Abstract
Under continuous, glucose-limited conditions, budding yeast exhibit robust metabolic cycles associated with major oscillations of gene expression. How such fluctuations are linked to changes in chromatin status is not well understood. Here we examine the correlated genome-wide transcription and chromatin states across the yeast metabolic cycle at unprecedented temporal resolution, revealing a “just-in-time supply chain” by which components from specific cellular processes such as ribosome biogenesis become available in a highly coordinated manner. We identify distinct chromatin and splicing patterns associated with different gene categories and determine the relative timing of chromatin modifications to maximal transcription. There is unexpected variation in the chromatin modification and expression relationship, with histone acetylation peaks occurring with varying timing and “sharpness” relative to RNA expression both within and between cycle phases. Chromatin modifier occupancy reveals subtly distinct spatial and temporal patterns compared to the modifications themselves.
Chromatin plays fundamental roles in DNA-related processes, including transcription, replication, recombination and repair1. For example, most histone acetylations and certain methylations (eg. H3K4me3, H3K36me3) are correlated with active transcription while deacetylation and other methylations are correlated with repression2,3. The pervasiveness of biological dynamics suggests that temporal interrogation of chromatin function is warranted. Compensatory and homeostatic effects almost certainly limit understanding of chromatin functions from steady-state studies4. The discovery of histone “readers” and “writers” further suggests the dynamics of chromatin5,6, as do studies of bulk acetylation upon depletion of acetyl-CoA using a temperature-sensitive mutant7. Examination of chromatin state and RNA expression level at single genes upon activation reveals many dynamic changes, such as the sequential appearance of H2BK123ub and H3K4me3 at GAL1 during galactose induction8. Genome-wide ChIP-seq studies reveal temporally dynamic chromatin patterns in circadian rhythm, heart development, yeast meiosis and other dynamic processes. These patterns have been correlated with transcription factor binding and RNA expression9-11 and support a generally dynamic role for chromatin in gene regulation. However, no prior studies have sufficient temporal resolution to dissect the specific roles of individual histone modifications during distinct steps in transcription in vivo, and the scope of genes regulated in these paradigms on a fast time scale has been limited.
Here we exploit a uniquely informative dynamical system, the yeast metabolic cycle (YMC), to examine regulation of >3,000 genes in Saccharomyces cerevisiae 12. Under continuous glucose-limited conditions, yeast exhibits a ~4-5 hour respiratory cycle with >3,000 transcripts and dozens of metabolites oscillating at the same pace13. Although nearly all cycling genes share the same period of oscillation, the YMC can be broadly divided into three phases based on the defining expression profile: OX (oxidative), RB (reductive-building) and RC (reductive-charging). The overall logic of the YMC is well-defined: growth genes, such as ribosomal and amino acid biosynthesis genes are activated in OX; mitochondria and cell cycle genes are expressed in RB; and genes responding to starvation, stress and survival are elevated in RC. These divergent expression profiles imply that the dynamic interaction between chromatin modifications and transcription across distinct biological processes can be queried in a single system. Additionally, acetyl-CoA, a most dynamically oscillating metabolite, is a critical metabolic signal driving the YMC via histone (and perhaps other proteins) acetylation14. Studies on acetyl-CoA, S-adenosylmethionine and other metabolites directly involved in histone modification indicate that chromatin could play key roles in coordinating metabolism and gene expression7,15. Given such prevalent orchestration of expression and metabolites in the YMC, investigating the dynamic patterns of chromatin state could be useful for gaining a systems-level understanding of the YMC and beyond, such as how cells respond to a wide variety of nutrient and other biological stimuli through chromatin modifications. Discoveries made in the YMC could also extend our understanding of other cycling systems, such as circadian clocks.
We set out to do time-course ChIP-seq of histone modifications and modifiers in the YMC. We reveal the genome-wide landscape of 7 histone modifications across the YMC and compare it to a high-resolution RNA-seq profile and dynamic localization of 3 critical chromatin modifiers, Gcn5p, Esa1p and Set1p. High sampling frequency allowed definition of clear-cut sub-phases of gene expression in each phase, such as the sequential activation of ribosome biogenesis (ribi) and ribosomal protein (RP) genes during the growth phase. We also defined 7 distinct chromatin patterns associated with genes of different functional classes, revealing the diversity of histone modification pathways during transcription. We also observed that binding of certain chromatin modifiers does not always correlate with the appearance of the corresponding histone modifications, suggesting sophisticated temporal regulation. Inspired by the pervasiveness of the changes on the H3 and H4 N-termini during the YMC, we evaluated mutants in H3 and H4 modifiable lysines and showed that only certain multi-lysine mutants abolish the YMC, implying high system redundancy. Together, these data provide a comprehensive and high temporal resolution view of the dynamics of transcription and chromatin states. As such, they reveal how transcription and chromatin modifications are coordinated for different classes of genes during distinct phases of the life of a yeast cell.
Results
Gene expression patterns in the YMC
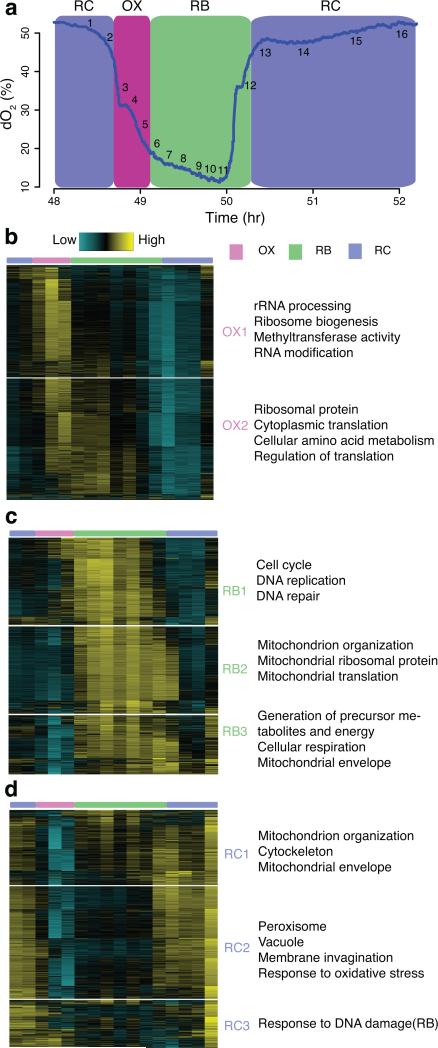
Microarray-based transcriptome analysis revealed three superclusters of genes defined by expression pattern in the YMC12. Because of the short duration of the OX growth phase (~0.5 h), the previous sampling strategy used, (evenly-spaced every ~25 min and spread over 3 cycles) may have missed important transient regulatory events. To improve temporal resolution, we collected 16 samples across one cycle for RNA-seq by increasing the density of time points taken in the OX phase (Fig. 1a). The same three superclusters of genes were observed in excellent agreement with the prior data (Supplementary Fig. 1a, Supplementary Table 1). We also identified an additional ~500 genes with a clear expression peak in the OX phase from the “non-periodic” genes defined previously (Supplementary Fig. 1b, Supplementary Table 1). Gene Ontology (GO) analysis indicated enrichment of ribosome biogenesis genes. Many showed hints of periodic expression in the microarray dataset but evaded detection by the periodicity algorithm due to less frequent sampling.
Figure 1. High temporal resolution analysis of gene expression reveals meticulous temporal compartmentalization in yeast.
(a) shows the oscillation of oxygen in the YMC and 16 time points of one cycle for RNA-seq. The same color scheme is used for labeling the three phases throughout the paper: Magenta = OX phase, Green = RB phase, Blue = RC phase. (b-d) K-means clustering of OX, RB and RC phase genes separately reveals subtly distinct temporal patterns. Enriched GO terms are listed on the right of each sub-cluster. Each row of the heat maps represents one gene and columns represent the 16 consecutive time points from left to right. 3-color bar on top marks the three phases. The blue-yellow gradient represents RNA signals normalized to read depth using Cuffdiff and centered to a mean of zero across 16 time points. (b) shows 2 subclusters of OX phase genes; (c) shows 3 subclusters of RB phase genes; (d) shows 3 subclusters of RC phase genes.
Thanks to high temporal resolution, we resolved several temporal subclusters associated with distinct biological functions within previously defined OX, RB and RC super-clusters (Fig. 1b-d). OX phase genes were subdivided into OX1, defining an earlier and more transient peak of expression compared to OX2 (Fig. 1b, Supplementary Fig. 1c). OX1 contained genes associated with ribosome biogenesis (ribi) and rRNA processing. Ribosomal subunit (RP), amino acid metabolism (aa) and cytoplasmic translation genes were enriched in OX2, and peaked ~5 min later. The subtle differences between ribi and RP genes were confirmed by reverse transcription followed by quantitative real-time PCRs (RT-qPCRs) (Supplementary Fig. 2f). This observation suggested precise temporal control of the energetically-demanding process of ribosome biogenesis. Ribosomal assembly factors and RNA processing factors were expressed just before ribosomal proteins and translational factors, providing a possible example of “just-in-time” orchestration of a critical process in the YMC leading to increased translational capacity. Three temporal subclusters of RB and RC genes were also observed (Fig. 1c, d, Supplementary Fig. 1c). Cell cycle and mitochondrial genes were readily distinguished although the two processes both occur under similar metabolic states. Collectively, the RNA-seq results not only recapitulated known gene expression patterns, but also displayed elaborate and fine-grained temporal control of coordinated sequential processes.
Accumulation of pre-mRNAs in the OX phase
Besides the temporal compartmentalization of gene expression, RNA-seq analysis also revealed an unexpected pattern in intron sequence accumulation. As discussed previously, RP genes are activated in OX, peaking at t5. Surprisingly, we observed a dynamic intron signal in several intron-containing RP genes consistently peaking at t4 (Fig. 2a). Several other intron-containing OX genes showed similar patterns (Fig. 2a).
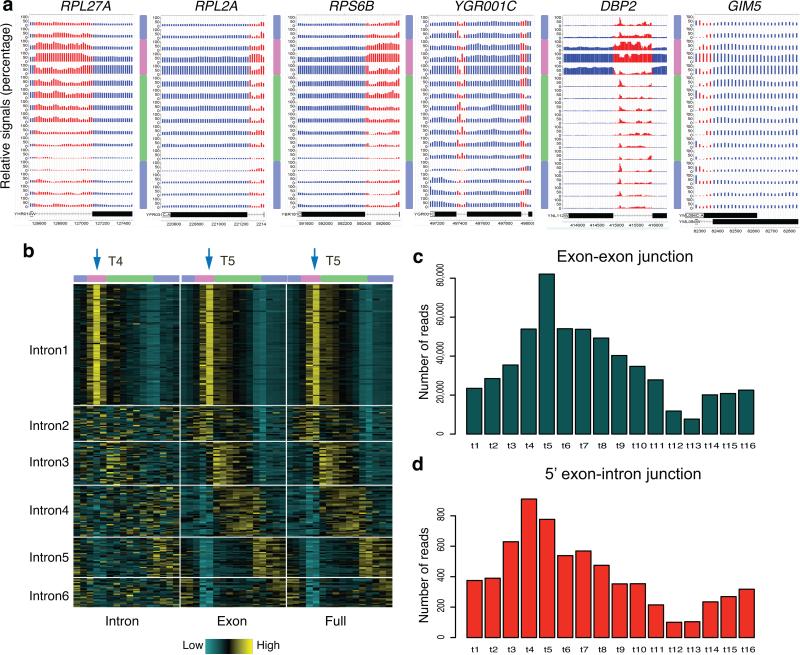
Figure 2. RNA-seq analysis at introns reveals transient accumulation of pre-mRNAs during OX phase.
(a) Relative RNA signals are displayed at intron-containing genes. Each track represents relative RNA levels at one of 16 time points, ordered sequentially from top to bottom. Signals are displayed as percentage of maximum value of the 16 time points at each 20 bp window. Red regions represent introns and blue regions represent exons. 3-color bars represent the 3 YMC phases vertically. Examples include genes in this cluster with the longest (DBP2), shortest (GIM5) intron and two introns (YGR001C). (b) A total of 280 intron-containing genes were divided into 6 groups based on their temporal RNA signals at introns, exons, and intron+exon (full) regions, ordered by the phase of peak full-length RNA expression. 16 time-point signals (individual columns) of each region were grouped together consecutively as one joint column. Three regions were ordered as “Intron”, “Exon”, “Full” and separated by white lines. Each single row represents signals of one gene standardized to a mean of zero. Arrows mark the peak time point. (c) The total number of reads spanning the exon-exon junctions of RP genes from Intron1 cluster are displayed across 16 time points in the bar plot. (d) The total numbers of junction reads spanning the 5’ end of RP introns from Intron1 cluster are displayed across 16 time points in the bar plot.
We evaluated RNA-seq reads across exon and intron regions separately for all intron-containing genes. Intron, exon and full-length signals were displayed in a heat map, identifying 6 clusters (Fig. 2b and Supplementary Table 2). Among them, only intron1 showed temporal enrichment of intron signals in the OX phase. 89 of the 103 genes in intron1 are RP genes. Both the intron signals and the ratio of intron/exon signals peaked at t4 (Fig. 2b and Supplementary Fig. 2a).
During RNA splicing, intron 5’ ends are cleaved and join to the branchpoint in the intron, forming a lariat. To distinguish intron accumulation as lariats or pre-mRNA, we examined exon exon and intron-exon junctional reads of the 89 RP genes. As expected, exon-exon junctional reads were highest at t5, consistent with the timing of exon signals (Fig. 2c). However, 5’ intron junctional reads, representing the pre-mRNA form, peaked at t4 (Fig. 2d). Additionally, we examined individual RP genes and discovered that the majority of the RP genes exhibited the same pattern (Supplementary Fig. 2b-d). RT-qPCR of 4 RP genes showed that the peaks of introns were consistently ahead of the peaks of exon signals by a mean time of 7.3 min (p value = 0.0286) (Supplementary Fig. 2f).
Interestingly, t4 marks a strikingly reproducible inflection point in the O2 curve of unknown functional importance occuring near mid-OX (Fig. 1a). These data suggested exquisite temporal control of RP gene splicing – RP genes appear to be transcribed at t4, but the corresponding mature spliced RP gene transcripts do not emerge until 5-7 min later at t5. Moreover, prior studies demonstrated accumulation of RP pre-mRNAs under stress conditions in yeast16,17. This delay in RP gene mRNA splicing in the YMC suggests that yeast might suffer a certain level of amino acid or other stresses during early OX (i.e., t4). Alternatively, RP mRNAs may be spliced only on cue, following proper assembly of the ribosome biogenesis machinery. These may lead to a transient pause in O2 consumption and the transient accumulation of RP pre-mRNAs precisely at this time point. Casein Kinase 2 complex (CK2) is required for RP gene pre-mRNA accumulation16; we observed peak expression of CK2 components at t4, t5 (Supplementary Fig. 2e).
Dynamics of histone modifications across YMC
Given the transcriptome dynamics and close dependence of transcription on chromatin modification, we next investigated chromatin states across the YMC. A similar set of 16 samples across one cycle were collected for ChIP-seq (Fig. 3a) and 7 histone modifications were assessed, including H3K4me3, H3K36me3, H3K9ac, H3K14ac, H3K56ac, H4K5ac, H4K16ac, and H3 as control. Most of these are associated with active transcription3,18. H3K56ac also functions in DNA replication and repair19 and H4K16ac is involved in silencing maintenance and lifespan20-22. The two time series were computationally aligned by performing ChIP-seq for H3K9ac on both set of samples used for RNA and ChIP-seq (Online Methods).
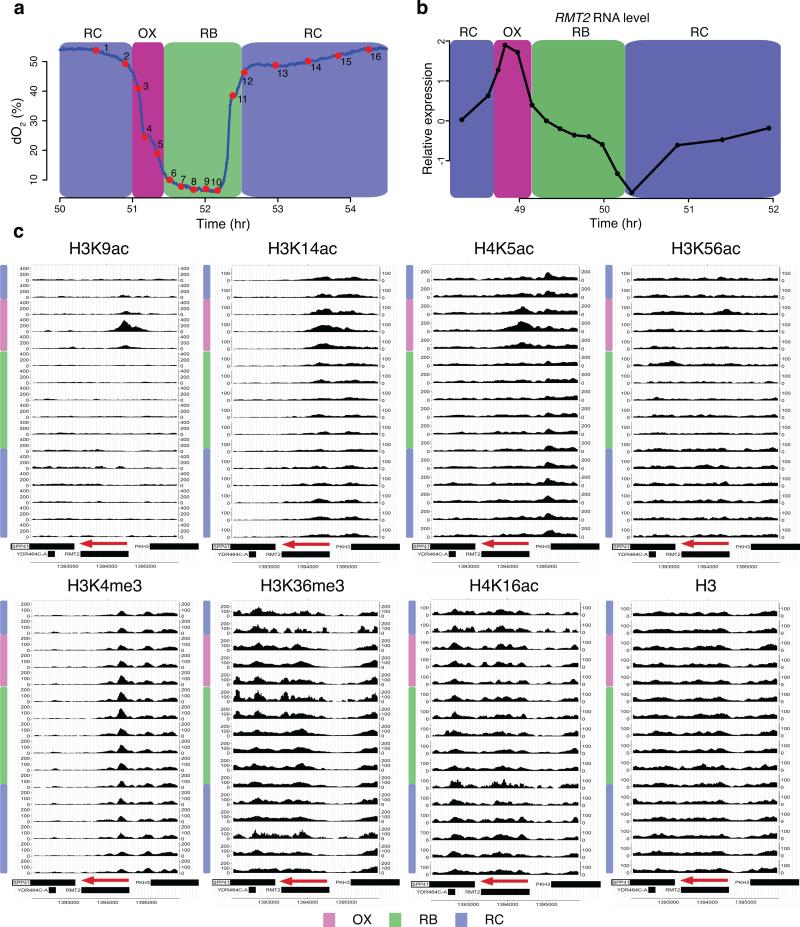
Figure 3. Dynamic chromatin states across the YMC.
(a) shows the oscillation of oxygen in one YMC. Cycling cells were collected at 16 intentionally uneven time points over one cycle for ChIP-seq. (b-c) Temporal relationship between RNA level and histone modifications at the RMT2 locus. (b) shows the RNA level of RMT2 from normalized RNA-seq data. (c) CisGenome browser views of ChIP-seq of H3K9ac, H3K14ac, H4K5ac, H3K56ac, H3K4me3, H3K36me3, H4K16ac and H3 at RMT2. 16 tracks represent 16 time points from top to bottom consecutively. RMT2 is encoded on the Crick strand, labeled by the red arrows pointing to the left.
Figure 3c shows the spatial and temporal signals at the RMT2 locus, encoding an arginine methyltransferase for ribosomal protein Rpl1223. Its expression peaked in mid-OX phase (Fig. 3b). Mapping of ChIP-seq signals across RMT2 revealed a typical spatial pattern5 where H3K9ac, H3K14ac, H4K5ac and H3K4me3 signals peaked in the 5’ end whereas the H3K36me3 signal was located in the coding region (Fig. 3c). Although all positively correlated with transcription, the kinetics and relative timing were quite distinct at this locus. The H3K9ac signal was most dynamic, with a transient peak at t4. H3K14ac and H4K5ac signals also increased in OX with temporally broader peaks than H3K9ac. Instead, the H3K4me3 signal was much more stable than these acetylation marks, with clear-cut peaks across all 16 time points. It started to increase only in late OX at a modest level and peaked in early RB, well after acetylation peaks. No other modifications were obviously dynamic at RMT2.
Expanding examination to broader regions revealed another unexpected difference between modifications. Most H3K9ac peaks were observed in OX, despite the presence of nearby RB and RC genes, consistent with previous study showing that H3K9ac appeared preferentially at OX growth genes (Supplementary Fig. 3). RC/stress response genes were less dependent on H3K9ac14. Instead, H3K14ac, H4K5ac and H3K4me3 peaks were found at most genes indiscriminately. Strikingly, H3K9ac peaks seemingly appeared and disappeared completely coupled with transcription of neighboring genes while H3K14ac, H4K5ac and H3K4me3 only displayed an adjustment of amplitude. The data suggests that H3K9ac is a very active modification that regulates mainly OX growth genes, possibly by directly sensing metabolic stimuli. Others, like H3K14ac, H4K5ac and H3K4me3, have a more general role in transcription regulation. For more examples see supplementary Fig. 4.
Next, we sought to determine average genome-wide chromatin patterns for various gene classes. First, we averaged the spatial signals of these modifications relative to the transcription start site (TSS) across the genome (Supplementary Fig. 5a). Gene-specific signals were calculated based on the their localization. A heat map was used to display the genome-wide interactive temporal patterns (Supplementary Fig. 5b, Supplementary Table 3). Generally, acetylation signals were more dynamic and less noisy than methylation signals, consistent with acetylation shifts reflecting the immediate chromatin response under continuous glucose-limitation. Fourteen clusters were defined, suggesting complex dynamic relationships among these modifications across the YMC.
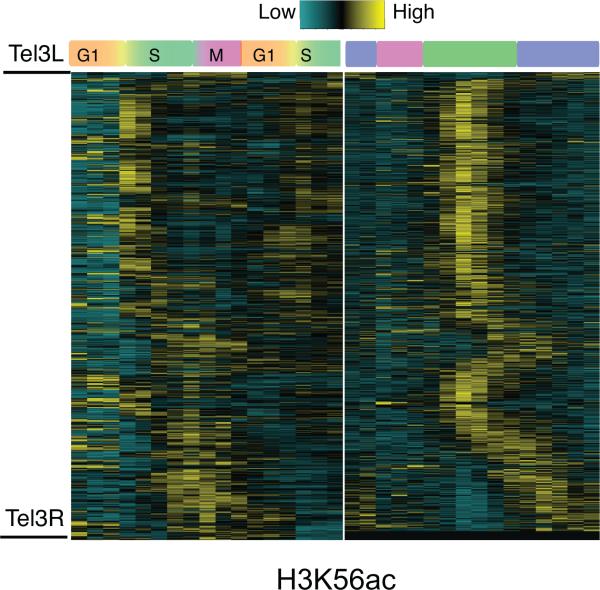
Intriguingly, H3K56ac exhibited very dynamic but noticeably different temporal patterns compared to other histone modifications. By comparing the results to previous work24, we clearly observed cell cycle-related dynamics of H3K56ac (Fig. 4). H3K56ac was strongly elevated in early S phase; similarly, YMC signals peaked at t8, the middle of RB phase, corresponding to S-phase entry. Correspondingly, DNA replication genes (RNA_RB1) were also expressed highest at t8 (Fig. 1c). The expression level of H3K56 acetyltransferase RTT109, peaked at t8 whereas RNA levels of H3K56 deacetylase HST3, began to increase after t8 (Supplementary Fig. 1d). These data corroborated known involvement of H3K56ac in DNA replication in RB and supported the relationship between cell cycle and YMC.
Figure 4. Temporal patterns of H3K56ac.
Comparison of H3K56ac incorporation between the standard cell cycle and YMC. The left panel shows the ChIP-chip data of H3K56ac across the cell cycle from the study of Kaplan et al. 2008, and the right panel is the new ChIP-seq data of H3K56ac across the YMC. The H3K56ac signal was calculated for every nucleosome on chromosome III, sorted by chromosomal location and standardized to a mean of zero.
Temporal association between transcription and chromatin
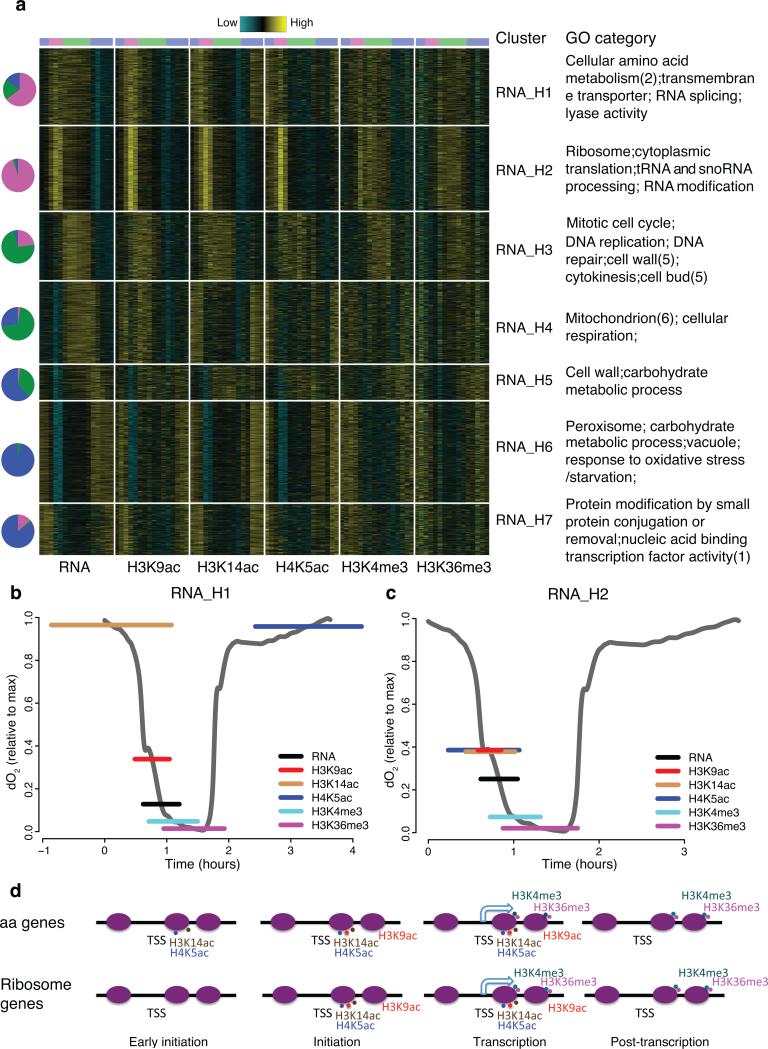
An important function for histone modifications is transcriptional regulation2. With this in mind, we examined the genome-wide temporal relationships between each chromatin modification and transcription. Given the correlation analysis (Online methods) and previous studies3, we focused on five modifications most clearly linked to transcription (Fig. 5). We discovered 7 clusters of cycling genes (Fig. 5a, Supplementary Table 4). Are there different histone modification pathways associated with transcription of different gene clusters? To tackle this, we estimated relative timing of each modification peak relative to its corresponding RNA peak and examined GO enrichment on all clusters.
Figure 5. Temporal combinatorial patterns of chromatin states and gene expression define functionally related genes.
(a) is a heat map showing 7 clusters of cycling genes (combining OX, RB, RC phase genes ordered by the time of peak gene expression. RNA, H3K9ac, H3K14ac, H4K5ac, H3K4me3 and H3K36me3 are included in this analysis and each is displayed in a joint column, separated by white lines. The data is standardized to a mean of zero. Pie charts on the left indicate composition of OX (Magenta), RB (Green), RC (Blue) phase genes in each cluster. Enriched GO categories are listed on the right. (b-c) Estimation of time in YMC when RNA and histone modifications in “RNA_H1” (b) and “RNA_H2” (c) reach the maximum. The middle of the horizontal colored lines on the O2 curves represent the mean peak value of genes in each cluster and the lengths represent s.d. for 464 and 516 genes in (b) and (c). (d) Models of histone modification pathways during the transcription of aa and ribosomal genes. Purple ovals represent −1, +1 and subsequent ORF nucleosomes for the indicated gene. Hooked arrows represent transcription and spots represent histone modifications.
In clusters RNA_H1 and 2, the H3K9ac peak notably appeared just minutes before gene expression peak (9, 6.8 minutes in cluster 1, 2) whereas the H3K4me3 and H3K36me3 peaked 10-30 minutes after the peak of RNA (Fig. 5b,c). Surprisingly, H3K14ac and H4K5ac signals appeared concomitantly with peak gene expression in RNA_H 2 (6.2 minutes of H3K14ac and 10.8 minutes of H4K5ac), but in RNA_H 1 they were advanced by one hour relative to RNA (48 minutes for H3K14ac and 75 minutes for H4K5ac). Aa genes were enriched in RNA_H 1, whereas RNA_H 2 contained mostly ribi and RP genes. Combined with the RNA-seq analysis (Fig. 1), we could define three distinct chromatin-transcriptional “programs” among OX growth genes. Ribosome biogenesis (ribi) and ribosomal protein (RP) genes shared the same chromatin pattern, distinct from that of aa genes. However, peak mRNA levels of RP genes and amino acid genes appeared later and lasted longer than those of the ribi genes. As discussed earlier, regulated splicing of RP genes might cause a delay of the peak level of mature RP mRNA (Fig. 2).
Among the RB clusters, RNA_H 3 contained primarily cell cycle genes and exhibited similar chromatin patterns to OX clusters relative to RNA dynamics (Fig. 5a). In contrast, the genes encoding mitochondrial proteins (RNA_H 4) behaved quite differently. Although they had a similar peak expression time, timing of the modification signals was out of phase. H3K14ac and H4K5ac increased in late RC and H3K9ac increased in late RC through OX. A motif sequence recognized by RNA-binding protein Puf3 was found in the 3’ untranslated regions (3’ UTR) of nearly all mitochondrial ribosomal genes12. This discrepancy between RNA level and chromatin states was consistent with the hypothesis that this gene class may be subject to post-transcriptional rather than transcriptional regulation.
The quantification of the relative timing of chromatin signals summarized different histone modification programs during transcription in yeast. In general, acetylation marks appear first, followed by RNA and then methylation marks, consistent with an overall initiation role for acetylation and an elongation role for methylation. Intriguingly, the fine-grained timing of different histone acetylation modifications could be a key factor to distinguish different transcription initiation mechanisms (Fig. 5d). In RNA_H 1, H4K5ac appeared at first, followed by H3K9ac and then RNA. This general sequence suggests that H4K5ac functions at an early stage of transcription initiation in these sets of genes, perhaps to “pre-set” their activation, while H3K9ac, which peaks much more sharply, may play a critical role in triggering phase transitions, and may regulate a later initiation step or a transition to elongation. In RNA_H 2, H4K5ac and H3K9ac peaked together immediately before RNA peak, suggesting dispensability of a presetting stage during transcription of these genes.
The distinct patterns between chromatin and gene expression raised the question whether specific transcription factors (TFs) are associated with these clusters. TF motif analysis (Supplementary Table 4) revealed that binding sites for TFs involved in amino acid metabolism, like Gcn4p, Bas1p, Cbf1p, Leu3p, were enriched in RNA_H 1, whereas in RNA_H 2 we observed enrichment primarily of TF motifs related to RP gene expression, such as Stb3p, Sfp1p, Rap1p, Pbf1p, Pbf2p, Dot6p, Tod6p. RNA_H 3 contained several common cell-cycle related TF motifs including those for Xbp1p, Mbp1p, Fkh1/2p, Swi4p. Many TF motifs associated with stress response were enriched in RNA_H 6, a typical RC cluster, such as Gis1p, Msn2/4p, Adr1p, Crz1p. Collectively, these results revealed a complex, dynamic and interactive regulatory network of chromatin states, TF binding sites, and post-transcriptional regulation.
Landscape of chromatin-modifying enzyme occupancy in the YMC
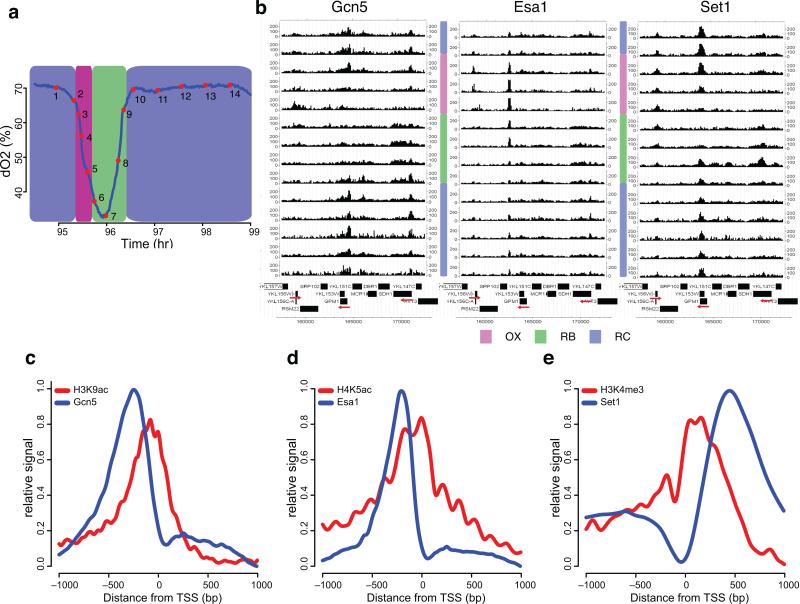
To further understand the dynamic patterns of histone modifications, we analyzed genome-wide occupancy of several chromatin-modifying enzymes in the YMC, specifically Gcn5p, Esa1p and Set1p5. Gcn5p is the major histone H3 tail acetyltransferase and Esa1p acetylates H4 tails, although other HATs also acetylate H3 and H4. Set1p is the catalytic subunit of COMPASS complex H3K4 methyltransferase.
To simultaneously evaluate all three enzymes, we constructed a doubly-tagged GCN5-FLAG, SET1-3HA strain employing an anti-native Esa1p antibody. This allowed ChIP-seq of Gcn5p, Esa1p and Set1p in one set of samples (Fig. 6a). First, we examined spatiotemporal patterns of these enzymes by displaying ChIP-seq signals at sample regions; temporal patterns were quite distinct (Fig. 6b). All three enzymes were recruited to RP gene RPS27A (YKL156W) in OX but with subtly distinct kinetics. Gcn5p occupancy was detected only at t5, corresponding to t4 in the 16 time point ChIP-seq of histone modifications. However, Esa1p and Set1p signals preceded this peak, spanning mostly t2, t3, although Set1p occupancy lasted much longer, extending through early RB. Only Gcn5p and Set1p signals were detected at GPM1(RC) and SDH1(RB) loci, in a manner correlated with RNA levels (Fig. 6b). Interestingly, the signal of Set1p appeared 1-2 time points (5~20 minutes) before Gcn5p signal at GPM1, similar to RPS27A.
Figure 6. Spatiotemporal patterns of chromatin modifiers and corresponding modifications.
(a) shows the oscillation of O2 in one YMC and 14 intentionally unevenly spaced time points for ChIP-seq of Gcn5p, Set1p and Esa1p. (b) CisGenome browser views of the temporal ChIP-seq patterns of Gcn5p, Set1p and Esa1p at chromosome XI: 157000-173000. The 14 tracks represent 14 consecutive time points from top to bottom. Red arrows label the direction of interesting genes: from left to right are YKL156W (RPS27A), YKL152C (GPM1), and YKL148C (SDH1). (c-e) Spatial distributions of the chromatin modifiers and the corresponding modifications. ChIP signals were averaged from −1000 to +1000 bp of TSSs (transcription start site) derived from MACS detected peaks. Signals of each modifier are plotted together with its corresponding modification normalized by H3.
Given the complex spatiotemporal relationships between the three enzymes, we sought to assess the interactive patterns and their relationship to corresponding histone modifications and gene expression from a genome-wide perspective. We found 2369 Gcn5p peaks, 2225 Esa1 peaks and 2279 Set1p peaks, including 1874, 1810 and 1629 with neighboring annotated genes (Supplementary Fig. 6a). Among these, ~1750 genes were bound by at least two modifiers and 1035 genes were bound by all three modifiers, suggesting that these enzymes were not necessary for expression of every cycling gene. Spatial analysis showed that Gcn5p and Esa1p were localized 200-300 bp upstream of the TSS while Set1p was localized within the transcribed regions (Fig. 6c,d,e). This supported the well-accepted hypothesis that Gcn5p and Esa1p function in transcriptional initiation whereas Set1p is associated with elongation25. Intriguingly, the locations of the modifiers did not overlap completely with corresponding histone modifications. Although Gcn5p and Esa1p located upstream of the TSS, H3K9ac and H4K5ac were shifted downstream. This suggested that Gcn5p and Esa1p can “reach across” the TSS to act on the downstream nucleosome. The Set1p signal was slightly shifted downstream of the H3K4me3 peak, presumably because Set1p also produces mono- and di-methylation on H3K4 during transcriptional elongation.
Next, we assessed temporal patterns of chromatin-modifiers, focusing on annotated genes associated with all three modifiers. We defined 4 superclusters of temporal patterns by K-means clustering (Supplementary Fig. 6b, Supplementary Table 5). The results suggested that all three modifiers are functional in all three phases and recruitment of modifiers is roughly in phase with corresponding gene expression.
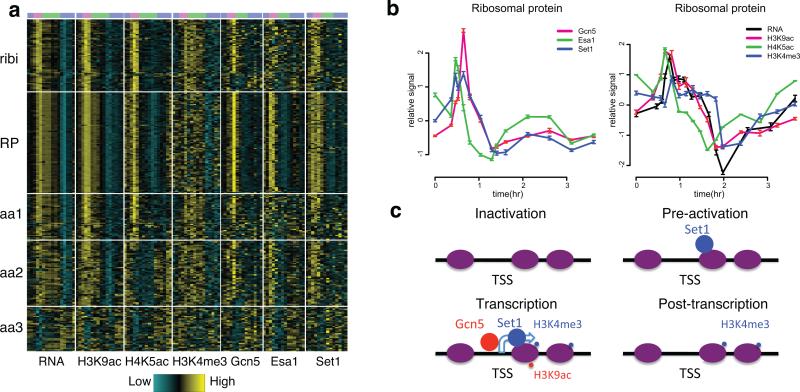
However, chromatin modifiers, chromatin states and gene expression were not simply correlated. Here we focused on cell growth genes, including ribi, RP and aa genes26. Ribi and RP genes individually exhibited one pattern while aa genes showed three distinct patterns (Fig. 7a, Supplementary Table 5). Since RP genes showed the cleanest signal, we examined relative timing within this group (Fig. 7b). As described above, H3K9ac and H4K5ac appeared with the RNA peak whereas H3K4me3 exhibited a delay. Surprisingly, Set1p was recruited ~10 minutes prior to the binding of Gcn5p, and remained bound until dissociation of Gcn5p. This pattern was consistent at RPS27A, GPM1 (described above) and many other loci. The relative timing of histone modifications and modifiers at ribosome and aa genes were further confirmed by ChIP-qPCR in similar YMC experiment using Gcn5-3FLAG, 6HA-Set1 (supplementary Fig. 6d,e). The temporal discrepancy between modifiers and modifications suggested that Set1p is recruited prior to transcription in a state that is non-functional, at least for tri-methylation, until binding of Gcn5p occurs (Fig. 7c), whereas Esa1p is also recruited early but is immediately active.
Figure 7. Similar but not identical temporal patterns of chromatin modifiers and corresponding modifications.
(a) is a heat map of the temporal patterns of RNA, histone modifications and corresponding modifiers for ribosome biogenesis genes, ribosomal protein genes and amino acid metabolism genes. (b) Averaged ChIP-seq signals of histone modifications and modifiers and RNA-seq signals for RP genes across the YMC. Error bars represent s.d. of 83 RP genes at each time point. (c) A model of Set1p/H3K4me3 and Gcn5p/H3K9ac during transcription. Purple ovals represent −1, +1 and subsequent ORF nucleosomes for the indicated gene. The hooked arrow represents transcription and small and big spots represent histone modifications and histone modifiers.
Histone mutant analysis
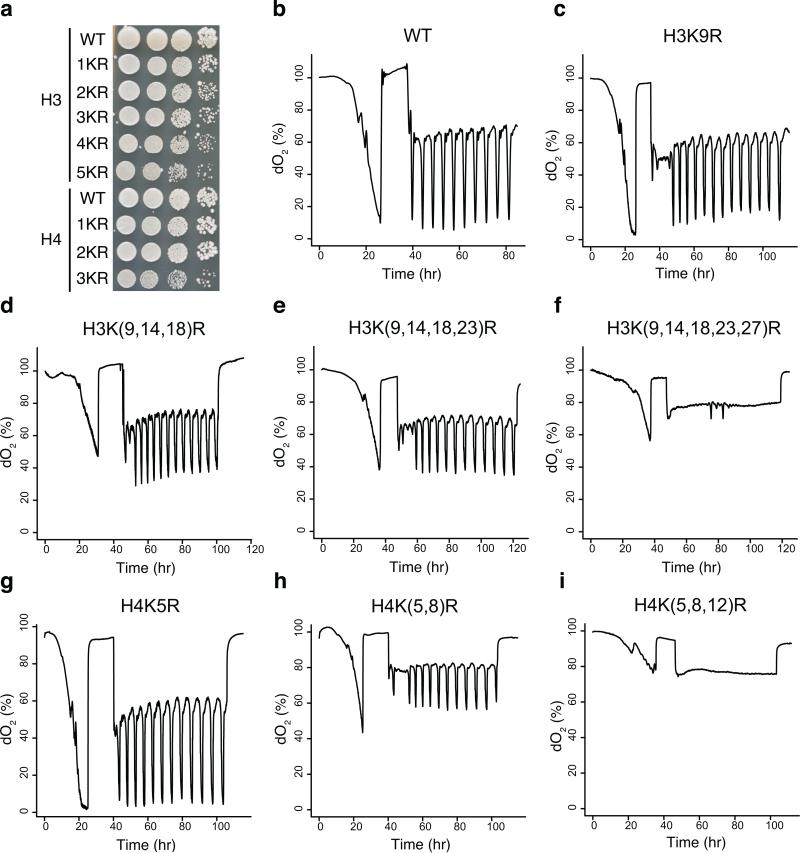
Given the complex patterns of histone modifications and modifiers in YMC, we investigated the criticality of individual lysine residue modifications. Gcn5p mediates entry into the growth phase, as a mutant lacking it does not exhibit metabolic cycles14. To broaden our understanding of histone modifications in the YMC, we examined the dO2 oscillation phenotype of all histone H3 and H4 lysine mutants corresponding to residues at which acetylation or methylation have been reported. All single lysine to alanine or arginine mutants, including H3K4, H3K9, H3K14, H3K18, H3K23, H3K27, H3K36, H3K56, H3K79, H4K5, H4K8, H4K12, H4K16, exhibited normal cycles, suggesting that no single lysine modification on H3 or H4 is critical for the YMC (data not shown). Next, we combined lysine to arginine mutations in H3 or H4 tails, including H3K(9,14)R, H3K(9,14,18)R, H3K(9,14,18,23)R, H3K(9,14,18,23,27)R, H4K(5,8)R, H4K(5,8,12)R. Notably, we observed decreased growth on YPD with increasing numbers of lysine to arginine mutations, consistent with published reports (Fig. 8a)27. As the number of K to R mutations increased, the amplitude of the dO2 oscillation decreased (Fig. 8b-i). Strikingly, H3K(9,14,18,23,27)R, and H4K(5,8,12)R mutants abolished dO2 oscillation. These results supported the hypothesis that PTMs such as acetylation on the H3 and H4 tails are required for normal cycling and support a role for acetyl-CoA in establishing the YMC14, although there is no essential role in the YMC for any individual lysine residue.
Figure 8. O2 consumption traces of histone point mutants.
(a) Strains were plated (1:10 serial dilutions) on YPD plates and grown at 30°C. H3 mutants represent H3K9R, H3K(9,14)R, H3K(9,14,18)R, H3K(9,14,18,23)R, H3K(9,14,18,23,27)R. H4 mutants represent H4K5R, H4K(5,8)R, H4K(5,8,12)R. (b-i) show O2 curves of histone mutants.
Discussion
In this study, we described the dynamic landscape of transcription and chromatin modifications across distinct metabolic states in yeast. We uncovered distinct combinatorial patterns associated with different functional groups of genes, emphasizing high-resolution timing of expression of distinct groups of genes and relative timing of histone modifications and modifiers within each group.
A “Just in time” supply chain?
Cellular processes are well organized spatiotemporally to maximize fitness especially in the face of limited nutrients. One interesting example is the sequential activation of promoters in bacterial aa biosynthesis pathways28. YMC transcriptional profiling revealed several examples of staged biosynthesis consistent with a “just in time” program. Ribi transcripts appeared just before those encoding RP and aa (OX phase) genes (Fig. 1b), supporting a “just in time supply chain” that maximizes efficiency of ribosome biogenesis and translation. The short pulse of ribi genes may also play a critical role in helping them achieve an efficient ramp-down of protein synthesis at the end of OX. The observation of transient RP gene pre-mRNA accumulation suggested the possibility of elaborate control of splicing in regulation of ribosome biogenesis. Alternatively, this staging may reflect the fact that ribi genes can be much larger and therefore take longer to translate than RP proteins, which are universally short. The translation of the longest ribi gene might require 3-4 minutes more than the longest RP gene, consistent with the relative timing of ribi and RP expression. It is likely that the sequential activation of ribi and RP leads to concurrent synthesis of all ribosome biogenesis proteins. Likewise, cell cycle-related genes and mitochondrial genes, the two major groups that had an elevated RNA level in the RB phase, also exhibited subtle timing disparities (Fig. 1c), perhaps indicating two distinct YMC subpopulations – one that will enter the CDC, and one which will not29. Collectively, the evidence suggested a complex and precise orchestration of macromolecular biosynthesis processes in yeast during the YMC from a transcriptional perspective. It will be interesting to examine whether protein factors are also organized temporally.
Different histone modification programs at distinct genes
We defined seven different combinatorial clusters between histone modifications and gene expression, revealing several different regimes of chromatin states in transcription (Fig. 5a). Interestingly, different histone acetylation marks behaved distinctly in association with different groups of genes. H3K9ac, H3K14ac and H4K5ac all appeared coincident with the peak of gene expression of ribosomal genes. However, H3K14ac and H4K5ac were shifted earlier at aa (OX phase_aa2) genes (1 hour before the RNA peak). We hypothesized that H3K14ac and H4K5ac function in “pre-setting” the promoters, perhaps by chromatin remodeling. However, RP genes did not show evidence of this pre-activation phase from the perspective of chromatin states, perhaps because they are permanently pre-activated. RP genes could be immediate sensors of critical metabolic and environmental stimuli, e.g. intracellular acetyl-CoA levels14. Conversely, aa2 genes appear to have a preparatory phase.
Dynamics of histone modifiers increase the complexity of chromatin states. In many cases, Set1p was recruited earlier than Gcn5p, in seeming contradiction to the order of appearance of H3K4me3 and H3K9ac (Fig. 7c). We hypothesize that Set1p may catalyze mono- or dimethylation in promoter regions to preset TSSs for activation. It is known that H3K4me1 accumulates at the MET16 promoter prior to induction in yeast30. Additionally, H3K4me1/me2 precede transcription in cardiac development and T cell differentiation, suggesting that wiring of at least some chromatin-based programs are phylogenetically conserved31,32.
Roles of histone modifications in the YMC
Various histone modifications may function “solo” or in a combinatorial or cumulative manner33,34. These modifications either change the charge status of histone tails or provide platforms to recruit transcriptional regulatory complexes. The very different patterns between H3K9ac and H4K5ac suggested distinct functions during transcription. Histone mutant analysis revealed functional redundancy within each histone tail. The H3-5KR quintuple and H4-3KR triple mutant each totally abolished the oscillation, partially corroborating the evidence that the gcn5Δ strain does not support oscillation14. Surprisingly however, H3-5KA and H4-3KA mutants grew better than corresponding KR mutants and still exhibited O2 oscillation phenotypes (Supplementary Fig. 7), suggesting that charge status of H3 and H4 may play a fundamental role in the YMC21. Maintaining the positive charges on these histone tails may “lock” growth genes into a repressed state, resulting in inability to undergo bursts of growth characteristic of the YMC and cells in general.
Similar chromatin dynamics between YMC and circadian cycle
Remarkably, circadian rhythm in mammals and this yeast ultradian rhythm show several noteworthy similarities. Through systematic analysis of the transcriptome and chromatin state as a function of circadian clock in mouse liver, Koike et al. revealed similar temporal patterns of histone modifications9, although the genes under regulation are completely distinct and the time scales of the cycles are very different, suggesting phylogenetically conserved roles of histone modifications during transcription. More intriguingly, beyond the oscillatory phenomenon of transcriptome, metabolism and chromatin states in these two systems, the metabolism-epigenome-transcriptome loop is consistent in both systems, suggesting that the circular interactions among the three players actually sustain oscillations. Our findings on dynamics of chromatin modifications and temporal associations between epigenome and transcriptome suggest an active metabolism-epigenome-transcriptome loop in YMC. The observation that histone acetylation mutants abolished oscillatory respiration in yeast directly supports the indispensability of histone modification in sustaining metabolic cycles and by extension perhaps the circadian clock.
We have revealed a network of gene expression, chromatin state, and chromatin modifiers across the YMC as highly synchronized yeast cells transition between metabolic states akin to growth, division, and quiescence, at very high temporal resolution. Our study not only demonstrates how chromatin states are related to and directing the timing of gene expression, but also provides examples of how these processes are coordinated in a dynamic fashion to achieve optimal growth, metabolic efficiency, and presumably fitness. This yeast study may help illuminate the exquisite temporal control of various dynamic biological processes.
Online Methods
Plasmids and histone mutagenesis
Plasmids carrying histone mutations were obtained from the histone mutant library described previously35. pJD233 and pJD234, each containing a hygMX selectable marker cassette, were used for making HHT1 and HHF1 mutants respectively. pZK8 was generated from pJD154 by replacing the URA3 cassette with a natMX cassette for mutagenesis on HHT2 and HHF2. Plasmids carrying multiple lysine mutations were generated by site-directed mutagenic fusion PCR and subcloning in the indicated backbones, pJD233, pJD234 and pZK8. Briefly, primers carrying the desired mutations were used to generate two PCR fragments containing the overlapping mutation regions. Fusion PCR was used to generate a single fragment and the product was digested and ligated to the indicated backbones, pJD233, pJD234 and pZK8 and verified by DNA sequencing.
Metabolic cycles
Metabolic cycle experiments were performed as previously described12 except for the timing of sampling for RNASeq and ChIP-Seq; samples were intentionally taken unevenly to more deeply sample the very rapidly changing OX phase, and less densely out side the OX phase. Fermentors were from New Brunswick Scientific (BioFlo 110 or BioFlo 3000). 10 mL overnight saturated culture was inoculated to start each YMC run. Depending on the model, YMC runs were operated at an agitation speed of 400 rpm (Bioflo 110) or 475 rpm (Bioflo 3000), an aeration rate of 1 L/min, a temperature of 30°C, and a pH of 3.4 in 1 L YMC medium.
Once the batch culture was saturated, at least 4 hour of starvation was performed. After starvation, fresh medium was added continuously at a dilution rate of ~0.09~0.1 h−1.
RT-qPCR
2 OD BY5764 cells from the cycle were collected and flash frozen. RNA was extracted using the Qiagen RNeasy Mini kit (QIAGEN, 74104, Valencia, CA) with the standard protocol. First strand cDNA was synthesized using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, 18080-051, Grand Island, NY). Oligo(dT)20 primer was used for reverse-transcription. Fast SYBR Green Master Mix (Applied Biosystems, 4385612, Foster City, CA) was used for real-time PCR and experiments were done on the platform of StepOnePlus Real-Time PCR System (Applied Biosystems, 4385612, Foster City, CA). PCR primers are listed in supplementary Table 6.
RNA-seq
RNA was extracted across 16 time points of one cycle at the indicated time points and RNA-seq was performed. With this strategy, the shortest interval between two OX phase time points is < 5 minutes, and across the entire cycle the samples were taken at intentionally uneven intervals. 1 mL BY5765 cells from the cycle (about 15 OD/mL) were collected and flash frozen. The pellet was resuspended in 200 μl RNA stat-60 (Tel-Test, Friendswood, TX) and disrupted in a Mini-Beadbeater-16 (Biospec, Bartlesville, OK) broken up three times with 100 μl glass beads in 1.5 ml conical screw cap tube (USA SCIENTIFIC, 1415-8700, Ocala, FL). Supernatant was collected into a new microcentrifuge tube and 800 μl RNA stat-60 was added to repeat bead beating one more time. The supernatants were combined and sit at room temperature for 10 min. 200 μl chloroform was then added and the tube was vortexed for 15 sec and left at room temperature for 10 min. The tubes were centrifuged at 4°C, 16000 rcf for 15 min and the supernatant was transferred to a new tube. 500 μl isopropanol was added and mixed by inverting the tube several times. The mixture was incubated at room temperature for 10 min and centrifuged at 4°C, 16000 rcf for 15 min. The pellet was washed with 70% ethanol twice, air dried and resuspended in 100 μl H2O. Library construction and sequencing were performed using the Hiseq platform supervised by the UTSW Microarray Core Facility. Briefly, Samples were run on the Agilent 2100 Bioanalyzer to ensure that high quality RNA was used. 4 μg total RNA was then prepared with the TruSeq™ RNA Sample Prep Kit (Illumina, San Diego, CA). mRNA was purified and fragmented before cDNA synthesis. cDNA was then end repaired and A-tailed. After adapter ligation, samples were PCR amplified and purified with AmpureXP beads (Agencourt, A63880, Brea, CA), then validated again on the Agilent 2100 Bioanalyzer. Before being run on the Illumina Hiseq 2000 samples were quantified by qPCR. Primers were within the adaptor sequence, which are as below:
P1: 5’ AAT GAT ACG GCG ACC ACC GA 3’
P2: 5’ CAA GCA GAA GAC GGC ATA CGA 3’.
Differential gene expression analysis was performed with TopHat and Cuffdiff following a standard protocol36. Briefly, the TopHat parameters used was: --bowtie1 -i 40 --genome-read-mismatches 4 --no-coverage-search -G Saccharomyces_cerevisiae_Ensembl_EF2/Saccharomyces_cerevisiae/Ensembl/EF2/Annotation/ Archives/archive-2012-03-09-08-22-49/Genes/genes.gtf Saccharomyces_cerevisiae_Ensembl_EF2/Saccharomyces_cerevisiae/Ensembl/EF2/Sequence/Bo wtieIndex/genome. K-means clustering and heatmaps were produced using an R. Previously defined three super-clusters of genes were obtained from a previous study12. A subset of genes from genes previously identified as non-cycling show elevated expression in the OX phase by hierarchical clustering using the dChiP package37. Previously defined and newly-identified OX phase genes were combined. FPKM values were imported into R and the list was intersected with the three superclusters. The values of each superclusters were then subject to K-means clustering using the function “kmeans”. Genes inside each subcluster were further ordered based on the timing of peak expression. 10 clusters were calculated by K-means clustering and ordered by the timing of peak expression from visualization. Genes inside clusters were further ordered by weighted mean time of peak of expression. Weights were calculated as the percentage of RNA-seq reads at each time point over total reads across 16 time points. The weighted mean for each gene was calculated as the sum of weight x time at each time point. Intron-containing genes were defined from the SGD annotation file of 20100102 corresponding to the mapping index. (http://downloads.yeastgenome.org/curation/chromosomal_feature/ ). To display RNA signals in the CisGenome Browser38, RNAseq data was first mapped to Saccer2 by bowtie (-5 3 -3 8 –best saccer2). Results were imported into CisGenome browser package and were displayed in 20 bp windows in the browser. Signals at each window were first normalized by total number of mappable reads across 16 time points and then displayed as the percentage of the maximum value of the 16 time points. Exon-exon junction reads were directly called from the files of junctions.bed by TopHat. Intron junction reads were identified using mappable reads of 39 bp (50 bp-3 bp-8 bp) from bowtie. Each end of a junction read should extend at least 4 bp away from the junction site. Junction reads spanning the 5’ and 3’ ends of 89 RP gene introns at 16 time points were counted and hierarchical clustering and heatmaps were generated in R using the heatmap.2 function with default parameters. Boxplots were drawn using the boxplot function at every time point. The sums of the 16 time point junction reads were calculated and displayed using the barplot function.
ChIP-seq
Chromatin immunoprecipitation was performed as described previously14. ~5 OD ZKY329 cycling cells per time point were collected for ChIP of single histone modification and ~50 OD ZKY428 cycling cells (GCN5-FLAG::natMX, SET1-3HA::kanMX6) per time point were collected for ChIP of each chromatin modifying enzyme. Antibodies used are list as following: H3K9ac (Millipore 06-942), H3K14ac (Millipore 07-353), H3K56ac (07-677), H4K5ac (Millipore 07-327), H4K16ac (Millipore 07-329), H3 (Millipore 05-928), H3K4me3 (Millipore CS200580), H3K36me3 (Abcam ab9050), Esa1 (Abcam ab4466), FLAG M2 (Sigma, F1804), HA 12CA5 (Roche, 11583816001). Validation is provided on the manufacturer's website. 3 μg primary antibody was used per ChIP experiment. Briefly, cells were first fixed in 1% formaldehyde at 25°C for 15 min and quenched in 125mM glycine at 25°C for 10 min. Cells were pelleted and washed twice with buffer containing 100 mM NaCl, 10 mM Tris-Cl pH 8.0, 1 mM EDTA, 1 mM PMSF, 1 mM benzamidine•HCl before freezing. The frozen pellet was resuspended in 0.45 ml ChIP lysis buffer (50 mM HEPES•KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate (DOC), 0.1% SDS, 1 mM PMSF, 10 μM leupeptin, 5 μM pepstatin A, Roche protease inhibitor cocktail) and lysed by bead beating. Lysate from 50 OD cells were split into two tubes each containing 280 μl lysate and sonicated for 16 cycles (30 sec on, 1 min off, high output) using a Bioruptor (Diagenode, Denville, NJ or Tosho Denki, Japan). The supernatant of the sonicated lysate was pre-cleared. 50 μl lysate was saved as input. For ChIP with histone antibodies, 50 μl whole cell extract (WCE) was diluted 1:10 and used for each ChIP. For ChIP of histone modifier proteins, 500 μl WCE and 3 μg antibody was used. After incubation overnight, 50 μl protein G magnetic beads (Invitrogen, Grand Island, NY, 10003D) or protein A Sepharose beads (GE Healthcare Life Sciences, Piscataway, NJ, 17-5280-01) resuspended in ChIP lysis buffer was added and incubated for 1.5 h at 4 °C. Beads were washed twice with ChIP lysis buffer, twice with DOC buffer (10 mM Tris•Cl pH 8.0, 0.25 M LiCl, 0.5% deoxycholate, 0.5% NP-40, 1 mM EDTA) and twice with TE. 125 μl of TES buffer (TE pH8.0 with 1% SDS, 150 mM NaCl, and 5 mM dithiothreitol) was added to resuspend the beads. Supernatant was collected after incubation at 65°C for 10 min. A second round of elution was performed and the eluates were combined. Reverse crosslinking was performed by incubation for 6 h at 65°C. An equal volume of TE containing 1.25 mg/ml proteinase K and 0.4 mg/ml glycogen was added to the samples after reverse crosslinking and samples were incubated for 2 h at 37°C. Samples were extracted twice with an equal volume of phenol and once with 25:1 chloroform:isoamyl alcohol. DNA was precipitated in 0.1 volume 3.0 M sodium acetate (PH 5.3) and 2.5 volume of 100% ice-cold ethanol at -20°C overnight. Pellets were washed once with cold 70% ethanol and resuspended in 20 μl TE. Library construction and sequencing were performed following the Illumina protocol. Briefly, DNA was end repaired and A-tailed. Barcoded adaptors were ligated and DNA was run in 2% agarose gel. DNA fragments from 150 bp to 300 bp long were excised from gel and used for PCR. PCR products were gel-extracted again and quantified on an Agilent Bioanalyzer. Sequencing was performed on an Illumina GXII or Hiseq 2000 or Solid. Raw reads were mapped to the reference genome (sacCer2) by bowtie and peaks were visualized by the CisGenome Browser as described above.
Distribution of ChIP-seq signals relative to TSSs
Information on TSS locations was adopted from a previous study39. TSSs of genes of interest were aligned at zero of X-axis and ChIP-seq reads were counted for each 10 bp window from -1000 bp to 1000 bp relative to the closest TSS. Number of reads in each window were summed and plotted in R using the plot function.
ChIP-seq signals at genes
ChIP signals of histone modifications at genes were evaluated by counting reads overlapping defined regions of every gene using the CisGenome package. Briefly, a −100 bp to +400 bp window spanning TSS was used for H3K9ac, H3K14ac, H3K56ac, H4K5ac and H3K4me3. The H3K36me3, H4K16ac and H4 signals were calculated from TSS to TES. Peaks of chromatin modifiers were called by MACS peak-calling package with p value=10−02 40 and annotated by the nearest TSS. ChIP values of chromatin modifiers were calculated similarly by counting reads −250 bp to +250 bp windows spanning the center of peaks or −100 bp to +400 bp window spanning TSS. Ribosomal assembly (ribi) factors were described by Jorgensen et al.,26. The ribosomal protein (RP gene) list is from the website, http://ribosome.med.miyazaki-u.ac.jp/rpg.cgi?mode=orglist&org=Saccharomyces%20cerevisiae. Amino acid metabolism genes are from the GO term “cellular amino acid metabolic process” of the SGD annotation file. ChIP values were normalized according to the total number of aligned reads. Time point 13 of H3K14ac and H4K5ac, time point 1 of H4K16ac and time point 5/7 of H3K36me3 show certain level of fluctuation. We tried to delete these samples or average these samples with neighboring samples and observed no significant differences in the downstream analysis. Based on the continuity of time course experiments, we smoothed these samples by averaging the signals with neighboring samples for visualization. K-means clustering and heatmaps were performed in R as described above. Hierarchical clustering was performed by dChiP package37.
Data Pre-processing
For each sample, we normalized the observed value by the total number of aligned reads. For each modification, we then standardized the 16 observed values (log2 of read counts plus 1) within each gene. We were primarily interested in the trend of change over time, rather than the relative magnitude of expression among genes. So we standardized the 16 values within each gene, i.e. we subtract the mean from the 16 values and then divide by their standard deviation. Then we pooled the data for all RNA-seq and ChIP-seq datasets together to get a data matrix, in which each row is a gene and each column is a time point.
Notations
For a given marker, we denote the matrixes as Y = [Ygj] as processed gene expression levels, where g = 1,...,N is the gene index, and j = 1,...,16 is the time point index. We denote tj as observation times.
Correlation between chromatin modifications and gene expression
A gene-specific correlation coefficient was calculated between 16-time-point gene expression values and each histone modification value. A 2-time-point shift was allowed to achieve the maximum correlation value. Basically, we calculated gene-specific correlation coefficient with time point shift of -2, -1, 0, 1, 2 and took the maximum correlation value as the output correlation coefficient. We note that some modifications are shifted forward or backward relative to RNA level and this could help get the accurate correlation value. A density distribution of correlation values was plotted across the whole genome or across subsets of interest (OX, RB, RC). We observed a positive correlation in the distribution of H3K4me3, H3K36me3, H3K9ac, H3K14ac and H4K5ac with RNA level (data not shown). Interestingly, the OX phase genes always showed the best correlation with expression, while RB phase genes showed a relatively poor correlation. We also noticed that H4K16ac and total H3 exhibit a slight negative correlation and H3K56ac shows no obvious correlation (data not shown).
Curve fitting
For each gene, we observed gene expression levels at 16 time points. To convert these discrete data into a continuous profile or function, we fit every set of 16 points into a smooth curve using a penalized B-spline41. We denote function fg(t) as fitted continuous profile from data [Yg1, ..., Yg16] and [t1, ..., t16]. fg(t) is a linear combination of B-spline basis functions (a definition of B-spline basis can be found in Eilers and Marx, 1996). Estimation of the coefficient of each basis function is obtained by a penalized optimization problem.
Time alignment (Batch effect removal)
We assume the dynamics of gene expression levels changes according to a biologically specified clock or time system. Such time systems can be affected by unknown or uncontrolled fluctuations in experimental conditions such as minute variations in medium composition, temperature, etc. When data are generated in multiple batches, the results can be biased by the different time systems.
RNA-seq and ChIP-seq of histone modifications were generated in two batches. To minimize batch to batch variations, the histone modification H3K9ac, which is the most periodic, was independently measured in both batches. We used this information to align or “time warp” the two batches into a single me time system. We denote and as the fitted gene profiles for two different batches. Theoretically, and should be the same under the identical time system. Thus we seek a mathematical transformation s:[t1, ..., t16] -> [s(t1), ..., s(t16)], such that and are most alike for all genes. Mathematically, we need to minimize
We obtain the final result [s(t1), ..., s(t16)] using a simulated annealing algorithm.
As a result, the observation times of the raw RNA-seq data are replaced by [s(t1), ..., s(t16)], and we assume that the batch effect is minimized as much as possible given the current data.
Clustering genes
We cluster the genes using the processed gene expression levels. The K-means algorithm is used for its speed and robustness of dealing with large data. The K-means algorithm is affected by the choice of initial cluster centers. To minimize the dependency on initial cluster number, we repeated the K-means algorithm 50 times using different randomly chosen initial cluster centers, and kept the result with the smallest total within-cluster sum of squares.
To choose the number of clusters, we tried different options range from 1 to 30. Finally we selected 7 clusters since it had the best Bayesian Information Criterion (BIC) value: BIC is one of the most popular criteria for model selection42. Similarly we selected 14 clusters of 8 histone modifications in supplementary Figure 5.
Finding the profile peak location of each cluster
For each cluster of genes we want to know when the expression profile reaches its peak value. For each gene, we can find the peak (highest point) of profile from the fitted smooth curve. The cluster peak location is defined as the average of peak locations for all genes in a cluster. Similarly we can calculate the standard deviation of the cluster peak location.
GO analysis
Gene ontology analysis was performed by the hypergeometric test using the SGD annotation file (http://www.yeastgenome.org/download-data/curation#.UQFqKhyLFXY). P values were adjusted by the False Discovery Rate (FDR).
Analysis of H3K56ac
The analysis of H3K56ac in the cell cycle was performed following procedure similar to one described previously24. We adopted the same regions of nucleosomes in chromosome III and calculated H3K56ac signals at these nucleosomes as described above. We sorted nucleosomal H3K56ac signals by genomic location and fit our data with those of the previous study by generating heat maps in R.
Supplementary Material
Acknowledgements
We dedicate this paper to the memory of our colleague, Y.Y.L.. We thank L.H. for general data analysis advice; J.D. for advices on histone mutants; L.S. and S.U., and G.R. for help with fermentor and YMC experiments; and S.T., B.C., J.B, P.M., Y.Y.L and members of the Boeke lab for helpful discussion. This work was supported by NIH grants R01HG006841 to H.J., R01GM094314 to B.P.T. and U54GM103520 to J.D.B.
Footnotes
Author Contributions
Z.K., L.C., B.P.T and J.D.B designed experiments; Z.K. and L.C. collected ChIP-seq and RNA-seq data; Z.K., X.Z. and H.J. performed the analysis; Z.K. made histone mutants and analyzed growth and YMC phenotypes; Z.K. wrote the manuscript with help from L.C., X.Z., H.J., B.P.T and J.D.B; all authors reviewed the manuscript.
Accession codes
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 3.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Weiner A, et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS biology. 2012;10:e1001369. doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nature Reviews Molecular Cell Biology. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 6.Brownell JE, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 8.Krebs JE. Moving marks: dynamic histone modifications in yeast. Mol. Biosyst. 2007;3:590–597. doi: 10.1039/b703923a. [DOI] [PubMed] [Google Scholar]
- 9.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paige SL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011;21:875–884. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 13.Tu BP, et al. Cyclic changes in metabolic state during the life of a yeast cell. Proceedings of the National Academy of Sciences. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaochar S, Tu BP. Gatekeepers of chromatin: Small metabolites elicit big changes in gene expression. Trends Biochem. Sci. 2012 doi: 10.1016/j.tibs.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergkessel M, Whitworth GB, Guthrie C. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA. 2011;17:1461–1478. doi: 10.1261/rna.2754011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Avvakumov N, Nourani A, Côté J. Histone chaperones: modulators of chromatin marks. Mol. Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moazed D. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 2001;13:232–238. doi: 10.1016/s0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 23.Chern MK, et al. Yeast ribosomal protein L12 is a substrate of protein-arginine methyltransferase 2. J. Biol. Chem. 2002;277:15345–15353. doi: 10.1074/jbc.M111379200. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan T, et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen P, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Megee PC, Morgan BA, Smith MM. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 28.Zaslaver A, et al. Just-in-time transcription program in metabolic pathways. Nat. Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 29.Laxman S, Sutter BM, Tu BP. Behavior of a Metabolic Cycling Population at the Single Cell Level as Visualized by Fluorescent Gene Expression Reporters. PloS one. 2010;5:e12595. doi: 10.1371/journal.pone.0012595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 34.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 35.Dai J, et al. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C, Wong WH. DNA-chip analyzer (dChip). The analysis of gene expression data: methods and software. 2003:120–141. [Google Scholar]
- 38.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eilers PH, Marx BD. Flexible smoothing with B-splines and penalties. Statistical science. 1996:89–102. [Google Scholar]
- 42.Schwarz G. Estimating the dimension of a model. The annals of statistics. 1978;6:461–464. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.