Abstract
Orderly termination of sister-chromatid cohesion during mitosis is critical for accurate chromosome segregation. During prophase, mitotic kinases phosphorylate cohesin and its protector sororin, triggering Wapl-dependent cohesin release from chromosome arms. The shugoshin (Sgo1)–PP2A complex protects centromeric cohesin until its cleavage by separase at anaphase onset. Here, we report the crystal structure of a human cohesin subcomplex comprising SA2 and Scc1. Multiple HEAT repeats of SA2 form a dragon-shaped structure. Scc1 makes extensive contacts with SA2, with one binding hotspot. Sgo1 and Wapl compete for binding to a conserved site on SA2–Scc1. Mutations of SA2 residues at this site that disrupt Wapl binding bypass Sgo1 requirement in cohesion protection. Thus, besides recruiting PP2A to dephosphorylate cohesin and sororin, Sgo1 physically shields cohesin from Wapl. This unexpected, direct antagonism between Sgo1 and Wapl augments centromeric cohesion protection.
The ring-shaped cohesin complex maintains genome integrity through regulating sister-chromatid cohesion, DNA repair, and transcription in eukaryotes1–6. Dysregulation of cohesin and its regulators has been implicated in human cancers and developmental diseases7,8. Cohesin consists of an Smc1–3 heterodimer and a non-Smc heterodimer1,9. The Smc1 and Smc3 ATPases heterodimerize through their hinge domains (see Fig. 1a). Scc1 binds to the ATPase domains of Smc1 and Smc3 through its C- and N-terminal winged helix domains, respectively, forming a ring. In human somatic cells, Scc1 binds through its central region to either SA1 or SA2, two homologous HEAT (Huntingtin, Elongation factor 3, A subunit, and TOR) repeat-containing proteins.
Figure 1.
Structure and binding interface of human SA2–Scc1. (a) Cartoon diagram of the crystal structure of human SA2 in complex with the SA2-binding region of Scc1, with SA2 colored blue and Scc1 colored pink. The N-terminal helical domain and the 17 HEAT repeats (R1-R17) of SA2 are labeled. The N- and C-termini of SA2 and Scc1 are indicated. The four major contact sites (I–IV) between SA2 and Scc1 are boxed in dashed lines. A schematic drawing of the cohesin architecture is shown in lower right corner. (b–e) Zoomed in views of sites I–IV, with SA2 and Scc1 residues in yellow and gray sticks, respectively. Scc1 residues are labeled in red.
The dynamics and mode of cohesin association with chromatin are tightly regulated by a set of cohesin regulators2,3,10,11. In telophase, cohesin is loaded onto chromatin by the cohesin loader Scc2–Scc4 (refs. 12–15). In G1, cohesin association with chromatin remains dynamic, as the chromatin-bound cohesin can be released by the cooperative actions of the adaptor protein Pds5 and the cohesin inhibitor Wapl16–18. During S phase, the cohesin protector sororin binds cohesin in part through Pds5, and antagonizes Wapl to establish cohesion19–21. During prophase in human somatic cells, the mitotic kinases Cdk1, Plk1, and Aurora B collaborate to phosphorylate cohesin and sororin, triggering Wapl-dependent cohesin release from chromosome arms22–24. A pool of cohesin at centromeres is protected by the Sgo1–PP2A complex25–27, which keeps cohesin and sororin in a hypophosphorylated state and maintains centromeric cohesion28. At the metaphase–anaphase transition, the proper kinetochore–microtubule attachment creates tension across sister kinetochores and silences the spindle checkpoint29,30, leading to separase activation. Kinetochore tension also triggers redistribution of Sgo1 from centromeres to kinetochores31,32, which is thought to inactivate Sgo1 and allow cohesin cleavage by active separase. Inactivation of Sgo1-mediated protection of centromeric cohesion leads to premature sister-chromatid separation and spindle-checkpoint-dependent mitotic arrest33,34.
The SA2–Scc1 heterodimer mediates the binding between cohesin and its regulators, including Scc2–Scc4, Pds5, Wapl, and Sgo1 (refs. 28,35,36). The mechanisms by which this non-Smc cohesin subcomplex coordinates these crucial molecular interactions are poorly understood. To gain structural insights into these interactions, we have determined the crystal structure of human SA2 bound to the SA2-binding region of Scc1, and mapped the binding sites of Sgo1 and Wapl. Our results establish a competition between Sgo1 and Wapl in cohesin binding, and implicate a role for this direct antagonism in centromeric cohesion protection.
RESULTS
Structure of human SA2 bound to Scc1
We co-expressed human SA2 and the SA2-binding region of Scc1 in insect cells, purified the resulting cohesin subcomplex, and determined its crystal structure (Fig. 1 and Table 1). SA2 and Scc1 form a simple 1:1 heterodimer. SA2 contains a helical N-terminal domain (N-domain) followed by 17 HEAT repeats (termed R1-R17). The SA2 structure is shaped like an oriental dragon. Two long helices of R1 and R2 resemble the snout of the dragon. The N-domain comprising 7 helices packs against R1 and resembles the head of the dragon. The body of the dragon bends sharply at R9 and R10. This bend divides SA2 into N- and C-halves. Scc1 consists of four short helices (termed αA–D) and two long extended segments. These elements interact extensively with SA2 at four major sites, in a way reminiscent of a dragon rider riding a dragon (Fig. 1 and Fig. 2a). As expected, many residues at the SA2–Scc1 interface are highly conserved in metazoans.
Table 1.
Data collection and refinement statistics
| Native SA2–Scc1–MES | SeMet SA2–Scc1 | SeMet SA2–Scc1–MES | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 78.455, 107.275, 180.103 | 78.606, 108.752, 181.791 | 78.733, 108.045, 180.835 |
| α,β,γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.97918 | 0.97918 | 0.97924 |
| Resolution (Å) | 2.95 | 3.05 | 2.85 |
| Rsym | 0.111 (0.959) | 0.102 (1.00) | 0.098 (1.00) |
| I / σI | 19.0 (1.8) | 19.9 (1.5) | 20.1 (1.5) |
| Completeness (%) | 100.0 (99.8) | 100.0 (99.9) | 99.9 (99.9) |
| Redundancy | 6.2 (6.2) | 10.0 (8.9) | 8.8 (8.2) |
| Refinement | |||
| Resolution (Å) | 47.68–2.95 | 45.45–3.05 | 41.71–2.85 |
| No. reflections | 32,803 | 30,393 | 36,729 |
| Rwork / Rfree (%) | 18.8 / 25.1 | 19.5 / 23.8 | 20.9 / 22.5 |
| No. atoms | |||
| Protein | 7,974 | 7,909 | 7,982 |
| Ligand/ion | 12 | 0 | 12 |
| Water | 0 | 22 | 14 |
| B-factors (Å2) | |||
| Protein | 88.1 | 80.6 | 59.5 |
| Ligand/ion | 82.9 | – | 60.3 |
| Water | – | 50.3 | 29.8 |
| r.m.s deviations | |||
| Bond lengths (Å) | 0.009 | 0.007 | 0.005 |
| Bond angles (°) | 1.21 | 1.03 | 0.87 |
Data sets were collected with one native crystal with MES in the crystallization buffer, one selenomethionine crystal without MES, and one selenomethionine crystal with MES.
Values in parentheses are for highest-resolution shell.
Figure 2.
Identification of a binding hotspot between SA2 and Scc1. (a) Cartoon diagram of the structure of human SA2–Scc1, with SA2 and Scc1 colored blue and pink, respectively, in an orientation that is rotated 180° relative to that in Fig. 1a. The N- and C-termini of both proteins and the four helices of Scc1 are labeled. The four SA2–Scc1 contact sites are boxed. (b) Anti-Myc and anti-Scc1 immunoblots of lysates and anti-Myc immunoprecipitates (IP) of HeLa cells transfected with the indicated Myc-SA2 plasmids. WT, wild type. Uncropped blots are included in Supplementary Fig. 6. (c) A zoomed-in view of contact site IV, with SA2 and Scc1 residues in yellow and gray sticks, respectively. Scc1 residues are labeled in red. The dashed red lines indicate hydrogen bonds. (d) Anti-SA2 and anti-tubulin immunoblots of lysates of HeLa cells transfected with the indicated siRNAs and plasmids. WT, wild type. Uncropped blots are included in Supplementary Fig. 6. (e) Quantification of the mitotic indices (defined as the percentage of MPM2-positive, 4N cells) of cells in d. Error bars, s.d. (n = 4 independent experiments). (f) Four major types of metaphase spreads of cells in d. The spreads were stained with DAPI (blue) and the kinetochore marker CREST (red). Selected sister chromatids were magnified and shown in insets. Scale bar, 5 μm. (g) Quantification of the percentage of cells in d with type III and IV chromosome morphologies as in f. Error bars, s.d. (n = 4 independent experiments).
At site I, the N-terminal loop of Scc1 makes mainly electrostatic interactions with SA2 residues in the N-domain and at the ridge of R1 and R2 (Fig. 1b). There are four salt bridges formed between R322, D327, K330, and E331 of Scc1 and E154, R213, D209, and R298 of SA2, respectively. In addition, Scc1 L324 packs against T149, F152, and V212, forming hydrophobic interactions. At site II, the αA helix of Scc1 packs against R2-R4 of SA2 (Fig. 1c). The sidechain packing between Scc1 and SA2 at this site appears to be sub-optimal. For example, even though I337 and L341 of Scc1 are located in the vicinity of Y297 and W334 of SA2, there is minimal contact between their van der Waals surfaces. At site III, the loop connecting αA and αB of Scc1 interacts with the edge of R5 and R6 of SA2 (Fig. 1d). Scc1 αB contacts R7, R9, and the R7–R8 loop in SA2. Specifically, L351, L353, A354, P355, and P356 of Scc1 develop hydrophobic interactions with L416, L473, H474, H476, and Y479 of SA2. Site IV is close to the bend, with αB, αC, and αD of Scc1 interacting extensively with R11-R13 of the C-half of SA2 (Fig. 1e). In particular, L385, L388, and F389 located in αD of Scc1 make hydrophobic contacts with L742, W743, L745, M796, and I797 of SA2. Thus, Scc1 interacts with SA2 through an extensive interface that spans nearly the entire length of SA2.
Identification of a binding hotspot between SA2 and Scc1
To ascertain the functional importance of the observed SA2–Scc1 interface, we created about 50 SA2 and Scc1 mutants targeting conserved, surface-exposed residues at or near the SA2–Scc1 interface. Strikingly, among these mutants, only one, SA2 D793K, abolished the binding between SA2 and Scc1 (Fig. 2b and Supplementary Fig. 1a–d). SA2 D793 is located at site IV. Its sidechain forms hydrogen bonds with the backbone amides of A377 and Q378 of Scc1, which reside in the loop connecting αC and αD (Fig. 2c). Consistently, deletion of P376 and A377 in this loop, deletion of αD, or mutation of the three hydrophobic residues in αD abolished Scc1 binding to SA2 (Supplementary Fig. 1e). Moreover, ectopic expression of Myc-SA2 wild type (WT), but not D793K, in HeLa cells rescued the premature sister-chromatid separation and mitotic arrest phenotypes caused by co-depletion of SA1 and SA2 (Fig. 2d–g). Therefore, SA2 and Scc1 interact through an extensive interface, with site IV being a binding hotspot. Interestingly, in the pre-fusion state of hemagglutinin (HA) of the influenza virus, an aspartate sidechain of the fusion peptide forms similar hydrogen bonds to the backbone amides of residues in a pocket on HA37. Disruption of this interaction is a rate-limiting step in viral membrane fusion.
Identification of a functional Sgo1-binding site on cohesin
Human Sgo1 associates with cohesin during mitosis, and phosphorylation of Sgo1 at T346 enhances this association28. It is unclear, however, whether the cohesin–Sgo1 interaction is direct. Through systematic deletion mutagenesis (data not shown), we mapped the cohesin-binding region of Sgo1 to a 41-residue conserved motif containing T346. Both the unphosphorylated Sgo1 and phospho-T346 Sgo1 (pSgo1) peptides bound purified recombinant SA2–Scc1 (Fig. 3a). pSgo1 bound more tightly than unphosphorylated Sgo1 did. As determined by isothermal titration calorimetry (ITC), unphosphorylated Sgo1 bound SA2–Scc1 with a dissociation constant (Kd) of 618 ± 80 nM (mean and standard deviation of three independent measurements), whereas pSgo1 had a Kd of 109 ± 35 nM (Fig. 3b). Both peptides bound SA2–Scc1 with a stoichiometry of 1:1, indicating that SA2–Scc1 has a single Sgo1-binding site. Thus, Sgo1 binds directly to the SA2–Scc1 cohesin subcomplex, and Sgo1 phosphorylation enhances, but is not required for, this binding.
Figure 3.
A conserved, functional Sgo1-binding site of SA2–Scc1. (a) Coomassie stained SDS-PAGE gel of recombinant SA2–Scc1 (input) and the same complex bound to the indicated beads. Uncropped images are included in Supplementary Fig. 6. (b) Isothermal titration calorimetry (ITC) curves of the binding between SA2–Scc1 and unphosphorylated or phospho-T346 Sgo1 peptides, with the dissociation constant (Kd) and binding stoichiometry (N) indicated. (c) Autoradiograph of 35S-labeled SA2–Scc1 proteins (input) and the same proteins bound to beads coupled to unphosphorylated or phospho-T346 Sgo1 peptides. WT, wild type. Uncropped images are included in Supplementary Fig. 6. (d) The Sgo1-binding site of SA2–Scc1. Color and labeling schemes are the same as in Fig. 1c. A MES molecule bound at this site is shown in sticks. (e) Anti-SA2 and anti-tubulin immunoblots of lysates of HeLa cells transfected with the indicated siRNAs and Myc-SA2 plasmids. The positions of the endogenous and Myc-SA2 are indicated. WT, wild type. Uncropped blots are included in Supplementary Fig. 6. (f) Quantification of the mitotic indices (defined as the percentage of MPM2-positive, 4N cells) of cells in e. Error bars, s.d. (n = 4 independent experiments). (g) Quantification of the percentages of mitotic cells in e with separated sister chromatids (types III/IV metaphase spreads). Error bars, range (n = 2 independent experiments).
Despite numerous attempts, we failed to obtain diffracting crystals of the SA2–Scc1–pSgo1 complex. The structural basis of phosphorylation-enhanced Sgo1 binding to cohesin remains to be established. We thus sought to define the Sgo1-binding site on SA2–Scc1 through mutagenesis. Among the SA2 and Scc1 mutations, only SA2 Y297A, R298E, D326K, K330E, Y331A, W334A, D793K, and K870E greatly diminished Sgo1 or pSgo1 binding in vitro (Fig. 3c and Supplementary Fig. 2a,b). The SA2 D793K mutant lost binding to Sgo1, because it could not bind Scc1. Another mutant K870E also had weakened Sgo1 binding. K870 is located at site IV and in close proximity to D793. Mutation of this residue likely affects Sgo1 binding indirectly through affecting Scc1 binding. Aside of D793 and K870, all other residues critical for Sgo1 binding, including Y297, R298, D326, K330, Y331, and W334, are clustered near site II of the SA2–Scc1 interface (Fig. 3d). When co-expressed with GFP-Sgo1 in HeLa cells, mutants targeting these residues also exhibited weaker binding to GFP-Sgo1 (Supplementary Fig. 2c). Moreover, the Sgo1-binding-deficient SA2 mutants, Y297A, R298E, Y331A, and W334A, failed to rescue the mitotic arrest phenotypes of HeLa cells depleted of SA1 and SA2 (Fig. 3e,f and Supplementary Fig. 2d,e). Consistently, Y331A and W334A also failed to prevent premature sister-chromatid separation in these cells (Fig. 3g). Collectively, these results establish site II of SA2–Scc1 as a functional Sgo1-binding site. The notion that Sgo1 binds near the SA2–Scc1 interface is also consistent with the fact that Sgo1 only binds to the SA2–Scc1 complex, but does not associate with either SA2 or Scc1 alone (data not shown).
We noticed that a 2-(N-morpholino)ethanesufonic acid (MES) molecule from the crystallization solution bound at the Sgo1-binding site (Fig. 3d and Supplementary Fig. 3a,b). In particular, the sulfate group of MES makes hydrogen bonds and favorable electrostatic interactions with Y297 and R298, and is in the vicinity of Y331. Y297 and R298 belong to a signature FVHRYRD motif conserved in SA proteins from yeast to man. SA2 Y297F and Y331F were defective in Sgo1 binding (Supplementary Fig. 2c), implicating their hydroxyl groups in engaging non-hydrophobic interactions with Sgo1. Thus, one intriguing possibility is that MES might mimic phospho-T346 of Sgo1. Future structural studies are needed to rigorously test this possibility.
The conserved FVHRYRD motif is involved in Scc2–Scc4 binding in the fission yeast35. Mutations of this motif cause a partial defect in topological cohesin loading to circular DNA in vitro and cohesion defect in yeast cells35. We tested whether this motif in human SA2 was also required for cohesin loading in human cells. GFP-SA2 Y297A and R298E localized to chromatin in telophase HeLa cells as efficiently as GFP-SA2 WT (Supplementary Fig. 3c,d). As a negative control, the Scc1-binding-deficient D793K mutant did not associate with chromatin. Therefore, we do not have evidence to indicate a role for the FVHRYRD motif of human SA2 in cohesin loading. Because Scc2–Scc4 interacts with multiple cohesin subunits35, disruption of a single interaction surface might not block cohesin association with chromatin. Our results thus do not rule out a role of this motif in cohesin loading. Nevertheless, as SA2 Y297A and R298E do not show gross chromosome loading defects, their functional defects in human cells are likely due to their inability to bind Sgo1.
Sgo1 competes with Wapl for cohesin binding
SA2–Scc1 also interacts with Wapl. In particular, several FGF motifs in the N-terminal region of Wapl have been implicated in SA2–Scc1 binding36. In addition, we have previously shown that a middle region of human Wapl (Wapl-M, residues 510–570) is critical for SA2–Scc1 binding in vitro38 (Fig. 4a). We further tested whether Wapl-M was required for Wapl function in human cells. As expected, depletion of Wapl greatly increased the percentage of mitotic cells with arm-closed sister chromatids (Fig. 4b,c), as cohesin was not effectively removed from chromosome arms and sister chromosomes were not completely decatenated in Wapl-depleted cells39–41. Expression of Wapl WT, but not the mutant with Wapl-M deleted (Δ510–570), rescued this arm-resolution deficiency. Consistent with the well established genetic antagonism between Sgo1 and Wapl in centromeric cohesion protection17,18, depletion of Wapl from HeLa cells rescued the premature sister-chromatid separation and the resulting mitotic arrest caused by Sgo1 depletion (Supplementary Fig. 4a,b). Restoring functional Wapl through increasing expression of Wapl WT restored mitotic arrest in cells depleted of both Sgo1 and Wapl. When expressed at similar levels, Wapl Δ510–570 was less effective in restoring Wapl function and mitotic arrest in these cells. Taken together, the middle region of Wapl is critical for its function in human cells.
Figure 4.
Competition between Wapl and Sgo1 for cohesin binding. (a) Schematic drawing of domains and motifs of human Wapl. (b) Quantification of the percentages of mitotic HeLa cells (transfected with the Wapl siRNA and increasing concentrations of the indicated plasmids) that had arm-closed chromosomes. WT, wild type. Error bars, range (n = 2 independent experiments). (c) Representative metaphase spreads of cells in b with arm-closed or arm-separated chromosomes. Selected sister chromatids were magnified and shown in inset. Scale bar, 5 μm. (d) Coomassie stained SDS-PAGE gel of recombinant SA2–Scc1 (input) and the same complex bound to GST or GST-Wapl, in the presence of increasing concentrations of the indicated Sgo1 peptides. (e) Quantification of the relative SA2 band intensities of the binding reaction shown in d. Error bars, range (n = 2 independent experiments). (f) Surface diagram of SA2, with Scc1 shown in ribbons and MES shown in sticks. The SA2 residues conserved in metazoans are colored yellow. The conserved patch at or near the Sgo1-binding site is boxed. (g) Ribbon drawing of the boxed region in f, with conserved, surface-exposed SA2 residues shown in sticks. Residues critical for binding to Wapl or Sgo1 or both are colored purple, green, and yellow, respectively. (h) Anti-Myc and anti-Wapl immunoblots of the anti-Myc IP of HeLa cells transfected with the indicated plasmids. WT, wild type. Uncropped blots are included in Supplementary Fig. 6.
Because both Sgo1 and Wapl interact with SA2–Scc1, we tested whether Sgo1 and Wapl-M competed for binding to SA2–Scc1. A GST-Wapl fragment containing Wapl-M pulled down recombinant SA2–Scc1 purified from insect cells (Fig. 4d). Addition of the pSgo1 peptide reduced SA2–Scc1 binding to GST-Wapl-M in a dose-dependent manner (Fig. 4d,e). As expected, the unphosphorylated Sgo1 peptide was less effective in the competition. As a control, a shorter pSgo1 peptide (with five residues flanking pT346 on either side) showed no competition.
We then mapped the Wapl-binding site on SA2–Scc1. We focused our analysis on the conserved patch of residues at the Sgo1-binding site (Fig. 4f,g). Indeed, mutations of K290, D326, and K330 abolished Myc-SA2 binding to endogenous Wapl in human cells (Fig. 4h). Mutations of Y331 and W334 in SA2 diminished (but did not abolish) Wapl binding in human cells. Four of the five Wapl-binding-deficient SA2 mutants, including D326K, K330E, Y331A, and W334A were also defective in Sgo1 binding. Mutation of SA2 Y328 had no effect on Wapl or Sgo1 binding. We further confirmed that the K290E, D326K, and K330E mutations greatly reduced the binding of SA2–Scc1 to GST-Wapl-M in vitro (Supplementary Fig. 4c,d). Therefore, Sgo1 and Wapl compete with each other for cohesin binding, and they bind to overlapping sites on SA2–Scc1.
SA2 mutations or high concentrations of pSgo1 do not completely block Wapl binding to SA2–Scc1, suggesting that Wapl might bind SA2–Scc1 through multiple interfaces. In addition, the Wapl/Sgo1-binding residues on SA2 are highly conserved in all metazoans, whereas the cohesin-binding motifs of Sgo1 and Wapl are only conserved in vertebrates, but not in Drosophila or C. elegans. It is possible that certain conserved structural features of Sgo1 and Wapl cannot be easily gleaned from amino acid sequences alone. Alternatively, these SA2 residues are conserved for other purposes, such as binding Scc2–Scc4.
Direct Sgo1–Wapl antagonism strengthens cohesion protection
An established function of cohesin-bound Sgo1 is to recruit PP2A to cohesin and enable PP2A to protect sororin from phosphorylation by Cdk1 (ref. 28). Hypophosphorylated sororin remains bound to Pds5 to counteract Wapl, thereby maintaining centromeric cohesion. Consistent with this notion, expression of the non-phosphorylatable sororin 9A mutant with all Cdk1 sites mutated bypasses the requirement for Sgo1 in cohesion protection during normal mitotic progression24,28. As shown above, pSgo1 binds to SA2–Scc1 with high affinity (around 100 nM) and directly competes with Wapl for cohesin binding. We thus examined whether the strong Sgo1 binding to SA2–Scc1 had additional roles, aside from enabling sororin dephosphorylation by PP2A.
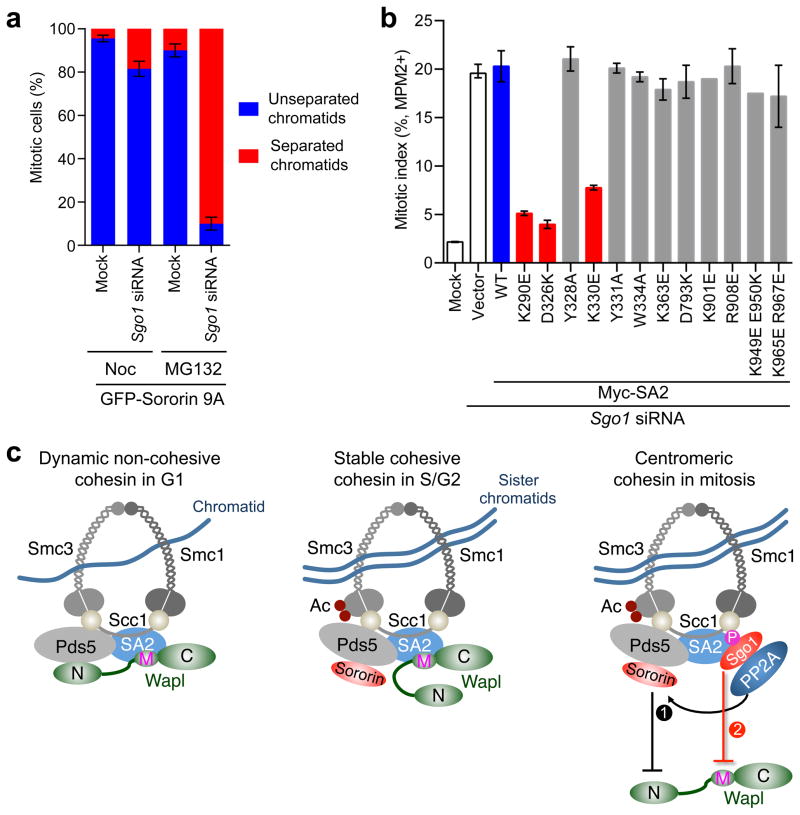
We first revisited the phenotypes of sororin 9A-expressing cells depleted of Sgo1. Consistent with our previous findings28, in the presence of nocodazole (which arrested cells in prometaphase, with all kinetochores not experiencing spindle-pulling force or tension), expression of sororin 9A in HeLa cells rescued the premature sister-chromatid separation phenotypes caused by Sgo1 depletion (Fig. 5a). In stark contrast, in cells treated with the proteasome inhibitor MG132 (which arrested cells at metaphase, with persistent spindle-pulling force and kinetochore tension), Sgo1 depletion caused massive premature sister-chromatid separation even in cells expressing sororin 9A (Fig. 5a). Thus, in the absence of the spindle-pulling force at kinetochores, hypophosphorylated sororin alone presumably suffices to protect centromeric cohesion. When kinetochores are under persistent tension, both Sgo1 and hypophosphorylated sororin are required to maintain sister-chromatid cohesion. This result suggests that, in addition to promoting sororin dephosphorylation, Sgo1 has other roles in cohesion protection.
Figure 5.
Sgo1 prevents Wapl from accessing a functional site on cohesin. (a) Quantification of the percentages of mitotic GFP-sororin 9A-expressing HeLa cells (transfected with the indicated siRNA and arrested in prometaphase with nocodazole or at metaphase with MG132) that had unseparated chromatids (types I/II) and separated chromatids (types III/IV). Error bars, range (n = 2 independent experiments). (b) Quantification of the mitotic indices (defined as the percentage of cells with 4N DNA content and positive MPM2 staining) of HeLa cells transfected with Sgo1 siRNA and the indicated Myc-SA2 plasmids. WT, wild type. Error bars, range (n = 2 independent experiments). The K901E and K949E E950K mutants were tested only once. (c) Model for sororin- and Sgo1-dependent cohesion protection during the cell cycle. Sgo1 protects centromeric cohesion in two ways: enabling sororin dephosphorylation by PP2A and directly shielding SA2–Scc1 from Wapl. Ac, acetyl group.
We hypothesized that the direct competition between Sgo1 and Wapl for cohesin binding might allow Sgo1 to shield cohesin from Wapl and account for the sororin-independent function of Sgo1 in cohesion protection. A strong prediction of this hypothesis was that cohesin containing Wapl-binding-deficient SA2 mutants should alleviate the need for Sgo1 protection. Indeed, overexpression of SA2 K290E, D326K, or K330E mutants that lost Wapl binding in HeLa cells partially rescued the premature sister-chromatid separation (Supplementary Fig. 5a–c) and mitotic arrest (Fig. 5b) caused by Sgo1 depletion. As controls, expression of SA2 mutants that retained partial or full Wapl binding, including Y328A, Y331A, and W334A, failed to bypass Sgo1 requirement in cohesion protection. Expression of other SA2 mutants targeting a conserved patch of residues in the C-half or the Scc1-binding-deficient D793K mutant also had no effect. Finally, overexpression of SA2 D326K or K330E even rescued the gradual loss of cohesion in the presence of prolonged MG132-induced metaphase arrest, a phenotype termed cohesion fatigue42 (Supplementary Fig. 5d). These results strongly suggest that a critical function of Sgo1 in cohesion protection is to directly shield cohesin from Wapl through binding to SA2–Scc1.
DISCUSSION
In this study, we have determined the crystal structure of the SA2–Scc1 cohesin subcomplex, which is the interaction hub for cohesin regulators. Further biochemical and functional analyses have uncovered direct competition between the cohesion protector Sgo1 and the cohesion inhibitor Wapl for cohesin binding, and demonstrated the relevance of this competition in cohesion protection.
Sgo1 forms a homodimer through its N-terminal coiled coil domain that binds PP2A27. One Sgo1 dimer binds to one PP2A complex43. We have shown that the pSgo1 peptide binds with 1:1 stoichiometry to SA2–Scc1. In principle, the two monomers of the Sgo1 dimer can each bind one cohesin complex, thus bridging two cohesin rings. We do not have evidence that this type of Sgo1-dependent cohesin crosslinking occurs in human cells. More importantly, the SA2 D326K and K330E mutants that are defective in Sgo1 binding can bypass the requirement for Sgo1 in cohesion, by virtue of their inability to interact with Wapl. Because these mutants cannot physically link cohesin rings, the putative Sgo1-mediated cohesin crosslinking (even if it does occur) is unlikely to directly contribute to sister-chromatid cohesion.
The results presented herein, along with previously published results, suggest the following model for cohesion establishment and maintenance in human cells (Fig. 5c). During telophase and G1, cohesin is loaded on chromosomes, but undergoes Wapl-dependent dynamic release from chromosomes. This release involves an opening of the cohesin ring at the Smc3–Scc1 juncture44,45, and requires an interaction between Pds5 and the N-terminal region of Wapl (Wapl-N), and interactions between cohesin and the middle region (Wapl-M) and C-terminal domain of Wapl (Wapl-C). The mechanism by which Wapl–Pds5 disrupts the Smc3–Scc1 interface is unknown, but is unlikely due to direct competition for binding38,46. One possibility is that Wapl–Pds5 allosterically stimulates the intrinsic ATPase activity of cohesin, possibly by stabilizing the transition state. ATP hydrolysis transiently disrupts the Smc3–Scc1 interface, and triggers the opening of the cohesin ring and its release from chromatin.
During S phase, Smc3 acetylation enables the binding of sororin to Pds5. Sororin blocks the Pds5–Wapl-N interaction, but does not completely displace Wapl from cohesin21,28, because Wapl-M and Wapl-C can still associate with cohesin. This form of cohesin stably associates with sister chromatids and establishes cohesion. During mitosis, phosphorylation of sororin by mitotic kinases disrupts the sororin–Pds5 interaction21, and Wapl-N reengages Pds5 to trigger cohesin release from chromosome arms.
At centromeres, however, phosphorylated Sgo1 binds SA2–Scc1 to protect cohesin from Wapl in two ways. First, Sgo1 recruits PP2A to cohesin28, which keeps sororin hypophosphorylated and bound to Pds5. Pds5-bound sororin antagonizes Wapl-N. Second, Sgo1 directly competes with Wapl-M for binding to SA2–Scc1. As a result, cohesin bound to both Sgo1 and sororin (indirectly through Pds5) is completely shielded from Wapl28. These two mechanisms collaborate to protect centromeric cohesion to the fullest extent. Strong centromeric cohesion is required to resist the spindle pulling force at sister kinetochores, and enables the generation of kinetochore tension necessary for spindle checkpoint inactivation and accurate chromosome segregation.
Another major function of shugoshin proteins is to protect meiotic cohesin at centromeres from separase-mediated cleavage during meiosis I47,48. One mechanism by which shugoshin protects meiotic cohesin from separase is likely through PP2A-dependent dephosphorylation of Rec8 (the meiotic counterpart of Scc1)43. Human Sgo1 appears to be capable of inhibiting separase-dependent removal of mitotic cohesin from centromeres, when the Sgo1–cohesin interaction is not disrupted properly31,32. Interestingly, the two separase-cleavage sites in human Scc1 flank the central SA2-binding region of Scc1 delineated in our structure49. In the future, it will be interesting to test whether SA2 contributes to Scc1 cleavage by providing a docking site for separase, and if so, whether Sgo1 binding to SA2–Scc1 also shields cohesin from separase.
ONLINE METHODS
Protein expression and purification
The cDNAs of human SA2 (residues 80–1060) and Scc1 (residues 281–420) were separately cloned into the FseI and AscI sites of a modified pFastBac HT vector (Invitrogen). The final constructs encoded an N-terminal His6-Flag tagged SA2 and an N-terminal His6-Strep tagged Scc1, with a tobacco etch virus (TEV) protease cleavage site between the Flag tag and SA2, and a PreScission protease cleavage site between the Strep tag and Scc1. Recombinant SA2 and Scc1 baculoviruses were constructed with the Bac-to-Bac system (Invitrogen) according to manufacturer’s protocols. For large-scale production of recombinant proteins, Hi5 insect cells (Sigma-Aldrich) were infected with both the SA2 and Scc1 baculoviruses, and harvested at about 50 h post-infection. Cells were resuspended in the lysis buffer (50 mM Tris-HCl pH 7.7, 150 mM KCl, 0.1 % [v/v] Triton X-100, and a protease inhibitor cocktail). After sonication and centrifugation, the supernatant was applied onto Ni2+-NTA resin (Qiagen) equilibrated with the lysis buffer and incubated at 4°C overnight. The resin was washed with wash buffer I (50 mM Tris-HCl pH 7.7, 1.2 M KCl, and 10 mM imidazole) and then washed with wash buffer II (20 mM HEPES pH 7.4, 100 mM NaCl, and 20 mM imidazole). The SA2–Scc1 complex was eluted with the elution buffer (20 mM HEPES pH 7.4, 100 mM NaCl, and 50–200 mM imidazole), and incubated with the TEV and PreScission proteases at 4°C overnight. The complex with the tags removed was applied onto a HiTrap Q HP column (GE Healthcare) equilibrated with the QA buffer (50 mM Tris-HCl pH 8.5) using an AKTA chromatography system (GE Healthcare). SA2–Scc1 was eluted with a linear salt gradient from 0 to 600 mM NaCl. SA2–Scc1 was further applied onto a HiLoad 16/60 Superdex 200 prep-grade column (GE Healthcare) equilibrated with the purification buffer (20 mM Tris-HCl pH 7.7, 300 mM NaCl, and 5 mM TCEP). Purified SA2–Scc1 was concentrated to 7 mg/ml, flash frozen in liquid nitrogen, and stored at −80°C for crystallization or binding assays.
The selenomethionine labeled (Se-Met) SA2–Scc1 complex was expressed in Hi5 cells according to manufacture’s protocols. Briefly, Hi5 cells cultured in the ESF921 medium (Expression Systems) were infected with both SA2 and Scc1 baculoviruses. The cells were incubated for 16 h and then supplied with 100 mg/l L-selenomethionine. After an additional 48 h, the cells were harvested, and the Se-Met SA2–Scc1 complex was purified as described above.
Crystallization and data collection
All crystallization experiments were performed at 20°C. Initial screening for the SA2–Scc1–MES complex was carried out by sitting-drop vapor-diffusion with the Phoenix crystallization robot (Art Robbins Instruments), using commercially available screening kits from Hampton Research, Qiagen, and Molecular Dimensions. Conditions were further optimized with the hanging-drop vapor-diffusion method. SA2–Scc1–MES crystals suitable for X-ray diffraction experiments appeared within a week with a reservoir solution consisting of 0.12 M Morpheus Divalents Mix, 0.1 M Morpheus Buffer System 1, and 27–30% (v/v) Morpheus EOD_P8K (Molecular Dimensions). Similarly, the SA2–Scc1 crystals were grown in one week with a reservoir solution consisting of 0.12 M Morpheus Divalents Mix, 0.1 M imidazole-HCl pH 6.5, and 27–30% (v/v) Morpheus EOD_P8K. All crystals were cryo-protected with a reservoir solution including 30% (v/v) Morpheus EOD_P8K before flash-frozen.
All X-ray diffraction data were collected at 100 K at the Structural Biology Center (Beamline 19ID) at Argonne National Laboratory. SA2–Scc1–MES (Native-MES), selenomethionine-labeled SA2–Scc1–MES (SeMet-MES), and selenomethionine-labeled SA2–Scc1 without MES (SeMet) crystals diffracted to 2.95 Å, 2.85 Å, and 3.05 Å, respectively (Table 1). Diffraction data were processed using the HKL software package50.
Structure determination and model refinement
Initial phases for the selenomethionine-labeled SA2–Scc1 (SeMet) were obtained by single-wavelength anomalous dispersion (SAD) using AutoSol in the PHENIX software package51. Model building of the SA2 N-terminal HEAT repeats and Scc1 was done with AutoBuild in PHENIX. Subsequent model building was carried out with the program COOT52, and the structure was refined with PHENIX. Structures of the Native-MES and SeMet-MES complexes were solved by molecular replacement with PHENIX, using the SeMet structure (without MES) as the search model. Statistics for refinement are summarized in Table 1. The final model of native SA2–Scc1–MES complex (Native-MES) contains residues 83–254, 261–438, 455–505, 512–543, 547–748, 752–805, 807–836, 853–933, 936–959, 965–986, 988–990, 993–1035, and 1037–1047 from SA2, residues 321–396 from Scc1, and one MES molecule. The final model of the selenomethionine SA2–Scc1–MES (SeMet-MES) contains residues 83–91, 93–255, 260–438, 455–505, 513–543, 548–748, 753–836, 853–959, 965–991, 994–1035, and 1037–1048 from SA2, residues 321–395 from Scc1, and one MES molecule. The final model of selenomethionine SA2–Scc1 complex (SeMet) contains residues 83–253, 261–438, 456–505, 513–543, 546–674, 677–748, 753–804, 808–836, 852–932, 938–959, 966–986, 988–990, and 993–1049 from SA2, residues 321–394 from Scc1. Remaining residues of SA2 and Scc1 are disordered. All structure drawings in this study were created using Pymol (DeLano Scientific LLC) and depicted the SeMet-MES complex.
In vitro binding assays
The cDNAs encoding full-length human SA2, full-length human Scc1, and the Scc1 fragment (residues 211–420) were cloned into the FseI and AscI sites of a modified pCS2-Myc vector. The SA2 and Scc1 mutants were constructed with the QuikChange Site-directed Mutagenesis Kit (Stratagene). The pCS2-Myc-SA2 (wild-type and mutants) or pCS2-Myc-Scc1 (wild-type and mutants) vectors were added either alone or as a mixture of 3:1 ratio to the TNT Quick Coupled Transcription Translation Systems (Promega), and incubated in the presence of 35S-methionine at 30°C for 90 min to produce 35S-labeled Myc-SA2, Myc-Scc1, or the Myc-SA2–Scc1 complex.
To assay the binding between SA2 and Scc1, GST-SA2 (residues 80–1060) and GST-Scc1 (residues 211–420) were expressed in bacteria and purified with the glutathione-Sepharose 4B resin (GE Healthcare). The glutathione-Sepharose beads bound to GST-SA2 or GST-Scc1 were incubated with 35S-labeled Myc-Scc1 (wild-type and mutants) or Myc-SA2 (wild-type and mutants), respectively, at 4°C overnight, and washed four times with TBS containing 0.05% Tween 20. Beads bound to GST were used as negative controls in both cases. The bound proteins were separated by SDS-PAGE. The gels were stained with Coomassie blue, dried, and analyzed with a phosphorimager (Fujifilm). Intensities of bound proteins were quantified with the program Image J.
To assay the interaction between Sgo1 and SA2–Scc1, synthetic Sgo1 (residues 313–353) and phospho-T346 Sgo1 (pSgo1) peptides (each with an extra C-terminal cysteine) were coupled to beads with the SulfoLink Immobilization Kits and Coupling Resin (Thermo Scientific) according to the manufacturer’s instructions. Recombinant purified SA280–1060–Scc1281–420 or the 35S-labeled (wild-type and mutant) Myc-SA2–Scc1211–420 complexes were incubated with Sgo1- or pSgo1-coupled beads at 4°C overnight, and washed four times with TBS containing 0.05% Tween 20. The bound proteins were separated by SDS-PAGE and stained with Coomassie blue. For the assays with radioactive proteins, the gels were dried and analyzed with a phosphorimager (Fujifilm). Intensities of bound proteins were quantified with the program Image J.
To assay the binding between Wapl and SA2–Scc1, GST-Wapl401–600 and GST-Wapl410–590 (wild-type and mutants) were expressed in bacteria and purified with glutathione-Sepharose 4B beads. Beads bound to GST or GST-Wapl proteins were incubated with recombinant purified SA280–1060–Scc1281–420 or the 35S-labeled wild-type and mutant Myc-SA2–Scc1211–420 complexes, in the presence of increasing concentrations of Sgo1 or pSgo1 peptides. The beads were washed with TBS containing 0.05% Tween 20. The bound proteins were analyzed and quantified as described above.
Isothermal titration calorimetry (ITC)
ITC was performed with a MicroCal iTC200 (GE Healthcare) at 20°C. Calorimetric measurements were performed with purified SA2–Scc1 and synthetic unphosphorylated or phospho-T346 Sgo1 peptides containing residues 313–353. For each titration, 300 μl of 23 μM SA2–Scc1 in a buffer containing 20 mM Tris-HCl pH 7.7, 100 mM NaCl, and 5 mM TCEP was added to the calorimeter cell. The Sgo1 or pSgo1 peptide (350 μM) in the same buffer was injected with 18 portions of 2 μl with an injection syringe. Binding parameters were evaluated using the Origin software package provided with the instrument.
Cell culture, transfection, and synchronization
HeLa Tet-On cells were grown in DMEM (Invitrogen) supplemented with 10% FBS and 2 mM L-glutamine. Plasmid transfection was performed when cells reached a confluency of 50% using the Effectene reagent (Qiagen) according to the manufacturer’s protocols. For siRNA transfection, cells were transfected with Lipofectamine RNAiMAX (Invitrogen) at 20%–40% confluency according to the manufacturer’s protocols, and analyzed at 24–48 h after transfection. The siRNA oligonucleotides targeting human Wapl (siWapl; 5′-CGGACTACCCTTAGCACAA-3′), Sgo1 (siSgo1; 5′-GAGGGGACCCUUUUACAGATT-3′), SA1 (siSA1; 5′-GAAUAGAGAUGUUUCGAAA-3′), and SA2 (siSA2; 5′-CCACUGAUGUCUUACCGAA-3′) were synthesized by Thermo Scientific. The siRNAs were transfected at a final concentration of 5 nM.
For mitotic synchronization, cells were treated with 2 mM thymidine for 16–18 h, released into fresh medium for 9 h, and blocked at mitosis with the addition of 5 μM nocodazole (Sigma) for 2 h. For the cohesin-loading assay, cells were treated with 2mM thymidine for 16–18 h, and released into fresh medium for 11 h before fixation. For the cohesion fatigue assay, cells were released into medium containing 10 μM MG132 for additional 2 h after nocodazole treatment.
For stable cell lines, HeLa Tet-On cells were transfected with pTRE2-GFP vector encoding wild-type human Sgo1. Clones were selected with 200 μg/ml hygromycin B. Inducible expression of GFP-Sgo1 was screened in the absence or presence of 1 μg/ml doxycycline (Invitrogen).
Antibodies, immunoblotting, and immunoprecipitation
The anti-Wapl antibody was generated against a C-terminal fragment of human Wapl (residues 601–1190) as described5. Rabbit polyclonal anti-GFP antibodies were raised against purified recombinant eGFP at Yenzym Antibodies, LLC. The following antibodies were purchased from the indicated commercial sources: anti-Myc (Roche, 11667203001), anti-SA2 (Santa Cruz, Biotechnology, sc-81852), anti-Scc1 (Bethyl Laboratories, A300–080A), and MPM2 (Millipore, 05–368). The Myc and Scc1 antibodies have been validated for immunoblotting and immunoprecipitation applications. The MPM2 antibody has been validated for flow cytometry. The relevant validation information and references can be found at the manufacturers’ websites. The SA2 antibody is validated for immunoblotting in this study (Fig. 2d), and the anti-GFP antibody is validated for both immunoblotting and immunoprecipitation (Supplementary Fig. 2c).
For immunoblotting, cells were lysed in the SDS sample buffer (pH 6.8), sonicated, and boiled. The lysates were separated by SDS-PAGE and blotted with the desired primary antibodies. The primary antibodies in the form of crude sera were used at 1:1000 dilutions whereas purified antibodies were used at a final concentration of 1 μg/ml. Horseradish peroxidase-linked donkey anti-rabbit or sheep anti-mouse IgGs (GE healthcare) were used as secondary antibodies. Immunoblots were developed with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) according to the manufacturer’s protocols. Original images of gels, autoradiographs, and blots in this study can be found in Supplementary Fig. 6.
For immunoprecipitation, the anti-Myc or anti-GFP antibodies were coupled to Affi-Prep Protein A beads (Bio-Rad) at a concentration of 1 mg/ml. Cells were lysed with the lysis buffer containing 25 mM Tris-HCl (pH 7.7), 75 mM NaCl, 0.1% (v/v) Nonidet P-40, 2 mM MgCl2, 10% (v/v) glycerol, 5 mM NaF, 0.3 mM Na3VO4, 10 mM β-glycerophosphate, 1 mM DTT, protease inhibitor mixture (Roche), and 50 units/ml Turbo Nuclease (Accelagen). After a 1-hr incubation on ice and a 10-min incubation at 37°C, all lysates were centrifuged at 4°C at 20,817 g for 20 min. The supernatants were incubated with the desired antibody beads for 3 h at 4°C. The beads were then washed three times with the lysis buffer containing 200 mM NaCl. Proteins bound to beads were dissolved in SDS sample buffer, separated by SDS-PAGE, and blotted with the appropriate antibodies.
Flow cytometry
Cells were harvested with trypsinization and fixed in 70% ice-cold ethanol overnight. After washing with PBS, cells were permeabilized with PBS containing 0.25% Triton X-100 on ice for 5 min. Then, cells were incubated with the MPM2 antibody in PBS containing 1% BSA for 3 h at room temperature followed by an incubation with a fluorescent secondary antibody (Invitrogen) for 30 min. After washing with PBS, cells were resuspended in PBS containing 0.1% Triton X-100, RNase A, and propidium iodide, and then analyzed with a flow cytometer. Data were processed with the program FlowJo.
Immunofluorescence and metaphase spreads
After synchronization, mitotic HeLa Tet-On cells were collected by shake-off. Cells were washed once with PBS, treated with 55 mM KCl hypotonic solution at 37°C for 15 min, and spun onto microscope slides with a Shandon Cytospin centrifuge. Cells on the slides were first permeabilized with the PHEM buffer (25 mM HEPES pH 7.5, 10 mM EGTA pH 8.0, 60 mM PIPES pH 7.0, 2 mM MgCl2) containing 0.3% Triton X-100 for 5 min and then fixed in 4% paraformaldehyde for 10 min. Fixed cells were washed three times with PBS containing 0.1% Triton X-100 for 2 min each time, and incubated with CREST in PBS containing 3% BSA and 0.1% Triton X-100 at 4°C overnight. Cells were then washed three times with PBS containing 0.1% Triton X-100 for 2 min each time, and incubated with fluorescent secondary antibodies (Molecular Probes) in PBS containing 3% BSA and 0.1% Triton X-100 for 1 h at room temperature. Cells were again washed three times with PBS containing 0.1% Triton X-100 and stained with 1 μg/ml DAPI for 2 min. After the final washes, the slides were sealed with nail polish and viewed with a 100X objective on a DeltaVision fluorescence microscope (GE Healthcare). Image processing and quantification were performed with Image J.
For the cohesin-loading assay, cells were plated in four-well chamber slides (LabTeck). Cells were transfected with GFP-SA2 plasmids for 8 h, followed by siSA1 and siSA2 transfection for another 36 h. Cells were first extracted with PHEM buffer containing 0.5% Triton X-100 for 5 min and then fixed in 2% paraformaldehyde for 15 min. After washing with PBS, cells were blocked in PBS containing 2% BSA for 1 h, and incubated with the anti-GFP and anti-tubulin antibodies in PBS containing 0.2% Triton X-100 and 3% BSA overnight at 4°C. After washing three times with PBS containing 0.05% Tween 20, cells were incubated with fluorescent secondary antibodies in PBS containing 0.2% Triton X-100 and 3% BSA for 1 h at room temperature. The cells were again washed three times with PBS containing 0.05% Tween 20 and stained with 1 μg/ml DAPI in PBS for 2 min. After final washes, slides were mounted and viewed with a 100X objective on a DeltaVision deconvolution fluorescence microscope. A series of z-stack images were captured at 0.2 μm intervals, deconvolved, and projected. Image processing and quantification were performed with Image J.
Supplementary Material
Acknowledgments
We thank C. Brautigam for assistance with isothermal titration calorimetry. Use of Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US Department of Energy under contract DE-AC02-06CH11357. H.Y. is supported as an investigator with the Howard Hughes Medical Institute. This work is supported by grants from the Cancer Prevention and Research Institute of Texas (RP110465-P3 to H.Y.) and the Welch Foundation (I-1441 to H.Y.).
Footnotes
Accession codes. The coordinates and structure factors of SeMet-SA2–Scc1, SeMet-SA2–Scc1–MES, and native SA2–Scc1–MES complexes have been deposited to the Protein Data Bank with the accession codes 4PJU, 4PJW, and 4PK7, respectively.
AUTHOR CONTRIBUTIONS
K.H. crystallized SA2–Scc1 and determined its structure with the help of Z.C. and D.R.T., and performed the in vitro binding assays with SA2 and Scc1 mutants. G.Z. performed the functional cellular assays. Q.Q. performed the cohesin binding assay that showed competition between Sgo1 and Wapl, and the in vitro binding assay between SA2–Scc1 and Wapl. Z.O. first observed competition between Sgo1 and Wapl in cohesin binding. H.L. performed the metaphase spread assay of sororin-9A-expressing cells depleted of Sgo1. H.Y. supervised the project, and wrote the paper with input from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 2.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 3.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–29. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 4.Wu N, Yu H. The Smc complexes in DNA damage response. Cell Biosci. 2012;2:5. doi: 10.1186/2045-3701-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu N, et al. Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 2012;26:1473–85. doi: 10.1101/gad.193615.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–97. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Solomon DA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–43. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remeseiro S, Cuadrado A, Losada A. Cohesin in development and disease. Development. 2013;140:3715–8. doi: 10.1242/dev.090605. [DOI] [PubMed] [Google Scholar]
- 9.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–7. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 10.Sherwood R, Takahashi TS, Jallepalli PV. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 2010;24:2723–31. doi: 10.1101/gad.1976710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shintomi K, Hirano T. Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma. 2010;119:459–67. doi: 10.1007/s00412-010-0271-z. [DOI] [PubMed] [Google Scholar]
- 12.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–54. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 14.Watrin E, et al. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–74. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 15.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–41. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 16.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–41. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 17.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–17. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kueng S, et al. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–67. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 19.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci U S A. 2010;107:20364–9. doi: 10.1073/pnas.1011069107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyama T, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–49. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 22.Hauf S, et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreier MR, Bekier ME, 2nd, Taylor WR. Regulation of sororin by Cdk1-mediated phosphorylation. J Cell Sci. 2011;124:2976–87. doi: 10.1242/jcs.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishiyama T, Sykora MM, Huis In ‘t Veld PJ, Mechtler K, Peters JM. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci U S A. 2013;110:13404–9. doi: 10.1073/pnas.1305020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitajima TS, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 26.Riedel CG, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 27.Tang Z, et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–85. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Rankin S, Yu H. Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nat Cell Biol. 2013;15:40–9. doi: 10.1038/ncb2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci. 2013;38:302–11. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, et al. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Jia L, Yu H. Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr Biol. 2013;23:1927–33. doi: 10.1016/j.cub.2013.07.078. [DOI] [PubMed] [Google Scholar]
- 33.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16:387–97. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 34.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–71. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009;23:2224–36. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanovic T, Choi JL, Whelan SP, van Oijen AM, Harrison SC. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. eLife. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang Z, et al. Structure of the human cohesin inhibitor Wapl. Proc Natl Acad Sci U S A. 2013;110:11355–60. doi: 10.1073/pnas.1304594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tedeschi A, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–8. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haarhuis JH, et al. WAPL-mediated removal of cohesin protects against segregation errors and aneuploidy. Curr Biol. 2013;23:2071–7. doi: 10.1016/j.cub.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Yu H. Chromosome biology: Wapl spreads its wings. Curr Biol. 2013;23:R923–5. doi: 10.1016/j.cub.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 42.Daum JR, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21:1018–24. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z, et al. Structure and function of the PP2A-shugoshin interaction. Mol Cell. 2009;35:426–41. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3-Scc1 gate. EMBO J. 2013;32:666–76. doi: 10.1038/emboj.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichinger CS, Kurze A, Oliveira RA, Nasmyth K. Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis. EMBO J. 2013;32:656–65. doi: 10.1038/emboj.2012.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee A, Zakian S, Hu XW, Singleton MR. Structural insights into the regulation of cohesion establishment by Wpl1. EMBO J. 2013;32:677–87. doi: 10.1038/emboj.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–7. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 48.Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–72. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–3. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.