Abstract
Background
Atrial fibrillation (AF) affects over 30 million individuals worldwide and is associated with an increased risk of stroke, heart failure, and death. AF is highly heritable, yet the genetic basis for the arrhythmia remains incompletely understood.
Methods & Results
To identify new AF-related genes, we utilized a multifaceted approach, combining large-scale genotyping in two ethnically distinct populations, cis-eQTL mapping, and functional validation. Four novel loci were identified in individuals of European descent near the genes NEURL (rs12415501, RR=1.18, 95%CI 1.13 – 1.23, p=6.5×10−16), GJA1 (rs13216675, RR=1.10, 95%CI 1.06 – 1.14, p=2.2×10−8), TBX5 (rs10507248, RR=1.12, 95%CI 1.08 – 1.16, p=5.7×10−11), and CAND2 (rs4642101, RR=1.10, 95%CI 1.06 – 1.14, p=9.8×10−9). In Japanese, novel loci were identified near NEURL (rs6584555, RR=1.32, 95%CI 1.26–1.39, p=2.0×10−25) and CUX2 (rs6490029, RR=1.12, 95%CI 1.08–1.16, p=3.9×10−9). The top SNPs or their proxies were identified as cis-eQTLs for the genes CAND2 (p=2.6×10−19), GJA1 (p=2.66×10−6), and TBX5 (p=1.36×10−05). Knockdown of the zebrafish orthologs of NEURL and CAND2 resulted in prolongation of the atrial action potential duration (17% and 45%, respectively).
Conclusions
We have identified five novel loci for AF. Our results further expand the diversity of genetic pathways implicated in AF and provide novel molecular targets for future biological and pharmacological investigation.
Keywords: atrial fibrillation, genetics, epidemiology, expression, functional analysis, zebrafish
Introduction
Atrial fibrillation (AF) is a common arrhythmia with major public health implications due to its high prevalence, significant morbidity and considerable associated healthcare costs.1 Currently, there are nearly 3 million individuals in the United States and over 8.8 million individuals in Europe affected by AF. With an aging population, the prevalence of AF is expected to dramatically increase. In addition to conventional risk factors,2 a genetic predisposition has been shown to contribute to AF risk.3 Over the last several years, numerous AF associated mutations, candidate genes, and risk loci have been identified; however, much of the heritability of AF remains unexplained.
Genome wide association studies (GWAS) have identified thousands of genetic loci associated with a wide range of conditions and traits. Most studies employ a stringent threshold of genome wide significance which, while minimizing false-positive associations, often fails to identify many disease associated loci. Increasing the sample size of a GWAS will enhance power, but for many diseases large numbers of affected individuals are unavailable and genotyping remains expensive. Since we have a limited understanding of the pathophysiology of AF, genetic discovery provides an important tool to identify novel pathways and therapeutic targets for the arrhythmia. Given these challenges, we sought to identify AF susceptibility loci using a combination of genotyping, eQTL mapping, and functional validation.
Methods
Overall study design
We have used available genome-wide association datasets for AF in Europeans and Japanese, respectively, and have identified selected genetic variants for additional replication in independent individuals. Following separate analyses in each replication cohort, we meta-analyzed the novel findings with the respective prior derivation stages. Variants that reached genome-wide significance for association with AF were subjected to additional analyses. First, we performed eQTL mapping in publicly available domains and left atrial tissue samples to identify gene expression changes depending on the identified genotypes. Second, we applied implicated loci pathway and gene enrichment analyses to better characterize novel candidate genes. Third, we performed candidate gene knockdown in an embryonic zebrafish model to test for morphologic and functional changes due to gene expression changes. Fourth, we conducted co-immunoprecipitation of candidate genes to inform protein-based interactions of our novel candidate genes. Last, we looked-up our association findings in a large consortial dataset of patients with ischemic stroke, a major consequence of AF. The study design including main results is summarized in Figure 1.
Figure 1.
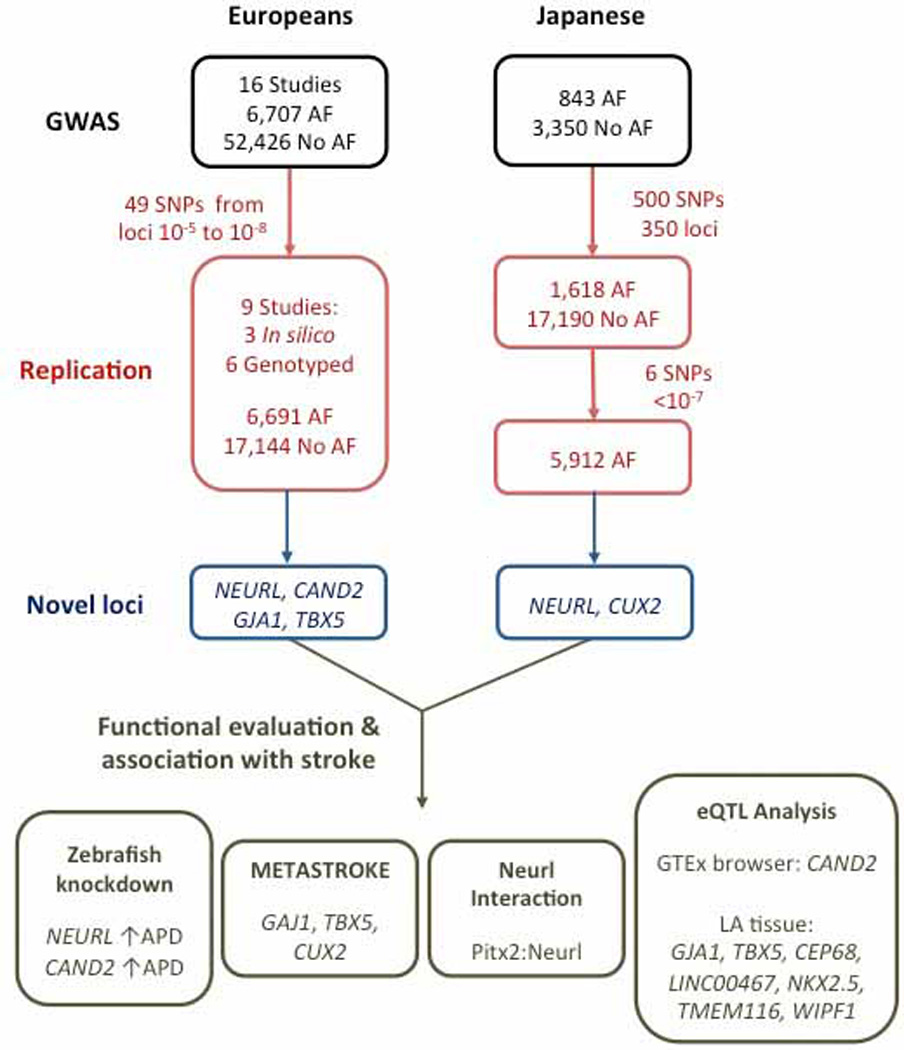
Flow-chart illustrating the study design and major results. Novel chromosomal loci associated with AF were identified independently in cohorts of European and Japanese descent by means of GWAS and subsequent replication. Signals in or around NEURL, TBX5, CAND2, GJA1, and CUX2 were detected. Additional studies revealed increased atrial action potential durations after knockdown of NEURL and CAND2, an interaction between NEURL and PITX2, an association of GJA1, TBX5, and CUX2 with stroke, and eQTL associations with CAND2, GJA1, TBX5, CEP68, LINC00467, NKX2.5, TMEM116, and WIPF1 in left atrial and other tissues. APD – action potential duration.
Study samples
Potential novel AF susceptibility signals in Europeans and Japanese were selected from a discovery sample consisting of cohorts with incident and prevalent AF, which has been previously described.4 To replicate variants from the discovery sample, we recruited additional samples and cohorts with available DNA for direct genotyping, or existing GWAS data for in-silico analysis. European replication samples included 6,691 independent AF cases and 17,144 controls. In Japanese, an additional 1,618 AF cases and 17,190 controls were analyzed; in a second replication stage, another 5,912 AF cases were added, totaling 8,373 AF cases. A detailed description of replication cohorts is available in the Supplemental Methods. Institutional Review Boards or Ethics Committees approved each contributing site. All participants provided written informed consent for participation in the cohorts, particularly allowing the analysis of DNA for genetic studies.
Selection of SNPs for replication
To identify SNPs for replication analyses in Europeans, we used the meta-analysis dataset from the GWAS performed by the AFGen consortium,4 and performed several selection steps: (1) We selected all SNPs (n=195) that demonstrated suggestive associations with the arrhythmia as defined by a meta-analysis p-value <5×10−5. This significance threshold for SNP inclusion was based on the expected power given an estimated independent validation sample size. (2) We then subjected SNPs within 1 Mb of the published genome-wide significant loci to further selection: all SNPs with a linkage disequilibrium measure r2≥0.1 with the published top-signals were omitted to avoid the inclusion of SNPs tagging the published results. (3) Finally we selected all SNPs with a minor allele frequency (MAF) ≥5%. SNPs with a MAF <5% were included if they were located in exons or the 3’ untranslated region (3’UTR) of known genes. Finally, we selected 49 variants. Given the smaller initial sample size of the GWAS in Japanese, a more extensive list of SNPs was considered for replication based on genotyping platform availability and cost. Balancing our statistical power and genotyping considerations, we thus selected the top 500 SNPs at 350 independent loci from a prior meta-analysis for successive rounds of genotyping as described in the supplemental methods.4
Genotyping
Cohorts of European descent were directly genotyped using the iPlex matrix assisted laser desorption / ionization time-of-flight (MALDI-TOF) mass spectrometry technique based on Sequenom platforms. All genotypes were analyzed using dedicated calling software applying the manufacturer’s recommendations. In Ottawa, TaqMan assays (Applied Biosystems, Inc., Foster City, CA) were used. For in-silico replication cohorts, genotypes from commercially available Affymetrix and Illumina genotyping arrays were used. Each cohort used genotyping results imputed to >2.5 million HapMap SNPs based on the HapMap CEU panel. Cohort-specific details are described in Supplemental Table 1. For genotyping in Japanese cases, the multiplex PCR-based Invader Assay (Third Wave Technologies) was used according to the manufacturer’s recommendations. Quality control for all genotyping results required a call rate ≥99% in both cases and controls, and deviations from the Hardy-Weinberg equilibrium were accepted to a p-value >1.0×10−6 in controls.
Statistical methods in Europeans
For genetic associations, studies from the GWAS discovery stage were calculated as described earlier.4 In the replication cohorts, we used logistic regression models to assess the associations between SNPs and AF; to achieve higher statistical power in smaller replication cohorts, we combined prevalent and incident AF cases. All models were adjusted for age at DNA draw and sex. Cohorts with multiple study centers further adjusted for site. Associations derived from GWAS datasets were also adjusted for principle components to account for population structure. Each cohort contributing in-silico replicated SNPs used significant principle components specific to their dataset. We assumed an additive model of inheritance. Associations were restricted to SNPs selected according to the description above. Directly genotyped SNPs were used following standard genotyping quality control. For imputed SNPs from GWAS cohorts, we used the observed to expected variance of the imputed SNP genotype count (r2) to adjudicate the imputation quality, and we only included SNPs with r2≥0.3 (range 0–1; 0: random imputation; 1: perfect imputation).
We meta-analyzed study-specific association results using METAL, applying a fixed effects approach weighted for the inverse of variance. Association effects are presented as relative risks (RR). For significant SNPs, we also computed tests of heterogeneity among the study effects; the p-values for the 4 SNPs in Table 1 were all >0.05 and thus not significant. We considered novel loci significantly associated when they exceeded the commonly accepted threshold of genome-wide significance at p=5×10−8 after meta-analyzing our GWAS discovery cohorts with the replication cohorts. For novel loci, we drew regional association plots using LocusZoom considering up to ±1000kb around the respective topSNP.
Table 1.
Meta-analyses of SNP-Associations with AF, by origin of study
| SNP | Chr | AF Risk allele |
Closest gene |
Relative location |
Original GWAS dataset4 | Replication | Overall Meta-analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF | RR (95% CI) |
p | RAF | RR (95% CI) |
p | RAF | RR (95% CI) |
p | |||||
| Europeans | |||||||||||||
| rs12415501 | 10q24 | T | NEURL | Intronic | 0.16 | 1.15 (1.10–1.22) |
9.0×10−8 | 0.16 | 1.22 (1.14–1.29) |
6.0 × 10−10 | 0.16 | 1.18 (1.13–1.23) |
6.5×10−16 |
| rs10507248 | 12q24 | T | TBX5 | Intronic | 0.73 | 1.13 (1.08–1.18) |
8.5×10−8 | 0.73 | 1.11 (1.05–1.17) |
0.0001 | 0.73 | 1.12 (1.08–1.16) |
5.7×10−11 |
| rs4642101 | 3p25 | G | CAND2 | Intronic | 0.65 | 1.11 (1.06–1.15) |
4.2×10−6 | 0.65 | 1.09 (1.04–1.15) |
0.0006 | 0.65 | 1.10 (1.06–1.14) |
9.8×10−9 |
| rs13216675 | 6q22 | T | GJA1 | Intergenic | 0.69 | 1.10 (1.05–1.15) |
5.0×10−5 | 0.68 | 1.10 (1.05–1.16) |
0.0001 | 0.69 | 1.10 (1.06–1.14) |
2.2×10−8 |
| Japanese | |||||||||||||
| rs6584555 | 10q24 | C | NEURL | Intronic | 0.12 | 1.33 (1.14–1.55) |
2.8×10−4 | 0.12 | 1.32 (1.25–1.39 |
1.6×10−22 | 0.12 | 1.32 (1.26–1.39) |
2.0×10−25 |
| rs6490029 | 12q24 | A | CUX2 | Intronic | 0.65 | 1.22 (1.09–1.37) |
6.3×10−4 | 0.64 | 1.11 (1.07–1.16) |
5.0×10−7 | 0.64 | 1.12 (1.08–1.16) |
3.9×10−9 |
In both the discovery and replication stages, each cohort provided cohort-specific results, which were subsequently meta-analyzed. In the overall meta-analysis, the summary results of each stage were meta-analyzed treating each stage as a cohort. In Ottawa, we used rs3825214 as a proxy SNP for rs12415501 (r2=0.76). SNP – single nucleotide polymorphism; AF – atrial fibrillation; Chr – chromosome; RAF – risk allele frequency; RR – relative risk; CI – confidence interval.
Statistical methods in Japanese
The associations of all SNPs were assessed with the Cochran-Armitage trend test. To further validate the results of the discovery-stage analysis, we selected the 500 SNPs with the most significant Cochran-Armitage trend p values for follow-up analyses in additional 1,618 Japanese AF cases and 17,190 AF-free controls. Of the selected 500 SNPs, 150 showed evidence of strong linkage disequilibrium with other selected markers as assessed by the Haploview software. We thus selected 350 SNPs for further genotyping. We combined the genotype data of both the first and second stage for meta-analysis using the Mantel-Haenszel method. We also assessed heterogeneity of our results for all significantly associated SNPs calculating Breslow-Day tests. All tests yielded p-values >0.05 and were thus non-significant: rs6584555, p=0.90; rs6490029, p=014; rs639652, p=0.46; rs1906599, p=0.77; rs6466579, p=0.27; rs12932445, p=0.98.
Analysis of eQTLs
We performed eQTL analyses from two sources: the Cleveland Clinic Atrial Tissue Bank and the publicly available Genotype-Tissue Expression Portal (GTEx) of the Broad Institute of Harvard and MIT. We first searched for all 49 SNPs considered for replication analysis in Europeans as well as the 2 SNPs identified in Japanese. Second, for those SNPs exceeding or approaching genome-wide significance after replication (Table 1), we additionally searched for all proxy SNPs defined as those with at least moderate linkage disequilibrium (r2≥0.5) with the sentinel SNPs. Detailed methods are provided in the supplement.
Implicated loci pathways
We also performed gene enrichment analyses at our implicated loci to determine known functional interactions between the 5 newly discovered loci and the 9 previously reported AF loci,4 in addition to 6 genes from eQTL analysis. The web-based tool GRAIL analyzes the connectivity between genetic loci using information retrieved from text mining.5 Here, we combined 20 loci, including the 14 AF loci and 6 eQTL genes as both the query and seed regions. The search was performed on the abstracts in PubMed published before August 2012. Out of the 20 queried loci, 10 showed an excess of connectivity (PGRAIL<0.05 after multiple testing correction). These loci were connected by keywords such as "cardiac", "heart", "channels", "atrial", or similar. In addition, we used the Ingenuity Pathway Analysis (IPA) tool to examine functional enrichment of the 14 AF loci. For each locus, we searched genes within 1Mb of the top SNP. A total of 275 genes were found. These genes were then analyzed by IPA, and the most significant canonical pathways were reported.
Knockdown of candidate genes in zebrafish
Zebrafish of the Tübingen/AB strain were maintained according to standard methods. Morpholino oligonucleotides (MOs) designed to disrupt the proper splicing or translation of zebrafish genes neurla, cand2, cand1, and cux2b were obtained from Genetools LLC (Corvallis, OR, USA). Measurements of heart rate, contractile function and optical mapping were obtained as previously described;6 detailed methods are provided in the supplement.
Co-immunoprecipitation in COS7 cells
For co-immunoprecipitation in COS7 cells, we transfected an expression plasmid of Myc- or FLAG-tagged target genes into COS7 cells (HSRRB; JCRB9127) using Fugene 6 (Roche). At 24h post transfection, immunoprecipitations were performed in lysis buffer (20mM Tris pH 7.5, with 150mM NaCl, 0.4% Nonidet P-40 containing 5µg/ml of MG-132 and protease inhibitor tablet EDTA- Roche) using anti-Myc tagged (Santa Cruz) or anti-FLAG tagged M2 agarose (Sigma). We visualized targets using HRP-conjugated anti-FLAG (Sigma) or anti-Myc antibodies (Santa Cruz).
Results
Study design
The overall design of the study is illustrated in Figure 1. In Europeans, the AFGen discovery sample comprised 16 studies that included 6,707 AF cases and 52,426 AF free controls.4 There were 195 single nucleotide polymorphisms (SNPs) with p values between 1×10−5 and 5×10−8 in the AFGen discovery sample. Based on a priori power calculations, we then selected 49 SNPs that were not in strong linkage disequilibrium with previously identified loci (r2<0.1). The SNPs were directly genotyped in 6 studies and in-silico replication was performed in 3 studies together consisting of 6,691 independent AF cases and 17,144 controls (Supplemental Tables 1 & 2). The mean age in the AF cases was 64.2±8.3 years versus 66.1±7.9 years in controls. Approximately two thirds of cases and half of the controls were male.
Following meta-analysis of the replication cohorts with the discovery stage results from the AFGen Consortium, four SNPs exceeded the threshold of genome-wide significance in Europeans; three further signals were near genome-wide significance (p<5×10−8). Results for the top 4 variants are shown in Table 1; full results for all 49 SNPs are provided in Supplemental Tables 3 & 4. Regional association plots for the top four associations in Europeans are shown in Figure 2.
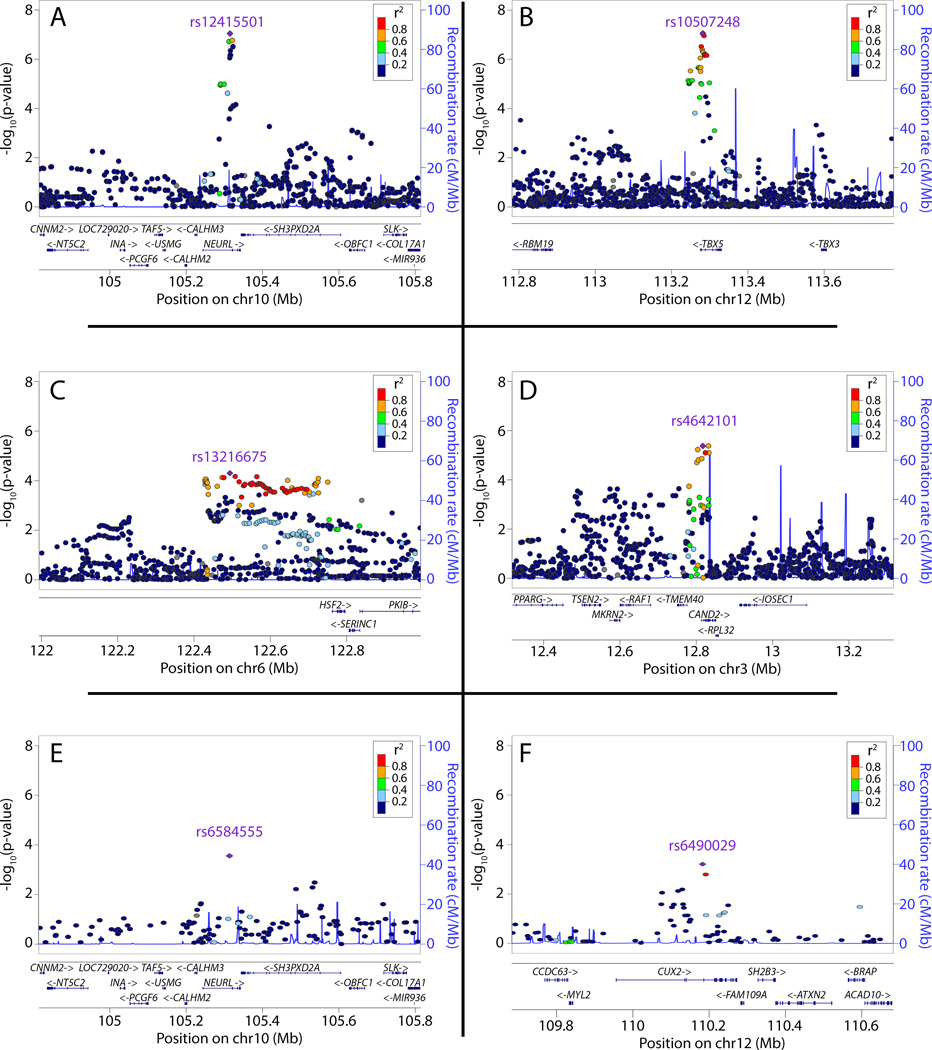
Figure 2.
Regional plots for novel atrial fibrillation susceptibility loci in Europeans and Japanese. Panels A-D (A: NEURL B: TBX5 C: GJA1 D: CAND2) show 4 novel loci detected in Europeans, panels E (NEURL) and F (CUX2) show 2 novel loci detected in Japanese. At each novel locus (p≤5×10−8), SNPs are plotted using the genomic position (NCBI Build 36) and discovery stage P values. In each panel, the sentinel SNP is labeled in purple. Each dot represents a SNP. The strength of the linkage disequilibrium of SNPs with the sentinel-SNP is indicated by a color gradient according to the legend in each panel, where red indicates strong, and blue indicates weak linkage disequilibrium. Estimated recombination rates are shown by the blue line, and spikes indicate locations of frequent recombination. Below each panel, the chromosomal positions of the SNPs and regional candidate genes are annotated. Linkage disequilibrium and recombination rates in panels A-F are based on the CEU HapMap release 22 (European) and JPT + CHB HapMap release 22 (Japanese), respectively. All regional association plots prepared using LocusZoom.
In Japanese, the GWAS discovery sample consisted of 843 AF cases and 3,350 AF free controls.4 A total of 500 SNPs from 350 loci were genotyped in a replication sample consisting of 1,618 AF cases and 17,190 controls, and the results were meta-analyzed with the Japanese GWAS discovery data. Six novel SNPs reaching p<1×10−7 were genotyped in 5,912 additional AF cases of Japanese ancestry, expanding the total number of AF cases to 8,373 (Supplemental Table 5); two SNPs remained significantly associated with AF (Table 1). Regional association plots for the two novel variants in Japanese are shown in Figure 2.
Five novel AF risk loci in Europeans and Japanese
The most significantly associated novel variants in both Europeans and Japanese were intronic to the gene NEURL on chromosome 10q24.33 (Europeans: rs12415501, relative risk for the AF risk allele (RR) 1.18, 95% confidence interval (CI) 1.13–1.23, p=6.5×10−16; Japanese: rs6584555, RR 1.32, 95% CI 1.26–1.39, p=2.0×10−25). Fine mapping of ten additional SNPs at the NEURL locus in the Japanese population did not reveal any independent susceptibility signals for AF at this locus (Supplemental Table 6).
The second locus identified in Europeans is intronic to TBX5 on chromosome 12q24 (rs10507248, RR 1.12, 95% CI 1.08–1.16, p=5.7×10−11). The third locus identified in Europeans is on chromosome 3p25.2 intronic to CAND2 (rs4642101, RR 1.10, 95% CI 1.06–1.14, p=9.8×10−9). The SNP rs4642101 is in moderate to strong linkage disequilibrium (r2=0.64) with the non-synonymous SNP rs2305398 that results in an amino acid substitution from glutamine to arginine (p.Q315R). The fourth locus identified in Europeans is on chromosome 6q22.31 in a large intergenic region (rs13216675, RR 1.10, 95% CI 1.06–1.14, p=2.2×10−8). The closest gene is GJA1; rs13216675 is located approximately 670kb downstream of the gene. Interestingly, each of the variants identified in Europeans at the TBX5, CAND2, and GJA1, were also associated with AF in Japanese (Supplemental Table 7). The fifth locus which, was identified only in Japanese individuals, is located intronic to CUX2 (rs6490029, RR 1.12, 95% CI 1.08–1.16, p=3.9×10−9) on chromosome 12q24.11–12; we did not observe evidence of an association at the CUX2 locus in Europeans (Supplemental Figure 1 and Supplemental Table 7).
Expression quantitative trait loci mapping
We assessed the influence of novel susceptibility signals on the expression of candidate genes by investigating eQTLs using two sources. First, accessing the publicly available Genotype-Tissue Expression Portal (GTEx), we found several significant associations between gene expression and novel susceptibility loci (Supplemental Table 8). The AF risk allele of the top SNP at the CAND2 locus, rs4642101, was significantly associated with a higher expression of CAND2 in skeletal muscle (p=2.6× 10−9). A proxy SNP for rs4642101 also had a significant eQTL with CAND2 (rs9877049, p=2.6×10−19, r2=0.64). No eQTLs were identified in the GTEx database at the four other novel loci.
Second, we associated SNP genotypes with gene expression levels in a large repository of left atrial tissue samples (n=289; Supplemental Table 8). AF was present at the time of tissue acquisition in 136 patients, 70 had no history of AF, and 80 patients were women. Among SNPs at the novel loci for AF, we found significant cis-eQTL associations where the AF risk allele correlated with a decreased expression of GJA1 (rs13216675, p=9.84×10−5) and the AF risk allele correlated with an increased expression of TBX5 (rs10507248, p=2.14 ×10−4). At both loci, we identified SNPs in linkage disequilibrium with the index SNPs, but with statistically stronger effects on gene expression: rs2176990 (r2=0.54 with rs13216675, p=2.66×10−6, 0.93 fold (0.90–0.95) decreased expression per AF risk allele) and rs1946295 (r2=0.87 with rs10507248, p=1.36×10−5, 1.12 fold (1.08–1.18) increased expression per AF risk allele).
Among the 49 SNPs initially tested for an association with AF in Europeans, we also observed significant eQTLs for SNPs at five other genes. These loci were only marginally associated with AF, but exceeded the threshold of significance at p<2.03×10−4 for eQTL analyses. The respective loci were found for SNPs in or around the candidate genes CEP68, LINC00467, NKX2.5, TMEM116, and WIPF1. In more detail, rs2723065 (association with AF p=7.6×10−8), and in particular rs2540950 (r2=0.93 with rs2723065) were strongly associated with the expression of CEP68 (p=9.70×10−17). The four other SNPs had a weaker association with AF, but a significant cis-eQTL association with the candidate genes LINC00467 (rs12733930, p value for association with AF = 8.2×10−4, p value for eQTL =1.59×10−24), NKX2.5 (p for AF=1.0×10−6, p for eQTL=8.78×10−6), TMEM116 (rs6490029, p for AF=3.9×10−9, p for eQTL=4.28×10−06), and WIPF1 (rs2358891, p for AF=2.0×10−6, p for eQTL=8.87×10−10) (Supplemental Table 4).
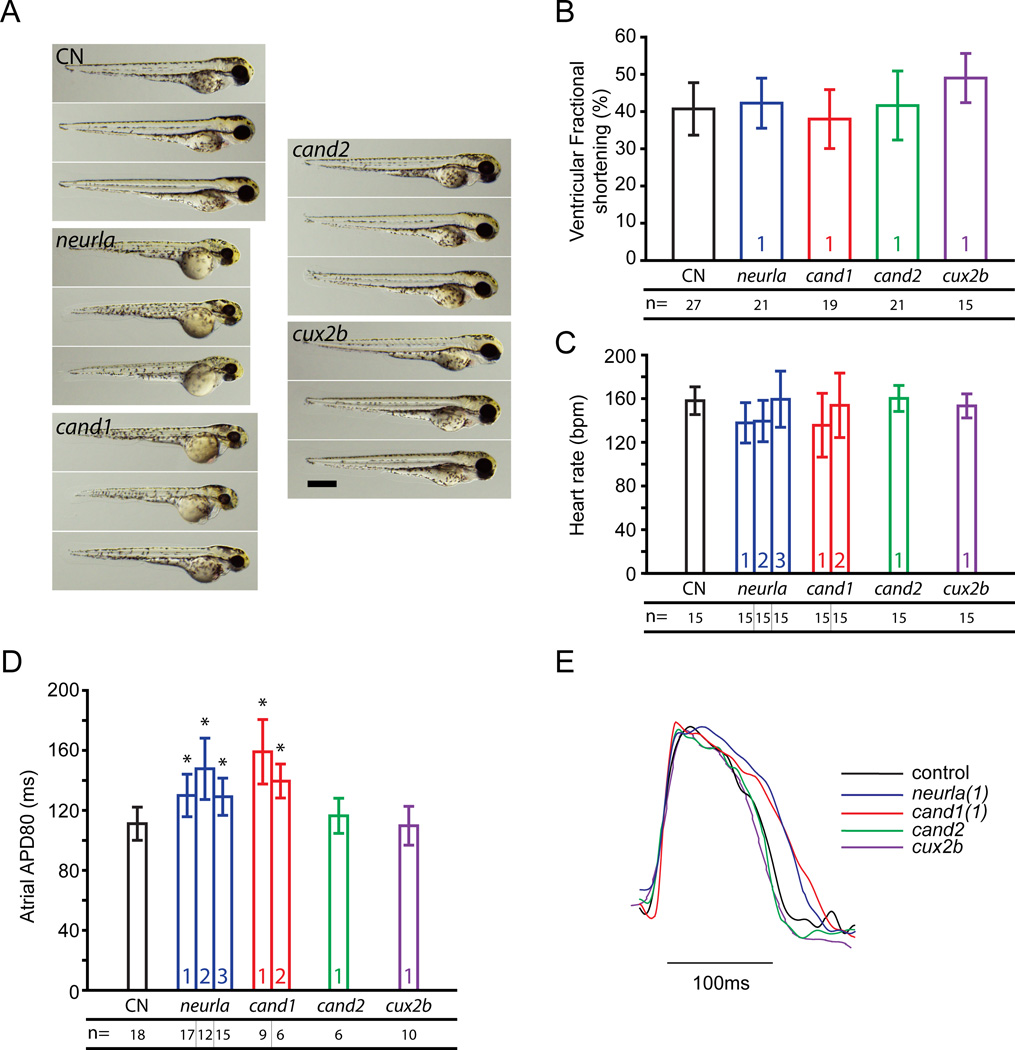
Zebrafish knockdown studies of NEURL, CAND2 and CUX2
For the novel AF risk loci identified in our genetic analyses, we sought to determine the potential role of these genes in cardiovascular function through morpholino-mediated knockdown of orthologues in zebrafish embryos (Supplemental Table 9). Since TBX5 and GJA1 have well-described roles in cardiovascular physiology, our zebrafish studies focused on the three novel candidate genes: NEURL, CAND2, and CUX2.
Zebrafish have a single ortholog of the NEURL and CUX2 genes, neurla and cux2b, but have two putative orthologs for the CAND2 gene, cand1 and cand2. We assessed the efficacy and morphologic consequences of gene knockdown, and the effect on resting heart rate, ventricular contractility, and atrial action potential duration (APD80). Knockdown efficacy was sufficient for all four genes (Supplemental Table 9). Morphologically, embryonic development was only slightly affected by knockdown of neurla and cand1, which showed mild developmental delay, whereas cand2 and cux2b morphants were indistinguishable from controls (Figure 3A). There were no significant effects on resting ventricular contractile function (Figure 3B) or heart rate (Figure 3C) for any knockdowns. We determined the atrial APD80 by analyzing optical mapping data as described earlier.6 For neurla knockdown embryos, the atrial APD80 was significantly lengthened by 17%, 34% and 19% for the three neurla-targeting morpholinos (Figure 3D, Supplemental Table 10). Knockdown of the zebrafish cand1 gene resulted in a prolongation of the atrial APD80 by 45% (Replication morpholino=31% APD80 increase) Knockdown of cand2 or cux2b did not significantly alter the APD80 (Figure 3D, Supplemental Table 10). Representative optical mapping recordings for all four gene knockdowns are presented in Figure 3E.
Figure 3.
Analysis of neurla, cand1, cand2, and cux2b knockdown in zebrafish. A: Brightfield micrographs of anesthetized 72hpf embryos injected with morpholinos. Scale bar = 500µm. B: Measurement of ventricular fractional shortening. C: Analysis of resting heart rate. D: Atrial action potential durations as assayed by optical mapping in zebrafish hearts. *Represents p<0.05 when compared to control. E: Representative traces of atrial action potentials from optical mapping. All numbers within bars indicate which morpholino was used for the presented data. Where no labels are shown, data represent pooled data obtained from all effective morpholinos. CN – control. n=number of biological replicates for a given experiment.
Interaction between Neurl and Pitx2
NEURL encodes an E3 ubiquitin ligase with a putative RING finger domain.7 E3 ubiquitin ligases have been shown to interact with several types of transcription factors.8 Since a number of AF GWAS loci reside at or near transcription factors (PITX2, ZFHX3, PRRX1, TBX5, and CUX2), we tested the direct interaction between NEURL and AF-associated transcription factors. NEURL was co-expressed in COS7 cells with each transcription factor using myc- or FLAG-tagged NEURL and myc-tagged PRRX1, ZFHX3, and TBX5 or FLAG-tagged PITX2 and CUX2. By co-immunoprecipitation, we demonstrated a NEURL-PITX2 protein interaction (Supplemental Figure 2a). We did not find evidence of a direct interaction between NEURL and PRRX1, CUX2, or TBX5; studies on ZFHX3 were unsuccessful (Supplemental Figure 2b, 2c, 2d).
Implicated loci pathways
To integrate our novel SNP and eQTL findings with the previously described 9 susceptibility loci for AF,4 we employed systems biology based gene enrichment analyses. Using the web-based tool GRAIL, 10 of the total 20 loci showed an excess of connectivity (p<0.05) involving keywords such as "cardiac", "heart", "channels", and "atrial" (Supplemental Figure 3). The most significantly enriched pathways by an Ingenuity analysis were those involving “calcium signaling” (p=5.3×10−5), “L-serine degradation” (p=4.1×10−4), and “geranylgeranyldiphosphate biosynthesis” (p=8.1×10−4).
Relation between novel AF risk loci and stroke
AF is strongly associated with an increased risk of stroke. We therefore determined whether the top 5 novel loci from our genetic analyses were associated with ischemic stroke in the METASTROKE collaboration of the International Stroke Genetics Consortium, a meta-analysis of GWAS combining 12,389 ischemic stroke patients and 62,004 controls (Table 2).9 For rs6490029, we detected an association with any type of ischemic stroke (CUX2, odds ratio 0.95, 95% CI 0.91–0.98, p=0.0034). Interestingly, the coded allele was hazardous for AF, but protective for ischemic stroke. Restricting our analyses to 2365 individuals with cardioembolic stroke, we also found associations for rs13216675 (GJA1; odds ratio 1.11, 95% CI 1.04–1.19, p=0.002) and rs10507248 (TBX5; odds ratio 1.13, 95% CI 1.05–1.21, p=0.0013). Consistent with findings from the METASTROKE collaboration, different subtypes of stroke show limited overlap in genetic associations.9
Table 2.
Association of Novel AF Loci in the METASTROKE Consortium9
| Phenotype | Cases | Controls | SNP | AF Risk Allele |
Closest gene |
RAF | OR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|
| Overall ischemic stroke | 12,389 | 62,004 | rs12415501 | T | NEURL | 0.16 | 1.03 (0.99 – 1.08) | 0.20 |
| rs10507248 | T | TBX5 | 0.73 | 1.05 (1.01 – 1.08) | 0.01 | |||
| rs4642101 | G | CAND2 | 0.65 | 0.95 (0.93 – 0.99) | 0.95 | |||
| rs13216675 | T | GJA1 | 0.69 | 1.04 (1.01 – 1.08) | 0.02 | |||
| rs6490029 | A | CUX2 | 0.23 | 0.95 (0.91 – 0.98) | 0.0034 | |||
| Cardioembolic stroke | 2,365 | 56,140 | rs12415501 | T | NEURL | 0.16 | 1.10 (1.01 – 1.20) | 0.03 |
| rs10507248 | T | TBX5 | 0.73 | 1.11 (1.04 – 1.19) | 0.0027 | |||
| rs4642101 | G | CAND2 | 0.65 | 1.04 (0.97 – 1.11) | 0.23 | |||
| rs13216675 | T | GJA1 | 0.69 | 1.13 (1.05 – 1.21) | 0.0013 | |||
| rs6490029 | A | CUX2 | 0.23 | 0.98 (0.91 – 1.05) | 0.55 |
Discussion
In the present study, we sought to integrate multiple parallel techniques to identify novel AF susceptibility loci. Large-scale genotyping in Europeans and Japanese identified novel AF risk loci at or near the genes NEURL, TBX5, CAND2, GJA1, and CUX2. Expression quantitative trait loci mapping in left atrial tissue analyses identified associations between AF SNPs at the CAND2, TBX5, GJA1, CEP68, LINC00467, NKX2.5, TMEM116, and WIPF1 loci. Functional characterization of NEURL and CAND2 orthologs in embryonic zebrafish demonstrated that knockdown of these genes resulted in a significant lengthening of the atrial action potential duration. Further, we found that NEURL and PITX2c physically interacted in a cellular overexpression model. Finally, AF-associated SNPs at the GJA1, TBX5, and CUX2 loci were also significantly associated with ischemic stroke.
The most significantly associated novel AF locus that we identified is intronic to the gene NEURL, which encodes an E3 ubiquitin ligase. NEURL has been reported to be a tumor-suppressor gene in malignant astrocytic tumors, and rat and mouse homologs of the gene are highly expressed in muscle tissue.10 The most consistent cellular abnormalities noted in AF are a calcium overload state and shortening of the atrial action potential duration.11 Using embryonic zebrafish, we found that knockdown of the NEURL ortholog specifically altered atrial action potential duration without affecting cardiac contractile function or heart rate. While it is unclear whether the AF-associated SNPs at the locus are associated with an increase or decrease in NEURL expression, our results provide compelling support for the role of NEURL in atrial repolarization and in turn, AF. 12
In 2007, a genetic locus was described for AF on chromosome 4q25, upstream from the gene encoding the transcription factor PITX2;22 in the ensuing years, the association between AF and variants at this locus has been widely replicated. Although the role of PITX2 in AF has not yet been fully understood, it is critical for the left-right symmetry of the heart during embryogenesis and the formation of myocardial sleeves in the pulmonary veins.13 Further, loss of one isoform, PITX2c, has been associated with an increased susceptibility to AF in murine models. Given the in vitro interaction between NEURL and PITX2 that we observed, it is interesting to speculate that NEURL may mediate a susceptibility to AF by ubiquitin–mediated alteration of PITX2 activity.
The second novel locus we identified resides at TBX5, a transcription factor that is critically involved in the development of the cardiac conduction system.14 We also found that SNPs at this locus modulate the expression of TBX5 in human atrial tissue. Mutations in TBX5 underlie Holt-Oram syndrome, features of which include atrial and ventricular septum secundum defects and conduction abnormalities including atrioventricular node block. In an atypical form of Holt-Oram syndrome with a high prevalence of AF, a TBX5 gain-of-function mutation was identified, findings that are consistent with our eQTL results.15 Two recent GWAS associated the electrocardiographic PR interval with variants intronic to or in proximity with TBX5.16, 17 In the study by Holm et al., the top SNP (rs3825214, r2=0.76 with rs10507248) also showed association with AF (p=4.0×10−5), but failed to reach genome-wide significance.16 In the study by Pfeufer et al., rs1896312 is independent of rs10507248 (r2=0) and showed no association with AF (p=0.72).17 Interestingly, we also found expression levels of NKX2.5 vary by SNP genotypes in our dataset. Together, TBX5 and NKX2.5 are known to play critical roles in both the differentiation of cardiomyocytes and the specialization of conduction and nodal tissue.14
At the third novel locus, CAND2 encodes a TATA-binding protein, TIP120b, which is muscle-specific and critical for myogenesis.18 We found that the AF associated SNP at this locus is associated with reduced CAND2/TIP120b expression in striated muscle tissue. While the specific role of CAND2/TIP120b in AF is currently unclear, we observed atrial action potential prolongation by morpholino-mediated gene knockdown in the zebrafish. Additionally, our eQTL analyses indicate that the risk allele is associated with increased expression of CAND2. Extrapolating our findings in the zebrafish, increased CAND2 levels would be predicted to shorten the atrial action potential duration, as has been widely observed in AF.
GJA1, a strong candidate gene at our fourth AF locus, encodes the gap junction protein connexin 43 on chromosome 6q22.31 which is abundantly expressed in the heart.19 We found that AF-associated SNPs influenced the transcription of GJA1 in both left atrial tissue and the whole heart. Connexin 43 is the predominant cardiac gap junction protein and facilitates coordinated electrical activity between adjacent myocytes. Germline mutations in GJA1 have been associated with syndromic diseases such as hypoplastic left heart syndrome, atrioventricular canal defects, or oculo-dento-digital dysplasia. Interestingly, a somatic, loss-of-function mutation in connexin 43 has been found to underlie AF in humans.20 Further, mice with 60% reduced atrial Gja1 expression showed an increased susceptibility to induced AF and atrial tachycardia.21 Two independent swine models with an AF induced reduction of GJA1 expression demonstrated that restoration of GJA1 expression ameliorated AF burden.22 More recently, SNPs in proximity of GJA1 have been reported to be associated with resting heart rate;23 however, the AF variants appear to be unrelated to both (r2=0.02 for each).
At the fifth locus, CUX2, cut-like homeobox 2, is a transcription factor implicated in cell-cycle progression relevant for spinal cord development,24 and has been investigated for its contribution to bipolar disorder. More recently, the Wellcome Trust Case Control Consortium identified variants at CUX2 as a significant susceptibility marker for type 1 diabetes.25 Yet, the reported SNP rs1265564 only displays weak linkage disequilibrium (r2=0.17) with the AF SNP rs6490029. In another GWAS of Koreans and Japanese for coronary artery disease, CUX2 was suggested as a susceptibility locus, but failed to replicate.26 The CUX2 association was Japanese specific as we did not find evidence for an association in the region among Europeans (Supplemental Figure 1). The specificity of the CUX2 association in Japanese was in contrast to other four loci that were all associated with AF to varying degrees (Table 1 and Supplemental Table 7). The variability in the association between individuals of European and Japanese ancestry may be due to differences in allele frequency, sample size or another intrinsic difference between the populations.
Clinically, AF confers a five-fold increased risk of stroke. We found that the AF SNPs at the CUX2, GJA1, and TBX5 loci were associated with ischemic stroke in the METASTROKE collaboration. Interestingly, we found that the AF risk allele at the CUX2 locus was associated with a decreased risk of ischemic stroke, whereas the AF risk alleles at the two other loci conferred an increased risk of cardioembolic stroke. Given that two of the strongest associations for stroke are at the PITX2 and ZFHX3 loci for AF,27, 28 it is possible that the associations we observed at the GJA1 and TBX5 loci are due to occult AF among the stroke cases. At present, it remains unclear why variants at CUX2 would be associated with a decreased risk of ischemic stroke.
Strengths of our work include the investigation of two large samples of AF cases in Europeans and Japanese, eQTL analyses in atrial tissue, functional studies supporting the role for NEURL and CAND2 in AF pathophysiology, and the association of three of the novel AF loci with stroke. However, our study was also subject to a number of limitations. We studied individuals of European and Japanese ancestry, thus extrapolation of our findings to other races and ethnicities may be limited. Although AF often occurs in association with other risk factors, we included all individuals with AF both to increase the generalizability and the statistical power of the current analyses. We acknowledge that the NEURL:PITX2 interaction that we observed was in vitro and further in vivo studies will be necessary. As with other GWAS, the AF associated SNPs are unlikely to be the causal variants; rather they are likely to be a marker of disease risk. Although we believe that our eQTL, co-immunoprecipitation, and zebrafish studies were important initial analyses, ultimately, further fine mapping, sequencing, and functional studies will be required to identify the specific role of these genes in the pathogenesis of AF.
In summary, using a combination of genetic association, eQTL analyses and functional mapping of novel genes, we have identified 5 susceptibility loci for AF. Functional analyses of NEURL and CAND2 via zebrafish knockdown resulted in alterations in atrial electrophysiology, and protein interaction analysis demonstrated an in vitro interaction between NEURL and PITX2. Finally, our findings indicate that the novel AF signals at GJA1, TBX5, and CUX2 were significantly associated with ischemic stroke or its subtypes. In aggregate, our studies further expand our understanding of the molecular pathways and clinical implications of this common and morbid arrhythmia.
Supplementary Material
Acknowledgments
Funding Sources: This work was funded by the following: AGES: NIH contract N01- AG-12100, NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). AFNET/KORA S4: German National Genome Research Network NGFN 01GS0838, 01GR0803, BMBF-01EZ0874, 01GR0803, NGFN 01GI0204, 01GR0103, NGFN-2, NGFNPlus 01GS0823, and NGFNPlus 01GS0834; German Federal Ministry of Education and Research (BMBF) 01EZ0874; German Competence Network on AF (AFNET) 01 GI 0204/N; Leducq Foundation 07-CVD 03, BMBF Spitzen cluster personalized medicine m4 (01 EX1021E), LMU Excellence Initiative (42595-6); Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. The KORA platform is funded by the BMBF and by the State of Bavaria. German Heart Foundation (Sinner). Supported by the DZHK (German Centre for Cardiovascular Research) and by the BMBF (German Ministry of Education and Research). ARIC: NHLBI contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01- HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367, R01HL086694 and U10HL054512; NHGRI contract U01HG004402; and NIH contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Additional support by grants RC1-HL-099452 from the NHLBI and 09SDG2280087 from the AHA. BioBank Japan: This study was supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan and by SENSIN Medical Research Foundation, Japan. DANFIB: Danish National Research Foundation Centre for Cardiac Arrhythmia, The John and Birthe Meyer Foundation, The Research Foundation of the Heart Centre Rigshospitalet, Lægernes Forsikringsforening af 1891, The Arvid Nilsson Foundation, and the Director Henrik Ibsens Foundation, the Villadsen Family Foundation, the Stock Broker Henry Hansen Wife Karla Hansen, Born Westergaard, Grant and The Danish Heart Foundation (grant no. 07-10-R60-A1815-B573-22398). Cleveland Clinic: R01 HL090620 from the NHLBI (Chung, Barnard, J. Smith, Van Wagoner); NIH/NCRR, CTSA 1UL-RR024989 (Chung, Van Wagoner); Heart and Vascular Institute, Department of Cardiovascular Medicine, Cleveland Clinic (Chung); Leducq Foundation 07-CVD 03 (Van Wagoner, Chung); Atrial Fibrillation Innovation Center, State of Ohio (Van Wagoner, Chung). CHS: NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the NINDS. Additional support was provided through AG023629 from the NIA. A full list of CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. FHS: NHLBI and Boston University School of Medicine based on analyses by Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. NHLBI’s Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No.N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Other support came from 1R01 HL092577 1RO1 HL076784, 1R01 AG028321, Evans Center for Interdisciplinary Biomedical Research ARC on Atrial Fibrillation at Boston University (Benjamin), 6R01-NS 17950 and AHA 09FTF2190028. Health ABC: NIA contracts N01AG62101, N01AG62103, and N01AG62106. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the NIH to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, NIA. HVH: NHLBI R01 HL 068986, R01 HL085251, and R01 HL073410. MAC: The Malmö Diet and Cancer study was made possible by grants from the Malmö city council. J. Gustav Smith was supported by the Swedish Heart-Lung foundation and the Thorsten Westerström Foundation. Olle Melander was supported by the Swedish Heart-Lung foundation, the Swedish Medical Research Council, the Medical Faculty of Lund University, Malmö University Hospital, the Albert Påhlsson Research Foundation, the Crafoord Foundation, the Region Skane, the Hulda and Conrad Mossfelt Foundation, the King Gustaf V and Queen Victoria Fund, the Lennart Hanssons Memorial Fund, and the Wallenberg Foundation. MGH: NIH grants K23HL114724 (Lubitz), T32HL007208 (Tucker), R01HL092577 (Ellinor and Benjamin), R01HL104156 and K24HL105780 (Ellinor), and an American Heart Association Established Investigator Award 13EIA14220013 (Ellinor). Ottawa: Heart and Stroke Foundations of Ontario (Gollob), and the Canadian Institutes for Health Research (Gollob). Dr. Gollob is supported by the Early Researcher Award program from the Government of Ontario and from the Heart and Stroke Foundation of Ontario Clinician-Scientist Award. PROSPER: Supported by an investigator initiated grant obtained from Bristol-Myers Squibb. Prof. Dr. J. W. Jukema is an Established Clinical Investigator of the Netherlands Heart Foundation (grant 2001 D 032). Support for genotyping was provided by the seventh framework program of the European commission (grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging grant 050-060-810). RS-I / RS-II: RS-I is supported by the Erasmus Medical Center and Erasmus University Rotterdam; The Netherlands Organization for Scientific Research; The Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by The Netherlands Organization for Scientific Research (NWO) (175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was supported by The Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project nr. 050-060-810. SHIP: SHIP is part of the Community Medicine Research Network of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania. This work supported partly by the research project Greifswald Approach to Individualized Medicine (GANI_MED). The GANI_MED consortium is funded by the BMBF and the Ministry of Cultural Affairs of the Federal State of Mecklenburg-West Pomerania (03IS2061A). Generation of genome-wide data has been supported by the BMBF (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG. WGHS: NHLBI HL 043851 (Buring) and HL69757 (Buring) and NCI CA 047988 (Buring), the Donald W. Reynolds Foundation and the Fondation Leducq (Ridker), with collaborative scientific support and funding for genotyping provided by Amgen. Atrial fibrillation endpoint confirmation was supported by HL-093613 (Albert) and a grant from the Harris Family Foundation (Tedrow). METASTROKE: Dr. Dichgans is supported by the Vascular Dementia Research Foundation. Dr. Rosand is supported by the National Institute of Neurological Disorders and Stroke U01-NS069208. Role of the Sponsors: None of the funding agencies had any role in the study design, data collection or analysis, interpretation of the data, writing of the manuscript, or in the decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Wann LS, Curtis AB, January CT, Ellenbogen KA, Lowe JE, Estes NA, 3rd, Page RL, Ezekowitz MD, Slotwiner DJ, Jackman WM, Stevenson WG, Tracy CM, Fuster V, Ryden LE, Cannom DS, Le Heuzey JY, Crijns HJ, Olsson SB, Prystowsky EN, Halperin JL, Tamargo JL, Kay GN, Jacobs AK, Anderson JL, Albert N, Hochman JS, Buller CE, Kushner FG, Creager MA, Ohman EM, Ettinger SM, Guyton RA, Tarkington LG, Yancy CW. 2011 accf/aha/hrs focused update on the management of patients with atrial fibrillation (updating the 2006 guideline): A report of the american college of cardiology foundation/american heart association task force on practice guidelines. Heart Rhythm. 2011;8:157–176. doi: 10.1016/j.hrthm.2010.11.047. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin EJ, Levy D, Vaziri SM, D'Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The framingham heart study. Jama. 1994;271:840–844. [PubMed] [Google Scholar]
- 3.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. Jama. 2010;304:2263–2269. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: Predicting genes at pathogenic snp associations and rare deletions. PLoS genetics. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kaab S, Roden DM, MacRae CA. Drug-sensitized zebrafish screen identifies multiple genes, including gins3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. Neuralized1 activates cpeb3: A function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–1383. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou W, Chen X, Shim JH, Huang Z, Brady N, Hu D, Drapp R, Sigrist K, Glimcher LH, Jones D. The e3 ubiquitin ligase wwp2 regulates craniofacial development through mono-ubiquitylation of goosecoid. Nat Cell Biol. 2011;13:59–65. doi: 10.1038/ncb2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, Thorsteinsdottir U, Nalls MA, Longstreth W, Wiggins KL, Yadav S, Parati EA, Destefano AL, Worrall BB, Kittner SJ, Khan MS, Reiner AP, Helgadottir A, Achterberg S, Fernandez-Cadenas I, Abboud S, Schmidt R, Walters M, Chen WM, Ringelstein EB, O'Donnell M, Ho WK, Pera J, Lemmens R, Norrving B, Higgins P, Benn M, Sale M, Kuhlenbaumer G, Doney AS, Vicente AM, Delavaran H, Algra A, Davies G, Oliveira SA, Palmer CN, Deary I, Schmidt H, Pandolfo M, Montaner J, Carty C, de Bakker PI, Kostulas K, Ferro JM, van Zuydam NR, Valdimarsson E, Nordestgaard BG, Lindgren A, Thijs V, Slowik A, Saleheen D, Pare G, Berger K, Thorleifsson G, Hofman A, Mosley TH, Mitchell BD, Furie K, Clarke R, Levi C, Seshadri S, Gschwendtner A, Boncoraglio GB, Sharma P, Bis JC, Gretarsdottir S, Psaty BM, Rothwell PM, Rosand J, Meschia JF, Stefansson K, Dichgans M, Markus HS. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmusk T, Palm K, Belluardo N, Mudo G, Neuman T. Dendritic localization of mammalian neuralized mrna encoding a protein with transcription repression activities. Mol Cell Neurosci. 2002;20:649–668. doi: 10.1006/mcne.2002.1148. [DOI] [PubMed] [Google Scholar]
- 11.Nattel S. New ideas about atrial fibrillation 50 years on. Nature. 2002;415:219–226. doi: 10.1038/415219a. [DOI] [PubMed] [Google Scholar]
- 12.Wakili R, Voigt N, Kaab S, Dobrev D, Nattel S. Recent advances in the molecular pathophysiology of atrial fibrillation. J Clin Invest. 2011;121:2955–2968. doi: 10.1172/JCI46315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. Pitx2c and nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 14.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including id2, tbx5, and nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 15.Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, Lam J, Wilde AA, Lekanne Deprez RH, Moorman AF. A gain-of-function tbx5 mutation is associated with atypical holt-oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102:1433–1442. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- 16.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U, Stefansson K. Several common variants modulate heart rate, pr interval and qrs duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 17.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WH, Kottgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BH, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Muller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann HE, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kaab S, Witteman JC, Alonso A, Benjamin EJ, Heckbert SR. Genome-wide association study of pr interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki T, Okada N, Ishida M, Yogosawa S, Makino Y, Tamura TA. Tip120b: A novel tip120-family protein that is expressed specifically in muscle tissues. Biochem Biophys Res Commun. 1999;261:911–916. doi: 10.1006/bbrc.1999.1147. [DOI] [PubMed] [Google Scholar]
- 19.Li JY, Hou XE, Dahlstrom A. Gap-43 and its relation to autonomic and sensory neurons in sciatic nerve and gastrocnemius muscle in the rat. J Auton Nerv Syst. 1995;50:299–309. doi: 10.1016/0165-1838(94)00101-o. [DOI] [PubMed] [Google Scholar]
- 20.Thibodeau IL, Xu J, Li Q, Liu G, Lam K, Veinot JP, Birnie DH, Jones DL, Krahn AD, Lemery R, Nicholson BJ, Gollob MH. Paradigm of genetic mosaicism and lone atrial fibrillation: Physiological characterization of a connexin 43-deletion mutant identified from atrial tissue. Circulation. 2010;122:236–244. doi: 10.1161/CIRCULATIONAHA.110.961227. [DOI] [PubMed] [Google Scholar]
- 21.Tuomi JM, Tyml K, Jones DL. Atrial tachycardia/fibrillation in the connexin 43 g60s mutant (oculodentodigital dysplasia) mouse. Am J Physiol Heart Circ Physiol. 2011;300:H1402–H1411. doi: 10.1152/ajpheart.01094.2010. [DOI] [PubMed] [Google Scholar]
- 22.Bikou O, Thomas D, Trappe K, Lugenbiel P, Kelemen K, Koch M, Soucek R, Voss F, Becker R, Katus HA, Bauer A. Connexin 43 gene therapy prevents persistent atrial fibrillation in a porcine model. Cardiovasc Res. 2011;92:218–225. doi: 10.1093/cvr/cvr209. [DOI] [PubMed] [Google Scholar]
- 23.Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, de Bakker PI, Muller M, Morrison AC, Smith AV, Isaacs A, Sanna S, Dorr M, Navarro P, Fuchsberger C, Nolte IM, de Geus EJ, Estrada K, Hwang SJ, Bis JC, Ruckert IM, Alonso A, Launer LJ, Hottenga JJ, Rivadeneira F, Noseworthy PA, Rice KM, Perz S, Arking DE, Spector TD, Kors JA, Aulchenko YS, Tarasov KV, Homuth G, Wild SH, Marroni F, Gieger C, Licht CM, Prineas RJ, Hofman A, Rotter JI, Hicks AA, Ernst F, Najjar SS, Wright AF, Peters A, Fox ER, Oostra BA, Kroemer HK, Couper D, Volzke H, Campbell H, Meitinger T, Uda M, Witteman JC, Psaty BM, Wichmann HE, Harris TB, Kaab S, Siscovick DS, Jamshidi Y, Uitterlinden AG, Folsom AR, Larson MG, Wilson JF, Penninx BW, Snieder H, Pramstaller PP, van Duijn CM, Lakatta EG, Felix SB, Gudnason V, Pfeufer A, Heckbert SR, Stricker BH, Boerwinkle E, O'Donnell CJ. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet. 2010;19:3885–3894. doi: 10.1093/hmg/ddq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development. 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 genomes-based imputation identifies novel and refined associations for the wellcome trust case control consortium phase 1 data. Eur J Hum Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JY, Lee BS, Shin DJ, Woo Park K, Shin YA, Joong Kim K, Heo L, Young Lee J, Kyoung Kim Y, Jin Kim Y, Bum Hong C, Lee SH, Yoon D, Jung Ku H, Oh IY, Kim BJ, Lee J, Park SJ, Kim J, Kawk HK, Lee JE, Park HK, Lee JE, Nam HY, Park HY, Shin C, Yokota M, Asano H, Nakatochi M, Matsubara T, Kitajima H, Yamamoto K, Kim HL, Han BG, Cho MC, Jang Y, Kim HS, Euy Park J, Lee JY. A genome-wide association study of a coronary artery disease risk variant. J Hum Genet. 2013;58:120–126. doi: 10.1038/jhg.2012.124. [DOI] [PubMed] [Google Scholar]
- 27.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, Kostulas K, Kuhlenbaumer G, Bevan S, Jonsdottir T, Bjarnason H, Saemundsdottir J, Palsson S, Arnar DO, Holm H, Thorgeirsson G, Valdimarsson EM, Sveinbjornsdottir S, Gieger C, Berger K, Wichmann HE, Hillert J, Markus H, Gulcher JR, Ringelstein EB, Kong A, Dichgans M, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in zfhx3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.