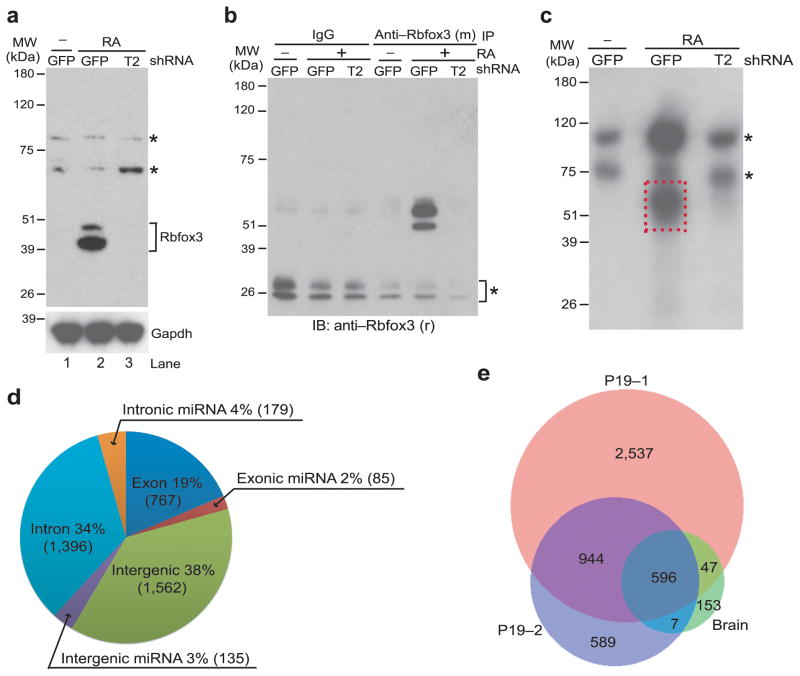
To identify endogenous binding sites of Rbfox3, we carried out PAR-CLIP experiments using neuronally differentiated P19 embryonal carcinoma cells and mouse neural tissues. We previously showed that P19 cells only express Rbfox3 during neuronal differentiation triggered by retinoic acid (RA)15. We confirm that observation here (Fig. 1a): Rbfox3 was expressed in RA-treated and neuronally differentiated P19 cells which were expressing the control shRNA against GFP (P19-GFP) but not in undifferentiated P19-GFP cells. RA-treatment did not increase Rbfox3 protein levels in P19 cells expressing T2 shRNA against Rbfox3 (P19-T2). P19-GFP and P19-T2 cells were incubated with photoactivatable 4SU and UV-irradiated. The crosslinked endogenous Rbfox3–RNA complexes were immunoprecipitated by mouse anti-Rbfox3 and radiolabeled in vitro with T4 polynucleotide kinase. Immunoprecipitated Rbfox3–RNA complexes were specifically detected using rabbit anti-Rbfox3 at the 50–60 kDa region after lithium dodecyl sulfate (LDS)-PAGE in the RA-treated P19-GFP cell sample but not in the untreated P19-GFP nor in the RA-treated P19-T2 cell samples (Fig. 1b). An autoradiogram of the equivalent gel showed the radiolabeled complexes at the 50–60 kDa region, which was detected only in the RA-treated P19-GFP cell sample (dashed rectangle, Fig. 1c). These data unambiguously demonstrates that the complexes with 50–60 kDa are specific for Rbfox3. These 50–60 kDa complexes have an approximately 10–20 kDa larger molecular mass compared to Rbfox3 itself (40 kDa), consistent with crosslinking with 30–60 nt RNAs. The crosslinked RNAs were converted into cDNA and sequenced. Two biological replicates generated 3.2 million reads (Supplementary Data Set 1) and 1 million reads uniquely mapped to the mouse genome, respectively. The mapped reads were analyzed with PARalyzer where the criteria call for at least 10 reads and one or more T-to-C conversion events, characteristic for crosslinked 4SU, in each cluster. Overall, we found 4,124 clusters (binding sites) from the 3.2 million reads, which were distributed as follows: 41% mapped to intergenic regions, 38% to intronic regions, and 21% to exonic regions (Fig. 1d, Supplementary Data Set 1). Strikingly, 9% (399 clusters) of Rbfox3-bound RNA clusters were mapped to miRNA hairpin loci. (The term of “miRNA hairpin loci” is used when RNA clusters were mapped to miRNA hairpins and does not distinguish pri-miRNA, pre-miRNA and mature miRNA. MiRNA hairpins are annotated in the miRBase (www.mirbase.org). Among the 1,977,090 reads that mapped to miRNA hairpin loci, 37,057 reads (1.9%) contained the T-to-C conversion. One of the reasons for the apparently low conversion ratio, which is still considered to be above the background, might be the low 4SU incorporation in neuronally differentiated P19 cells (0.208 ± 0.088%, mean ± s.d., n = 3 biological replicates, cell cultures), which is less than 1/10th compared to that reported for actively proliferating cells27. The target distributions of the data set from the 1 million reads were similar to those from the 3.2 million reads. Moreover, 72% unique clusters (intergenic, 75%; intronic, 72%; exonic, 66%; miRNA hairpin loci, 84%) were also in the list of clusters from the 3.2 million reads with a minimal 5-nt overlap, demonstrating the high reproducibility of the PAR-CLIP experiments.
Figure 1.
PAR-CLIP for endogenous Rbfox3 in neuronally differentiated P19 cells. (a) Immunoblot detecting Rbfox3. Samples are lysates from retinoic acid (RA)-treated or untreated (−) clonal P19 cells which were stably expressing shRNA targeting Rbfox3 (T2) or control shRNA for GFP. The bands indicated with * are unidentified proteins which cross-reacted with mouse anti-Rbfox3 but are unlikely Rbfox3-related proteins because of unresponsiveness to T2 shRNA. The lower panel is an immunoblot detecting Gapdh which serves as a loading control. (b) Immunoblot detecting Rbfox3 in crosslinked RNA–protein complexes. Samples are anti-Rbfox3 immunoprecipitates prepared from 4SU-incorporated and UV-irradiated P19 cells. RA-treatment and shRNAs applied to P19 cells are indicated. Anti-Rbfox3 (m) and anti-Rbfox3 (r) indicate mouse anti-Rbfox3 used for immunoprecipitation (IP) and rabbit anti-Rbfox3 used for immunoblotting (IB), respectively. The bands indicated with * are mouse IgG light chains cross-reacting to secondary goat antibodies against rabbit IgG. (c) Autoradiogram of radioactively phosphorylated RNA–protein complexes following LDS-PAGE. Samples are T4 polynucleotide kinase-phosphorylated anti-Rbfox3 immunoprecipitates prepared from 4SU-incorporated and UV-irradiated P19 cells. RA-treatment and shRNAs applied to P19 cells are indicated. The rectangle boxed with broken red lines was considered to be Rbfox3–RNA complexes and processed for further analysis. The bands indicated with * are unidentified complexes. (d) Genomic distribution of Rbfox3 binding clusters determined by PAR-CLIP analysis. Numbers in parenthesis indicate cluster numbers. A full list of PAR-CLIP clusters is provided in Supplementary Data Set 1. (e) Venn diagram of cluster numbers overlapping among three PAR-CLIP experiments.
To further investigate Rbfox3 targets in the mouse central nervous system (CNS), we developed an in vivo mouse PAR-CLIP protocol (Supplementary Fig. 1a–c). Although 4SU incorporation in total RNA in brain (0.026 ± 0.012%, mean ± s.d., n = 3 biological replicates, mice) was approximately 1/10th compared to RA-treated P19 cells, we identified 803 clusters. Of these clusters, 41% mapped to intergenic regions, 36% to intronic regions, and 23% to exonic regions (Supplementary Fig. 1d, Supplementary Data Set 2), similar to the data obtained from P19 cells. Moreover, 80% of the clusters (intergenic, 80%; intronic, 79%; exonic, 80%; miRNA hairpin loci, 90%) in the in vivo PAR-CLIP analysis were also in the list of clusters from P19 cells. Notably, the binding of Rbfox3 to miRNA hairpin loci (157 clusters) was also observed in the in vivo PAR-CLIP experiment (Supplementary Fig. 1d). The number of clusters overlapping among three PAR-CLIP experiments is shown in Figure 1e.
To our surprise, only a few clusters include the (U)GCAUG motif and our attempts using several motif discovery algorithms failed to find any motif including (U)GCAUG with statistically significant enrichment in the clusters. Despite high reproducibility of the detected target sites of Rbfox3 in intact cells, their primary sequences showed broad sequence specificity. The following study focuses on the potential role of Rbfox3 through its binding to non-canonical RNA targets.
Rbfox3 affects miRNA biogenesis in cells
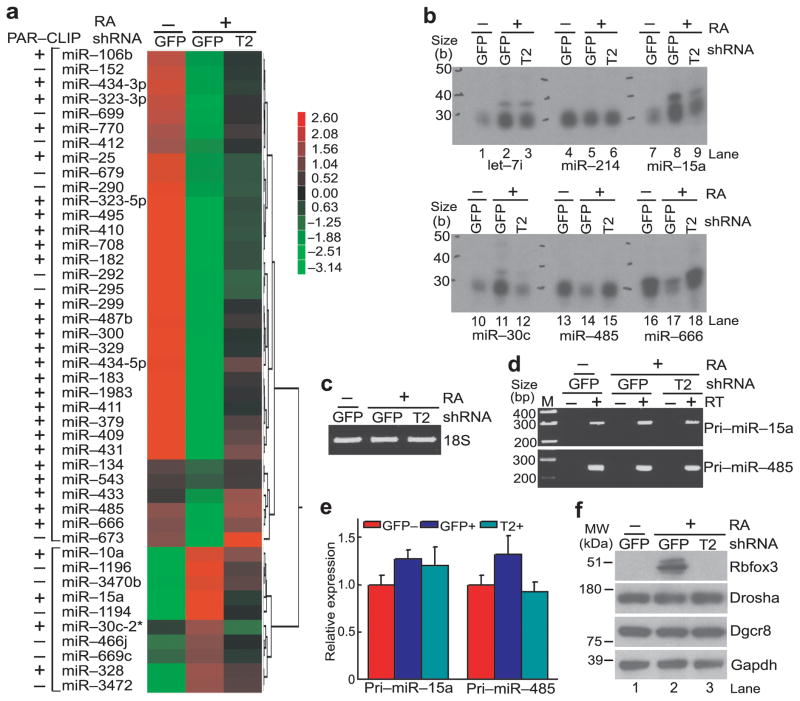
The observation that Rbfox3 binds to miRNA hairpin loci prompted us to investigate whether Rbfox3 could alter the expression level of miRNAs during neuronal differentiation of P19 cells. We performed miRNA microarray analyses using the RNAs extracted from untreated P19-GFP, RA-treated P19-GFP, or RA-treated P19-T2 cells. In total 97 miRNAs were differentially expressed between any two different culture conditions (Supplementary Data Set 3). Among them, the expression levels of 34 miRNAs decreased during differentiation and Rbfox3 knock-down prevented these changes. The expression levels of 10 miRNAs increased during differentiation and Rbfox3 knock-down inhibited these increases (Fig. 2a, Supplementary Data Set 3, n = 3 biological replicates, cell cultures, P < 0.01). Therefore expression of these 44 miRNAs appeared to correlate with Rbfox3 expression. Thirty miRNAs out of the 44 miRNAs (68%) were also identified as Rbfox3 targets by PAR-CLIP, suggesting that the Rbfox3 binding to miRNA hairpin loci correlates with the expression level of a subset of miRNAs.
Figure 2.
Rbfox3-regulated miRNAs in P19 cells. (a) Heat map showing comparative miRNA microarray analysis among three different conditions of P19 cell cultures. Samples are RNAs isolated from RA-treated (+) Rbfox3-specific T2shRNA-expressing (P19-T2) or control GFPshRNA expressing P19 cells (P19-GFP) or untreated (−) GFPshRNA-expressing P19 cells. Color intensities are shown as a log2 scale. The left-most column shows positive (+) and negative (−) detection in PAR-CLIP. Relative Rbfox3 expression levels in these cultures are shown in panel f. (b) Splinted ligation-mediated miRNA detection analysis. Autoradiograms following urea-PAGE of radiolabeled detection oligonucleotides which were ligated specifically to the indicated mature miRNAs from untreated (−) P19-GFP cells and RA-treated P19-GFP and P19-T2 cells are shown. (c) Agarose gel electrophoresis analysis of total RNAs. Ethidium staining of 18S ribosomal RNA (rRNA) from the indicated three P19 cell cultures is shown. (d) RT-PCR analysis of pri-miRNAs. PCR products for the indicated pri-miRNAs from the indicated P19 cell cultures following reverse transcription (RT, +) and without RT (−) are shown following agarose gel electrophoresis. RT (−) serves as a negative control for genomic DNA contamination. (e) Quantitative RT-PCR (qRT-PCR) of pri-miRNAs. No statistically significant difference in pri-miRNA abundance among the indicated three different P19 cell cultures was observed. n = 3 (biological replicates, cell cultures), average ± s.e.m., P > 0.05 (two-sided t-test). (f) Immunoblot analysis of Rbfox3 and the microprocessor complex.
In order to confirm the results obtained by microarray data, we analyzed expression of candidate miRNAs using a splinted-ligation-mediated miRNA detection method43. The expression level of miR-214, which was not detected by PAR-CLIP analysis, was unchanged under all three conditions (Fig. 2b, lanes 4–6). Although let-7i was induced by RA, Rbfox3 knock-down did not affect let-7i expression (Fig. 2b, lanes 1–3). In agreement with the microarray data, induction of Rbfox3 was accompanied by increased expression of miR-15a and miR-30c (Fig. 2b, lanes 8,11), whereas Rbfox3 knock-down (T2) canceled these increases (Fig. 2b, lanes 9,12). In contrast, miR-485 and miR-666 levels were decreased by Rbfox3 induction (Fig. 2b, lanes 14,17), whereas Rbfox3 knock-down restored their expression levels (Fig. 2b, lanes 15,18). We next asked whether the Rbfox3-associated changes in miRNA expression originated from the changes in the pri-miRNA expression level. Quantitative RT-PCR showed that pri-miR-15a and pri-miR-485 were not significantly altered in the various states of P19 cells (Fig. 2d, e). There was no obvious change in the protein levels of Drosha and Dgcr8 (Fig. 2f), which are essential for pri-miRNA processing to pre-miRNA44,45.
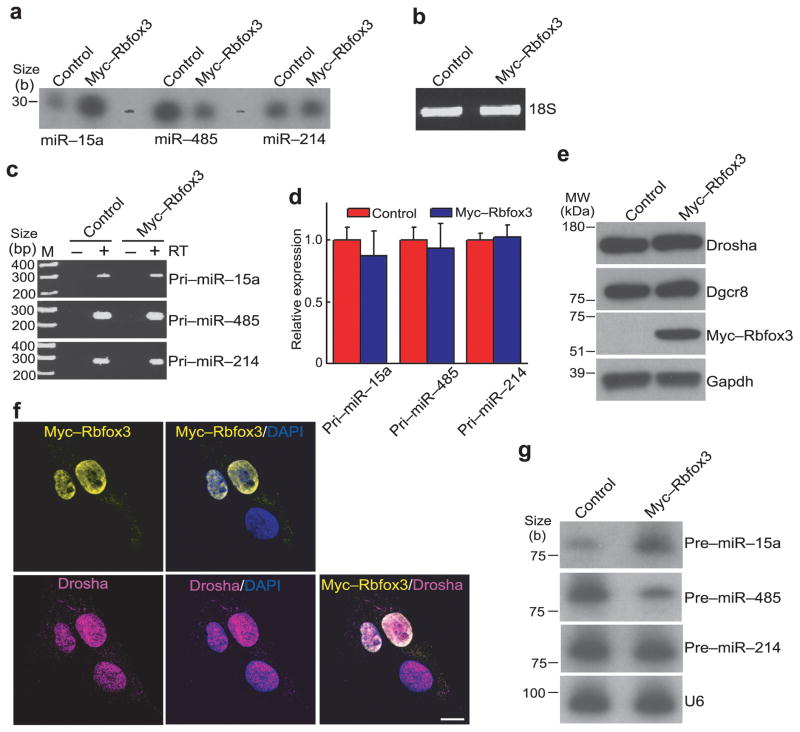
To further investigate the effect of Rbfox3 on miRNA expression, we measured the miRNA expression levels after exogenous Rbfox3 expression in P19 cells. Consistent with the knock-down experiments, exogenous Rbfox3 expression increased and decreased miR-15a and miR-485 levels, respectively (Fig. 3a). There was no significant change in the levels of the pri-miRNAs, Drosha and Dgcr8 (Fig. 3c–e). However the possibility remains that Rbfox3 might affect steps before the pri-miRNA processing. Therefore we analyzed the effect of Rbfox3 on the pri-miRNA levels in the Drosha knocked-down P19 cells, where conversion of pri-miRNA to pre-miRNA is prohibited. The pri-miR-15a and pri-miR-214 levels increased moderately but the pri-miRNA-485 level was unchanged by Drosha knock-down (Supplementary Fig. 2a). These observations reflect that only a small fraction of the pri-miRNAs (host transcripts) is processed by the Drosha complex. Exogenous Rbfox3 expression did not affect the levels of all three pri-miRNAs in either control or Drosha knock-down cells (Supplementary Fig. 2a). Therefore the Rbfox3-mediated change in mature miRNA abundance most likely occurs during posttranscriptional steps. Because Rbfox3 is predominantly localized in the nucleus with nucleolar exclusion (Fig. 3f), Rbfox3 might play a role in regulating miRNA biogenesis at the stage of pri-miRNA processing into pre-miRNA. To examine this possibility, the endogenous pre-miRNA levels were measured after exogenous Rbfox3 expression using RNA blot analysis. Consistent with our hypothesis, pre-miR-15a and pre-miR-485 levels were increased and decreased by exogenous Rbfox3 expression, respectively, whereas the pre-miR-214 level was unchanged (Fig. 3g). These results suggest that Rbfox3 has an effect on the processing of pri-miRNA into pre-miRNA for a subset of miRNAs in the nucleus.
Figure 3.
Rbfox3 effects on miRNA biogenesis in P19 cells. (a) Splinted ligation-mediated miRNA detection analysis. Indicated miRNAs are detected in total RNA from P19 cells which were transfected with an Rbfox3 expression construct (myc-Rbfox3) or empty vector (control). (b) Agarose gel electrophoresis analysis of total RNAs. Ethidium staining of 18S rRNA in total RNAs isolated from P19 cells transfected with the indicated vectors is shown. (c) RT-PCR analysis of pri-miRNAs. RT (−) serves a negative control for genomic DNA contamination. (d) qRT-PCR of pri-miRNAs. No statistically significant difference in pri-miRNA abundance was observed between myc-Rbfox3 transfected and control P19 cells. n = 3 (biological replicates, cell cultures), average ± s.e.m., P > 0.05 (two-sided t-test). (e) Immunoblot analysis of Rbfox3 and the microprocessor complex. (f) Immuno-fluorescent micrographs showing nuclear localization of Rbfox3 and Drosha. Exogenously expressed myc-Rbfox3 and endogenous Drosha in P19 cells were immunostained. DAPI stains nuclei. Bar, 10 μm. (g) RNA blots detecting pre-miRNAs. U6 serves as a loading control. Uncropped images of panels a and g are provided in Supplementary Figure 5.
Rbfox3 binds to pri-miRNAs lacking the UGCAUG motif
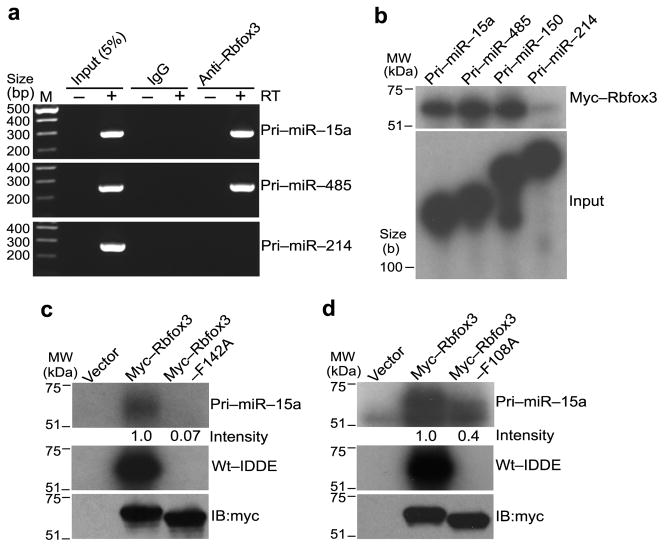
Although PAR-CLIP analysis indicated that Rbfox3 binds a subset of miRNA hairpin loci in P19 cells and in mouse brain, the cluster sequences determined by PAR-CLIP overlap with the mature and/or pre-miRNA sequences. Therefore we analyzed interactions of Rbfox3 with pri-miRNAs in neuronally differentiated P19 cells by RNA–protein complex immunoprecipitation (RIP) followed by RT-PCR using primers which detect only pri-miRNAs but not mature or pre-miRNAs. Rbfox3 interacted with pri-miR-15a and pri-miR-485 but not pri-miR-214 (Fig. 4a). These results confirm the interaction of Rbfox3 with specific pri-miRNAs in the cells and the specificity is consistent with the PAR-CLIP data.
Figure 4.
Binding of Rbfox3 to pri-miRNAs in P19 cells and in vitro. (a) RT-PCR detecting pri-miRNAs in RNA–Rbfox3 complexes from RA-treated P19 cells. RT-PCR products amplifying the indicated pri-miRNAs from RNA samples, which were recovered from anti-Rbfox3-immunocomplex from RA-treated P19 cell lysates, are shown following agarose gel electrophoresis. Immunoprecipitation with nonspecific IgG and PCR without RT (−) serve as negative controls. (b) In vitro pri-miRNA–Rbfox3 crosslinking analysis. Samples are RNase-treated anti-myc-immunoprecipitates of in vitro synthesized myc-Rbfox3 which was UV crosslinked with the indicated radiolabeled pri-miRNAs. Autoradiograms of myc-Rbfox3–RNA complexes following LDS-PAGE (upper panel) and input pri-miRNAs following urea-PAGE (lower panel) are shown. (c, d) In vitro UV crosslinking of Rbfox3 mutant with RNAs. Upper and middle panels are autoradiograms of the indicated wild-type or mutant (F142A, F108A) myc-Rbfox3 crosslinked with radiolabeled pri-miR-15a and wt-IDDE, respectively, following LDS-PAGE. Numbers under the upper panel indicate relative intensities of radioactive bands. The empty expression vector (vector) serves as a negative control. The wt-IDDE is a 201-nt intronic distal downstream enhancer region for alternative exon N30 of human nonmuscle myosin heavy chain II-B gene (MYH10) and contains two copies of UGCAUG. Lower panel shows an immunoblot (IB) with anti-myc. Uncropped images of panels b, c, and d are provided in Supplementary Figure 5.
To determine whether Rbfox3 itself binds directly to pri-miRNA, we performed in vitro UV crosslinking experiments using radiolabeled pri-miRNA and in vitro synthesized Rbfox3 protein. Rbfox3 directly interacted with pri-miR-15a, pri-miR-485 and pri-miR-150 but not with pri-miR-214 (Fig. 4b), consistent with the PAR-CLIP and RIP-RT-PCR results. Next, we addressed the mechanism of Rbfox3 binding to the pri-miRNA compared with its binding to the UGCAUG motif. Based on the solution structure of Rbfox1 and the UGCAUGU oligonucleotide46, two phenylalanine (F) residues F108 and F142 within the Rbfox3 RRM are critical for the recognition of the 5′-UGC nucleotides and the second 5′-UG nucleotides, respectively. We postulated that the critical amino acid residues for Rbfox3 binding to the pri-miRNA and to the UGCAUG motif might be different. We examined this possibility using mutant Rbfox3 where F108 or F142 was changed to alanine (A). The two mutant proteins with F108A and F142A completely lost their binding activities to the wild-type IDDE (wt-IDDE) which contains two copies of the UGCAUG element7 (Fig. 4c, d, middle panels). However, whereas the F142A mutant essentially did not bind at all to pri-miR-15a, 40% of the binding of the F108A mutant to pri-miR-15a was retained (Fig. 4c, d, upper panels). These results suggest that Rbfox3 may employ different binding mechanisms in a target-specific manner.
Rbfox isoforms have different binding activities to RNAs
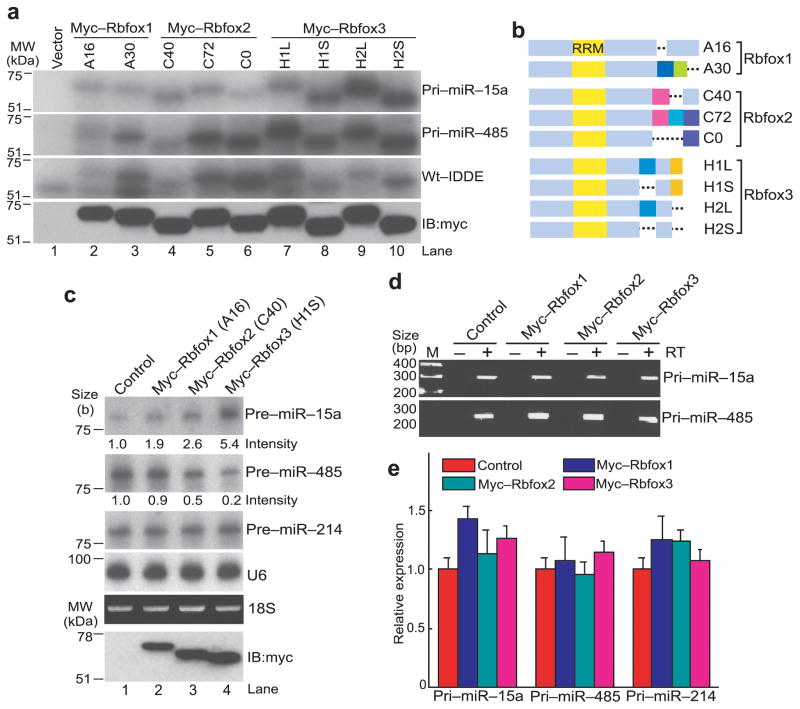
Extensive alternative splicing of all three Rbfox family genes generates C-terminal variants7,8,15,16. Interestingly, although all Rbfox family proteins contain an almost identical RRM, individual isoforms of the Rbfox family showed different binding activities to pri-miR-15a and pri-miR-485 as well as to wt-IDDE (Fig. 5a, b). All four isoforms of Rbfox3 efficiently bound to pri-miR-15a and pri-miR-485, and two Rbfox2 isoforms (C72, C0) also bound strongly to pri-miR-485 but not to pri-miR-15a (Fig. 5a). Two isoforms of Rbfox1 (A16, A30) and the C40 isoform of Rbfox2 showed weaker binding to pri-miR-485 compared with the C72 and C0 isoforms. Each isoform of Rbfox also had different binding activity to the wt-IDDE. Therefore the region outside of the RRM, specially the C-terminal domain, appeared to affect the binding activities of Rbfox proteins to the target RNAs, regardless of the presence or absence of the UGCAUG motif. We next examined whether Rbfox1 and Rbfox2 also have an effect on pri-miRNA processing. RNA blot analysis showed that exogenous expression of Rbfox1 or Rbfox2 increased or decreased the expression level of pre-miR-15a or pre-miR-485, respectively, although the extents of changes were smaller than Rbfox3 (Fig. 5c). The pre-miR-214 level was unchanged by exogenous Rbfox expression. There were no significant changes in the pri-miRNA levels by exogenous Rbfox expression (Fig. 5d, e). These results suggest that the C-terminal domain of the Rbfox family protein influences the strength of its RNA binding activity and thus its ability to promote or inhibit pri-miRNA processing.
Figure 5.
Differential RNA binding activities and pre-miRNA processing activities among Rbfox isoforms. (a) In vitro UV crosslinking of various Rbfox isoforms with RNAs. Upper three panels are autoradiograms showing the indicated myc-tagged Rbfox isoforms crosslinked with the indicated radiolabeled RNAs following immunoprecipitation with anti-myc, RNase-treatment and LDS-PAGE. Bottom panel shows an immunoblot (IB) with anti-myc. (b) Diagram of Rbfox isoforms. The amino acids in the same colors indicate the same amino acid sequences within each member of the Rbfox gene family but not among the three different Rbfox genes. (c) RNA blot analysis of pre-miRNAs after exogenous expression of various Rbfox isoforms in P19 cells. Samples for upper five panels are total RNA from P19 cells which were transfected with expression constructs for the indicated proteins or with the empty vector (control). Numbers under the upper two panels indicate relative intensities of radioactive bands. The RNA blot for U6 and ethidium staining of 18S rRNA serves as loading and quality controls. Bottom panel is an immunoblot (IB) with anti-myc. (d, e) RT-PCR detecting pri-miRNAs after exogenous expression of Rbfox isoforms in P19 cells. End products of gel electrophoresis are shown in d. Quantification by qRT-PCR is shown in e. No statistical difference was observed. n = 3 (biological replicates, cell cultures), average ± s.e.m., P > 0.05 (two-sided t-test). Isoforms A16, C40 and H1S were used (see panel b). Uncropped images of panels a and c are provided in Supplementary Figure 5.
Rbfox3 can bind to various regions of pri-miRNA structure
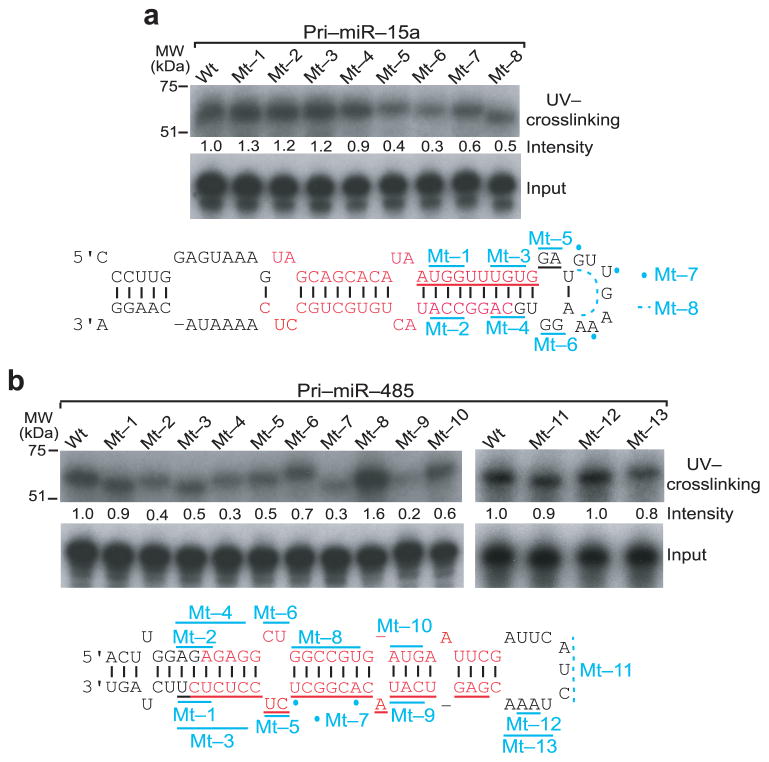
To determine the precise sequence on pri-miRNA bound by Rbfox3, we introduced nucleotide mutations in the stem-loop regions of pri-miR-15a and pri-miR-485. Mutant pri-miRNAs were used for in vitro UV crosslinking experiments. Mutation in the stem region of pri-miR-15a did not decrease Rbfox3 binding (Fig. 6a, mt-1–mt-4, Supplementary Fig. 3a). Mutation of the terminal loop in pri-miR-15a, which extends the stem base pairing (mt-5), changes nucleotides while maintaining the loop structure (mt-6, mt-7) or deletes nucleotides (mt-8), made the binding to Rbfox3 less efficient (Fig. 6a). These results suggest that Rbfox3 binds the terminal loop of pri-miR-15a and both the sequence and the loop structure are important for its binding. In the case of pri-miR-485, we detected an overall decrease of Rbfox3 binding by mutation in the stem region (Fig. 6b, mt-1–mt-7, mt-9, Supplementary Fig. 3b). Specifically, disruption of base paring in the stem region proximal to the terminal loop decreased Rbfox3 binding (mt-7, mt-9). Recovering base paring with a compensatory mutation restored the binding to Rbfox3 (mt-8, mt-10), suggesting that the stem structure is more important than the primary sequence of this region. On the other hand, various mutations in the terminal loop of pri-miR-485 did not affect Rbfox3 binding (Fig. 6b, mt-11–mt-13). These results suggest a possible mechanism for the Rbfox3 effect on miRNA processing: Rbfox3 enhances pri-miRNA processing by binding to the terminal loop region, whereas it represses pri-miRNA processing by binding to the stem region. To obtain further supports for this idea, we determined nucleotides crosslinked with Rbfox3 for additional pri-miRNAs by in vitro PAR-CLIP using in vitro transcribed RNA with 4S-UTP. In this analysis the conversion of U-to-C due to crosslinking to Rbfox3 was not necessarily detected in the exact binding sites as observed in the IDDE where the conversion occurred 3 nucleotides away from the UGCAUG element (Supplementary Fig. 4c). In the case of pri-miR-328, where mature miRNA increased in the presence of Rbfox3 (Fig. 2a), the crosslinked U was found very close to the terminal loop, similar to pri-miR-15a (Supplementary Fig. 4a). In contrast, in the case of pri-miR-300 and pri-miR-485 whose mature miRNAs decreased in the presence of Rbfox3 (Fig. 2a), the crosslinked U residues were located in the stem region far away from the terminal loop (Supplementary Fig. 4b). These results support the model that positive and negative effects of Rbfox3 on pri-miRNAs processing depend on its binding location on the stem-loop structure of pri-miRNAs.
Figure 6.
Determination of Rbfox3 binding regions in pri-miRNAs. (a, b) In vitro UV crosslinking of Rbfox3 with wild-type and mutant pri-miR-15a and pri-miR-485. Upper panels are autoradiograms of myc-Rbfox3 crosslinked with indicated radiolabeled pri-miRNAs following LDS-PAGE. The relative band intensities are indicated under the panel. Middle panels are autoradiograms of input radiolabeled pri-miRNAs following Urea-PAGE. Lower panels show stem-loop region of wild-type (wt) pri-miRNAs. Mature miR-5p and 3p are indicated in red. The region underlined with red and black corresponds to the Rbfox3-binding cluster detected by PAR-CLIP. Some of nucleotides marked with cyan lines and dots are mutated for the indicated mutants (mt-1–mt-13). Nucleotides marked with cyan broken lines are deleted for the indicated mutants. Detailed changes in nucleotide, position, and base pairing for individual mutants are provided in Supplementary Figure 3. Uncropped images are provided in Supplementary Figure 5.
Rbfox3 regulates pri-miRNA processing to pre-miRNA in vitro
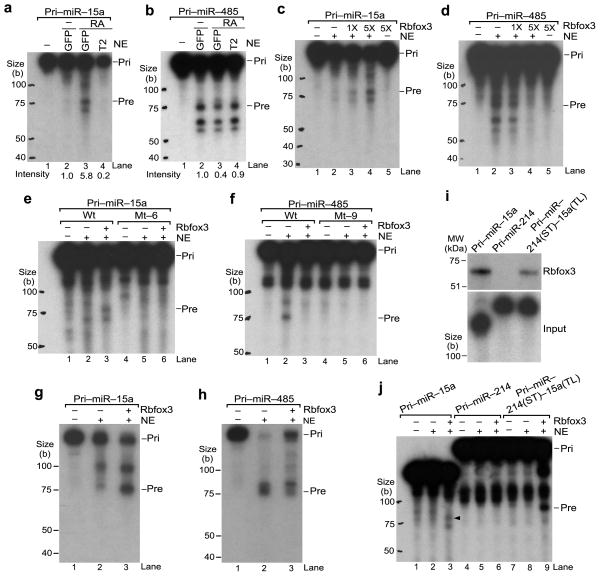
To test the direct involvement of Rbfox3 in the pri-miRNA processing into pre-miRNA, in vitro pri-miRNA processing assays were performed by incubating radiolabeled pri-miRNA substrates with nuclear extracts (NE) from undifferentiated P19-GFP, neuronally differentiated (RA-treated) P19-GFP, or RA-treated P19-T2 cells. The production of pre-miR-15a was readily detected in NE from neuronally differentiated P19-GFP cells but not from undifferentiated P19-GFP cells (Fig. 7a, lanes 2,3, quantified below). Moreover, the pre-miR-15a generation was remarkably reduced in NE from Rbfox3 knock-down P19-T2 cells (Fig. 7a, lane 4). In contrast, the pre-miR-485 production was reduced in NE from neuronally differentiated P19-GFP cells compared with those from undifferentiated P19-GFP cells (Fig. 7b, lanes 2,3, quantified below). Rbfox3 knock-down restored the pre-miR-485 production (Fig. 7b, lane 4). Because there is a possibility that compositions of NE from three different culture conditions might be different in not only the Rbfox3 level, we repeated the in vitro processing reactions using the same NE from undifferentiated P19-GFP cells with different amounts of in vitro synthesized Rbfox3. Increasing the amounts of Rbfox3 increased or decreased the production of pre-miR-15a or pre-miR-485, respectively, in a dose-dependent manner (Fig. 7c, d). RNA blot analysis after pri-miRNA processing assays using the complementary sequence to the mature miRNA as a probe, detected pre-miR-15a and pre-miR-485 with similar sizes to those detected in intact cells (see Figs. 3g, 5b), and thus verified the pre-miRNA products (Fig. 7g, h). These results demonstrate that Rbfox3 functions as a positive or negative regulator at the stage of pri-miRNA processing to pre-miRNA. Next we tested processing of mutant pri-miRNAs which bind poorly to Rbfox3. Mt-6 of pri-miR15a, in which the terminal loop is mutated, was not processed into pre-miRNA even in the presence of Rbfox3 (Fig. 7e, lane 6), consistent with the idea that Rbfox3 binding to the terminal loop is required to process pri-miR-15a into pre-miR-15a. Mt-9 of pri-miR-485, in which stem base pairs are disrupted, is not efficiently processed into pre-miRNA regardless of the presence or absence of Rbfox3 (Fig. 7f, lanes 5,6). This suggests that mt-9 is neither a substrate for the microprocessor nor a target for Rbfox3 binding. To test whether the terminal loop of pri-miR-15a can function as an Rbfox3-dependent enhancer element for pri-miRNA cleavage in other pri-miRNA contexts, we generated a chimeric pri-miRNA. The terminal loop of pri-miRNA-214 was replaced by the pri-miR-15a terminal loop and the stem and flanking regions remained as pri-miR-214 (pri-miR-214(ST)-15a(TL)). Both wild-type pri-miR-214 and the chimeric pri-miR-214(ST)-15a(TL) were hardly processed into pre-miRNAs in the NE in the absence of Rbfox3 (Fig. 7j, lanes 5,8). Remarkably, addition of Rbfox3 to the NE resulted in generation of pre-miRNA from the chimeric pri-miRNA (Fig. 7j, lane 9), whereas Rbfox3 did not show any effect on the pri-miR-214 cleavage (Fig. 7j, lane 6). Rbfox3 binding to the chimeric pri-miRNA but not to pri-miR-214 was confirmed (Fig. 7i). Thus, the terminal loop of pri-miR-15a confers Rbfox3 regulation on unregulated pri-miRNA processing.
Figure 7.
Direct effects of Rbfox3 on pri-miRNA processing into pre-miRNA. (a, b) In vitro pri-miRNA processing by NE from different conditions of P19 cell cultures. Samples are RNA products from radiolabeled pri-miRNA which was incubated with NE or without NE (−) from RA-treated P19-GFP, P19-T2 cells or untreated (−) P19-GFP cells. Autoradiograms following urea-PAGE are shown. Original pri-miRNA (pri) and processed pre-miRNA (pre) are indicated. Intensity numbers under panels indicate relative pre-miRNA abundance compared to that in untreated P19-GFP. (c, d) In vitro pri-miRNA processing by NE with Rbfox3. Samples are RNA products from radiolabeled pri-miRNAs incubated in NE from untreated P19-GFP cells with increasing amounts (1X, 5X) of in vitro synthesized myc-Rbfox3 or with control reticulocyte lysates (myc-Rbfox3,−). (e, f) In vitro processing of mutant pri-miRNAs. Mt-6 and mt-9 are the same as those in Figure 6. (g, h) RNA blot analysis of in vitro processed RNAs from pri-miRNAs. Samples are unlabeled pri-miRNA incubated with (+) or without (−) NE and myc-Rbfox3 as indicated. RNA blots using radiolabeled oligoDNAs complementary to the mature miRNA sequences are shown. (i) In vitro UV crosslinking of Rbfox3 with the chimeric pri-miRNA containing the terminal loop of pri-miR-15a. Upper panel shows an autoradiogram of myc-Rbfox3 crosslinked with radiolabeled pri-miRNAs following LDS-PAGE. Lower panel shows input pri-miRNAs following Urea-PAGE. (j) In vitro processing of the chimeric pri-miRNA containing the pri-miR-15a terminal loop in the presence of Rbfox3. An arrowhead indicates pre-miR-15a. Uncropped image of panels i is provided in Supplementary Figure 5.
Rbfox3 modulates the Drosha–Dgcr8 binding to pri-miRNAs
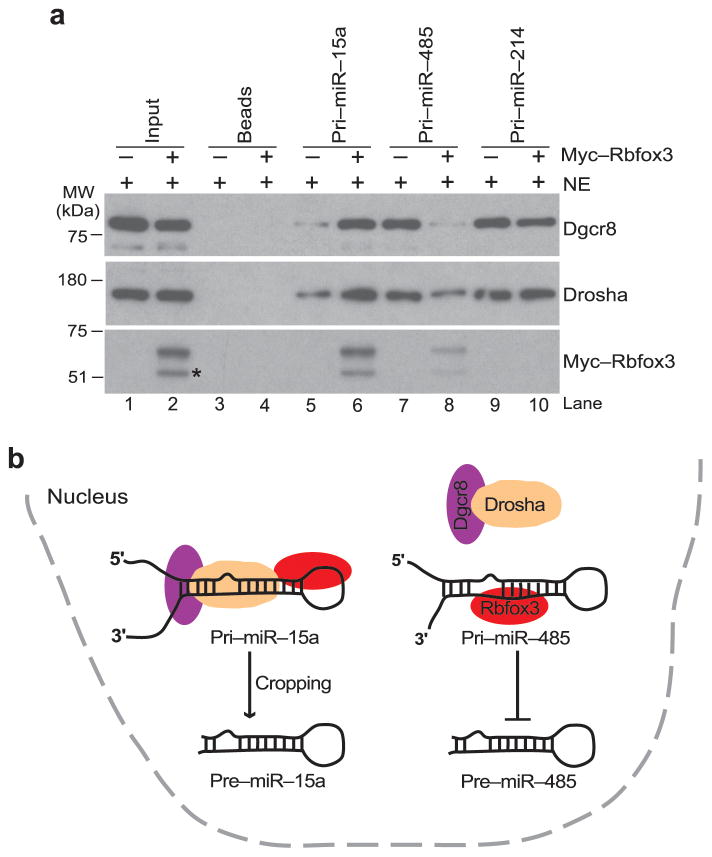
Next, we investigated how Rbfox3 exerts its activity as a negative or positive regulator in pri-miRNA processing. Because the Drosha–Dgcr8 complex is a critical factor for pri-miRNA processing in the nucleus, we explored the possibility that Rbfox3 strengthens or weakens the association of the Drosha–Dgcr8 complex with pri-miRNA. Pull-down experiments using immobilized pri-miRNA on agarose beads, the NE from undifferentiated P19 cells and in vitro synthesized Rbfox3 confirmed that Rbfox3 binds to pri-miR-15a and pri-miR-485 but not to pri-miR-214 (Fig. 8a, bottom panel, lanes 6,8,10). In the absence of Rbfox3, pri-miR-15a recruited Drosha and Dgcr8 to a small extent, while pri-miR-485 and pr-miR214 did so to a larger extent (Fig. 8a, top and middle panels, lanes 5,7,9). Strikingly, Rbfox3 addition enhanced the recruitment of the Drosha–Dgcr8 complex to pri-miR-15a, whereas it repressed the Drosha–Dgcr8 recruitment to pri-miR-485 (Fig. 8a, lanes 6,8). There was no change with pri-miR-214 (Fig. 8a, lane 10). These results suggest that Rbfox3 modulates pri-miRNA processing through the recruitment of the Drosha–Dgcr8 complex to pri-miRNA.
Figure 8.
Regulation of pri-miRNA processing by Rbfox3. (a) Immunoblots detecting pri-miRNA-recruited microprocessor complex in the presence and absence of Rbfox3. Samples are proteins associated with indicated pri-miRNAs immobilized to beads or beads alone following incubation with NE from untreated P19-GFP cells in the presence (+) or absence (−) of in vitro synthesized myc-Rbfox3. 5% of input was loaded in lanes 1, 2. The band indicated with * in lane 2 is most likely a truncated fragment of myc-Rbfox3. Uncropped images are provided in Supplementary Figure 5. (b) A model for Rbfox3-mediated regulation of pri-miR-15a and pri-miR-485 processing. In the case of pri-miR-15a, Rbfox3-binding to the terminal loop causes enhancement of the microprocessor (Drosha and Dgcr8) recruitment thereby increasing pre-miR-15a cropping. In the case of pri-miR-485, Rbfox3 binding to the stem region causes inhibition of the microprocessor recruitment thereby decreasing pre-miR-485 cropping.
DISCUSSION
We identified more than 4,000 binding sites for endogenous Rbfox3 in neuronally differentiated P19 cells using PAR-CLIP analysis that were reproducible in mouse CNS tissues. This study underscores the unexpected finding that Rbfox3 binds non-UGCAUG sequence containing RNAs including pri-miRNAs. Moreover, we provided extensive evidence that supports the direct involvement of Rbfox3 in miRNA biogenesis.
Although most RBPs have low sequence specificities for their RNA binding sites, Rbfox proteins have been thought to bind the UGCAUG sequence with extraordinarily high sequence specificity. This sequence was also identified as the most favored Rbfox2-binding sequence in embryonic stem cells by CLIP-seq and individual nucleotide-resolution CLIP-seq (iCLIP) and most recently in brain as a common target site for all three Rbfox family members13,47,48. However, these CLIP-seq studies showed that a substantial proportion of Rbfox-RNA crosslink sites do not overlap with the (U)GCAUG motif, suggesting that the RNA binding sites of Rbfox are not limited to the UGCAUG sequence in intact cells. Our analysis of 2,153 cluster sequences with a 5-nt extension in both sides from P19 cells, of which T-to-C conversion ratios are 0.9 and over (maximum 1.0), revealed that the top most over-represented pentamers were CAAGG, AAGGT, and CCAAG (z-scores: 35.39, 34.82, and 23.23, respectively), which overlap each other. Notably, the AAGGT sequence exists in the terminal loop of pri-miR-15a. However, a number of motif analyses of our PAR-CLIP clusters with a 20-nt or 200-nt extension in both sides failed to find a binding motif including the UGCAUG sequence. Overall Rbfox3 appears to bind a wide-range of sequences in neuronal cells. The reason why the UGCAUG sequence was not enriched in Rbfox3-binding clusters could be explained by a number of possibilities. First, the immunoprecipitated Rbfox3 isoforms in this study may have a different target binding preference compared with the isoform of Rbfox1 used for SELEX. As demonstrated in this study, Rbfox isoforms with C-terminal variations showed different binding activities to different target RNAs in vitro. Second, it has been reported that Rbfox binding partner proteins affect the binding strength of Rbfox to target RNAs10,14. Interaction of Rbfox3 with other RBPs might also result in targeting to different RNA sequences. Rbfox3 could be crosslinked to sequences which have a low affinity for Rbfox3 alone in intact cells. Third, the accessibility of anti-Rbfox3 antibody to Rbfox3 in large ribonucleoprotein complexes might be limited to certain complexes. Our analysis may not include all Rbfox3–RNA complexes. Fourth, our PAR-CLIP analyses are unlikely to be saturating. Low abundance RNA targets, such as short-lifetime intermediate products of RNA processing or metabolism might be missed.
Among Rbfox3-targeted non-UGCAUG RNAs in neuronal cells, we focused on pri-miRNAs in this study. The specific and direct binding of Rbfox3 to the pri-miRNAs is demonstrated by in vitro binding assays. Moreover, in vitro binding assays using mutant Rbfox3 suggest that the mode of the interaction between Rbfox3 and a pri-miRNA is different from that of the interaction between Rbfox3 and the UGCAUG sequence. The RRM has a β1α1β2β3α2β4 topology that forms a four-stranded β-sheet packed against two α-helices with loops connecting β-strands and α-helices49. A F142A mutation, which is located in the core ribonucleoprotein domain-1 in the β3-sheet, abolishes Rbfox3 binding to the pri-miRNA similar to its effect on binding to UGCAUG, suggesting that the β-sheet surface of the Rbfox3 RRM is used for binding to both target RNAs. On the other hand, a F108A mutation completely abolishes Rbfox3 binding to the UGCAUG but not to the pri-miRNA. A previous structural study using the mammalian Rbfox1 RRM with 5′-UGCAUGU-3′ RNA has shown that F126 located in the loop between β1 and α1 is critical for binding to the 5′-end UGC sequence46. Consistent with this study, F108 in Rbfox3 (equivalent to F126 in human RBFOX1) is crucial for binding to the UGCAUG sequence. However, F108 is not as critical to the pri-miRNA. Because the β-sheet does not always bind RNA with sequence-specificity and RRMs have the capacity to interact with RNAs using all the elements comprising their structure (i.e. β-sheets, loops, α-helices)49, it is conceivable that Rbfox3 binds various RNA sequences using different structural modes.
Multiple lines of evidence shown in this study point to a role for Rbfox3 in regulation of miRNA biogenesis. Our results revealed that Rbfox3 has a positive and negative influence on miRNA processing at the stage of Drosha cleavage through its direct binding to pri-miRNAs. Rbfox3 binds to the terminal loop of pri-miR-15a and promotes the recruitment of Drosha and Dgcr8, resulting in enhanced pri-miRNA cleavage. On the other hand, Rbfox3 binds to the stem region of pre-miR-485 and inhibits the recruitment of Drosha and Dgcr8, resulting in reduced pri-miRNA cleavage (Fig. 8b). Moreover there are similar correlations between the Rbfox3 binding location on pri-miRNA and the Rbfox3 effect on miRNA biogenesis for pri-miR-328 and pri-miR-300. Based on these data, we speculate that regulation of pri-miRNA processing by Rbfox3 may depend on its binding position within the stem-loop structure. In addition, Rbfox3 may affect the efficiency of stem-loop formation of pri-miRNA, changing the recruitment of the microprocessor complex to the stem-loop structure. To understand how Rbfox3 binding to the terminal loop or the stem region influences the stem-loop structure of pri-miRNA, precise structure probing of pri-miRNAs before and after Rbfox3 binding will be required, as demonstrated for Hnrnpa1 and Khsrp37,50.
It has been proposed that phylogenetic conservation of pri-miRNA terminal loops reflects the requirement for auxiliary factors that bind to these conserved loops to ensure optimal pri-miRNA processing50. A more recent study further identified a conserved primary sequence motif in terminal loops critical for cleavage of a set of pri-miRNAs51. Notably, pri-miR-15a and two more pri-miRNAs (10a and 30c-2) out of four pri-miRNAs whose processing is enhanced by Rbfox3 have phylogenetically conserved terminal loops. Previous studies have demonstrated that Hnrnpa1 and Khsrp bind to the terminal loops of pri-miRNAs. Hnrnpa1 can promote and inhibit pri-miRNA processing depending on the pri-miRNA context, while Khsrp promotes pri-miRNA processing35,37,50. Srsf1 and Smad proteins have been reported to bind the stem region of pri-miRNAs and activate Drosha cleavage36,38. Although these examples focused on regulation of individual miRNAs, a recent study has demonstrated that SRp20 (Srsf3) plays an important role in Drosha cleavage of a large number of pri-miRNAs through its binding to a phylogenetically conserved CNNC motif in the 3′ single-stranded flanking region of pri-miRNAs51. In the case of Rbfox3, pri-miRNA cleavage is regulated positively and negatively depending on the pri-miRNA context.
Rbfox3 is expressed in a wide-range of postmitotic neurons from early development to adulthood. Although Rbfox3 is expected to play a role in the differentiation of neuronal cells and to function in mature neurons, its biological function has just begun to be addressed in cultured cells and animals21. To understand the biological function of Rbfox3, it is essential to study unbiased transcriptome-wide Rbfox3–RNA interactions, which were previously limited to the bioinformatics analysis of the UGCAUG motif12. Our study does not deny the previous notion that Rbfox proteins regulate alternative splicing through their binding to the UGCAUG motif, but adds the idea that Rbfox3 can bind to non-UGCAUG RNAs and can regulate other RNA processing events such as miRNA biogenesis. As regards other members of the Rbfox family, we have shown that some isoforms of Rbfox1 and Rbfox2 are also capable of binding to pri-miRNAs and regulating their biogenesis. In addition, the fact that Rbfox family proteins can form homo- or hetero- dimers suggests that Rbfox1 and Rbfox2 might also participate in miRNA processing with Rbfox3. Future studies will elucidate the biological relevance of Rbfox-regulated miRNA biogenesis.
ONLINE METHODS
Cell culture, transfection, shRNAs, and siRNA
P19 mouse embryonal carcinoma cells were purchased from ATCC and maintained in MEMα containing 7.5% calf serum and 2.5% fetal bovine serum. Establishment of the clonal P19 cell lines expressing T2 shRNA for Rbfox3 and control shRNA for GFP was described previously15. Neuronal differentiation of P19 cells was induced by retinoic acid as described previously21. In brief, cells were cultured in a bacterial-grade Petri dish in a medium containing 5 × 10−7 M all-trans retinoic acid for 4 d. The cell aggregates were resuspended by mild pipetting and trypsin/EDTA treatment. The resuspended cells were then transferred to a poly-d-lysine-coated tissue culture dish and cultured for an additional 4 d. P19 cells were transfected with plasmids alone or together with siRNA by electroporation (Amaxa Nucleofector, Lonza). SiRNA against Drosha was purchased from Santa Cruz (sc-44812).
Preparation of cell extracts, immunoblot analysis and antibodies (Abs)
Cell extracts were prepared using radioimmune precipitation assay (RIPA) buffer (Sigma) supplemented with a protease inhibitor cocktail (Roche) as described previously15. For immunoblot analysis, the lithium dodecyl sulfate (LDS)-denatured and reduced protein samples were resolved on a 4–12% polyacrylamide NuPAGE Bis-Tris gel (Invitrogen) and transferred onto a nitrocellulose membrane. Binding of antibodies was detected by the SuperSignal system (Pierce). The primary Abs used in this study were rabbit polyclonal anti-Rbfox315 (1:3,000), mouse monoclonal anti-Rbfox3 (1:500, Millipore, MAB377), mouse monoclonal anti-myc (1:5,000, Invitrogen, 46–0603), mouse monoclonal anti-Gapdh (1:5,000, Biodesign, H86504M), rabbit polyclonal anti-Drosha (1:1,000, Abcam, ab12286), and goat polyclonal anti-Dgcr8 (1:1,000, Abcam, ab109098). The antibodies have been validated for immunoblot analysis of mouse samples on the manufactures’ websites.
Immunofluorescence microscopy
Cell staining and immunofluorescence microscopy were carried out as described previously15. In brief, P19 cells transfected with an Rbfox3 expression plasmid were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline. Fixed cells were incubated with primary mouse monoclonal anti-Rbfox3 (1:500, Millipore, MAB377) and rabbit polyclonal anti-Drosha (1:1,000, Abcam, ab12286). Alexa-488- and Alexa-594-conjugated goat Abs against mouse IgG and rabbit IgG (1:500, Molecular Probes, A11001, A11012) were used as secondary Abs. The antibodies have been validated for immunocytochemical and immunofluorescent analysis of mouse samples on the manufactures’ websites. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, 5 μg/ml). The specimens were examined using a Zeiss LSM 510 Meta confocal laser-scanning microscope.
Plasmids and DNA templates
The expression plasmids for Rbfox3 in the pCS3+MT vector, which contains the cytomegalovirus promoter, the SP6 promoter and a myc epitope, have been described previously15. Mouse Rbfox3 isoforms H1L, H1S, H2L, and H2S correspond to mRbfox3-L(374), mRbfox3-S(327), mRbfox3-L(361), and mRbfox3-S(314), respectively, in a previous publication21. Rbfox1 A16 and A30 and Rbfox2 C40 (same as F11) were described previously7. The cDNAs for Rbfox2 C72 and C0 were obtained from mouse brain total RNA by RT-PCR using the primers 5′-ctc agg cct cca cta gtt ATG GAG AAA AAG AAA ATG GTAA CTC-3′ and 5′-ctc agg cct cct cta gaa GTA GGG GGC AAA TCG GCT GTA-3′. Lower case letters represent adapter sequences including restriction enzyme sites. The F108A and F142A mutant clones of Rbfox3 were generated by GenScript. The template DNA containing the T7 promoter upstream of the IDDE, which was used for in vitro transcription, was described previously7. DNA templates containing the T7 promoter and encoding an approximately 200 nt-region of pri-miRNAs were generated by PCR from MicroRNA Expression Plasmids (ORIGENE) using the following primer sets: 5′-TTG TAA TAC GAC TCA CTA TAG GGA AGC CTT TTC TGT AAA TTA C-3′ and 5′-GCT ATC ATA GGA GCT ATG AAT AAA AAG-3′ for pri-miR-15a, 5′-TTG TAA TAC GAC TCA CTA TAG GGA GAG AGT CGA TGT GTT CTT C -3′ and 5′-ACA TGG GTT TAG CCA TCC AGA AAC CCA CC-3′ for pri-miR-485, 5′-TTG TAA TAC GAC TCA CTA TAG GGC TGG AAA ACA GGC TGA TTG T-3′ and 5′-GTA GCT CTT GGT GTA GAT GCT ATG TTG TG-3′ for pri-miR-214, and 5′-TTG TAA TAC GAC TCA CTA TAG GGC AGT GCT TTC CGC AGC ATC t-3′ and 5′-CCC ACT TAG GAA GGG ACC CAA GGC ATC CC-3′ for pri-miR-150. Underlined nucleotides represent the T7 promoter sequence. The plasmid constructs encoding the mutants of pri-miR-15a, pri-miR-485, and chimeric pri-miR-214(ST)-15a(TL) with the upstream T7 promoter were generated by GenScript.
RNA preparation, RT-PCR, quantitative RT-PCR (qRT-PCR)
Total RNA was extracted with the miRNeasy kit (Qiagen) or Trizol (Invitrogen) and reverse transcribed by the SuperScript III First-Strand Synthesis System (Invitrogen) with random hexamers. PCR was performed using FastStart PCR Master Premix (Roche). qRT-PCR was performed in triplicate with FastStart Universal SYBR Green Master Premix (Roche) on a 7500 Real-Time PCR System (Applied Biosystems). Relative differences in RNA levels were calculated according to the 2−ΔΔCt method and normalized against Gapdh mRNA. Statistical analysis from 3 biological replicates (minimum sufficient number) was performed by two-sided t-test or ANOVA followed by Bonferroni’s multiple comparisons test. The data met the assumptions of the tests. The PCR primers used were 5′-GGA AAT ACT TTT TAT TCT GCT GAA AGC C-3′ and 5′-GCT ATC ATA GGA GCT ATG AAT AAA AAG-3′ for pri-miR-15a, 5′-CCA ATG CTG TCC CAC CCC TTC TTC ACA GG-3′ and 5′-ACA TGG GTT TAG CCA TCC AGA AAC CCA CC-3′ for pri-miR-485, 5′-CTC CTT TCC CTT TAT CCC CCT GTC CTT C-3′ and 5′-GTA GCT CTT GGT GTA GAT GCT ATG TTG TG-3′ for pri-miR-214, and 5′-CCA TGG AGA AGG CTG GGG-3′ and 5′-CAA AGT TGT CAT GGA TGA CC-3′ for Gapdh mRNA.
Analysis for miRNA microarray
The microarray assay was performed and analyzed using the GeneChip miRNA 2.0 Array (Affymetrix) following the manufacturer’s instructions (Santa Clara). The minimum sufficient sample size (n = 3 biological replicates, cell cultures) was chosen for each condition. The total nine chips raw data were pre-processed by global background correction, quantile normalization and median polish summarization. The data distributions were approximate to normal distribution after log transformation and quantile normalization. We performed the principle component analysis to estimate the variation and detect the outlier for triplicated three groups. The mouse mature miRNA probesets were compared between three groups by the One-way ANOVA model in the MSCL Analysis’s Toolbox implemented in JMP software (http://abs.cit.nih.gov/MSCLtoolbox/). The differentially expressed miRNAs were selected by the filters of greater than 40% expression level change and P values less than 0.01.
Splinted-ligation-mediated miRNA detection
MiRNAs were detected using the miRtect-IT miRNA Labeling and Detection Kit (Affymetrix) according to the manufacturer’s instructions. In brief, miRNAs were captured from 4 μg of total RNA using a bridge oligonucleotide that is designed to specifically detect one miRNA variant, and ligated to a 32P-labeled detection oligonucleotide using T4 DNA ligase (New England Biolab). The following bridge oligonucleotides were used: let-7i, 5′-GAA TGT CAT AAG CGA ACA GCA CAA ACT ACT ACC TCA-3′; miR-214, 5′-GAA TGT CAT AAG CGG CAC AGC AAG TGT AGA CAG GCA-3′; miR-15a, 5′-GAA TGT CAT AAG CGC ACA AAC CAT TAT GTG CTG CTA-3′; miR-30c, 5′-GAA TGT CAT AAG CGG CTG AGA GTG TAG GAT GTT TAC A-3′; miR-485, 5′-GAA TGT CAT AAG CGG AAT TCA TCA CGG CCA GCC TCT-3′; and miR-666, 5′-GAA TGT CAT AAG CGG GCT CTC ACA GCT GTG CCC GCT-3′. The resulting products were separated by electrophoresis in a urea-15% polyacrylamide TBE gel (Invitrogen) with Dynamarker Small RNA plus (BioDynamics Laboratory, Tokyo, Japan) as a size marker and exposed to X-ray film (Kodak).
RNA–protein UV crosslinking
Preparation of RNA substrates and myc-tagged proteins were carried out as described previously21. In brief, the RNA substrates were synthesized from PCR-generated DNA templates by T7 RNA polymerase in the presence of [α-32P]UTP using a MAXIscript kit (Ambion). The myc-tagged proteins were synthesized in vitro from pCS3+MT constructs using the TNT Coupled Reticulocyte Lysate System with SP6 RNA polymerase (Promega). Binding reactions were carried out in a 25 μl mixture that contains 10 mM HEPES (pH 7.9), 2 mM MgCl2, 1 mM ATP, 20 mM creatine phosphate, 50 ng yeast tRNA, 2 mM DTT, 2% polyethylene glycol (M.W. 3,550), the reticulocyte lysate reaction mixture and 1 × 106 c.p.m. RNA probe for 20 min at 30 °C. Reaction mixtures were irradiated with 254 nm UV in a UV Stratalinker 2400 (Stratagene) for 20 min on ice, digested with 2 μl RNase-A/T1 (Ambion) and immunoprecipitated with 2 μg of anti-myc. Samples were subject to LDS-PAGE followed by autoradiography. The radioactive bands were quantified using NIH ImageJ.
RNA affinity purification
Pri-miRNAs were generated by in vitro transcription from PCR-generated DNA templates which contain the T7 promoter using a MAXIscript kit (Ambion). The RNAs were treated with sodium m-periodate (Sigma) and coupled to adipic acid dihydrazide agarose beads (Sigma) as described previously52. By this procedure, the 3′ end of the RNA molecule attaches to agarose. Affinity purification of pri-miRNA-binding proteins from nuclear extract of P19 cells was performed as described52. Proteins associated with the immobilized pri-miRNAs were analyzed by immunoblotting.
In vitro pri-miRNA processing assays
Pri-miRNA substrates were synthesized from PCR-generated DNA templates by T7 RNA polymerase in the presence of [α-32P]UTP using a MAXIscript kit (Ambion). Nuclear extracts of P19 cells were prepared by Nuclear Extract Kit (ACTIVE MOTIF) and dialyzed in a buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, 0.2 mM EDTA, 0.4 mM PMSF, 0.5 mM DTT and 20% (v/v) glycerol. In vitro processing assays was performed as described38 with some modifications. Briefly, reactions were carried out in 25 μl mixtures containing 50% (v/v) nuclear extract, 20% (v/v) reticulocyte lysate reaction mixture, 10 mM HEPES (pH 7.9), 2 mM MgCl2, 1 mM ATP, 20 mM creatine phosphate, 50 ng yeast tRNA, 2 mM DTT, 2% polyethylene glycol (M.W. 3,550), and 100,000 c.p.m. of each pri-miRNA probe for 30 min at 30 °C. The reaction mixtures were then extracted with phenol-chloroform and the RNAs were precipitated with ethanol. The RNA samples were resolved on a urea-6% polyacrylamide TBE gel (Invitrogen) with Dynamarker Small RNA plus as a size marker and exposed to X-ray film (Kodak) at −80 °C.
RNA–protein complex immunoprecipitation (RIP)
Whole cell extracts were prepared from RA-treated neuronally differentiated P19 cells. Approximately 300 μl of cell pellets were lysed in 1 ml of the RIPA buffer, and 500 μl of cell extracts were subjected to RIP with 10 μg mouse anti-Rbfox3 (Millipore) or control IgG using Magna RIP kit (Millipore). RNA recovered from immunoprecipitates was subjected to RT-PCR.
RNA blot analysis
RNAs were resolved on a urea-10% polyacrylamide TBE gel (Invitrogen) under denaturing conditions with Dynamarker Small RNA plus as a size marker. RNAs in the gel were transferred electronically onto a Zeta-probe GT membrane (Bio-Rad) and then crosslinked to the membrane by UV-light at 1,200 mJ in a UV Stratalinker 2400 (Stratagene). The oligonucleotide complementary to the target RNA was end-labeled with [γ-32P]ATP by T4 polynucleotide kinase and used as a probe for RNA blotting. The following oligonucleotides were used as probes: miR-15a, 5′-CAC AAA CCA TTA TGT GCT GCT A-3′; miR-485, 5′-GAA TTC ATC ACG GCC AGC CTC T-3′; miR-214, 5′-GCA CAG CAA GTG TAG ACA GGC A-3′; and U6 snRNA, 5′-ATA TGG AAC GCT TCA CGA ATT-3′. Hybridization was performed in Expresshyb solution (Clontech). The membrane was exposed to X-ray film (Kodak).
PAR-CLIP
After labeling with 100 μM 4-thiouridine (4SU, Sigma, T4509) for 16 h, P19 cells were processed according to the PAR-CLIP protocol as described27. In brief, 365 nm UV irradiated cells were lysed in the NP-40 lysis buffer and incubated with RNase T1. Immunoprecipitation was carried out with protein G magnetic beads (Invitrogen) coupled to Rbfox3 antibody (Millipore) for 2 h at 4 °C. Immunoprecipitated RNA–Rbfox3 complexes on the beads were treated with RNase T1 and calf intestinal alkaline phosphatase (New England Biolabs) and then phosphorylated by T4 polynucleotide kinase and ATP including [γ-32P]ATP. The crosslinked and radiolabeled protein–RNA complexes were solubilized in LDS loading buffer and resolved on a 4–12% polyacrylamide NuPAGE Bis-Tris gel (Invitrogen). The RNA–protein complexes migrating at the 50–60 kDa region corresponding to Rbfox3–RNA complexes were excised and electro-eluted from the gel. Following Proteinase K digestion and phenol-chloroform extraction, the recovered RNAs were processed for a cDNA library preparation using an adaptor set compatible with Solexa sequencing according to the standard small RNA protocol as described38.
For mouse in vivo PAR-CLIP, 2-month-old C57BL/6 male and female mice were intraperitoneally injected with 100 μl of 200 mM 4SU followed by a second injection with 100 μl of 400 mM 4SU 1 h later. After an additional 1 h, whole brain and the upper part of the spinal cord were dissected and passed through a 40 μm cell strainer to prepare a single-cell suspension. The cells were irradiated with 365 nm UV-light and processed according to the regular PAR-CLIP protocol. All mouse procedures were performed with approval from the National Heart, Lung, and Blood Institute Animal Care and Use Committee.
For PAR-CLIP data analysis, raw sequencing reads were first trimmed and collapsed to unique reads by fastx_clipper and fastx_collapser, respectively. The resulting sequence reads were then mapped to the reference mouse genome (mm9) with Bowtie with the following parameter (-v 2 –m10 –best –strata – all). PARalyzer was then used to identify PAR-CLIP peaks, each of which has to contain a minimum of 10 sequence reads and at least one T-C conversion event. The resulting PAR-CLIP peaks were then filtered with RepeatMasker to remove repetitive elements (e.g. SINE, LINE) followed by comparison with known gene annotations (RefSeq) to determine their relative location in the genome. Supplementary Data Set 4 summarized the numbers of reads at each of processing, mapping, and filtering step of PAR-CLIP experiments using P19 cells and the mouse CNS. Analysis of clusters overlapping (minimum 5 nts) among three PAR-CLIP experiments (two from P19 cells and one from the mouse brain) was performed and an area-proportional Venn diagram was generated using a web application BioVenn53.
In vitro PAR-CLIP
RNA was synthesized by T7 RNA polymerase from PCR-generated DNA template in the presence of 4S-UTP (at a ratio to UTP of 1:4) using a MAXIscript kit (Ambion). The myc-Rbfox3 was synthesized in vitro from pCS3+MT constructs using the TNT Coupled Reticulocyte Lysate System with SP6 RNA polymerase (Promega). Binding reactions were carried out in the reaction mixture as described in the section of “RNA–protein UV crosslinking” and irradiated with 365 nm UV for 5 min. Without RNase digestion, crosslinked RNA–protein complexes were immunoprecipitated with anti-myc. RNA was recovered from the complexes by proteinase K digestion followed by phenol-chloroform extraction and then subjected to RT-PCR. The cDNA from the entire length of RNA was cloned in the TOPO-T vector (Invitrogen) and sequenced.
Determination of 4SU incorporation rate into total RNA
After incubation with 4SU, total RNA was isolated from neuronally differentiated P19 cells and mouse brain and was subjected to enzymatic digestion with or without dephosphorylation to generate single nucleosides or nucleotides, respectively, for HPLC analysis54. Briefly, 40 μg of purified total RNA was incubated in a 28 μl volume for overnight with 0.09 U snake venom phosphodiesterase (Worthington Biochemical) alone or together with 0.4 U bacterial alkaline phosphatase (Worthington Biochemical). The resulting single nucleosides or nucleotides were analyzed by a modified ion-paring high-performance liquid chromatography (HPLC) using the Agilent 1100 HPLC (Agilent technologies) equipped with a reverse phase column, Supelco LC-18-T (150 × 4.6 mm, 3 μm particle size). Chromatography was performed at the flow rate 0.7 ml/min and the nucleoside or nucleotide detection was performed at 260 nm and 333 nm. The HPLC-reverse phase column was calibrated with AMP, CMP, GMP, UMP, uridine and 4-thiouridine obtained from Sigma-Aldrich.
Supplementary Material
Acknowledgments
We thank U. Ohler, D.L. Corcoran and N. Mukherjee (Duke Univ.) for help in PARalyzer analysis, H. Wu (Lawrence Berkeley National Laboratory) for advice on preparation of a sequence library for PAR-CLIP, D.-Y. Lee (Biochemistry Core Facility, National Heart, Lung, and Blood Institute) for HPLC analysis of ribonucleosides, and C.A. Combs (Light Microscope Core Facility, NHLBI) for advice on confocal microscopy. We also thank D.E. Saunders and A.F. Smith for technical assistance, S. Nakahata for reagents, and M.A. Conti for critical reading of the manuscript. This work was supported by Division of Intramural Research, NHLBI, US National Institutes of Health, USA.
Footnotes
ACCESSION CODES
The PAR-CLIP sequence data have been deposited in the NCBI Sequence Read Archive with accession number SRP032810. The microarray data have been deposited in NCBI Gene Expression Omnibus with accession number GSE52377.
AUTHOR CONTRIBUTIONS
K.K.K. and S.K. designed the study. K.K.K. performed the biological experiments, and K.K.K., R.S.A. and S.K. interpreted the data. Y.Y. and J.Z. performed bioinformatics analyses. K.K.K., R.S.A. and S.K. wrote the manuscript with input from Y.Y. and J.Z.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–12. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponthier JL, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–74. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 3.Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–74. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 4.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271:17613–6. [PubMed] [Google Scholar]
- 5.Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–45. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brudno M, et al. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–48. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–89. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–16. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–22. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol. 2007;27:8612–21. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–41. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–63. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–7. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2011;39:3064–78. doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–61. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–93. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher TL, et al. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359:251–61. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–11. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fogel BL, et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–86. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehman LT, et al. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–60. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim KK, Nam J, Mukouyama YS, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200:443–58. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pistoni M, et al. Rbfox1 downregulation and altered calpain 3 splicing by FRG1 in a mouse model of Facioscapulohumeral muscular dystrophy (FSHD) PLoS Genet. 2013;9:e1003186. doi: 10.1371/journal.pgen.1003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–7. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macias S, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19:760–6. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jungkamp AC, et al. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell. 2011;44:828–40. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 30.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 31.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–39. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 32.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 34.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 35.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–84. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–8. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, et al. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rau F, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840–5. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 40.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–8. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morlando M, et al. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31:4502–10. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Vogel G, Yu Z, Richard S. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol Cell Biol. 2013;33:1233–43. doi: 10.1128/MCB.01604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maroney PA, Chamnongpol S, Souret F, Nilsen TW. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA. 2007;13:930–6. doi: 10.1261/rna.518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 45.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–73. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637–51. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weyn-Vanhentenryck SM, et al. HITS-CLIP and Integrative Modeling Define the Rbfox Splicing-Regulatory Network Linked to Brain Development and Autism. Cell Rep. 2014;6:1139–52. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–8. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–58. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–7. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrus A, Kuimelis RG. Base composition analysis of nucleosides using HPLC. Curr Protoc Nucleic Acid Chem. 2001;Chapter 10(Unit 10):6. doi: 10.1002/0471142700.nc1006s01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.