Abstract
Homologous recombination is a conserved pathway for repairing double–stranded breaks, which are processed to yield single–stranded DNA overhangs that serve as platforms for presynaptic complex assembly. Here we use single–molecule imaging to reveal the interplay between Saccharomyce cerevisiae RPA, Rad52, and Rad51 during presynaptic complex assembly. We show that Rad52 binds RPA–ssDNA and suppresses RPA turnover, highlighting an unanticipated regulatory influence on protein dynamics. Rad51 binding extends the ssDNA, and Rad52–RPA clusters remain interspersed along the presynaptic complex. These clusters promote additional binding of RPA and Rad52. Together, our work illustrates the spatial and temporal progression of RPA and Rad52 association with the presynaptic complex, and reveals a novel RPA–Rad52–Rad51–ssDNA intermediate, which has implications for understanding how the activities of Rad52 and RPA are coordinated with Rad51 during the later stages recombination.
Introduction
DNA double–strand breaks (DSBs) are among the most toxic forms of DNA damage, and can lead to genomic rearrangements and other severe chromosomal abnormalities. Homologous recombination (HR) is a conserved pathway that can be used to repair these lesions through an error–free mechanism that relies upon the presence of an undamaged homologous chromosome to serve as a template for repair of the broken DNA1–4. Defects in HR are widely associated with genetic abnormalities and cancer, highlighting the importance of this pathway for maintaining genome integrity. During HR, the newly exposed DNA ends are processed through Exo–1 or Sgs1–dependent 5′→3′ resection pathways, yielding long 3′ single stranded DNA (ssDNA) overhangs5–9. These ssDNA overhangs are then paired with homologous sequence elsewhere in the genome, and any missing sequence information is restored using the homologous DNA as a template for replication. The replicated intermediate is then resolved, regenerating the continuity of the broken chromosome. HR requires a complex repertoire of proteins, which are responsible for sensing damage, repair factor recruitment, and processing and repairing the damaged DNA. Many of the eukaryotic proteins involved in HR were identified as Saccharomyces cerevisiae mutants defective in the repair of DNA damage caused by ionizing radiation and are collectively referred to as the RAD52 epistasis group, which includes Rad50, Rfa1, Rad52, Rad54, Rdh54 (Tid1), Rad55–57, Rad59, Mre11, and Xrs2. In addition to these original members, there are now known to be more than 30 different proteins or protein complexes involved in HR1–3.
Replication protein A (RPA) is an abundant protein that participates in all aspects of eukaryotic DNA metabolism involving ssDNA intermediates10,11. During the early stages of homologous recombination the processed ssDNA overhangs are bound by RPA, which is a heterotrimeric complex comprised of Rfa1, Rfa2, and Rfa35,8,10,12–14. RPA protects the ssDNA from enzymatic degradation, removes secondary structure, serves as a checkpoint signaling intermediate15,16, and recruits specific HR proteins5,8,11,17–20. Rad51 is a DNA recombinase that assembles into an extended helical filament on the RPA–coated ssDNA21,22, and the resulting presynaptic complex is a critical HR intermediate in all eukaryotes2,3,23–26. This presynaptic filament is responsible for aligning the processed ssDNA overhang with a homologous dsDNA template, and also performs strand invasion whereby the ssDNA is paired with the complementary DNA strand from the homologous duplex.
Many other accessory factors are also essential for the successful completion of HR, and these proteins promote numerous events during recombination. For example, RPA can outcompete Rad51 for ssDNA–binding, therefore presynaptic complex assembly is stimulated by mediator proteins that assist Rad51 loading2,23. Rad52 is a key mediator in S.cerevisiae27–30, and Rad52 is also required for the second strand capture and strand annealing reactions that take place during the later stages of recombination31–34. Rad52 co–localizes with RPA and Rad51 at induced DSBs in vivo17,35, and Rad52 also forms spontaneous foci during S–phase, reflecting HR–dependent repair of stalled replication forks35,36. The importance of Rad52 is revealed by the extreme susceptibility of yeast rad52 mutants to DNA damage24. Although unrelated in sequence, Rad52 is functionally similar to human Brca2 (refs. 37,38), and it has recently been shown that human cells deficient for both Rad52 and Brca2 exhibit extensive chromosome abnormalities37.
While there is a growing knowledge of the proteins involved in HR and the contribution that they make to the final outcome of the repair processes, there remains relatively little information regarding how the macromolecular complexes involved in HR are assembled and disassembled, and how the individual protein components influence one another during the course of a reaction. Here we use two–color single–molecule imaging and ssDNA curtains to reveal the interplay between RPA, Rad52, and Rad51 during presynaptic complex assembly. We show that individual Rad52 complexes bind tightly to an RPA–coated ssDNA, which mimics the physiologically relevant substrate present at the free ends of processed DSBs. The initial Rad52 complexes serve as nucleation sites allowing for association of additional Rad52 molecules, which spread along the RPA–ssDNA. RPA binds very tightly to ssDNA when free RPA is absent from the surrounding solution, but the bound proteins remain poised for very rapid concentration–dependent turnover through a mechanism involving a microscopically dissociated intermediate, enabling free RPA present in solution to compete for transiently exposed patches of ssDNA39,40. Remarkably, Rad52 suppresses RPA turnover through a mechanism requiring direct protein–protein contacts between RPA and Rad52. This finding highlights an unanticipated regulatory influence of Rad52 on RPA dynamics during the very early stages of HR. Rad51 binding cause srapid extension of the ssDNA, and small Rad52–RPA clusters remain interspersed between extended tracts of Rad51. These clusters serve as nucleation sites for the binding of additional molecules of RPA and Rad52. Together, our work illustrates the spatial and temporal progression of RPA and Rad52 association with the presynaptic complex throughout the early stages of HR, and reveals the existence and assembly pathway of a novel RPA–Rad52–Rad51–ssDNA presynaptic intermediate.
Results
Visualizing Rad52–RPA–ssDNA interactions
We utilized ssDNA curtains to mimic the temporal progression of events thought to take place during the early stages of recombination (Fig. 1a)41. S. cerevisiae RPA–mCherry (80 pM)was injected into a microfluidic sample chamber containing ssDNA curtains. The ssDNA substrates were generated using a biotinylated oligonucleotide and circular M13 ssDNA as a template for rolling circle replication. The resulting ssDNA was anchored to a lipid bilayer on the surface of a microfluidic sample chamber through a biotin–streptavidin linkage, and the anchored ssDNA molecules were then aligned along the leading edge of zig–zag shaped nanofabricated chromium (Cr) barriers through the application of hydrodynamic force41,42. The ssDNA remains compacted in the absence of RPA, but quickly unravels when incubated with RPA39–41. Once extended, the downstream ends of the RPA–ssDNA complexes become anchored to exposed Cr pedestals41. The resulting double–tethered RPA–ssDNA complexes could then be visualized with total internal refection fluorescence microscopy (TIRFM) even in the absence of buffer flow (Fig. 1b).
Fig. 1.
Single–stranded DNA curtain assay for presynaptic complex assembly. (a) Schematic of ssDNA curtains. (b) Wide–field images of RPA–mCherry (magenta) bound to ssDNA in the absence (upper panel) and presence of 50 pM SNAP488–Rad52 (green; lower panel). (c) Wide–field images of 50 pM SNAP488–Rad52 (upper panel) or 1 nM SNAP488–Rad52 (lower panels) bound to wild–type (unlabeled) RPA on ssDNA. The 5′→3′ orientation of the ssDNA is indicated in this and all subsequent figures.
RPA–ssDNA represents the first HR intermediate that is anticipated following resection of a DSB. The next stage of presynaptic complex was mimicked by injecting SNAP–tagged S. cerevisiae Rad52 labeled with either Alexa 488 or Alexa 546(SNAP488–Rad52 and SNAP546–Rad52; Supplemental Fig. 1–2) and then asking whether these fluorescent versions of Rad52 could bind to the RPA–ssDNA. Fluorescent Rad52 co–localized with both RPA–mCherry (Fig. 1b), and unlabeled wild–type (wt) RPA (Fig. 1c). At low concentrations (50 pM) individual fluorescent Rad52 complexes were readily resolved (Fig. 1c), whereas Rad52 coated the RPA–ssDNA at higher protein concentrations (1 nM)(Fig. 1c). Although Rad52 could bind to naked ssDNA in our assays, these ssDNA molecules remained highly compacted (not shown); characterization of the Rad52–ssDNA complexes in the absence of RPA was not pursued given that the RPA–ssDNA complex is the physiologically relevant substrate.
Mechanism of Rad52 assembly on RPA–ssDNA
When viewed in real–time Rad52 was found to bind to numerous different locations along the length of the RPA–ssDNA, the bound molecules of Rad52 remained at fixed positions, and Rad52 could accumulate along the entire length of the RPA–ssDNA (Fig. 2a, b). Control reactions confirmed that there was no Rad52 binding to regions of the sample chamber surface lacking RPA–ssDNA (Fig. 2c). There was no evidence that RPA was displaced from ssDNA upon association of Rad52 (Fig. 2d &Supplemental Fig. 3), in agreement with previous studies43–45. Rad52 forms a heptameric ring in solution46–48, and the individual Rad52 complexes displayed uniform signal intensities (x̄ = 59.2±10.3 a.u., n = 591; Fig. 2e), consistent with expectations for association events involving discrete, well–defined complexes. Moreover, quantitation of the Rad52 binding distributions revealed no discernable pattern of preferential binding sites (Fig. 2f).
Fig. 2.
Individual Rad52 complexes binding to RPA–ssDNA. (a) Kymographs showing the association of single SNAP488–Rad52 (50 pM) complexes with RPA–mCherry–ssDNA or (b) wt (dark) RPA–ssDNA. (c) Kymograph confirming that SNAP488–Rad52 does not associate with a control surface lacking an ssDNA molecule. (d) Example of Rad52 bound to an RPA–ssDNA complex highlighting that RPA is not selectively lost from sites bound by Rad52. (e) Histogram showing the uniform intensity distribution for individual SNAP–tagged Rad52 complexes bound to RPA–ssDNA (n = 591). (f)Distribution of Rad52 nucleation sites along the length of the RPA–ssDNA (n = 498). Error bars represent the standard deviation (s.d.) from n bootstrap samples55.
We next asked whether we could detect evidence of Rad52–Rad52 interactions on the RPA–ssDNA. For this we conducted two–color pulse–chase experiments to determine whether differentially labeled Rad52 complexes would preferentially associate with one another on the DNA (Fig. 3a). SNAP488–Rad52 (50 pM)was injected first and allowed to bind the RPA–ssDNA, and then chased immediately with an injection of SNAP546–Rad52 (50 pM; Fig. 3b). These experiments revealed a modest preference for incoming molecules of Rad52 to bind to the RPA–ssDNA at sites already occupied by pre–existing Rad52 (Pearson’s coefficient r = 0.43, P=6×10−13, n = 262; Fig. 3c), with 64% of binding events (n = 69) occurring at pre–existing Rad52 complexes (Fig. 3d & Supplemental Fig. 4). The remaining events represented new binding to regions of the RPA–ssDNA lacking detectable SNAP488–Rad52; for brevity we refer to these as novel binding events (Fig. 3d). The existence of these novel binding events is not surprising considering overall length of the RPA–ssDNA (~13–μm, ~36,000–nt), so even after an initial Rad52 nucleation event the amount of RPA–ssDNA still vastly exceeds the small number of bound Rad52 complexes. Together, these findings suggested that at low protein concentrations Rad52 could bind at random locations along the RPA–ssDNA and also had a modest preference for self–association after binding to the RPA–ssDNA.
Fig. 3.
Nucleation and growth of Rad52 on RPA–ssDNA. (a) Schematic illustration of two–color pulse–chase experiment used to determine spatial distribution of new Rad52 binding events relative to pre–existing Rad52 complexes (b) Kymograph showing the binding of SNAP546–Rad52 to a RPA–ssDNA molecule already bound to SNAP488–Rad52. (c) Correlation of normalized SNAP488–Rad52 and SNAP546–Rad52 signals (n = 262). (d) Relative number of co–localized Rad52 binding events compared to binding events at new (novel) sites on the RPA–ssDNA. (e) Potential models for Rad52 nucleation and growth. (f) Integrated SNAP488–Rad52 (625 pM) signal across entire ssDNA molecules over time. (g) Kymographs highlighting the nucleation and bidirectional growth of Rad52 (625 pM) along the RPA–ssDNA. Arrowheads (f) & (g) indicate when initial nucleation events are visually detected in the kymographs, and these are also defined as the zero time point in (f).
If Rad52 binding occurred through a cooperative mechanism, then at higher protein concentrations quantitation of the total Rad52 signal on an individual ssDNA molecule should reveal an initial slow phase, corresponding to slow nucleation events, followed by a more rapid growth phase (Fig. 3e). Moreover, for a cooperative binding mechanism the individual nucleation events might be expected to lead to lateral growth of the Rad52 complexes along the RPA–ssDNA. In contrast, if Rad52 bound through a non–cooperative mechanism then there should be a uniform increase in the Rad52 intensity all along the RPA–ssDNA. As shown in Fig. 3f, at higher concentrations of Rad52 (625 nM) binding appeared to begin with an initial nucleation phase followed by more rapid growth, suggesting assembly did involve cooperative association of Rad52. Note that due to their low signal intensity relative to saturation, the initial Rad52 binding events are not detected when the total Rad52 signal is integrated across the length of the ssDNA (Fig. 3f), however, the precise timing of initial binding events (i.e. nucleation) becomes readily apparent upon inspection of the corresponding kymographs (Fig. 3g), which reveal that the initial binding events precede the more rapid growth phase of the reaction. In addition, these kymographs also revealed that the Rad52 individual nucleation events were followed by rapid (124 ± 41 nm sec−1, n = 8)growth in both the 5′→3′ and 3′→5′ directions as revealed by wedge–shaped patterns emanating outward from the initial binding sites (Fig. 3g). We conclude that Rad52 could bind RPA–ssDNA through a mechanism involving an initial nucleation event followed by more rapid bi–directional growth along the RPA–ssDNA.
Rad52 binds tightly to RPA–ssDNA
We have previously shown that RPA exhibits a lifetime ≫2 hours on ssDNA curtains, but this long lifetime is only observed when there is no free RPA present in solution (i.e. at the infinite dilution limit; see below)39. We next examined the stability of Rad52 bound to the RPA–ssDNA. For these experiments, fluorescent Rad52 was incubated with RPA–ssDNA, the unbound proteins were then flushed away, and the resulting complexes were monitored to determine whether Rad52 remained bound or dissociated into free solution (Fig. 4a). The Rad52–RPA–ssDNA complexes were monitored over periods of either 10 minutes or 2 hours (Fig. 4b); importantly, the laser was shuttered between each acquired image and the image acquisition rates were adjusted such that the total time that each sample was exposed to laser illumination was identical for the 10 minute and 2 hour experiments. For experiments spanning either 10 minutes or 2 hours, the loss of Rad52 fluorescence occurred by a gradual decrease in fluorescence signal indicative of photo–bleaching of a multimeric Rad52 complex, rather than the abrupt signal loss that would be expected for protein dissociation. These findings indicated that Rad52 binds tightly to the RPA–ssDNA, and remained stably associated with the RPA–ssDNA for ≥ hours; note that experiments beyond 2 hours are intractable due to stage drift and spontaneous breakage of the tethered ssDNA.
Fig. 4.
Rad52 regulates RPA turnover. (a) Schematic for determining Rad52 binding lifetime on RPA–ssDNA. (b) Kymographs showing SNAP488–Rad52 dissociation over 10–minutes (upper panel) or 2–hours (lower panel); shuttering time was adjusted so that the total illumination time was identical for both experimental measurements 39. (c) Models for the potential influence of Rad52 on RPA turnover. (d) RPA–mCherry turnover after chasing with wt RPA, ±1 nM SNAP488–Rad52. Aligned images of ssDNA molecules showing persistent co–localization of SNAP488–Rad52 and RPA–mCherry clusters. (e) Examples showing quantitation of RPA–mCherry turnover on single ssDNA molecules. (f) Aligned images of RPA–mCherry and SNAP488–Rad52 on different ssDNA molecules. (g) Line graphs of SNAP488–Rad52 and RPA–mCherry co–localization. (h) Correlation analysis of RPA–mCherry and SNAP488–Rad52 after exchange with wt RPA (n = 255).
Rad52 regulates RPA turnover
RPA has an exceptionally slow off–rate at the infinite dilution limit, yet it remains poised for rapid macroscopic dissociation though an unusual, newly recognized mechanism for the turnover of DNA–binding proteins involving concentration–driven dissociation of a microscopically dissociated intermediate39,49,50. In brief, ssDNA–bound RPA undergoes constant microscopic dissociation under all conditions, but does not equilibrate with free solution39. These microscopic dissociation events are manifested as macroscopically detectable dissociation into free solution only when other ssDNA–binding proteins are present to compete with the transiently unbound species for exposed patches of ssDNA39,49,50.
The ability of RPA to exchange between free and bound states when free RPA is present in solution raises the question of what happens to Rad52 during RPA turnover. There are at least three possibilities: (i) RPA turn over might cause Rad52 dissociation; (ii) Rad52 could remain bound without affecting RPA turnover; or (iii) Rad52 might alter RPA turnover kinetics (Fig. 4c). To distinguish between these possibilities SNAP488–Rad52–RPA–mCherry–ssDNA complexes were chased with unlabeled (dark) wild–type RPA (Fig. 4c). In the absence of Rad52, the majority of the RPA–mCherry (75.8 ± 2.7%) exchanged with dark RPA within ~30 sec (Fig. 4d), as previously demonstrated39,40. In the presence of Rad52 a much smaller fraction of RPA–mCherry under went rapid turnover (38.8 ± 14.5%), and all of the Rad52 remained bound to the RPA–ssDNA (Fig. 4d, e). Remarkably, the RPA molecules bound by Rad52 did not undergo turnover, as revealed by highly persistent tracks of RPA–mCherry co–localized with Rad52, whereas the RPA molecules not bound by Rad52 underwent rapid turnover (Fig. 4d). Inspection of numerous ssDNA molecules confirmed that the Rad52 complexes always co–localized with persistent tracks of RPA (Fig. 4f, g) and Pearson analysis confirmed a strong correlation between Rad52 binding and the clusters of RPA resistant to turnover (r = 0.74, P=0, n = 255). Moreover, experiments with wild–type Rad52 (unlabeled) revealed similarly persistent tracks of RPA–mCherry, and only 43.7± 7.2% of the RPA–mCherry underwent rapid exchange (Supplemental Fig. 5), and control experiments using two different fluorescently labeled Rad52 constructs revealed identical results (Supplemental Fig. 1–2, and not shown), confirming that the outcome was not influenced by the Rad52 labeling strategy. Together, these results indicate that Rad52 bound tightly to RPA and suppressed its exchange between free and ssDNA–bound states, and this effect required direct protein–protein contact between Rad52 and RPA as revealed by the spatial correlation of Rad52 with the turnover–resistant RPA clusters.
Influence of Rad51 assembly on Rad52–RPA complexes
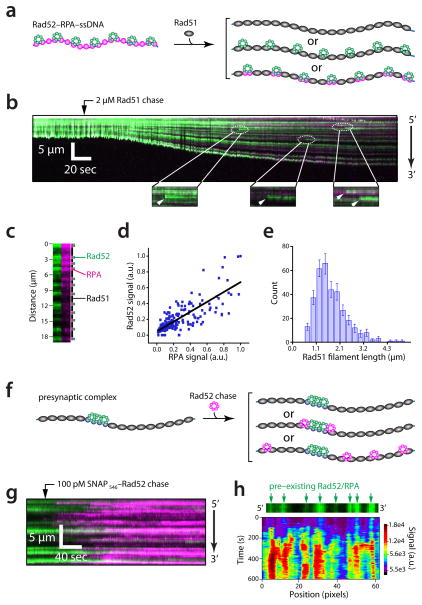
Assembly of the Rad51 filament coincides with the displacement of RPA from the ssDNA2,3,39,43. The ability of Rad52 to act as a mediator of Rad51 assembly is well–established27,29,30,35,43,51. However, the fate of Rad52 during and after the assembly of Rad51–ssDNA filaments remains unexplored. The fact that Rad51 physically displaces RPA from ssDNA raises questions regarding the fate of Rad52 bound to the RPA–ssDNA (Fig. 5a). This question is critical because in addition to its role as a mediator Rad52 is also required for second strand capture and stand annealing, which take place during the later stages of recombination31–34. The participation of Rad52 in these later stages of HR raises the possibility that it might remain bound to the presynaptic complex. Alternatively, Rad51 could provoke the dissociation of Rad52 from the RPA–ssDNA, in which case Rad52 would need to be recycled and re–associate with the HR machinery at a later step in the pathway. Therefore we next asked what happens to Rad52 when Rad51 binds the ssDNA.
Fig. 5.
Protein dynamics during presynaptic complex assembly. (a) Schematic illustration showing examples of the potential influence of Rad51 filament assembly on Rad52 bound to RPA–ssDNA. (b)Kymograph of wild–type (unlabeled) Rad51 (1 μM)binding to Rad52–RPA–ssDNA complexes containing RPA–mCherry and SNAP488–Rad52 in the presence of ATP (2.5 mM); new Rad52 binding events are highlighted. (c)Example of the spatial distribution of RPA (magenta), Rad52 (green), and Rad51 (dark) on a single ssDNA molecule. (d)Correlation analysis of RPA–mCherry and SNAP488–Rad52 within the Rad51 presynaptic filaments (n = 187). (e) Rad51 filament length distribution; filament length is defined as the distances between adjacent Rad52–RPA clusters (n = 346). Error bars represent the standard deviation (s.d.) from n bootstrap samples55.(f) Two–color experiment for testing whether newly added Rad52 can bind the presynaptic filaments. (g) Kymograph showing SNAP546–Rad52 re–binding to a presynaptic filament. (h) Spatially distinct re–binding kinetics of SNAP546–Rad52 to a presynaptic complex.
To determine the fate of Rad52 during presynaptic complex assembly we watched the binding of unlabeled wild–type Rad51 to single–tethered ssDNA curtains pre–bound by RPA–mCherry and SNAP488–Rad52 (ref. 39); single–tethered curtains allowed visual assessment of ssDNA extension by Rad51. As expected, Rad51 binding coincided with a rapid increase in the contour length of the ssDNA (~138%; Fig. 5b), which arises from physical extension of the ssDNA and the greater stiffness of the Rad51–ssDNA filament relative to RPA–ssDNA. Remarkably, Rad52 remained bound to the ssDNA during presynaptic complex assembly, yielding the appearance of small protein clusters separated by long tracts of dark Rad51–ssDNA (Fig. 5b). The remaining Rad52 clusters always co–localized with small patches of fluorescent RPA (r = 0.79, P=0, n = 187; Fig. 5b–d), revealing the continued presence of both proteins within the presynaptic complex. The distances between Rad52–RPA clusters followed a Poisson distribution (x̄= 1.6 ± 0.79 μm, n= 346), reflecting the stochastic nature of the underlying molecular event(s) dictating the lengths of the Rad51 filaments or the dispersion of the Rad52–RPA clusters within the filaments (Fig. 5e). Assuming that the Rad51–ssDNA molecules are extended to near their full contour lengths, and that they are stretched by ~50% relative to an equivalent length of B–form dsDNA, then a 1.6 μM Rad51–ssDNA filament would correspond to ~1,000 molecules of Rad51. We conclude that small clusters of Rad52–RPA remained embedded between long Rad51–ssDNA filaments after assembly of the presynaptic complex.
Rad52–RPA clusters are nucleation sites for protein binding
Interestingly, close inspection of the Rad51 chase experiments revealed that numerous new Rad52 binding events occurred during presynaptic complex assembly, and these new binding events occurred exclusively at sites already occupied by Rad52–RPA (Fig. 5b); the likely source of these new proteins were Rad52 complexes that had dissociated from ssDNA upstream of the viewing area. These observations suggested the possibility that the Rad52–RPA clusters embedded within the Rad51 presynaptic filaments might serve as nucleation sites allowing the binding of additional Rad52. Therefore, we next asked whether newly added Rad52 could bind pre–assembled presynaptic complexes. To address this issue we assembled presynaptic filaments comprised of Rad51–RPA–SNAP488–Rad52, and then chased these complexes with an injection of SNAP546–Rad52 (Fig. 5f). These experiments confirmed that additional Rad52 could indeed associate with the presynaptic complex, revealing this pathway as a secondary entry way for recruitment of Rad52 to the presynaptic filament (Fig. 5g, h). Moreover, the spatial distribution of the newly bound Rad52 coincided primarily with the positions of the pre–bound Rad52–RPA clusters, indicating that these locations served as sites for more extensive association of Rad52(Fig. 5h). There was more limited accumulation of new Rad52 at locations lacking detectable Rad52–RPA clusters, suggesting the possibility that Rad52 might be able to bind directly to Rad51–ssDNA, albeit to a lesser extent than the binding observed at the site of pre–existing Rad52–RPA. This differential binding pattern suggested the existence of two kinetically distinct pathways, as might be expected for association mechanisms involving distinct sets of protein–protein contacts (e.g. lower–affinity Rad52–Rad51 interactions versus higher–affinity Rad52–Rad52 interactions).
We have previously shown that the Rad51 filaments are highly stable when ATP is present in the surrounding buffer, and that RPA does not bind to these Rad51–ssDNA filaments39. If ATP is removed from the buffer, then Rad51 is quickly replaced with RPA, which coincides with a decrease in the ssDNA extension39. Together, these findings show that RPA alone does not bind extensively to the pre–assembled Rad51 filaments. We next asked whether co–injection of Rad52 and RPA could lead to more extensive binding to the pre–assembled Rad51–Rad52–RPA–ssDNA complexes, and if so whether the newly bound molecules exhibited a distinct spatial association pattern (Fig. 6a). Remarkably, co–injection of both Rad52 and RPA lead to extensive association of both proteins with the presynaptic complexes (Fig. 6b); bead pull down assays provide additional evidence supporting the notion that Rad51, Rad52, and RPA can all co–occupy ssDNA (Supplementary Fig. 6). Moreover, inspection of the spatial and temporal distribution of the binding events revealed that Rad52 binding initiated at the pre–existing clusters of Rad52–RPA and then spread outward along the length of the presynaptic complex (Fig. 6b–d). Interestingly, the newly added RPA bound to the pre–existing Rad52–RPA cluster, and spread outward along the presynaptic complex, although association and spreading of RPA was slightly delayed relative to Rad52 (c.f. Fig. 6c, d). These observations suggested a temporal order of association beginning with the binding of Rad52 at pre–existing Rad52–RPA clusters, followed by the more gradual association of RPA. Importantly, the observed contour length of the presynaptic complexes did not change significantly (<4%) even upon extensive association of the newly added Rad52 and RPA (Fig. 6b–d). Taken together, our results strongly suggest that Rad51 remained bound directly to the ssDNA as an extended helical filament within the context of these higher–order macromolecular complexes.
Fig. 6.
Assembly of Rad51–Rad52–RPA–ssDNA presynaptic intermediates. (a) Experimental schematic for detecting Rad52 and RPA binding to a pre–assembled Rad51–Rad52–RPA–ssDNA complex. (b) Kymographs showing wild-type Rad51 filament assembly on Rad52–RPA–ssDNA, followed by co–injection of additional Rad52 and RPA (as indicated). The upper panel shows a two–color overlay, and the lower panels show the individual red and green channels, as indicated. (c) & (d) show the spatially distinct binding kinetics of newly added RPA and Rad52, respectively, along the lengths of the pre–assembled Rad51–Rad52–RPA–ssDNA presynaptic filament. Arrowheads indicate the locations of the pre–existing Rad52–RPA clusters within the Rad51 filaments.
Discussion
Here we have used ssDNA curtains to visualize the spatial and temporal progression of the molecular events accompanying the assembly of S. cerevisiae presynaptic complexes. Together, our experiments reveal the existence and assembly pathway of a novel Rad51–Rad52–RPA–ssDNA presynaptic intermediate (Fig. 7a). We demonstrate that Rad52 binds directly to the RPA–ssDNA as discrete units consistent with expectations for a well–defined oligomer, and at higher protein concentrations Rad52 spreads along the RPA–ssDNA. We have previously shown that in the absence of free RPA, ssDNA–bound RPA undergoes continuous microscopic dissociation without equilibrating into solution39. Microscopic dissociation from the ssDNA is only manifested as macroscopic dissociation into bulk solution when free RPA is available to compete for partially exposed patches of ssDNA; the importance of this mechanism is that allows for both extremely high binding affinities as well as rapid exchange kinetics39,49,50. Remarkably, Rad52 suppresses the concentration–driven turnover of RPA. The ability of Rad52 to restrict RPA exchange implies that Rad52 restricts microscopic dissociation of RPA from the ssDNA, which in turn prevents macroscopic dissociation into free solution. When considering that Rad52 acts as a mediator of Rad51 assembly, one might intuitively expect that it should destabilize the RPA–ssDNA complex, as opposed to the stabilizing effect that is revealed in our assays. However, an important consideration is that the concentration–driven exchange of RPA between free and bound states is not specific to RPA, as is revealed by the finding that E. coil SSB can readily drive the dissociation of RPA from ssDNA, and vice versa39. We propose that the suppression of RPA turnover by Rad52 may be necessary to prevent inappropriate access of non–HR proteins to ssDNA recombination intermediates, while at the same time directing Rad51 to the RPA–coated ssDNA.
Fig. 7.
Model for RPA and Rad52 dynamics during presynaptic complex assembly. (a) Assembly pathway for the Rad51–Rad52–RPA–ssDNA presynaptic complexes. (b) Influence of Rad52–RPA on strand invasion and second strand capture during the later stages of homologous recombination. Details of the models are presented in the Discussion.
Rad51 assembles into extended filaments on the RPA–Rad52–ssDNA, and these long Rad51 filaments are punctuated by small clusters of Rad52–RPA, which are retained as a stable component embedded within the Rad51 presynaptic filaments. These clusters serve as nucleation sites enabling more extensive binding of Rad52 and RPA (Fig. 7a). Importantly, the Rad51–ssDNA remains in a highly extended configuration, even after extensive association of late arriving Rad52 and RPA. This finding is only consistent with a model in which Rad51 remains bound to the ssDNA, suggesting that the newly bound RPA and Rad52 are associated with the surface of the Rad51–ssDNA filament through protein–protein contacts; additional studies will be essential for establishing the molecular details of the protein–protein and protein–ssDNA contacts responsible for assembly of these macromolecular complexes. The resulting Rad52–RPA–Rad51–ssDNA presynaptic intermediate has not previously been identified, but its existence has important mechanistic implications for how the activities of RPA and Rad52 are coordinated with those of Rad51 during the downstream steps in homologous recombination. Namely, this novel intermediate places Rad52 and RPA in the right location to immediately stabilize the non–complementary ssDNA strand as it is being displaced by Rad51 during strand invasion (Fig. 7b)31–34. Moreover, the continued presence of RPA and Rad52 within the presynaptic complex may also help coordinate strand invasion with the capture and annealing of the second processed DNA end31–34. Finally, the finding that Rad52 and RPA can both stably co–exist within the Rad51 presynaptic complex provides a possible explanation for in vivo observations showing that RPA and Rad52 arrive at DSBs prior to Rad51, and remain at these sites even during and after the arrival of Rad5135,52–54. Future studies using ssDNA curtains may help reveal how other HR proteins interact with and influence the properties of presynaptic complexes, and may also begin shedding new light on later stages of homologous DNA recombination.
Online Methods
Proteins and DNA
RPA and Rad51 were purified as described39. RPA–mCherry was cloned, expressed in E. coli, and purified over Ni–resin (Qiagen) and a MonoQ column (GE Healthcare). Concentrations of RPA–mCherry were determined based on absorbance at 587 nm (ε587 nm=72,000 cm−1M−1).
S. cerevisiae Rad52 has multiple potential start codons, but the protein expressed in yeast corresponds to codons 34–505, yielding a 472 amino acid protein56. Therefore, Rad52 codons 34–504 were cloned with an N–terminal 6xHis and SNAP–tag, and a C–terminal intein/chitin–binding domain; for brevity we refer to this construct as SNAP–Rad52. As an alternative, Rad52 codons 1–504 were cloned with an N–terminal 6xHis and SNAP–tag, and a C–terminal intein/chitin–binding domain. Use of this longer reading frame provides an additional 34 amino acids placed between the SNAP tag and the N–terminus of Rad52; we refer to this construct as SNAP–34–Rad52. Bacteria (E. coli Rossetta™) were grown at 37°C, induced overnight at 18°C with 0.5 mM IPTG, and lysed by sonication. The clarified lysate was purified over a chitin column (NEB), washed with buffer A (50 mM Tris–HCl [pH 7.5], 600 mM NaCl) plus 1 mM EDTA, and eluted overnight in buffer A plus 1 mM EDTA and 50 mM DTT. The eluate was dialyzed into buffer A plus 0.1 mM DTT and 10 mM imidazole, loaded onto a Ni–NTA column, washed sequentially with buffer A containing 60 mM and 125 mM imidazole, and eluted in buffer A plus 250 mM imidazole. Proteins were concentrated with a Vivaspin concentrator (50 kDa MWCO; GE Healthcare), exchanged into storage buffer (40% glycerol, 40 mM Tris–HCl [pH 7.4], 600 mM NaCl, and 1 mM DTT), and stored at −80°C. SNAP–Rad52 was labeled with SNAP–Surface Alexa Fluor 488 (ε495 nm=71,000 cm−1M−1) or SNAP–Surface Alexa Fluor 546 (ε556 nm=104,000 cm−1M−1) overnight at 4°C, as per the manufacturers instructions (NEB). Unreacted dye was removed with a Sephadex G20 spin column, and labeling efficiency (~0.55–0.60 dyes per Rad52 monomer) was determined by comparing protein UV absorbance to the absorbance of the Alexa dye.
As an additional alternative fluorescent tagging strategy, Rad52 was labeled at the N–terminus with a succinimidyl ester of 5(6) carboxytetramethylrhodamine (TAMRA; Sigma Cat# 21955), essentially as described57. Rad52 (codons 34–504)was cloned with an N–terminal 6xHis and a C–terminal intein/chitin–binding domain, and purified from bacteria as described above for the SNAP–tagged proteins. After elution from the chitin column the purified protein was dialyzed extensively against 50 mM phosphate buffer [pH 7.0], containing 500 mM NaCl, 0.1 mM DTT, and 10% glycerol. The purified protein was mixed with a 12–times molar excess of TAMRA succinimidyl ester and incubated for 4 hours at 4°C. The labeling reactions were then terminated with the addition of 50 mM Tris–HCl [pH 7.5], and free dye was removed by gel filtration using a Sephacryl S–300HR column equilibrated with buffer containing 50 mM Tris–HCl [pH 7.4], 500 mM NaCl, 0.5 mM EDTA, 10% glycerol, and 0.1 mM DTT. The labeled protein was then concentrated by dialysis against dry poly (ethylene glycol) (PEG; 20,000 MW) and stored at −80°C.
Bulk biochemical assays
The oligomeric state of SNAP–Rad52 was assessed by gel filtration chromatography58. Rad52 and SNAP–Rad52 (~2 mg ml−1; 100 μl each) were resolved on a Superose 6 GL 30/100 column (GE Healthcare) in buffer containing 50 mM phosphate [pH 7.2] and 150 mM NaCl, and the column was calibrated using a set of defined molecular mass standards (blue dextran 2000, thyroglobulin (669 KDa), ferritin (440 KDa), aldolase (158 kDa), conalbulmin (75 kDa), and ovalbumin (43 kDa); GE Healthcare). The elution positions were monitor by UV absorbance, and confirmed by SDS–PAGE and Coomassie staining. Wt Rad52 and SNAP–Rad52 eluted at volumes consistent with the formation of a protein oligomer (not shown), confirming that the SNAP tag did not disrupt the assembly of Rad52 heptameric rings in solution.
The strand annealing and mediator activities of the fluorescent Rad52 constructs were tested using radio labeled ssDNA oligonucleotides. Strand annealing activities were conducted as previously described58, using 83–nucleotide ssDNA substrates (oligo–1: 5′–AAA TGA ACA TAA AGT AAA TAA GTA TAA GGA TAA TAC AAA ATA AGT AAA TGA ATA AAC ATA GAA AAT AAA GTA AAG GAT ATA AA–3′; and oligo–2: 5′–TTT ATA TCC TTT ACT TTA TTT TCT ATG TTT ATT CAT TTA CTT ATT TTG TAT TAT CCT TAT ACT TAT TTA CTT TAT GTT CAT TT–3′). 83–mer ssDNA (oligo–1; 60 nM molecules, 5 μM nucleotides) was incubated in the absence or presence of RPA (370 nM) in 12 μl annealing buffer (40 mM Tris–HCl [pH 7.5], 5 mM MgCl2, 0.1 μg ml−1 BSA, 1 mM DTT, 50 mM KCl) at 25 °C for 5 min, followed by a 5 min–incubation with 200 nM Rad52 at 25 °C. In a separate tube 32P–labeled complementary ssDNA (Oligo–2, 60 nM) was incubated with RPA and Rad52 under these same reaction conditions. The separate two mixtures were then combined and incubated for the indicated time periods at 25 °C. The reaction was stopped by treatment of 8 μl of the reaction with 1 μl of Oligo–2 (7 μM molecules) and then deproteinated with the addition of 0.5% SDS and 0.5 μg μl−1 proteinase K. The samples were resolved on 8% polyacrylamide gel in TAE buffer (30 mM Tris acetate [pH 7.4], 0.5 mM EDTA) and subjected to phosphorimaging analysis.
Strand exchange assays were conducted as previously described59,60, using a 150–nucleotide ssDNA (5′– TCT TAT TTA TGT CTC TTT TAT TTC ATT TCC TAT ATT TAT TCC TAT TAT GTT TTA TTC ATT TAC TTA TTC TTT ATG TTC ATT TTT TAT ATC CTT TAC TTT ATT TTC TCT GTT TAT TCA TTT ACT TAT TTT GTA TTA TCC TTA TCT TAT TTA–3′) and a 32P–labeled 40–bp dsDNA (5′–TAA TAC AAA ATA AGT AAA TGA ATA AAC AGA GAA AAT AAA G–3′). The 150–mer ssDNA oligonucleotide (3 μM nucleotides) was incubated with a combination of RPA (1 μM), Rad51 (1 μM), and Rad52 (20, 40, or 80 nM) in 11.5 μl of strand exchange buffer (35 mM Tris–HCl [pH 7.5], 5 mM MgCl2, 1 mM ATP, 100 ng μl−1 BSA, 1 mM DTT, 36 mM KCl) for 15 min at 37°C. Then 0.5 μl each of 32P –labeled homologous 40–bp dsDNA (20 μM base pairs; 32P–labeled at the 5′ end of the top strand) and spermidine (50 mM) were added to the reactions, followed by an incubation for 30 min at 37°C. Reactions were stopped with 0.5% SDS and 0.5 μg μl−1 proteinase K and the samples were resolved on 8% polyacrylamide gel in TAE buffer (30 mM Tris acetate [pH 7.4], 0.5 mM EDTA) and subjected to phosphorimaging analysis.
The relative strand annealing and mediator activities of the fluorescently tagged versions of Rad52 were determined by comparing the percent of product formed to reactions using unlabeled Rad52 at the same protein concentrations (Supplementary Fig. 1–2). Biochemical assays for strand annealing activity revealed that TAMRA–Rad52, SNAP–Rad52, and SNAP–34–Rad52 displayed an average of ~69%, ~70%, and ~72% of the strand annealing activity of unlabeled Rad52, respectively (Supplementary Fig. 1). The biochemical assays for mediator activity revealed that TAMRA–Rad52, SNAP–Rad52, and SNAP–34–Rad52 displayed an average of ~93%, ~41%, and ~67% mediator activity relative to reactions with unlabeled Rad52, respectively (Supplementary Fig. 2). Note that SNAP–Rad52 was used for detailed quantitation and figure preparation, however, all three different fluorescently tagged versions of Rad52 gave qualitatively similar results for the single molecule assays (not shown).
Bead pull down assays
Pull down assays were used to verify co–occupancy of Rad52 and RPA on ssDNA (Supplementary Fig. 3). Each reaction sample contained 20 μl Streptavidin magnetic beads (Life Technologies, Dynabeads M–280 Streptavidin), which were bound to excess of ssDNA oligonucleotide (5′ biotin–TTT TTT GTA AAA CGA CGG CCA GTG CCA AGC TTG CAT GCC TGC AGG TCG ACT CTA GAG GAT CCC CGG GTA CCG AG) and then washed with assay buffer (30 mM Tris–Acetate pH 7.5, 150 mM KCl, 5 mM Mg–Acetate, 2 mM ATP, 1 mM DTT, 0.01% Tween20) containing 20 mg ml−1 BSA. The beads where then incubated with 400 nM RPA for 20 minutes at 30 °C. The unbound sample was removed and replaced with fresh buffer containing 400 nM RPA to which Rad52 (0, 80, 240, 400, 800 nM) was added and incubated for 20 minutes at 30°C. Unbound protein was recovered and the beads were washed with assay buffer lacking BSA. Bound protein was recovered with the addition of 1% SDS. Samples were resolved on 8–16% Tris–Glycine gradient gels and proteins were detected by Coomassie staining.
Pull down assays using either ssDNA cellulose or magnetic beads were used to confirm co–occupancy of Rad51, Rad52, and RPA on ssDNA (Supplementary Fig. 6). Reactions contained either 10 μl ssDNA cellulose resin or 20 μl Streptavidin magnetic beads (Life Technologies, Dynabeads M–280 Streptavidin) bound to an ssDNA oligonucleotide (as above) rinsed with assay buffer plus 20 mg ml−1 BSA. A 10 μl sample of pre–mixed protein components (Rad51 10 μM, Rad52 1 uM, RPA 0–400 nM) were added to the beads and incubated at 37°C for 10 minutes. The unbound protein was removed and saved, and the ssDNA cellulose was then washed with assay buffer lacking BSA. Bound protein was recovered by the addition of 1% SDS. Samples were resolved on 8–16% Tris–Glycine gradient gels and proteins were detected by Coomassie staining.
TIRFM experiments data analysis
Single–molecule experiments were conducted with a custom–built total internal reflection fluorescent microscope and ssDNA curtains, as described41,61. Single–stranded substrates were generated by rolling circle replication of circular M13mp18 ssDNA (7,249–nucleotides; New England Biolabs). All single molecule experiments began with the addition of either 100 pM wt RPA or 100 pM RPA–mCherry, as indicated. The ssDNA substrates remain highly compacted in the absence of RPA–mCherry and cannot be visualized by TIRF microscopy41. Experiments using just RPA and Rad52 were conducted with double–tethered DNA curtains at flow rates of 0.1 – 0.2 ml min−1; experiments involving Rad51 utilized single–tethered DNA curtains and continuous buffer flow (0.1 ml min−1)41. Reactions were conducted at 30°C in buffer containing 30 mM Tris–acetate [pH 7.5], 5 mM Mg–Acetate, 50 mM KCl, 1 mM DTT, and 200 μg ml−1 BSA43. The reaction buffer was supplemented with 2.5 mM ATP for all experiments involving Rad51. Three fluorescently tagged versions of Rad52 were tested to ensure that experimental outcomes were not influenced by the labeling strategy: SNAP–Rad52, in which the SNAP domain was fused to the N–terminus of Rad52; SNAP–34–Rad52, which bears an additional 34 amino acid linker between the SNAP domain and Rad52; and TAMRA–Rad52, in which Rad52 was labeled at the N–terminus with 5(6) carboxytetramethylrhodamine (Supplemental Fig. 1). Use of the SNAP–tagged Rad52 constructs permits ready use of different color fluorophores, therefore SNAP488–Rad52 or SNAP546–Rad52 was used for detailed quantitation and figure preparation, however, all three different versions of Rad52 gave qualitatively similar results in the single molecule assays.
Image processing, fluorescence intensity and signal distribution measurements were made using NIH Image J 1.48c. Images (100–msec exposures) were collected from 10–minute time–lapse experiments at 1 second intervals and the lasers where shuttered in between frame acquisition to minimize photo–bleaching. For longer time–lapse experiments images (100–msec exposures) were collected at 24–second intervals for 2–hours, and the resulting data were corrected for stage drift. All data analysis was restricted to individual molecules of ssDNA identified based upon visual inspection of the RPA–mCherry or RPA–eGFP signal; overlapping ssDNA molecules were excluded from any further analysis or quantitation; and all conclusions were based on data collected from at least three separate experiments.
DNA curtains
Chromium (Cr) barriers were fabricated by e–beam lithography, as described41,61. Bilayers were prepared with 91.5% DOPC, 0.5% biotinylated–DPPE, and 8% mPEG 550–DOPE41,61. DNA substrates were generated using M13mp18 (7,249–bp; Invitrogen) as a template for rolling circle replication41. A biotinylated primer was annealed to the template, and reactions were initiated with addition of ϕ29 DNA polymerase (100 nM)in buffer containing 50 mM Tris [pH 7.4], 2 mM DTT, 10 mM MgCl2, 10 mM ammonium sulfate, and 200 μM dNTPs. The ssDNA molecules were aligned at the barriers with buffer flow, as described16.
TIRF microscopy
Experiments were performed using a prism–type TIRF microscope (Nikon) with two back–illuminated iXon EMCCDs (Andor Technology)61. Illumination was provided by a 200 mW, 488–nm laser and a 150 mW, 561–nm laser (Coherent, Inc.). Intensity at prism face was ~14 mW and ~25 mW for the 488–nm and 561–nm lasers, respectively. Fluorescence signals were separated by a filter cube equipped with a dichroic mirror (ZT561rdc), band pass filter (ET525/50m), and long pass filter (ET575lp)(Chroma Technology Corp.).
Rad52 binding to RPA–ssDNA
All RPA and Rad52 single–molecule experiments were conducted at 30°C in buffer containing 30 mM Tris–acetate (pH 7.5), 5 mM Mg–Acetate, 50 mM KCl, 1 mM DTT, and 200 μg ml−1 BSA43. Rad52 binding measurements were made using pre–assembled RPA (wild–type or mCherry–tagged, as indicated)/ssDNA filaments in a double–tethered DNA curtain format; unless otherwise indicated, free RPA was flushed from the sample chamber prior to the injection of Rad52. Single Rad52 particle fluorescence intensity measurements were made from wide–field images collected from reactions performed in the presence of 100 pM wt RPA and 50 pM SNAP488–Rad52 after free proteins had been flushed from the sample chamber. Rad52 nucleation site distributions were based upon data collected from ssDNA bound by RPA–mCherry after the injection of 50 pM SNAP488–Rad52. Two–color cooperative Rad52 binding measurements were conducted by using wt RPA/ssDNA complexes and an initial injection of 50 pM SNAP488–Rad52. Unbound protein was flushed from the sample chamber, and followed by a second injection of 50 pM SNAP546–Rad52. Co–localization of the two differentially labeled proteins was determined by Pearson correlation analysis of the overlap between pre–existing SNAP488–Rad52 clusters with the binding distributions of the newly injected SNAP546–Rad52. New SNAP546–Rad52 binding events that occurred at sites occupied by pre–existing molecules of SNAP488–Rad52 was manually scored based on inspection of the kymographs, and new SNAP546–Rad52 binding events were considered overlapping if they were occurred within ±1 pixel of the center position of a pre–existing peak of SNAP488–Rad52.
Rad52 nucleation and growth measurements were made by injecting 625 nM SNAP488–Rad52 into sample chambers containing pre–assembled wt RPA–ssDNA complexes. Nucleation sites and bidirectional growth were identified by visual inspection of the resulting data, and growth rates were estimated from the linear expansion of the accumulating Rad52 fluorescence moving outward from the initiating site of nucleation. Analysis of the growth rates was restricted to nucleation events that were well resolved from the nearest neighboring events, and the rates were only estimated up until the point the Rad52 fluorescence signal merged with the signal of adjacent complexes. Rad52 binding lifetime measurements were made using 100 pM RPA–mCherry (100 pM) and 50 pM SNAP488–Rad52. After initial binding, free proteins were flushed from the sample chamber and data were acquired for either 10–minute or 2–hour intervals frame acquisition frequencies of either 1 frame per second or 1 frame per 24 seconds, respectively; the lasers where shuttered in between frame acquisition so that the total time that each sample was exposed to laser illumination was identical for each of the two different time courses. RPA chase experiments were conducted using pre–assembled RPA–mCherry–ssDNA complexes or RPA–mCherry–SNAP488–Rad52–ssDNA complexes made with 100 pM RPA–mCherry and 1 nM SNAP488–Rad52 or 0.6 – 1.0 nM wild–type Rad52, as indicated. Free proteins were flushed from the sample chamber and then rapidly replaced with buffer containing 100 nM wt RPA. The kymographs in Fig. 4d were collected under identical illumination conditions and camera gain settings, and the image contrast of the RPA–mCherry signal for the wt RPA chase was adjusted identically for reactions in either the presence or absence of SNAP488–Rad52. RPA–mCherry and SNAP488–Rad52 co–localization analysis after chasing with 100 nM wt RPA was made using normalized pixel intensity profiles generated from separate kymographs for each to the two spectrally separated fluorophores, and the resulting data were quantified by Pearson correlation analysis.
Presynaptic complex assembly
Reactions involving Rad51 were conducted at 30°C in buffer containing 30 mM Tris–acetate [pH 7.5], 5 mM Mg–Acetate, 50 mM KCl, 1 mM DTT, 2.5 mM ATP and 200 μg ml−1 BSA43. RPA–mCherry/SNAP488–Rad52/ssDNA complexes were assembled onto single tethered ssDNA curtains, as described above with the exception that the reactions utilized 150 pM SNAP488–Rad52. Presynaptic complex assembly was then initiated by injection of 1–3 μM Rad51 under continuous buffer flow (0.2 ml min−1) while acquiring images at 1–second intervals for approximately 10 minutes. Control reactions confirmed that Rad51 filament formation on the ssDNA was ATP–dependent39. The length of the protein–bound ssDNA was measured as a function of time tracking the location of the most 3′ distal detectable complex of SNAP488–Rad52. The dark Rad51–bound ssDNA may extend beyond the most distal Rad52 complex, but would not be detectable because neither the ssDNA nor the Rad51 are fluorescently tagged; this possibility does not impact the interpretation of our results. The total relative amount of SNAP488–Rad52 bound to the ssDNA before and during the injection of Rad51 was calculated based upon the integrated signal intensity across the full length of each individual ssDNA molecule over the course of the reactions. The resulting data was then calculated as a percent decrease in each fluorescence signal over time. The apparent Rad51 filament length distributions are reported based upon the experimentally observed center–to–center distances between the small isolated clusters of RPA–mCherry that remained bound to the ssDNA after Rad51 filament formation.
Rad52 and RPA binding to pre–assembled presynaptic complexes
Presynaptic complexes were assembled exactly as described above using RPA–mCherry (100 pM), SNAP488–Rad52 (150 pM), and wt Rad51 (1–3 μM); and free proteins were then flushed out of the sample chamber using buffer containing 30 mM Tris–acetate (pH 7.5), 5 mM Mg–Acetate, 50 mM KCl, 1 mM DTT, 2.5 mM ATP and 200 μg ml−1 BSA. Control experiments demonstrated that the Rad51 filaments remained stable in the presence of ATP, and all subsequent buffers contained 2.5 mM ATP to prevent dissociation of the Rad51 filaments. The pre–assembled presynaptic complexes were then chased with SNAP546–Rad52 and data acquired at 1–second intervals for 10 minutes, as described above. SNAP546–Rad52 binding kinetics were quantified by spatially segregating sections of the presynaptic complex that were either bound by wt Rad51 or by SNAP488–Rad52/RPA–mCherry complexes based on visual inspection, and the increase in integrated SNAP546–Rad52 signal intensity (a.u.) for each region was determined as a function of time.
Supplementary Material
Acknowledgments
We thank L. Symington, and members of the Greene and Sung laboratories for comments on the manuscript. This research was funded by the US National Institutes of Health (USA) grants GM074739 (E.C.G), RO1ES007061 (P.S.), and CA146940 (E.C.G. and P.S.). This work was partially supported by the Nanoscale Science and Engineering Initiative of the US National Science Foundation (USA) under award No. CHE–0641523, and by the New York State Office of Science, Technology, and Academic Research (NYSTAR). E.C.G. is an Early Career Scientist with the Howard Hughes Medical Institute.
Footnotes
Author Contributions
B.G. and L.F.Y. cloned, expressed and purified RPA and Rad52, conducted the single–molecule experiments, and analyzed the resulting data. Y.K. and H.N. expressed and purified Rad51, and Y.K. conducted bulk biochemical experiments. E.C.G. supervised the project. B.G. and E.C.G. wrote the manuscript with input from all co–authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Mazon G, Mimitou EP, Symington LS. SnapShot: Homologous recombination in DNA double–strand break repair. Cell. 2010;142:646, 646 e1. doi: 10.1016/j.cell.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–71. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 4.Cromie GA, Connelly JC, Leach DR. Recombination at double–strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–74. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 5.Cejka P, et al. DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature. 2010;467:112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA double–strand break ends. Nature. 2012;489:576–80. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double–strand break processing. Nature. 2008;455:770–4. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu H, et al. Mechanism of the ATP–dependent DNA end–resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–11. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double–strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wold MS. Replication protein A: a heterotrimeric, single–stranded DNA–binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 11.Broderick S, Rehmet K, Concannon C, Nasheuer HP. Eukaryotic single–stranded DNA binding proteins: central factors in genome stability. Subcell Biochem. 2010;50:143–63. doi: 10.1007/978-90-481-3471-7_8. [DOI] [PubMed] [Google Scholar]
- 12.Mimitou EP, Symington LS. DNA end resection––unraveling the tail. DNA Repair (Amst) 2011;10:344–8. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 2009;8:1068–76. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Symington LS, Gautier J. Double–strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–71. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 15.Choi JH, et al. Reconstitution of RPA–covered single–stranded DNA–activated ATR–Chk1 signaling. Proc Natl Acad Sci U S A. 2010;107:13660–5. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marechal A, et al. PRP19 Transforms into a Sensor of RPA–ssDNA after DNA Damage and Drives ATR Activation via a Ubiquitin–Mediated Circuitry. Mol Cell. 2014;53:235–46. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–21. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hays SL, Firmenich AA, Massey P, Banerjee R, Berg P. Studies of the interaction between Rad52 protein and the yeast single–stranded DNA binding protein RPA. Mol Cell Biol. 1998;18:4400–6. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plate I, et al. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J Biol Chem. 2008;283:29077–85. doi: 10.1074/jbc.M804881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–6. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–9. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 23.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–39. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double–strand break repair. Microbiol Mol Biol Rev. 2002;66:630–70. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 26.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 27.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–7. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 28.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51–mediated recombination. Nature. 1998;391:404–7. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 29.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–10. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 30.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–4. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 31.Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double–strand–break repair. Mol Cell. 2008;29:517–24. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52–mediated second–end capture during DSB repair. Mol Cell. 2008;29:510–6. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 33.Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second–end DNA capture in double–stranded break repair to form complement–stabilized joint molecules. Proc Natl Acad Sci U S A. 2009;106:3077–82. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52–mediated DNA annealing after Rad51–mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–48. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–82. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Z, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108:686–91. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51–mediated recombination. Nature. 2010;467:678–83. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibb B, et al. Single–molecule imaging reveals dynamic behavior of RPA during assembly of the Saccharomyces cerevisiae Rad51 presynaptic complex. PLOS One. 2014;9:e89722. [Google Scholar]
- 40.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology–mediated repair of DNA double–strand breaks. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2786. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibb B, Silverstein TD, Finkelstein IJ, Greene EC. Single–stranded DNA curtains for real–time single–molecule visualization of protein–nucleic acid interactions. Anal Chem. 2012;84:7607–12. doi: 10.1021/ac302117z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visnapuu ML, Fazio T, Wind S, Greene EC. Parallel arrays of geometric nanowells for assembling curtains of DNA with controlled lateral dispersion. Langmuir. 2008;24:11293–9. doi: 10.1021/la8017634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)–single–stranded DNA to accelerate Rad51–mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–72. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 44.Sugiyama T, Kantake N. Dynamic regulatory interactions of rad51, rad52, and replication protein–a in recombination intermediates. J Mol Biol. 2009;390:45–55. doi: 10.1016/j.jmb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single–stranded DNA. Proc Natl Acad Sci U S A. 1998;95:6049–54. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co–operates with RPA in single–strand DNA annealing. Genes Cells. 1998;3:145–56. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 47.Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single–strand annealing domain of human RAD52 protein. Proc Natl Acad Sci U S A. 2002;99:13492–7. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stasiak AZ, et al. The human Rad52 protein exists as a heptameric ring. Curr Biol. 2000;10:337–40. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- 49.Graham JS, Johnson RC, Marko JF. Concentration–dependent exchange accelerates turnover of proteins bound to double–stranded DNA. Nucleic Acids Res. 2011;39:2249–59. doi: 10.1093/nar/gkq1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sing CE, Olvera de la Cruz M, Marko JF. Multiple–binding–site mechanism explains concentration–dependent unbinding rates of DNA–binding proteins. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–50. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Haber JE. Role of Saccharomyces single–stranded DNA–binding protein RPA in the strand invasion step of double–strand break repair. PLoS Biol. 2004;2:E21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyazaki T, Bressan DA, Shinohara M, Haber JE, Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double–strand break repair. EMBO J. 2004;23:939–49. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mine–Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14:510–7. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 55.Efron B, Tibshirani R. An Introduction to the Bootstrap. Champman and Hall, Inc; New York: 1993. [Google Scholar]
- 56.Antunez de Mayolo A, et al. Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res. 2006;34:2587–97. doi: 10.1093/nar/gkl280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–8. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 58.Krejci L, et al. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J Biol Chem. 2002;277:40132–41. doi: 10.1074/jbc.M206511200. [DOI] [PubMed] [Google Scholar]
- 59.Busygina V, et al. Hed1 regulates Rad51–mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–95. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon Y, Zhao W, Sung P. Biochemical studies on human Rad51–mediated homologous recombination. Methods Mol Biol. 2011;745:421–35. doi: 10.1007/978-1-61779-129-1_24. [DOI] [PubMed] [Google Scholar]
- 61.Greene EC, Wind S, Fazio T, Gorman J, Visnapuu ML. DNA curtains for high–throughput single–molecule optical imaging. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.