Abstract
We propose a nonparametric approach for cumulative incidence estimation when causes of failure are unknown or missing for some subjects. Under the missing at random assumption, we estimate the cumulative incidence function using multiple imputation methods. We develop asymptotic theory for the cumulative incidence estimators obtained from multiple imputation methods. We also discuss how to construct confidence intervals for the cumulative incidence function and perform a test for comparing the cumulative incidence functions in two samples with missing cause of failure. Through simulation studies, we show that the proposed methods perform well. The methods are illustrated with data from a randomized clinical trial in early stage breast cancer.
Keywords: competing risks, cumulative incidence function, missing at random, multiple imputation, two-sample tests
1. Introduction
In studies of time to event data, competing risks, where individuals may fail from one of several mutually exclusive causes, are frequently present. For example, after cancer diagnosis and treatment, patients may die of cancer but also may die of other causes prior to death from cancer. In such a case, medical investigators may be interested in predicting the probability of dying of cancer by a particular time t in the presence of death from other causes. The probability is defined by the cumulative incidence function
| (1) |
where T is the time to failure and ε ∈ {1, 2, … K} is the cause of failure. This quantity is useful in the analysis of competing risks in the sense that it reflects the risk of the cause of interest without ignoring the presence of other competing events. Another useful quantity in the analysis of competing risks is the cause-specific hazard function defined by
which is the instantaneous rate of cause k at time t in the presence of other competing risks. The cumulative incidence function given in (1) can be expressed as a function of all cause-specific hazards as
where is the overall survival function and is the cumulative cause-specific hazard function for cause k.
In competing risk studies, it may be known that a failure has occurred but the failure type may be unknown. For example in the cancer example, the cause of death may be unknown or missing for some patients due to incomplete reporting or documentation, or other reasons. The simplest way to deal with missing causes of failure may be to exclude subjects with missing causes from the analysis. However, such an approach will result in information loss and may yield biased results. Goetghebeur and Ryan [1] and Lu and Tsiatis [2] proposed methods for estimating regression parameters under the cause-specific proportional hazards model for competing risk data with missing cause of failure. Gao and Tsiatis [3] and Lu and Liang [4] proposed inference procedures for estimation of regression parameters under a linear transformation model and an additive hazards model for competing risks with missing cause of failure, respectively. Bakoyannis et al. [5] used multiple imputation to estimate regression parameters for the proportional subdistribution hazards model. Moreno-Betancur and Latouche [6] proposed a general framework for regression modeling of the cumulative incidence function with missing causes of failure using pseudo-values.
However, methods for estimating the cumulative incidence function in the presence of missing cause of failure have not been widely studied. Recently, Lee et al. [7] proposed multiple imputation methods for estimating the cumulative incidence function with missing cause of failure under the cause-specific proportional hazards model. Nicolaie et al. [8] used vertical modeling to estimate the cumulative incidence function with missing cause of failure. In this paper, we propose nonparametric inferences for the cumulative incidence function with missing cause of failure using methods in Lin [9] and Lu and Tsiatis [2]. Assuming missingness is random conditional on the observed data, we impute missing cause of failure multiple times and estimate the cumulative incidence function by averaging nonparametric cumulative incidence estimators obtained from each of several imputed data sets. We prove the asymptotic normality of the cumulative incidence estimators obtained from multiple imputation methods and derive a consistent variance estimator for the cumulative incidence estimators. We describe how to construct confidence intervals for the cumulative incidence function and perform a test for comparing two cumulative incidence functions in the presence of missing cause of failure. The performance of the proposed methods are extensively evaluated by simulation studies. Finally, we apply the methods to study the influence of tamoxifen treatment on mortality from breast cancer, using data from a randomized clinical trial in early stage breast cancer.
2. Inference procedures
For simplicity, we consider two competing events, ε ∈ {1, 2}. Define X = min(T, C), where T and C are the failure time and censoring time, respectively. We assume that the censoring time C is independent of (T, ε). Let δ = I(T ≤ C)ε, where I(·) is an indicator function. When causes of failure are known for all subjects, the observed data consist of (Xi, δi) (i = 1, …, n).
A nonparametric estimator of the cumulative incidence function (called the Aalen-Johansen estimator [10]) is given by
| (2) |
where Ŝ(t−) is the left-continuous Kaplan-Meier estimator [11] and Λ̂k(t) is the Nelson-Aalen estimator [12] for Λk(t). Aalen [13] first established the uniform consistency and weak convergence of the Aalen-Johansen estimator using a non-homogeneous Markov process formulation. Recently, variance estimation of the Aalen-Johansen estimator in small samples has caused some discussion [14, 15, 16]. Aalen’s variance formula [13] is generally smaller than the so-called Greenwood estimator in small samples, resulting in narrow confidence intervals and inflated type I error rates (more power). Building off Aalen’s work [13], Lin [9] presented a martingale-based estimator for the asymptotic variance of the Aalen-Johansen estimator. Lee and Fine [17] note that Lin’s martingale-based variance estimator equals the Greenwood type estimator given in Equation (7) of Allignol et al. [18] in the absence of ties. As an alternative to Aalen’s work [13], we use Lin’s result for variance estimation of the Aalen-Johansen estimator.
Define the counting process notation Nki(t) = I(Xi ≤ t, εi = k), Yi(t) = I(Xi ≥ t), , and . Let . Lin (1997) showed that
where
by the martingale theory, converges weakly to a zero-mean Gaussian process, and the variance can be consistently estimated by
When cause of failure is missing for some patients, the cumulative incidence estimator and its variance may be obtained from the above after excluding those with the missing cause information. However, such an approach may lead to biased inferences. We propose multiple imputation methods (referred to as improper multiple imputation) for nonparametric estimation of the cumulative incidence function with missing cause of failure.
2.1. Multiple imputation method
Lu and Tsiatis [2] proposed multiple imputation methods to estimate the proportional hazards regression parameters for competing risks data with missing cause of failure. In this section, we propose a nonparametric approach for estimating cumulative incidence with missing cause of failure using imputation methods described in Lu and Tsiatis [2].
Define the missingness indicator Ri, that is, Ri = 1 if the cause of failure is known and Ri = 0 if the cause of failure is missing or unknown. When the failure time is censored, we set Ri = 1 since this is known and it is not related to missing cause of failure. The observed data consist of (Ri, Xi, Riδi) (i = 1, …, n). We assume that the cause of failure is missing at random [19], which implies that given δi(> 0) and Xi, the probability that the cause of failure is missing depends only on the observed information Xi, not on the unobserved δi. That is,
Let D1i = I(δi = 1) and π(Xi) = Pr(δi = 1 | Ri = 0, δi > 0, Xi). When the cause of failure is missing, we impute missing D1i values from the conditional distribution of D1i given the observed data as in Lu and Tsiatis [2]. D1i has a Bernoulli distribution with success probability π(Xi) which can be specified by a parametric model with an unknown parameter γ. It is natural to fit the logistic regression model for π(Xi), logitπ(Xi, γ) = γ1 + γ2Xi, where γ = (γ1, γ2). Let γ0 = (γ01, γ02) be the true value of γ. The assumption that the cause of failure is missing at random implies
Thus we can infer π(Xi, γ0) from the complete cases for whom (Ri = 1, δi > 0). Maximizing the likelihood function for the complete cases
yields the maximum likelihood estimator γ̂ of γ. If the model π(X, γ) for π(X) is correctly specified, γ̂ is consistent and asymptotically normal under the regularity conditions. The asymptotic normality follows from the influence function expansion given in Appendix A. Using the maximum likelihood estimator γ̂, the missing value can be imputed as
where D1ij(Ri, γ̂) is the imputed value of D1i from the jth imputed data set, j = 1, …, m, and is randomly selected to be 1 or 0 from a Bernoulli distribution with success probability π(Xi, γ̂) when Ri = 0. Let F̂kj(t) be a nonparametric estimator, estimated as in (2), of cumulative incidence for cause k from the jth imputed data set. The multiple imputation estimator for Fk(t) is given by averaging nonparametric estimators obtained from each of m imputed data sets as
As noted in Lu and Tsiatis [2], Rubin [20]’s variance calculation is not applicable here; since we generate imputations from the conditional distribution of missing data (D1i) given the observed data evaluated at the fixed γ̂ across imputations, our imputation is not proper in the sense of Rubin [20]. Wang and Robins [21] indicates that under these conditions, Rubin’s variance will yield an inconsistent estimator for the sampling variance. We derive variance estimators for F̂kj(t) and F̂k(t) directly as in Lu and Tsiatis [2] (see Appendix A).
To establish the asymptotic properties for F̂1j(t) and F̂1(t), we assume that the parametric model for π(X) is correctly specified. Using the methods described in Lu and Tsiatis [2] or Tsiatis et al. [22], we show that U1j(t) = n1/2{F̂1j(t) − F1(t)} and U1(t) = n1/2{F̂1(t) − F1(t)} are asymptotically equivalent to (A.3) and (A.6) which converge weakly to mean zero Gaussian processes with respective variances V1j(t) and V1(t) given by equations (A.4) and (A.7) in Appendix A. The variances can be consistently estimated by V̂1j(t) and V̂1(t) in equations (A.5) and (A.8).
Note that we imputed missing causes of failure from the conditional distribution of missing data based on the maximum likelihood estimation of γ. The second term of V1j(t) in (A.4) accounts the variability of γ̂ from single imputation. When there are no missing causes of failure, the second term vanishes since and our inference procedures reduce to those in Lin [9]. Equation (A.7) shows that V1(t) ≤ V1j(t) for all m and V1(t) is decreased as the number of imputations m is increased, indicating an improvement in efficiency from multiple imputation over single imputation. Although more imputations provide more efficient estimators, the number of imputations should be determined based on the magnitude of V1j(t) and the second term in equation (A.7). In practice, a few imputations (such as 5–10) would be sufficient to achieve good efficiency.
2.2. Confidence intervals
Pointwise (1 − α) confidence intervals for F1(t) can be constructed based on a transformation of F1(t) to let F1 be bounded by 0 and 1 and improve the coverage probability. Denote K(t) = n1/2ϕ(t) [g{F̂1(t)} − g{F1(t)}], where g is a known function with non-zero continuous derivative g′ and ϕ is a weight function which converges to a non-negative bounded function. By the functional delta method, the process K(t) is asymptotically equivalent to ϕ(t)g′{F̂1(t)}U1(t), where U1(t) = n1/2{F̂1(t) − F1(t)}. Pointwise (1 − α) confidence intervals for F1(t) are given by
where zα/2 is an upper α/2 percentile of the standard normal distribution. With g(x) = log{− log(x)} and ϕ(x) = F̂1(t) log{F̂1(t)}V̂1(t)−1/2, pointwise (1 − α) confidence intervals for F1(t) are given by
| (3) |
2.3. Two-sample tests
We consider a test which compares the cumulative incidence functions in two samples. Let be the cumulative incidence function for cause 1 in group l (l = 1, 2). With two competing risk samples, we are interested in testing the null hypothesis for all t ≤ τ, where τ is the observed largest time point. Gray [23] proposed a test comparing weighted averages of the subdistribution hazards in several groups. Pepe [24] proposed a test based on the integrated weighted difference between the cumulative incidence estimates in two samples. Lin [9] proposed a Kolmogorov-Smirnov type statistic which compares the maximum difference between the cumulative incidence estimates in two samples. Pepe’s test and Lin’s test directly compare the cumulative incidence functions in two samples while Gray’s test compares subdistribution hazards in several groups. Pepe’s test is sensitive to stochastic ordering alternatives with for some t and can be used as an alternative to Gray’s test when the cumulative incidence functions cross [24, 25]. Lin’s test uses the Kolmogorov-Smirnov type statistic which is rank-based thus it may share some shortcomings of the rank statistic. As noted in Pepe and Fleming [26], the Kolmogorov-Smirnov type statistic may be sensitive to a large difference over a short period of time but can be very insensitive to a moderate difference over a long period of time. The latter case is of more interest in practice. Among the three tests, we focus on Pepe’s test to directly compare the cumulative incidence functions in two samples.
Let be the Aalen-Johansen estimator of in group l (l = 1, 2). Pepe [24] developed a test statistic for comparing two cumulative incidence functions, where
is a variance estimator of Q and nl is the number of subjects in group l. Here, W(·) is a data-dependent weight function used to ensure stability of a test statistic under the null hypothesis for all t ≤ τ. In practice, the weight function should be chosen based on alternative hypotheses and the choice of the weight function is analogous to that for weighted Kaplan-Meier statistics [26, 27]. A decreasing (increasing) weight function gives less weight to differences between two cumulative incidence estimates over later (early) time periods. Under the null hypothesis for all t ≤ τ, the test statistic has an asymptotic standard normal distribution. Pepe [24] notes that method of moment type estimator tends to be too small and underestimates the variance, resulting in larger type I errors. As an alternative to method of moment type estimator , Bajorunaite and Klein [28] proposed a martingale-based estimator for the variance of the statistic Q. Through simulation studies, they demonstrated that the method of moment type estimator underestimated the true variance and Pepe’s test with this variance estimator had larger type I errors, especially when the sample size was small, while the test with the martingale-based variance estimator performed well. Hence, we use Bajorunaite and Klein’s result for variance estimation of the statistic Q.
In the presence of missing cause of failure, we modify Pepe’s test statistic by using an imputation estimator for . Let be a nonparametric estimator of the cumulative incidence function for cause 1 from the jth imputed data set in group l and be the multiple imputation estimator obtained by averaging ’s from m imputed data sets in group l. To simplify the presentation, we let W(t) = 1. Then the statistic Q can be defined by
for single imputation and
for multiple imputation. To establish the asymptotic properties for QS and QM, we assume that a parametric model for π(X) is correctly specified. We derive variance estimators for QS and QM in Appendix B. Under the null hypothesis for all t ≤ τ, we show that QS and QM have asymptotic normal distributions with respective variances
and
where and are given in (B.2) and (B.5) in Appendix B. The variances can be consistently estimated by replacing and with their estimators and given in (B.3) and (B.6). Note that when there are no missing causes of failure, our test statistic reduce to Pepe’s [24] test statistic with the martingale-based variance estimator proposed by Bajorunaite and Klein [28] since in (B.1).
3. Numerical studies
Simulation studies were conducted to evaluate the performance of the proposed estimators for the cumulative incidence function when the cause of failure is missing. Cumulative incidence estimators and corresponding variance estimators were examined under various missingness mechanisms, followed by evaluations of performance for the two-sample tests.
3.1. Performance under correct imputation model
The cumulative incidence functions for cause 1 and 2 are assumed to be F1(t) = π1{1 − exp(−v1tθ1)} and F2(t) = π2{1 − exp(−v2tθ2)} with (π1, π2, v1, v2, θ1, θ2) = (2/3, 1/3, 1, 0.8, 1, 1) as in configuration I described in Section 3.1 of Peng and Fine [25]. The type 1 and 2 failures were generated from a Bernoulli distribution with success probability Pr(ε = 1) = π1. The failure times (T) were generated from the conditional distribution of T given ε = k, Pr(T ≤ t | ε = k) = Fk(t)/Pr(ε = k) for k = 1, 2. The censoring times (C) were generated from a uniform distribution U(0, 7.2). The observed times (X) were obtained by min(T, C) and the range of X was 4.568 on average. Under this setting, on average, we had 57% failures from cause 1, 28% failures from cause 2, and 15% censoring. We generated the missingness indicator R from logitPr(R = 0 | δ > 0, X) = η1 + η2X. With different choices of η’s, we generated 20, 30, and 40% missing causes of failure. Following Rubin’s taxonomy of missingness [19], given δ > 0 and X, if the probability that the cause of failure is missing does not depend on the unobserved cause of failure δ, the missingness mechanism is called missing at random (MAR), otherwise missing not at random (MNAR). If the missingness probability is constant, the missingness mechanism is called missing completely at random (MCAR). In Table 1, the first scenario indicates MCAR case and the second to fifth scenarios correspond to MAR case. Note that from the missing at random assumption, π(X) = Pr(δ = 1 | R = 0, δ > 0, X) = Pr(δ = 1 | δ > 0, X). Thus π(X) is related to the ratio of the cause-specific hazards for cause 1 and 2, that is, λ1(x)/λ2(x) = π(x)/{1 − π(x)}. By this relationship, the true model for π(X) is logitπ(X) = log(5/2) − 0.2X since for k = 1, 2. We fitted a correctly specified logistic regression model logitπ(X, γ) = γ1 + γ2X. We conducted 1000 simulations with sample sizes of 100 and 300. Table 1 shows the true value of F1(0.7), bias of F̂1(0.7), empirical variance (Var(F̂1)), averages of the variance estimate (E(V̂1)), mean square error (MSE), and empirical coverage probabilities (CP) for 95% confidence intervals given in (3) from single imputation (m = 1), 10 imputations (m = 10), and the complete case analysis (CC) obtained by excluding subjects with missing cause of failure. Under the correct imputation model, the imputation estimate shows that the bias is small and the variance estimate agrees with the empirical variance. The empirical coverage probabilities are close to the nominal level. Multiple imputation estimates have slightly smaller variance estimates than single imputation estimates and have the smallest MSEs. However, the complete case estimates result in larger biases and MSEs, and lower coverage probabilities compared to the imputation estimates. Supplemental analyses (see Figures 1–3 in Supplementary Materials) show that the complete case analysis can either underestimate or overestimate cumulative incidence, depending on whether deaths with missing cause occur early or late in follow-up. In either case, the imputation method performs well.
Table 1.
Simulation results under the correct imputation model.
| n | % missing | Method | F1(t = 0.7) = 0.3356 | ||||
|---|---|---|---|---|---|---|---|
| Bias(F̂1) | Var(F̂1) | E(V̂1) | MSE | CP | |||
| 100 | 20% | m=1 | −0.00142 | 0.00264 | 0.00264 | 0.00264 | 0.949 |
| (η1, η2) | m=10 | −0.00141 | 0.00248 | 0.00251 | 0.00248 | 0.951 | |
| = (−1.38, 0) | CC | −0.01065 | 0.00274 | 0.00273 | 0.00286 | 0.944 | |
| m*=10 | −0.00162 | 0.00242 | 0.00256 | 0.00243 | 0.953 | ||
| 30% | m=1 | −0.00043 | 0.00313 | 0.00311 | 0.00313 | 0.947 | |
| (η1, η2) | m=10 | −0.00067 | 0.00279 | 0.00286 | 0.00279 | 0.953 | |
| = (−1.38, 0.56) | CC | −0.06251 | 0.00290 | 0.00278 | 0.00681 | 0.770 | |
| m*=10 | −0.00167 | 0.00280 | 0.00296 | 0.00280 | 0.961 | ||
| 40% | m=1 | −0.00166 | 0.00340 | 0.00323 | 0.00340 | 0.949 | |
| (η1, η2) | m=10 | −0.00167 | 0.00294 | 0.00295 | 0.00294 | 0.950 | |
| = (−1.38, 1.1) | CC | −0.05030 | 0.00333 | 0.00322 | 0.00586 | 0.837 | |
| m*=10 | −0.00268 | 0.00296 | 0.00307 | 0.00297 | 0.954 | ||
| 30% | m=1 | 0.00100 | 0.00293 | 0.00273 | 0.00293 | 0.941 | |
| (η1, η2) | m=10 | 0.00183 | 0.00264 | 0.00259 | 0.00264 | 0.945 | |
| = (−0.1, −1) | CC | 0.01730 | 0.00337 | 0.00319 | 0.00367 | 0.936 | |
| m*=10 | 0.00122 | 0.00261 | 0.00262 | 0.00261 | 0.948 | ||
| 40% | m=1 | −0.00052 | 0.00308 | 0.00281 | 0.00308 | 0.942 | |
| (η1, η2) | m=10 | −0.00028 | 0.00283 | 0.00264 | 0.00283 | 0.946 | |
| = (−0.1, −0.36) | CC | 0.04804 | 0.00394 | 0.00377 | 0.00625 | 0.881 | |
| m*=10 | −0.00075 | 0.00285 | 0.00266 | 0.00285 | 0.949 | ||
|
| |||||||
| 300 | 20% | m=1 | −0.00073 | 0.00091 | 0.00089 | 0.00091 | 0.958 |
| (η1, η2) | m=10 | −0.00074 | 0.00082 | 0.00085 | 0.00082 | 0.960 | |
| = (−1.38, 0) | CC | −0.00985 | 0.00093 | 0.00093 | 0.00102 | 0.933 | |
| m*=10 | −0.00078 | 0.00082 | 0.00086 | 0.00082 | 0.961 | ||
| 30% | m=1 | 0.00090 | 0.00102 | 0.00105 | 0.00102 | 0.948 | |
| (η1, η2) | m=10 | 0.00070 | 0.00090 | 0.00096 | 0.00090 | 0.952 | |
| = (−1.38, 0.56) | CC | −0.06132 | 0.00094 | 0.00094 | 0.00470 | 0.482 | |
| m*=10 | 0.00037 | 0.00091 | 0.00100 | 0.00091 | 0.953 | ||
| 40% | m=1 | −0.00079 | 0.00106 | 0.00110 | 0.00106 | 0.949 | |
| (η1, η2) | m=10 | −0.00068 | 0.00092 | 0.00100 | 0.00092 | 0.959 | |
| = (−1.38, 1.1) | CC | −0.04953 | 0.00107 | 0.00110 | 0.00352 | 0.686 | |
| m*=10 | −0.00096 | 0.00093 | 0.00105 | 0.00093 | 0.965 | ||
| 30% | m=1 | −0.00003 | 0.00095 | 0.00092 | 0.00095 | 0.950 | |
| (η1, η2) | m=10 | 0.00015 | 0.00089 | 0.00087 | 0.00089 | 0.956 | |
| = (−0.1, −1) | CC | 0.01678 | 0.00115 | 0.00108 | 0.00143 | 0.926 | |
| m*=10 | 0.00009 | 0.00088 | 0.00089 | 0.00088 | 0.958 | ||
| 40% | m=1 | −0.00011 | 0.00099 | 0.00096 | 0.00099 | 0.939 | |
| (η1, η2) | m=10 | 0.00018 | 0.00091 | 0.00090 | 0.00091 | 0.949 | |
| = (−0.1, −0.36) | CC | 0.05030 | 0.00140 | 0.00129 | 0.00393 | 0.717 | |
| m*=10 | 0.00003 | 0.00091 | 0.00091 | 0.00091 | 0.948 | ||
m*=10, 10 proper imputations based on methods of [5].
To deal with missing cause of failure, Bakoyannis et al. [5] considered Rubin’s multiple imputation procedure under the MAR assumption, which is referred to as proper imputation. Their imputation methods can be applied to nonparametric estimation of the cumulative incidence function with missing cause of failure by using proper multiple imputation to impute missing cause of failure. The cumulative incidence function F1(t) can be estimated by averaging nonparametric estimators obtained from each of multiply imputed data sets. Its variance can be estimated by Rubin’s variance formula , where the within imputation variance Wvar is the mean of the variance estimates across m imputations and the between imputation variance Bvar is the sample variance of the m cumulative incidence estimates. Pointwise 95% confidence intervals for F1(t) can be computed by using (3) with replacing z0.025 by , where is the upper 2.5th percentile of a t-distribution with ψ degrees of freedom, ψ = (m − 1)[1 + Wvar/{(1 + m−1)Bvar}]2. We compared our imputation procedure (referred to as improper imputation) with that of Bakoyannis et al. [5] in the case of nonparametric estimation of the cumulative incidence function with missing cause of failure. As an imputation model for methods of Bakoyannis et al. [5], we fitted a correctly specified logistic regression model logitπ(X, γ) = γ1 + γ2X. As shown in Table 1, under the correct imputation model, the multiple imputation estimates from 10 proper imputations (m* = 10) show that the biases are small and the variance estimates agree with the empirical variance. The empirical coverage probabilities are close to the nominal level. However, the variance estimates from our imputation methods are smaller than those from Bakoyannis et al. [5]. This is because we derive variance estimators for imputation estimators directly. The MSEs of both methods are similar. This is because the proper multiple imputation estimators are consistent and unbiased, and its variances can be correctly estimated by Rubin’s variance formula when the imputation and analysis models agree (i.e., congenial as defined in [29]).
3.2. Performance under misspecified imputation model
Our imputation estimators are obtained based on the maximum likelihood estimation of γ from a parametric model specified for π(X). Therefore, the estimators may be biased if the parametric model is misspecified [30]. To justify the robustness of the imputation estimators, we fitted an incorrectly specified logistic regression model logitπ(X, γ) = γ1 + γ2 log(1 + exp(−X)), where we generated data from the same scenario as before. We conducted 1000 simulations with sample sizes of 100 and 300. In Table 2, the biases, variance estimates, and MSEs of the imputation estimates are small even if the parametric model is misspecified. The variance estimates agree with the empirical variances, and the empirical coverage probabilities of 95% confidence intervals are close to the nominal level. Supplemental analyses (see Supplemental Figures 4–6) confirm that the imputation method performs well under a misspecified parametric model.
Table 2.
Simulation results under the incorrect imputation model.
| n | % missing | Method | F1(t = 0.7) = 0.3356 | ||||
|---|---|---|---|---|---|---|---|
| Bias(F̂1) | Var(F̂1) | E(V̂1) | MSE | CP | |||
| 100 | 20% | m=1 | −0.00041 | 0.00276 | 0.00266 | 0.00276 | 0.945 |
| (η1, η2) | m=10 | −0.00048 | 0.00258 | 0.00254 | 0.00258 | 0.945 | |
| = (−1.38, 0) | CC | −0.00895 | 0.00290 | 0.00273 | 0.00298 | 0.935 | |
| m*=10 | −0.00096 | 0.00254 | 0.00256 | 0.00255 | 0.952 | ||
| 30% | m=1 | 0.00071 | 0.00333 | 0.00322 | 0.00333 | 0.950 | |
| (η1, η2) | m=10 | 0.00027 | 0.00305 | 0.00298 | 0.00305 | 0.953 | |
| = (−1.38, 0.56) | CC | −0.06176 | 0.00300 | 0.00277 | 0.00682 | 0.759 | |
| m*=10 | −0.00125 | 0.00305 | 0.00301 | 0.00305 | 0.953 | ||
| 40% | m=1 | 0.00193 | 0.00368 | 0.00336 | 0.00368 | 0.954 | |
| (η1, η2) | m=10 | 0.00137 | 0.00322 | 0.00309 | 0.00322 | 0.955 | |
| = (−1.38, 1.1) | CC | −0.04831 | 0.00350 | 0.00323 | 0.00583 | 0.846 | |
| m*=10 | −0.00002 | 0.00319 | 0.00310 | 0.00319 | 0.955 | ||
| 30% | m=1 | 0.00098 | 0.00283 | 0.00274 | 0.00284 | 0.955 | |
| (η1, η2) | m=10 | 0.00156 | 0.00263 | 0.00259 | 0.00263 | 0.956 | |
| = (−0.1, −1) | CC | 0.01797 | 0.00331 | 0.00318 | 0.00363 | 0.942 | |
| m*=10 | 0.00089 | 0.00262 | 0.00260 | 0.00262 | 0.954 | ||
| 40% | m=1 | 0.00160 | 0.00318 | 0.00281 | 0.00318 | 0.942 | |
| (η1, η2) | m=10 | 0.00061 | 0.00288 | 0.00264 | 0.00288 | 0.946 | |
| = (−0.1, −0.36) | CC | 0.05067 | 0.00399 | 0.00378 | 0.00655 | 0.882 | |
| m*=10 | 0.00008 | 0.00289 | 0.00267 | 0.00289 | 0.943 | ||
|
| |||||||
| 300 | 20% | m=1 | −0.00025 | 0.00085 | 0.00091 | 0.00085 | 0.959 |
| (η1, η2) | m=10 | 0.000002 | 0.00081 | 0.00087 | 0.00081 | 0.959 | |
| = (−1.38, 0) | CC | −0.00943 | 0.00087 | 0.00093 | 0.00096 | 0.942 | |
| m*=10 | 0.00002 | 0.00081 | 0.00087 | 0.00081 | 0.960 | ||
| 30% | m=1 | −0.00054 | 0.00111 | 0.00110 | 0.00111 | 0.940 | |
| (η1, η2) | m=10 | −0.00107 | 0.00098 | 0.00101 | 0.00098 | 0.950 | |
| = (−1.38, 0.56) | CC | −0.06315 | 0.00099 | 0.00094 | 0.00498 | 0.472 | |
| m*=10 | −0.00156 | 0.00098 | 0.00102 | 0.00098 | 0.945 | ||
| 40% | m=1 | 0.00135 | 0.00123 | 0.00115 | 0.00123 | 0.935 | |
| (η1, η2) | m=10 | 0.00131 | 0.00103 | 0.00105 | 0.00103 | 0.948 | |
| = (−1.38, 1.1) | CC | −0.04836 | 0.00113 | 0.00110 | 0.00347 | 0.702 | |
| m*=10 | 0.00040 | 0.00103 | 0.00106 | 0.00103 | 0.950 | ||
| 30% | m=1 | −0.00139 | 0.00099 | 0.00093 | 0.00099 | 0.944 | |
| (η1, η2) | m=10 | −0.00143 | 0.00091 | 0.00088 | 0.00091 | 0.944 | |
| = (−0.1, −1) | CC | 0.01495 | 0.00117 | 0.00108 | 0.00139 | 0.923 | |
| m*=10 | −0.00176 | 0.00089 | 0.00088 | 0.00089 | 0.944 | ||
| 40% | m=1 | 0.00061 | 0.00098 | 0.00095 | 0.00098 | 0.955 | |
| (η1, η2) | m=10 | 0.00075 | 0.00091 | 0.00090 | 0.00091 | 0.952 | |
| = (−0.1, −0.36) | CC | 0.05120 | 0.00140 | 0.00129 | 0.00402 | 0.710 | |
| m*=10 | 0.00048 | 0.00091 | 0.00091 | 0.00091 | 0.952 | ||
m*=10, 10 proper imputations based on methods of [5].
Table 2 shows results from proper multiple imputation of Bakoyannis et al. [5] under the misspecified imputation model logitπ(X, γ) = γ1 + γ2 log(1 + exp(−X)), where we did not use a non-parametric bootstrap method as suggested in [5] to avoid biases in the variance estimates obtained by Rubin’s variance formula under misspecification of an imputation model: because we found that the use of the bootstrap method for variance estimation was not necessary. The multiple imputation estimates show that the biases and variance estimates are small even if the imputation model is misspecified. The variance estimates agree with the empirical variances, and the empirical coverage probabilities for 95% confidence intervals are close to the nominal level. The variance estimates from our improper multiple imputation methods are slightly smaller than those from [5]. The MSEs of both approaches are similar.
3.3. Additional evaluations for two-sample tests
Additional numerical studies were conducted to evaluate the performance of the proposed test in two samples with missing cause of failure. As described in Section 3.1 of Peng and Fine [25], we considered configurations II, III, and IV along with configuration I; (π1, π2, v1, v2, θ1, θ2) = (2/3, 1/3, 1, 1.2, 1, 1) in configuration II, (2/3, 1/3, 1, 1.2, 0.5, 1) in configuration III, and (1/2, 1/2, 0.8, 1.2, 1, 1) in configuration IV. We generated the sample indicator Z from a Bernoulli distribution with success probability 0.5. When Z = 1 (denoted by group 1), we generated data from configuration I, while we generated data from configuration II, III, or IV when Z = 0 (denoted by group 2). The censoring times (Cl) were generated from a uniform distribution U(0, 7.2) for group 1 and from a uniform distribution U(c1, c2) for group 2, giving the censoring proportion of 15% for each group l (l = 1, 2). As noted in Peng and Fine [25], the cumulative incidence functions for cause 1 are identical under configurations I and II, there is a crossover of the cumulative incidence functions for cause 1 under configurations I and III, and the cumulative incidence function for cause 1 under configuration I is stochastically larger than that under configuration IV. For each group l (l = 1, 2), we generated the missingness indicator Rl from , where Xl = min(Tl, Cl). With different choices of η(l)’s, we generated 20, 30, and 40% missing causes of failure for group 1, and 10% missing causes of failure for group 2. The true model for π(X) is logitπ(X) = log(5/2) − 0.2X in configuration I, logitπ(X) = log(5/3) + 0.2X in configuration II, logitπ(X) = log(5/6) − 0.5 log(X) − X1/2 + 1.2X in configuration III, and logitπ(X) = log(2/3) + 0.4X in configuration IV. We fitted a correctly specified logistic regression model logitπ(X, γ) = γ1 + γ2X in configurations I, II, and IV, and logitπ(X, γ) = γ1 + γ2 log(X) + γ3X1/2 + γ4X in configuration III. We conducted 1000 simulations with sample sizes of 200 and 400. Table 3 shows that under the correct imputation model, the imputation tests have empirical sizes close to the nominal level 0.05 and high powers in detecting differences between the two cumulative incidence functions for cause 1, whereas the test obtained by the complete case analysis results in larger empirical size and less power compared to the imputation methods. Note that in configurations I vs. III and I vs. IV with , the complete case test has higher power than the imputation tests. This is because the complete case analysis can overestimate cumulative incidence when deaths with missing cause of failure occur late in follow-up.
Table 3.
Empirical rejection rates and powers of two-sample tests under the correct imputation model; The null hypothesis scenario corresponds to the first column (I vs. II). The second and third columns (I vs. III and I vs. IV) indicate powers. Configuration I is the generation model for group 1 (Z = 1), which is fixed, and configurations II, III, and IV correspond to the generation models for group 2 (Z = 0).
| n | % missing in configuration I | Method | 10% missing in configurations II, III, and IV | ||
|---|---|---|---|---|---|
| ( ) | ( ) | ( ) | |||
| = (−2.65, 0.5) | = (−2.6, 0.5) | = (−2.68, 0.5) | |||
| I vs. II | I vs. III | I vs. IV | |||
| 200 | 20% | m=1 | 0.056 | 0.085 | 0.634 |
| ( ) | m=10 | 0.050 | 0.088 | 0.659 | |
| = (−1.38, 0) | CC | 0.062 | 0.075 | 0.601 | |
| 30% | m=1 | 0.060 | 0.070 | 0.548 | |
| ( ) | m=10 | 0.054 | 0.075 | 0.578 | |
| = (−0.1, −1) | CC | 0.096 | 0.046 | 0.369 | |
| 40% | m=1 | 0.054 | 0.089 | 0.566 | |
| ( ) | m=10 | 0.046 | 0.092 | 0.585 | |
| = (−0.1, −0.36) | CC | 0.105 | 0.058 | 0.398 | |
| 30% | m=1 | 0.057 | 0.095 | 0.649 | |
| ( ) | m=10 | 0.054 | 0.079 | 0.663 | |
| = (−1.38, 0.56) | CC | 0.059 | 0.077 | 0.693 | |
| 40% | m=1 | 0.056 | 0.099 | 0.562 | |
| ( ) | m=10 | 0.049 | 0.090 | 0.599 | |
| = (−1.38, 1.1) | CC | 0.072 | 0.117 | 0.712 | |
|
| |||||
| 400 | 20% | m=1 | 0.053 | 0.126 | 0.896 |
| ( ) | m=10 | 0.050 | 0.128 | 0.924 | |
| = (−1.38, 0) | CC | 0.061 | 0.115 | 0.897 | |
| 30% | m=1 | 0.061 | 0.118 | 0.861 | |
| ( ) | m=10 | 0.050 | 0.121 | 0.886 | |
| = (−0.1, −1) | CC | 0.138 | 0.057 | 0.712 | |
| 40% | m=1 | 0.050 | 0.129 | 0.822 | |
| ( ) | m=10 | 0.049 | 0.119 | 0.868 | |
| = (−0.1, −0.36) | CC | 0.113 | 0.064 | 0.687 | |
| 30% | m=1 | 0.057 | 0.117 | 0.867 | |
| ( ) | m=10 | 0.054 | 0.118 | 0.878 | |
| = (−1.38, 0.56) | CC | 0.059 | 0.146 | 0.909 | |
| 40% | m=1 | 0.057 | 0.120 | 0.829 | |
| ( ) | m=10 | 0.056 | 0.128 | 0.856 | |
| = (−1.38, 1.1) | CC | 0.070 | 0.204 | 0.928 | |
To justify the robustness of the imputation test, we fitted an incorrectly specified logistic regression model logitπ(X, γ) = γ1 + γ2 log(1 + exp(−X)) in configurations I, II, III, and IV. We generated data from the same scenario as before. We conducted 1000 simulations with sample sizes of 200 and 400 (Table 4). Even with misspecified imputation model, the imputation tests result in empirical sizes close to the nominal level and high powers.
Table 4.
Empirical rejection rates and powers of two-sample tests under the incorrect imputation model; The null hypothesis scenario corresponds to the first column (I vs. II). The second and third columns (I vs. III and I vs. IV) indicate powers. Configuration I is the generation model for group 1 (Z = 1), which is fixed, and configurations II, III, and IV correspond to the generation models for group 2 (Z = 0).
| n | % missing in configuration I | Method | 10% missing in configurations II, III, and IV | ||
|---|---|---|---|---|---|
| ( ) | ( ) | ( ) | |||
| = (−2.65, 0.5) | = (−2.6, 0.5) | = (−2.68, 0.5) | |||
| I vs. II | I vs. III | I vs. IV | |||
| 200 | 20% | m=1 | 0.055 | 0.085 | 0.641 |
| ( ) | m=10 | 0.056 | 0.074 | 0.654 | |
| = (−1.38, 0) | CC | 0.064 | 0.064 | 0.593 | |
| 30% | m=1 | 0.064 | 0.084 | 0.590 | |
| ( ) | m=10 | 0.052 | 0.090 | 0.629 | |
| = (−0.1, −1) | CC | 0.107 | 0.053 | 0.430 | |
| 40% | m=1 | 0.054 | 0.111 | 0.555 | |
| ( ) | m=10 | 0.053 | 0.092 | 0.582 | |
| = (−0.1, −0.36) | CC | 0.105 | 0.065 | 0.392 | |
| 30% | m=1 | 0.060 | 0.084 | 0.604 | |
| ( ) | m=10 | 0.057 | 0.094 | 0.634 | |
| = (−1.38, 0.56) | CC | 0.071 | 0.107 | 0.660 | |
| 40% | m=1 | 0.053 | 0.088 | 0.582 | |
| ( ) | m=10 | 0.049 | 0.078 | 0.596 | |
| = (−1.38, 1.1) | CC | 0.085 | 0.105 | 0.700 | |
|
| |||||
| 400 | 20% | m=1 | 0.046 | 0.146 | 0.887 |
| ( ) | m=10 | 0.048 | 0.143 | 0.911 | |
| = (−1.38, 0) | CC | 0.060 | 0.120 | 0.881 | |
| 30% | m=1 | 0.054 | 0.122 | 0.840 | |
| Equation | m=10 | 0.052 | 0.112 | 0.863 | |
| = (−0.1, −1) | CC | 0.148 | 0.064 | 0.675 | |
| 40% | m=1 | 0.045 | 0.144 | 0.822 | |
| ( ) | m=10 | 0.045 | 0.138 | 0.856 | |
| = (−0.1, −0.36) | CC | 0.117 | 0.063 | 0.678 | |
| 30% | m=1 | 0.059 | 0.131 | 0.884 | |
| ( ) | m=10 | 0.060 | 0.128 | 0.906 | |
| = (−1.38, 0.56) | CC | 0.061 | 0.161 | 0.924 | |
| 40% | m=1 | 0.055 | 0.134 | 0.857 | |
| ( ) | m=10 | 0.045 | 0.131 | 0.887 | |
| = (−1.38, 1.1) | CC | 0.061 | 0.197 | 0.934 | |
4. Application: Effect of tamoxifen on breast-cancer specific mortality in women with early stage breast cancer
4.1. Background
We illustrate the proposed methods using data from a randomized clinical trial in early stage breast cancer, obtained from the National Surgical Adjuvant Breast and Bowel Project (NSABP). The NSABP is a U.S. National Cancer Institute sponsored multi-center clinical trials group that conducts research into the treatment and prevention of breast and colorectal cancers. Tamoxifen is an estrogen-like compound used in the treatment of breast cancer [31].
Between 1982 and 1988, 2892 women with estrogen receptor-positive breast tumors and no axillary node involvement were enrolled in NSABP Protocol B-14, a double-blind randomized trial comparing five years of tamoxifen (20 mg/day) with placebo. The primary trial endpoints were overall survival (time surviving after surgery, with an event being death from any cause) and disease-free survival (time free of tumor recurrence, new breast cancer in the opposite breast, other cancers, or death from any cause preceding these events). Findings indicated a strong positive effect of tamoxifen on disease-free survival [32], and with longer follow-up duration, a significant reduction in overall mortality [33]. The latter endpoint (overall mortality) is used as a conservative approach in that it avoids complexities of accurate ascertainment of cause of death and also requires that the net effect of treatment be a longer lifetime. However, most of the influence on improvement in overall survival arises from avoidance of death from breast cancer. Thus cause-specific mortality (especially, breast cancer-specific mortality) is of interest. While information on cause of death was desired and requested, because the endpoint was overall survival, missing cause of death data were not rigorously resolved, and so there is a substantial portion of cases with missing cause. Table 5 shows vital status for 2817 trial eligible patients included in this analysis. Of patients who died, 20.9% of patients assigned to placebo and 26.3% of patients assigned to tamoxifen had missing cause of death.
Table 5.
Vital status among women from the NSABP B-14 randomized trial.
| Status/Cause of Death | Treatment Arm | Total | |
|---|---|---|---|
| Placebo | Tamoxifen | ||
| Alive | 919 | 1001 | 1920 |
| Breast cancer | 241 | 155 | 396 |
| Other cause | 150 | 142 | 292 |
| Unknown | 103 | 106 | 209 |
|
| |||
| Total | 1413 | 1404 | 2817 |
4.2. Results
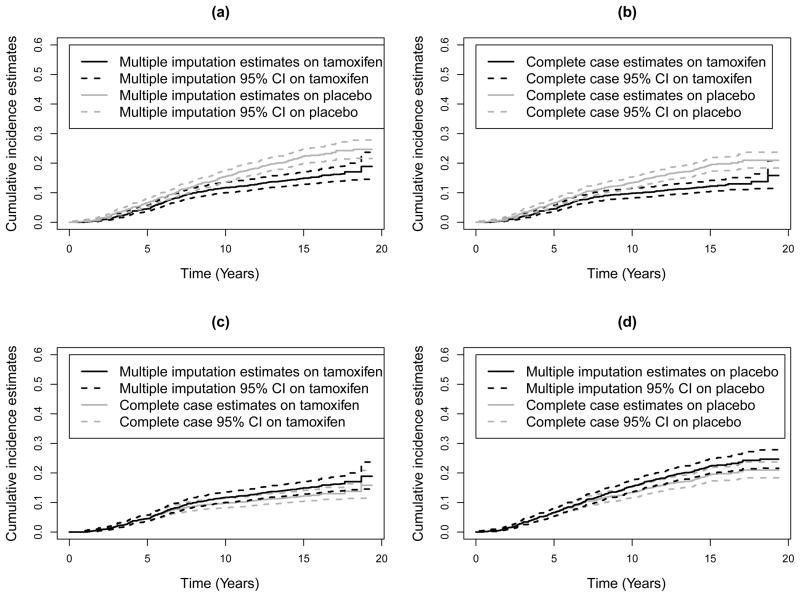
We applied our multiple imputation methods to estimate cumulative incidence of breast cancer-specific death on tamoxifen and placebo. In each group, the logistic imputation model for π(X) included time since diagnosis which was statistically significantly associated with breast cancer-specific mortality among those with complete cause of death information (R = 1, δ > 0). Figure 1 shows estimates of cumulative incidence of death due to breast cancer from 10 multiple imputations and the complete case estimates obtained by excluding patients with missing causes of death, along with their corresponding 95% pointwise confidence intervals (CI) given in (3). As shown in Figure 1(a) and (b), the estimates of cumulative incidence of breast cancer death in the tamoxifen group are lower than those in the placebo group in both multiple imputation and complete case analyses. In Figure 1(c) and (d), patients in the tamoxifen group from the multiple imputation methods have 10-year cumulative incidence of breast cancer death (with 95% confidence interval) of 11.7% (10.0–13.6%), while in the placebo group, 10-year cumulative incidence is 15.4% (13.5–17.4%). The complete case estimates are 9.8% (8.3–11.6%) for the tamoxifen group and 13.4% (11.6–15.3%) for the placebo group. The 10-year cumulative incidences of breast cancer death from the complete case analysis are 16% on tamoxifen and 13% on placebo lower than those from the multiple imputation methods. These differences continue to grow as follow-up lengthens.
Figure 1.
Estimates of cumulative incidence of breast cancer death with 95% pointwise confidence intervals (CI) by the multiple imputation method and complete case analysis. The upper panels show results by analysis and by group, whereas the lower panel shows results by group and by analysis.
In order to compare cumulative incidences of breast cancer death between tamoxifen and placebo groups, we computed the multiple imputation test based on 10 imputations and the complete case test. The test statistics (p-value) for breast cancer death are −3.746 (0.0002) for the multiple imputation test and −3.903 (0.00009) for the complete case test, both indicating that tamoxifen significantly reduces mortality from breast cancer. The result from the complete case test is very similar to that from the multiple imputation test. This may be because cumulative incidences of death due to breast cancer in both tamoxifen and placebo groups are underestimated to a similar extent by the complete case analysis (see Figure 1).
5. Discussion
We investigated multiple imputation methods for nonparametric inferences of the cumulative incidence function in the presence of missing causes of failure. Under the missing at random assumption, we develop asymptotic theory for multiple imputation methods to estimate the cumulative incidence function nonparametrically and perform a test for comparing the cumulative incidence functions in two samples. These procedures are straightforward to implement. When there are no missing causes of failure, our inference procedures reduce to those in Lin [9] and Pepe’s test with a martingale-based variance estimator [24, 28]. Simulation studies show that our multiple imputation methods perform well, even if the model for imputation is misspecified. The simulations also show that our improper imputation approach is slightly more efficient compared to the proper imputation [5]. In supplemental analyses, we show that the complete case analysis can either underestimate or overestimate cumulative incidence, depending on whether deaths with missing cause occur early or late in follow-up. In the breast cancer example, estimates of cumulative incidence of breast cancer death in both tamoxifen and placebo groups from the complete case analysis were lower than those from the multiple imputation methods. Since cumulative incidences of breast cancer death in both groups are underestimated to a similar extent by the complete case analysis, the complete case test provided a similar result to the multiple imputation test for comparing breast-cancer specific mortalities between the two groups. Thus, the complete case test is unlikely to be biased for comparing differences between the cumulative incidence functions in two samples where the missing cause of death mechanism is the same in each group.
We proposed nonparametric inferences of the cumulative incidence function with missing cause of failure under the assumption of missing at random when we observe (Xi, Ri, Riδi) (i = 1, …, n). In a practical setting, covariate information may be available and one may include auxiliary covariates in the imputation model to make the missing at random assumption more plausible, as the methods of [2]. In the case, our multiple imputation methods for nonparametric inferences of the cumulative incidence function with missing causes of failure are applicable incorporating auxiliary covariates. A nonparametric estimator of the cumulative incidence function can be obtained by imputing missing cause of failure from a Bernoulli distribution with success probability π(Wi, γ̂) and averaging nonparametric estimators across several imputed data sets, where Wi = (Xi, Zi), Zi is auxiliary covariates, and the success probability π(Wi, γ̂) can be obtained by fitting a logistic regression model, , under the missing at random assumption. The variance of the multiple imputation estimator can be estimated by V̂1(t) in equation (A.8) with replacing π(Xi, γ̂) by π(Wi, γ̂) and πγ(Xi, γ̂) by πγ(Wi, γ̂) in equation (A.5). The variance estimator of the multiple imputation test in two samples can be obtained by replacing πγl(Xil, γ̂l) by πγl(Wil, γ̂l) in (B.3) and π(Xil, γ̂l) by π(Wil, γ̂l) in equation (B.6).
We assumed that the cause of failure is missing at random. Unfortunately, assumptions about the missing mechanism such as the missing at random are not testable from the observed data [5, 34]. Thus, one may conduct sensitivity analyses to assess the robustness of inferences to departures from the missing at random assumption [5, 6, 35].
We used the Aalen-Johansen estimator for nonparametric estimation of the cumulative incidence function. This is based on the assumption that the censoring time C is independent of (T, ε). When the censoring time is only independent of (T, ε) conditional on covariates, the Aalen-Johansen estimator is unbiased if the censoring distribution does not depend on covariates (i.e., Pr(C > t | Z = z) = Pr(C > t) for covariates Z) and hence censoring is independent of covariates. However, the estimator may be biased if censoring depends on covariates [36]. In that case, Binder et al. [36] proposed an alternative estimator to the Aalen-Johansen estimator by using a regression model for the censoring time. Thus, when the censoring time is only independent of (T, ε) given covariates, our imputation methods to estimate nonparametrically the cumulative incidence function with missing cause of failure are valid if censoring is independent of covariates. However, if censoring depends on covariates, alternative estimators proposed by Binder et al. [36] may be used. Similar derivations used in our paper may be applicable to multiple imputation inferences for the alternative estimators.
In this paper, we discussed multiple imputation inferences for nonparametric estimation of the cumulative incidence function with two types of failure in the presence of missing causes of failure. When there are more than two causes of death, the missing cause can be imputed from a multinomial distribution with probabilities estimated by using a parametric modeling under the missing at random assumption. A multinomial logistic regression can be considered as a parametric imputation model. In that case, the variance formula can be obtained following asymptotic theory given in Appendix A along with the influence function expansion of the maximum likelihood estimators of parameters in a multinomial logistic imputation model.
When the imputation and analysis models agree (i.e., congenial as defined in [29]), results from the proper multiple imputation approach may be similar to those from our improper multiple imputation methods, as shown in the simulation studies. When covariate information is available and censoring depends on some covariates, one may include covariates (including auxiliary covariates) in the imputation model to make the missing at random assumption more plausible and use a regression model for the censoring time. In that case, the imputation and analysis models may then be uncongenial, and Rubin’s variance formula may produce some bias in the variance estimates of the multiple imputation estimates, as noted in [29]. Note that we derive variance estimators for the multiple imputation estimators directly. This may suggest that our multiple imputation method is more efficient.
Supplementary Material
Acknowledgments
The NSABP clinical trial in the example was supported by U.S. National Institute of Health grants U10-CA12027, U10-CA37377, U10-CA69651, and U10-CA69974. J. Dignam received support from U10-CA21661 and U10-CA180822. The authors thank the two reviewers and the associate editor for their helpful and constructive comments.
APPENDIX A
Define the counting process notation Nkij(t, Ri, γ) = I(Xi ≤ t, Dkij(Ri, γ) = 1), Yi(t) = I(Xi ≥ t), and . Let
Let Λ̂kj(t) be the cumulative cause-specific hazard estimate for cause k from the jth imputed data set that can be expressed by the counting process notation as
n1/2{Λ̂kj(t) − Λk(t)} is asymptotically equivalent to
| (A.1) |
Using the consistency of Ŝ(·), Taylor expansion and integration by parts as described in Lin [9], U1j(t) = n1/2{F̂1j(t) − F1(t)} from the jth imputed data set can be expressed as
Using (A.1) and S(t) = 1 − F1(t) − F2(t), U1j(t) = n1/2{F̂1j(t) − F1(t)} is given by
| (A.2) |
where
To establish the asymptotic theory for single imputation and multiple imputation estimates, we use the methods described in Lu and Tsiatis [2] or Tsiatis et al. [22]. Consider the centered sum , where μf (γ, t) = E{fij(γ, t)}. Then n−1/2{Hj(γ̂, t) − Hj(γ0, t)} converges in probability to zero [37]. Using the convergence of the centered sum and Taylor series expansion,
Using arguments similar to those in Tsiatis et al. [22],
which is estimated by
where subscript j means that the corresponding estimate is obtained from the jth imputed data set and .
The asymptotic normality of γ̂ follows from the influence function expansion
where Oi = (Ri = 1, δi > 0, Xi),
and J(γ0) is the information matrix
Applying the approximation for n1/2(γ̂ − γ0), U1j(t) is equal to
| (A.3) |
This is a sum of independent and identically distributed mean zero random variables, and the central limit theorem proves asymptotic normality. Its variance is given by
| (A.4) |
where E{fij(γ0, t)⊗2} is the asymptotic variance of n1/2{F̂1(t) − F1(t)} when all causes of failure are observed and is estimated by
and D2ij(Ri, γ̂) is the imputed value of D2i from the jth imputed data set as
For any j (j = 1, …, m), the variance V1j(t) is estimated by
| (A.5) |
For multiple imputation, U1(t) = n1/2{F̂1(t) − F1(t)} is asymptotically equivalent to
| (A.6) |
This is a sum of independent and identically distributed mean zero random variables, and the central limit theorem yields the asymptotic normality. Its variance is given by
| (A.7) |
The variance V1(t) is estimated by
| (A.8) |
APPENDIX B
Suppose that there are two independent competing risk samples with missing cause of failure. In group l (l = 1, 2), the observed data consist of (Ril, Xil, Rilδil) (i = 1, …, nl), where Ril is the missingness indicator in group l, Xil = min(Til, Cil), and δil = I(Til ≤ Cil)εil. For the ith individual from the jth imputed data set in group l, define the counting process notation Nkijl(t, Ril, γ) = I(Xil ≤ t, Dkijl(Ril, γ) = 1), Yil(t) = I(Xil ≥ t), and . Let
The test statistic for single imputation can be written as
Under the null hypothesis for all t ≤ τ,
Using (A.2) and changing the order of integration, is given by
where γ̂l is the maximum likelihood estimator of γ in group l and
To establish the asymptotic theory for single imputation and multiple imputation tests, we use the methods described in Lu and Tsiatis [2] or Tsiatis et al. [22]. We consider the centered sum for in group l, , where . Let γ0l be the true value of γl. Then converges in probability to zero [37]. Using the convergence of the centered sum and Taylor series expansion, and applying the approximation for in group l, is equal to
| (B.1) |
where using arguments similar to those in Tsiatis et al. [22],
This is a sum of independent and identically distributed mean zero random variables, and the central limit theorem yields the asymptotic normality. Its variance is given by
| (B.2) |
where is the asymptotic variance of when all causes of failure are observed and is estimated by
For any j (j = 1, …, m), the variance is estimated by
| (B.3) |
where
Therefore, the variance estimate of the test statistic QS is given by
Under the null hypothesis H0, the test statistic for multiple imputation is written as
Using (B.1), is asymptotically equivalent to
| (B.4) |
This is a sum of independent and identically distributed mean zero random variables, and the central limit theorem yields the asymptotic normality. Its variance is given by
| (B.5) |
The variance is estimated by
| (B.6) |
We obtain the variance estimate of the test statistic QM as
References
- 1.Goetghebeur E, Ryan L. Analysis of competing risks survival data when some failure types are missing. Biometrika. 1995;82:821–834. doi: 10.1093/biomet/82.4.821. [DOI] [Google Scholar]
- 2.Lu K, Tsiatis AA. Multiple imputation methods for estimating regression coefficients in the competing risks model with missing cause of failure. Biometrics. 2001;57:1191–1197. doi: 10.1111/j.0006-341X.2001.01191.x. [DOI] [PubMed] [Google Scholar]
- 3.Gao G, Tsiatis AA. Semiparametric estimators for the regression coefficients in the linear transformation competing risks model with missing cause of failure. Biometrika. 2005;92:875–891. doi: 10.1093/biomet/92.4.875. [DOI] [Google Scholar]
- 4.Lu W, Liang Y. Analysis of competing risks data with missing cause of failure under additive hazards model. Statistica Sinica. 2008;18:219–234. [Google Scholar]
- 5.Bakoyannis G, Siannis F, Touloumi G. Modelling competing risks data with missing cause of failure. Statistics in Medicine. 2010;29(30):3172–3185. doi: 10.1002/sim.4133. [DOI] [PubMed] [Google Scholar]
- 6.Moreno-Betancur M, Latouche A. Regression modeling of the cumulative incidence function with missing causes of failure using pseudo-values. Statistics in Medicine. 2013;32:3206–3223. doi: 10.1002/sim.5755. [DOI] [PubMed] [Google Scholar]
- 7.Lee M, Cronin KA, Gail MH, Dignam JJ, Feuer EJ. Multiple imputation methods for inference on cumulative incidence with missing cause of failure. Biometrical Journal. 2011;53(6):974–993. doi: 10.1002/bimj.201000175. [DOI] [PubMed] [Google Scholar]
- 8.Nicolaie MA, Houwelingen HC, Putter H. Vertical modeling: analysis of competing risks data with missing causes of failure. Statistical Methods in Medical Research. 2011 doi: 10.1177/0962280211432067. [DOI] [PubMed] [Google Scholar]
- 9.Lin DY. Non-parametric inference for cumulative incidence functions in competing risks studies. Statistics in Medicine. 1997;16:901–910. doi: 10.1002/(SICI)1097-0258(19970430)16:8<901::AID-SIM543>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 10.Aalen OO, Johansen S. An empirical transition matrix for non-homogeneous Markov chains based on censored observations. Scandinavian Journal of Statistics. 1978;5:141–150. [Google Scholar]
- 11.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 12.Nelson W. Theory and applications of hazard plotting for censored failure data. Technometrics. 1972;14:945–965. [Google Scholar]
- 13.Aalen OO. Nonparametric inference for a family of counting processes. Annals of Statistics. 1978;6:701–726. doi: 10.1214/aos/1176344247. [DOI] [Google Scholar]
- 14.Betensky RA, Schoenfeld DA. Nonparametric estimation in a cure model with random cure times. Biometrics. 2001;57:282–286. doi: 10.1111/j.0006-341X.2001.00282.x. [DOI] [PubMed] [Google Scholar]
- 15.Choudhury JB. Non-parametric confidence interval estimation for competing risks analysis: application to contraceptive data. Statistics in Medicine. 2002;21:1129–1144. doi: 10.1002/sim.1070. [DOI] [PubMed] [Google Scholar]
- 16.Braun TM, Yuan Z. Comparing the small sample performance of several variance estimators under competing risks. Statistics in Medicine. 2007;26 :1170–1180. doi: 10.1002/sim.2661. [DOI] [PubMed] [Google Scholar]
- 17.Lee M, Fine JP. Inference for cumulative incidence quantiles via parametric and nonparametric approaches. Statistics in Medicine. 2011;30:3221–3235. doi: 10.1002/sim.4349. [DOI] [PubMed] [Google Scholar]
- 18.Allignol A, Schumacher M, Beyersmann J. A note on variance estimation of the Aalen-Johansen estimator of the cumulative incidence function in competing risks, with a view towards left-truncated data. Biometrical Journal. 2010;52:126–137. doi: 10.1002/bimj.200900039. [DOI] [PubMed] [Google Scholar]
- 19.Rubin DB. Inference and missing data. Biometrika. 1976;63:581–592. doi: 10.1093/biomet/63.3.581. [DOI] [Google Scholar]
- 20.Rubin DB. Multiple Imputation for Nonresponse in Surveys. Wesley; New York: 1987. [Google Scholar]
- 21.Wang N, Robins JM. Large sample inference in parametric multiple imputation. Biometrika. 1998;85:935–948. doi: 10.1093/biomet/85.4.935. [DOI] [Google Scholar]
- 22.Tsiatis AA, Davidian M, Mcneney B. Multiple imputation methods for testing treatment differences in survival distributions with missing cause of failure. Biometrika. 2002;89:238–244. doi: 10.1093/biomet/89.1.238. [DOI] [Google Scholar]
- 23.Gray RJ. A class of k-sample tests for comparing the cumulative incidence of a competing risk. Annals of Statistics. 1988;16:1141–1154. doi: 10.1214/aos/1176350951. [DOI] [Google Scholar]
- 24.Pepe MS. Inference for events with dependent risks in multiple endpoint studies. Journal of the American Statistical Association. 1991;86:770–778. doi: 10.1080/01621459.1991.10475108. [DOI] [Google Scholar]
- 25.Peng L, Fine JP. Nonparametric quantile inference with competing risks data. Biometrika. 2007;94(3):735–744. doi: 10.1093/biomet/asm059. [DOI] [Google Scholar]
- 26.Pepe MS, Fleming TR. Weighted Kaplan-Meier statistics: a class of distance tests for censored survival data. Biometrics. 1989;45:497–507. [PubMed] [Google Scholar]
- 27.Pepe MS, Fleming TR. Weighted Kaplan-Meier statistics: large sample and optimality considerations. Journal of the Royal Statistical Society, Series B. 1991;53:341–352. [Google Scholar]
- 28.Bajorunaite R, Klein JP. Two-sample tests of the equality of two cumulative incidence functions. Computational Statistics and Data Analysis. 2007;51 :4269–4281. doi: 10.1016/j.csda.2006.05.011. [DOI] [Google Scholar]
- 29.Meng XL. Multiple imputation inferences with uncongenial sources of input (with discussion) Statistical Science. 1994;9:538–573. [Google Scholar]
- 30.Satten GA, Datta S, Williamson JM. Inference based on imputed failure times for the proportional hazards model with interval-censored data. Journal of the American Statistical Association. 1998;93:318–327. doi: 10.1080/01621459.1998.10474113. [DOI] [Google Scholar]
- 31.Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351:1451–1467. doi: 10.1016/S0140-6736(97)11423-4. [DOI] [PubMed] [Google Scholar]
- 32.Fisher B, Costantino J, Redmond C, et al. A randomized clinical trial evaluating tamoxifen in the treatment of patients with node-negative breast cancer who have estrogen-receptor-positive tumors. New England Journal of Medicine. 1989;320:479–484. doi: 10.1056/NEJM198902233200802. [DOI] [PubMed] [Google Scholar]
- 33.Fisher B, Dignam J, Bryant J, et al. Five versus more Than five years of tamoxifen therapy for breast cancer patients with negative lymph nodes and estrogen receptor positive tumors. Journal of the National Cancer Institute. 1996;88:1529–1542. doi: 10.1093/jnci/88.21.1529. [DOI] [PubMed] [Google Scholar]
- 34.Molenberghs G, Beunckens C, Sotto C, Kenward MG. Every missingness not at random model has a missingness at random counterpart with equal fit. Journal of the Royal Statistical Society, Series B. 2008;70:371–388. doi: 10.1111/j.1467-9868.2007.00640.x. [DOI] [Google Scholar]
- 35.Molenberghs G, Kenward MG. Missing Data in Clinical Studies. Wesley; Chichester: 2007. [Google Scholar]
- 36.Binder N, Gerds TA, Andersen PK. Pseudo-observations for competing risks with covariate dependent censoring. Lifetime Data Analysis. 2013;20:303–315. doi: 10.1007/s10985-013-9247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Vaart AW. Asymptotic Statistics. Cambridge University Press; Cambridge: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.